Abstract
BACKGROUND
Variant RHCE alleles with diminished expression of C, c, E, and e antigens have been described and indicate the genetic diversity of this gene locus in several populations. In this study the molecular background of variant RhCE antigens identified by standard serologic routine testing in German blood donors and patients was determined.
STUDY DESIGN AND METHODS
Samples from blood donors and patients were routinely analyzed for RhCE phenotype using the PK7200 analyzer with two sets of monoclonal anti-C, -c, -E, and -e reagents. Samples with confirmed variant RhCE antigens were analyzed by nucleotide sequencing of the 10 RHCE exons. A multiplex polymerase chain reaction with sequence-specific priming (PCR-SSP) method was established for rapid typing of the rare RHCE alleles.
RESULTS
We identified 43 samples with serologic RhCE variants. Molecular analysis revealed variant RHCE alleles in 34 samples. Altogether 22 RHCE alleles were detected; 10 have not been published before. Twenty alleles harbored distinct single-nucleotide substitutions, 18 of which encoded amino acid changes and 2 of which occurred in noncoding regions. Two samples represented RHCE-D-CE hybrid alleles involving different segments of the RHCE Exon 5. A multiplex PCR-SSP screening for 17 RHCE alleles was negative in 1344 samples of the DNA bank GerBS. The cumulative phenotype frequency was estimated between 1 in 488 (0.20%) and 1 in 8449 (0.012%).
CONCLUSION
Single-amino-acid substitutions were the molecular basis for variant RhCE antigen expression in most samples. Nucleotide substitutions in RHCE exons were excluded as possible mechanism of diminished RhCE antigen expression in one-fifth of the serologically identified samples.
Rh is the most polymorphic of all 30 blood group systems.1 The Rh blood group system is encoded by the two highly homologous genes, RHD and RHCE. The two polymorphic genes encode the five major antigens: RHD encodes the D antigen and RHCE the C, E, c, and e antigens. Forty-five additional low- and high-prevalence Rh antigens are defined to date. Many RHCE alleles were identified by molecular analyses in different populations.2,3 Variant RhCE phenotypes may be caused by single- or multiple-nucleotide substitutions in the RHCE gene or by RHCE-D-CE hybrid alleles at the RHCE gene locus. Rare RhCE phenotypes were found either by antibodies against low-prevalence4-7 and high-prevalence8,9 RhCE antigens or by weak and partial expression of the major RhCE antigens.5,10-12
In early studies the serologic patterns established by monoclonal anti-E allowed categorization of four variants of the E antigen (cat EI to EIV).13 The molecular basis in RHCE alleles are the single-amino-acid substitution M167K (500T>A) for cat EI,13 later recognized to express the low-prevalence antigen Ew;14 the RHcE-D(2-3)-cE hybrid allele for cat EII;13 the two D-specific amino acid substitutions Q233E and M238V (697C>G, 712A>G) for cat EIII;13 and the single D-specific amino acid substitution R201T (602G>C) for cat EIV,15 which is occurring in a cE haplotype and located suspiciously close to the amino acid 223 encoding the E/e antigens.
Another way to discover RhCE variants is the expression of low-prevalence antigens, some of which occur in RhCE proteins whose major antigens are altered. For example, the Rhce-D(5)-ce hybrid allele encodes an RhCE protein expressing the low-prevalence antigens RH33 (Har) and RH50 (FPTT) and a weakened e antigen.5 An RHCe-D(4)-Ce hybrid allele encodes an RhCE protein expressing the low-prevalence antigens RH32 (RN) and RH54 (DAK) and weakened C and e antigens, but lacking the high-prevalence antigen RH46 (Sec).10 The ceCF variant expresses the low-prevalence RH43 (Crawford) antigen that reacted with a polyclonal antibody inadvertently occurring in former commercial polyclonal anti-D reagents and is cross-reactive with a currently available monoclonal anti-D.16 Similarly, the RhCE variants ceRT and ceSL17,18 associated with the ce haplotype were found by the unexpected expression of D epitopes. If the same ceSL amino acid substitution occurs in the Ce haplotype, it induces the expression of the low-prevalence antigen RH53 (JAHK) and is associated with significantly weakened C and e antigens.19
Hence, minor modifications, like single-amino-acid substitutions typically occurring in the extracellular part of the RhCE protein but also in transmembraneous or intracellular segments, may induce the expression of neoantigens, recognized as low-prevalence antigens, or weakened epitope and antigen expression, which may involve conformational changes of the RhCE protein, reduce membrane integration, or hamper the interaction with other proteins of the Rh complex in the red blood cell (RBC) membrane.
In this study we investigated the RhCE variants encountered in the routine serologic workup of blood donors and patients in four reference laboratories of a large blood donor service in Southwestern Germany. Our screening procedure allowed detecting the weakened expression of the major RhCE antigens. We found 22 RHCE alleles among which 10 have not been published before.
MATERIALS AND METHODS
Blood samples and immunohematology
Blood samples were collected from blood donors in Southwestern Germany, which comprises a population of 16.5 million people. The first- and second-time donors were typed for ABO, D, CcEe, and K with an automated analyzer (PK7200; Olympus, Hamburg, Germany) as a routine serologic method. Two sets of monoclonal antibodies (MoAbs) diluted in 0.9% NaCl with 0.1% bromelin were used: anti-C (MS24 at a final dilution of 1 in 300, Ortho, Neckargmünd, Germany; and 392/P3x25513G8 at 1 in 175, Biotest, Dreieich, Germany), anti-E (MS258/906 and MS260/MS12, both at 1 in 150, Biotest), anti-c (MS33 at 1 in 100, Immucor, Rödermark, Germany; and MS42 at 1 in 100, Ortho), and anti-e (MS63/MS16/MS21 at 1 in 30, Ortho; and BS260/267 at 1 in 8, Biotest). The incubation time was 60 minutes at room temperature.
We examined samples in tube or column agglutination techniques that showed discrepant, weak, or unclear results for any of the antigens CcEe with the PK7200 analyzer and blood samples from patients with aberrant CcEe phenotypes that had been sent to our reference laboratories. Tube tests were performed for 15 minutes at room temperature with the same monoclonal reagents used for the PK7200 analyzer (MS24, 392/P3x25513G8; MS258/906, MS260/MS12; MS33, MS42; MS63/MS16/MS21, BS260/267). CcEe typing with column agglutination techniques was performed using the same MoAbs at room temperature (ID-Card, DiaMed, Ottobrun, Germany; and ScanGel, BioRad, München, Germany) or polyclonal (ID-Card Rh-subgroups plus K; DiaMed) RHCE typing cards. For the polyclonal typing gel card (DiaMed) the RBCs were suspended in Diluent 1 (bromelin solution, DiaMed) and in Diluent 2 for monoclonal typing cards. If a serologically variant RhCE phenotype was confirmed, the sample was subjected to molecular investigation of the RHCE gene.
RHCE nucleotide sequencing
For specific polymerase chain reaction (PCR) amplification and nucleotide sequencing of the 10 RHCE exons we used primers and PCR conditions as previously described20 except for the RHCE-specific reverse primer in Exon 5. This primer was replaced by the RHD/RHCE-specific primer rb15.21
RHD zygosity
The presence of the hybrid Rhesus box was determined by PCR (Ready Gene D neg, inno-Train, Kronberg, Germany).
Multiplex PCR for rare RHCE alleles
A PCR with sequence-specific priming (PCR-SSP) with five multiplex reactions was established for the rapid detection of the observed rare RHCE alleles. All primers were used at a final concentration of 1 mmol/L, except the control primers HBB-F and HBB-R, which were added to each multiplex PCR mix at 0.4 mmol/L (Table 1). The total volume of each PCR-SSP was 10 mL containing 10 ng of sample DNA. The cycling conditions were 2 minutes at 95°C, followed by 10 cycles with 15 seconds at 95°C and 1 minute at 65°C, followed by 20 cycles with 15 seconds at 95°C, 1 minute at 61°C, and 30 seconds at 72°C. The total thermocycling time was 1.5 hours. Amplification products were examined in 2% agarose gels with 0.5 ng/mL ethidium bromide (ultraviolet documentation device with charged coupled device camera, UVP, Upland, CA).
TABLE 1.
Primers for PCR-SSP typing of rare RHCE alleles
| Name | Nucleotide sequence (5′-3′) | Genomic region | Direction | Specificity |
|---|---|---|---|---|
| F-10T | TGGAACCCCTGCACAGAGAT | 5′-UTR Exon 1 | Sense | RHCE(−10T) |
| F28T | AAGTACCCGCGGTCTGTCT | Exon 1 | Sense | RHCE(28T) |
| RIVS1 | acactgttgrctgaatttcggtgc | Intron 1 | Antisense | RHD/RHCE |
| F341A | ccttctcacccccagTATTCA | Exon 3 | Sense | RHCE(341A) |
| F344C | tctcacccccagTATTCGGCC | Exon 3 | Sense | RHCE(344C) |
| F344G | tctcacccccagTATTCGGCG | Exon 3 | Sense | RHCE(344G) |
| F364C | GGCCACCATGAGTGCTATGC | Exon 3 | Sense | RHCE(364C) |
| F374A | GAGTGCTATGTCGGTGCTGAA | Exon 3 | Sense | RHCE(374A) |
| R455C | TACTGATGACCATCCTCAGGG | Exon 3 | Antisense | RHCE(455C) |
| FIVS3-5G | ctctactgctcttactgggttttatg | Intron 3 | Sense | RHCE(IVS3-5G) |
| F497T | ggttttattgcagACAGACTACCT | Exon 4 | Sense | RHCE(497T) |
| F500A | tttattgcagACAGACTACCACAA | Exon 4 | Sense | RHCE(500A) |
| F506A | gACAGACTACCACATGAACCA | Exon 4 | Sense | RHCE(506A) |
| F602C | GAACGGAGGATAATGATCAGAC | Exon 4 | Sense | RHCE(602C) |
| RIVS4 | gaggtccctaaaaggagtgc | Intron 4 | Antisense | RHCE |
| F649C | ccagGCGCCCTCTTCTTGC | Exon 5 | Sense | RHCE(649C) |
| F722T | GGAAGAATGCCATGTTCAACAT | Exon 5 | Sense | RHCE(722T) |
| F800A | CCCCAAAGGAAGATCAGCAA | Exon 5 | Sense | RHCE(800A) |
| RIVS5 | agctccaccacccggcatgt | Intron 5 | Antisense | RHCE |
| FIVS5 | gccccaacacaggggagag | Intron 5 | Sense | RHD/RHCE |
| R890G | CCCAGCCACAAGACCCG | Exon 6 | Antisense | RHCE(890C) |
| R908T | GCTCCCCCGATGGAGATCT | Exon 6 | Antisense | RHCE(908A) |
| HBB-F | GGTTGGCCAATCTACTCCCAGG | 5′-UTR Exon 1 | Sense | HBB * |
| HBB-R | GCTCACTCAGTGTGGCAAAG | Exon 2 | Antisense | HBB * |
Internal control primers specific for conserved sequences of the β-hemoglobin gene (HBB).
Using the multiplex PCR-SSP method, the DNA bank GerBS (German Blood Service) control series22 was screened. GerBS consists of 1344 DNA samples of healthy, unrelated blood donors from the southwestern area of Germany, which corresponds to the geographical origin of the blood donors and patients of the current study. Equal numbers of female and male subjects were investigated with a median age of 50 years (range, 18-68 years); their ABO, Rh, and Kell blood group phenotypes were random (Table 2).23
TABLE 2.
Blood group distribution among the samples of the GerBS control series
| Blood group | Blood donors | ||
|---|---|---|---|
|
|
|
||
| System | Phenotype | Number | % |
| ABO | O | 527 | 39.2 |
| A | 556 | 41.4 | |
| B | 177 | 13.2 | |
| AB | 84 | 6.2 | |
| Rh | DCcee | 487 | 36.2 |
| DCCee | 267 | 19.9 | |
| DCcEe | 156 | 11.6 | |
| DccEe | 156 | 11.6 | |
| DccEE | 22 | 1.6 | |
| Dccee | 16 | 1.2 | |
| DCcEE | 1 | 0.1 | |
| ddccee | 227 | 16.9 | |
| ddCcee | 7 | 0.5 | |
| ddccEe | 5 | 0.4 | |
| Kell | kk | 1245 | 92.6 |
| Kk | 99 | 7.4 | |
| KK | 0 | 0 | |
| Total | 1344 | 100 | |
Nomenclature
Novel RHCE alleles were named after the involved amino acid substitution, such as RHcE(R10W). In case of variations in noncoding regions, the allele was named after the involved nucleotide change. Hence, Rhce(-10C>T) represents a single-nucleotide substitution in the untranslated region (UTR) located 10 nucleotides upstream of (5′ to) the start codon, and RHCe(IVS3-5G) represents a substitution in Intron 3 located five nucleotides 5′ to the first nucleotide of Exon 4.
RESULTS
We identified 43 samples with weak expression of one or two major RhCE antigens (Table 3) by testing blood donors and patients with routine serologic methods. For automated blood donor testing, diluted monoclonal antisera were used. In approximately four of five samples, nucleotide sequencing revealed a variant RHCE allele, while nine samples showed no deviation from one of the regular RHCE exon nucleotide sequences. Between June 2005 and October 2007, a total of 1,208,162 whole-blood donations were collected in Baden-Württemberg (Southwestern Germany) at our DRK Blood Donor Service, of which 143 donations were from the 34 blood donors (Table 3).
TABLE 3.
Blood donor and patient samples in this study
| Blood donors | Patients | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Rhesus phenotype | Variant RHCE allele (n) | Regular RHCE allele (n) | Variant RHCE allele (n) | Regular RHCE allele (n) | Total |
| DCcEe | 17 | 6 | 1 | 1 | 25 |
| DCcee | 3 | 0 | 4 | 1 | 8 |
| DccEe | 7 | 0 | 2 | 0 | 9 |
| DCcEE | 0 | 1 | 0 | 0 | 1 |
| Total | 34 | 9 | 43 | ||
TABLE 4.
RHCE alleles identified in samples with aberrant RhCE antigen expression
| Allele | Trivial name | Nucleotide change |
Genomic region |
Amino acid change |
Membrane localization* |
Original report† |
Observed by Doescher et al.24 |
Accession number |
|---|---|---|---|---|---|---|---|---|
| Single-nucleotide polymorphisms | ||||||||
| RHce(5′-UTR-10C>T) | −10C>T | 5′-UTR Exon 1 | none | NA | 2008 | Yes | FM866412 | |
| RHcE(R10W) | 28C>T | Exon 1 | R10W | IC | This study | No | FJ486155 | |
| RHce(W16C) | ce(C48) | 48G>C | Exon 1 | W16C | TM | Westhoff et al., 200112 |
Yes | DQ266400 |
| RHCe(A36T) | Cx | 106G>A | Exon 1 | A36T | EF | Mouro et al., 19954 |
No | NA |
| RHCe(Q41R) | CW | 122A>G | Exon 1 | Q41R | EF | Mouro et al., 19954 |
Yes | NA |
| RHce(R114Q) | ce JAL+ | 341G>A | Exon 3 | R114Q | TM | 2003 | No | AJ548432 |
| RHCe(L115R) | 344T>G | Exon 3 | L115R | TM | 2004 | Yes | AJ867774 | |
| RHcE(L115P) | 344T>C | Exon 3 | L115P | TM | This study | Yes | FJ486156 | |
| RHCe(S122P) | 364T>C | Exon 3 | S122P | TM | This study | No | FJ486157 | |
| RHcE(l125N) | 374T>A | Exon 3 | I125N | TM | This study | No | FJ486158 | |
| RHCe(IVS3-5G) | 487-5T>G | Intron 3 | none | NA | 2008 | Yes | FM866415 | |
| RHCe(H166L) | 497A>T | Exon 4 | H166L | EF | This study | No | FJ486159 | |
| RHcE(M167K) | cat El (Ew) | 500T>A | Exon 4 | M167K | EF | Strobel et al., 200414 |
Yes | NA |
| RHcE(L169Q) | 506T>A | Exon 4 | L169Q | TM | this study | No | FJ486160 | |
| RHcE(R201T) | cat EIV | 602G>C | Exon 4 | R201T§ | IC | 199915 | Yes | FJ486161 |
| RHce(W217R) | 649T>C | Exon 5 | W217R | TM | This study | No | FJ486162 | |
| RHCE(T241I) | 722C>T | Exon 5 | T241I | TM | This study | No | FJ486163 | |
| RHCe(M267K) | 800T>A | Exon 5 | M267K§ | TM | This study | No | FJ486164 | |
| RHCe(L297P) | CE-OL10 | 890T>C | Exon 6 | L297P | TM | 2006 | Yes | AM295501 |
| RHcE(L303Q) | 908T>A | Exon 6 | L303Q | TM | This study | No | FJ486165 | |
| Gene conversions | ||||||||
| RHCe-D(5)-Ce | RHCeVA | 667G>T; 697G>C; 712A>G; 733C>G; 744T>C; 787A>G; 800T>A |
Exon 5 | V223F; Q233E; M238V; L245V; R263G; M267K |
TM | Noizat-Pirenne et al., 200211 |
Yes NA |
|
| RHCe-D(5667-712)-Ce | RHCeVF | 667G>T; 697G>C; 712A>G |
Exon 5 | V223F; Q233E; M238V |
TM | 2005 | No AJ867777 |
Amino acid changes located in the intracellular (IC), exofacial (EF), or one of the 12 transmembrane (TM) regions of the RhCE protein.
Reference for original publication or year of GenBank nucleotide sequence database submission.
NA = not applicable.
RHCE alleles
Among 34 samples (Table 3), we encountered 22 different RHCE alleles, 10 of which have previously not been published (Table 4). In a parallel study by Doescher and colleagues24 describing RhCE variants in Northern Germany, only 10 of these 22 RHCE alleles were observed, although the variety of alleles was similar. However, none of our donors and patients overlapped with those found in the Northern German study, published in this issue of TRANSFUSION.24
Immunohematology
The pattern of serologic reactivity with the major RhCE antigens confirmed the haplotype association for known RHCE alleles and allowed to identify the probable haplotypes for the 10 new RHCE alleles (Table 5). The RHCe(S122P) allele in the Ce haplotype had diminished C and e antigens but was not found to encode RH53 (JAHK), although RH53 is known to be encoded by the RHCe(S122L) allele in the Ce haplotype with significantly diminished antigens C and e.19
TABLE 5.
Serologic characteristics of samples with RHCE alleles
| Trivial name or allele | Rhesus antigen expression* |
RHD zygosity | Phenotype† | Probable variant haplotype |
Number of probands |
||||
|---|---|---|---|---|---|---|---|---|---|
| D | C | c | E | e | |||||
| RHce(5′-UTR-10C>T) | ++++ | − | ++++ | ++++ | ++ | Dd | DccEe | dce | 1 |
| RHcE(R10W) | ++++ | ++++ | +++/++ | +++/+ | ++++ | DD | DCcEe | DcE | 1 |
| ce(C48) | ++++ | − | ++++ | ++++ | ++/+ | Dd | DccEe | dce | 1 |
| ++++ | ++++ | ++++ | ++++ | ± | DD | DCcEe | Dce | 1 | |
| CX | ++++ | ++++/+++ | ++++ | ++++ | ++++/+++ | DD | DCxCcEe | DCxe | 1 |
| CW | ++++ | ++++/+++ | ++++ | ++++ | ++++ | Dd | DCwCcEe | DCwe | 1 |
| RHce(R114Q)‡ | ++++ | − | ++++ | ++++ | + | Dd | DccEe | dce | 1 |
| RHCe(L115R) | ++++ | ++/− | ++++ | − | ++++ | DD | DCcee | DCe | 1 |
| RHcE(L115P) | ++++ | ++++ | +++/++ | +++/− | ++++ | DD | DCcEe | DcE | 1 |
| RHCe(S122P) | ++++ | ++++/+++ | ++++ | ++++ | +++/++ | DD | DCcEe | DCe | 1 |
| ++++ | ++++/+++ | ++++ | − | ++++ | Dd | DCcee | DCe | ||
| RHcE(I125N) | ++++ | ++++ | ++ | +/− | ++++ | DD | DCcEe | DcE | 2 |
| RHCe(IVS3-5G) | ++++ | +++ | ++++ | ++++ | + | DD | DCcEe | DCe | 1 |
| ++++ | +++ | ++++ | − | ++++ | Dd | DCcee | DCe | ||
| RHCe(H166L) | ++++ | ++/− | ++++ | − | ++++ | DD | DCcee | DCe | 1 |
| cat El [Ew] | ++++ | − | ++++ | +++/+ | ++++ | DD | DccEe | DcE | |
| RHcE(L169Q) | ++++ | ++++ | ++++ | ++++/++ | ++++ | DD | DCcEe | DcE | 1 |
| cat EIV [RHcE(R201T)] | ++++ | − | ++++ | ++++/++ | ++++ | Dd | DccEe | DcE | 4 |
| ++++ | ++++ | ++++ | ++++/++ | ++++ | DD | DCcEe | DcE | 5 | |
| RHce(W217R) | ++++ | − | ++++ | ++++ | +++/++ | Dd | DccEe | dce | |
| RHCE(T241I) | ++++ | ++/+ | ++++ | +++/− | ++++ | Dd | DCcEe | DCE | |
| RHCe(M267K) | ++++ | +++/+ | ++++ | − | ++++ | Dd | DCcee | DCe | |
| RHCe(L297P) | ++++ | ++++/++ | ++++ | ++++ | ++/± | DD | DCcEe | DCe | |
| RHcE(L303Q) | ++++ | ++++ | ++ | ++ | ++++ | DD | DCcEe | DcE | 1 |
| RHCeVA | ++++ | +++ | ++++ | − | ++++ | Dd | DCcee | DCe | 1 |
| RHCeVF | ++++ | ++++/+++ | ++++ | − | ++++ | Dd | DCcee | DCe | 1 |
| Regular RHCE | ++++ | ++++/+++ | ++++ | ++++ | ++++ | Dd | DCcEe | DCE | 3 |
| ++++ | +++ | ++++ | ++++ | − | DD | DCcEE | DCE | 1 | |
| Regular RHCe | ++++ | ++++/++ | ++++ | ++++ | +++/± | DD | DCcEe | DCe | 3 |
| ++++ | ++++/+++ | ++++ | ++++ | ++++/+++ | Dd | DCcEe | DCe | 1 | |
| Regular RHce | ++++ | ++++ | ++++/+++ | − | ++++ | DD | DCcee | Dce | 1 |
Some reactions patterns, indicated by two agglutination strengths like +++/−, varied among different undiluted routine monoclonal and polyclonal antisera in gel matrix antiglobulin technique.
Antigens that are underlined indicate weak or absent reactions with some undiluted routine monoclonal antisera.
RHce(R114Q) represents a second sample and independent observation of the same allele reported by Hustinx et al.32 It carried W16C heterozygously and lacked L245V. Although RHCE Exon 3 sequences were deposited only, both samples of the initial documentation of the molecular bases of JAL in 2003 were heterozygous for C48 (GenBank Accession Number AJ548431 Ce JAL+ for RHCe(R114W)31,32 and AJ548432 ce JAL+ for RHce(R114Q)32); heterozygous W16C was expected for Ce JAL+ but unusual for ce JAL+.
Population frequencies
We devised a multiplex PCR-SSP method for detection of 17 of the 20 rare RHCE alleles with single-nucleotide polymorphisms (Table 6). The variant alleles ce(C48), CX, and CW were not included. This multiplex PCR facilitated the rapid screening of DNA samples (Fig. 1). We validated our multiplex PCR-SSP method by testing of the applicable DNA samples (Table 5), which corroborated their nucleotide sequencing data (data not shown). All 1344 blood donor samples of the GerBSDNA bank,22 which represented regular RhCE phenotypes (Table 2), were negative for any of the tested RHCE alleles.
TABLE 6.
Multiplex PCR-SSP typing for rare RHCE alleles
| Multiplex primer mix* | Allele specificity | Primers | PCR product size (bp) | |
|---|---|---|---|---|
|
| ||||
| Forward | Reverse | |||
| A | RHce(−10C>T) | F-10T | RIVS1 | 341 |
| RHcE(R10W) | F28T | 303 | ||
| B | RHce(R114Q) | F341A | R455C | 155 |
| RHcE(L115P) | F344C | 152 | ||
| RHCe(L115R) | F344G | 152 | ||
| RHCe(S122P) | F364C | 131 | ||
| RHcE(I125N) | F374A | 122 | ||
| C | RHCe(IVS3-5G) | FIVS3-5G | RIVS4 | 371 |
| RHCe(H166L) | F497T | 355 | ||
| RHcE(M167K) | F500A | 352 | ||
| RHcE(L169Q) | F506A | 343 | ||
| RHcE(R201T) | F602C | 248 | ||
| D | RHce(W217R) | F649C | RIVS5 | 291 |
| RHCE(T241I) | F722T | 221 | ||
| RHCe(M267K) | F800A | 141 | ||
| E | RHCe(L297P) | FIVS5 | R890G | 254 |
| RHcE(L303Q) | R908T | 274 | ||
Each multiplex primer mix contained primers HBB-F and HBB-R to amplify a 536-bp fragment of the HBB gene as an internal control.
Fig. 1.
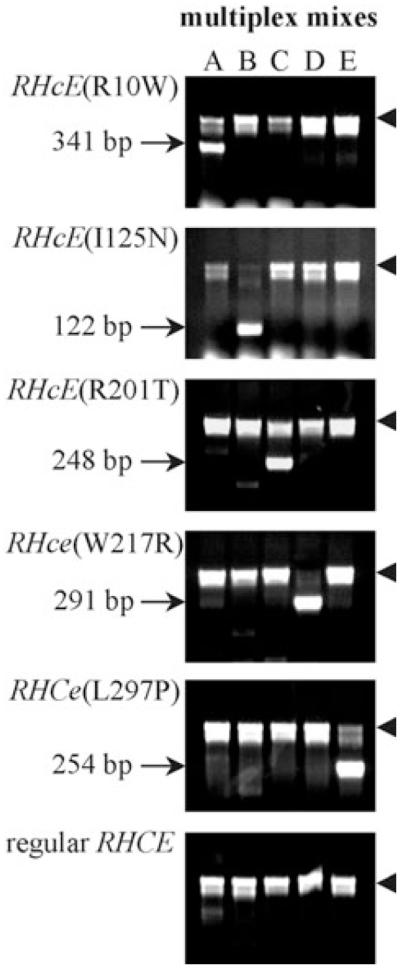
Multiplex PCR for rare RHCE alleles. Representative results are shown for the multiplex PCR-SSP typing of samples with five rare RHCE alleles and a sample with the regular RHCE genotype that is positive for antigens CcEe. The multiplex PCR-SSP reactions in the lanes comprised the Primer Mixes A to E (see Table 6). In all PCR-SSP procedures a 536-bp fragment of the HBB gene was coamplified as internal control (arrowhead).
The frequency estimate for each rare RHCE allele is less than 1 in 488 donors (0.2%; upper limit of 95% confidence interval, one-sided Poisson distribution). Because none of the 17 rare RHCE alleles in our assay (Table 6) were found in the 1344 samples, even the cumulative population frequency estimate for these rare RHCE alleles may be less than 1 in 488. The documented cumulative phenotype frequency was approximately 1 in 8449 (0.012%) for the RhCE variants detected in our serologic routine testing.
Samples carrying the regular RHCE allele
The DCcEe and DccEE phenotypes may be encoded by DCE/dce and DCE/cDE genotypes, respectively. The DCE haplotype is known to be associated with a diminished C antigen.25-27 Four samples with regular RHCE alleles had diminished C but, if any, a normal e antigen; all four samples could be explained by a DCE haplotype, as shown by RHD zygosity analysis (Table 5). With this observation we confirmed that this long recognized DCE haplotype is composed of a regular RHD and a regular RHCE allele with normal exon and splice site nucleotide sequences. Five samples also harbored regular RHCE alleles but carried diminished C antigen or e antigen or both, which cannot be explained by the mechanism of this DCE haplotype. Our study allowed identifying these unusual samples for further investigation toward their underlying molecular cause(s) that did not fit any known mechanism.
DISCUSSION
We identified 43 samples with variant RhCE phenotypes in a serologic routine setting. Among these, 34 carried variant RHCE alleles detectable by nucleotide sequencing of the 10 RHCE exons. Ten of the encountered alleles have not been published previously. Nine samples showed regular RHCE nucleotide sequences within the 10 exons and their adjacent intron regions. The occurrence of the DCE haplotype underlay the diminished C antigen in four samples with regular RHCE alleles, which left five samples in our study carrying a regular RHCE allele whose molecular basis for variant RhCE expression remained unexplained. In contrast to RHCE, a diminished D antigen expression is rarely associated with an apparently regular RHD allele.21,28
The proportion of novel RHCE alleles was unexpected. A large number of alleles have been discovered in previous population screens. Furthermore, a parallel study in Northern Germany24 revealed a similar proportion of novel alleles, but barely half of them were congruent with the alleles found in this study in Southwestern Germany. Our results otherwise correlated well with the observations in Northern Germany (Table 4). The differing Ew phenotype frequencies may be explained by the use of distinct serologic routine methods or by actual frequency variations.
Because most samples were from Caucasian donors and patients, two approaches allowed us to gauge the prevalence of variant RHCE alleles in the Southwestern German population. The calculated cumulative frequency of approximately 1 in 8500 (0.012%) may well represent a lower limit for variant RhCE phenotypes in the population. The cumulative frequency of 1 in 488 (0.20%) was the estimated upper limit for the distinct RHCE alleles as determined by screening 1344 samples of a DNA bank. The molecular screening that we applied was more powerful than our serologic routine screen, because the rare alleles may be detected in heterozygous fashion even when the variant phenotype is masked by a regular RhCE antigen. Almost all identified RHCE alleles will neither be novel mutations nor limited to isolated cases or families. Hence, the presented multiplex PCR tool (Table 6 and Fig. 1) may be used for population screening.
The accumulated evidence indicates a huge variety of RHCE alleles in populations, much of which remains to be explored. Although data on definite frequencies for variant RHCE alleles are missing to date, the available estimates are surprisingly similar to population frequencies of sporadically occurring alleles in other blood group genes. For example, the Bombay allele frequency was estimated to be 1 in 347 (0.28%) by a population genetics approach.29 For the D antigen, the cumulative phenotype frequency of weak D is approximately 1 in 227 (0.44%)23 and the most prevalent weak D type 1 allele frequency is approximately 1 in 277 (0.29%).21
It will be interesting to explore whether the variant RhCE proteins express novel low-prevalence antigens, which may cause immunizations in transfusion recipients. Examples of variant RhCE proteins are known that express low-prevalence antigens of appreciable immunogenicity. Cw,4 Cx,4 Ew,14 JAHK,19 Crawford,16 and JAL30-32 antigens are all caused by single-amino-acid substitutions in the RhCE protein. For example, the S122L substitution (365C>T) in the Ce haplotype encodes the JAHK antigen and is associated with a suppression of the C and e antigens. An RhCE protein with the S122P substitution (364T>C) in the Ce haplotype may lack JAHK expression and is associated with a suppression of the C and e antigens (Table 5).
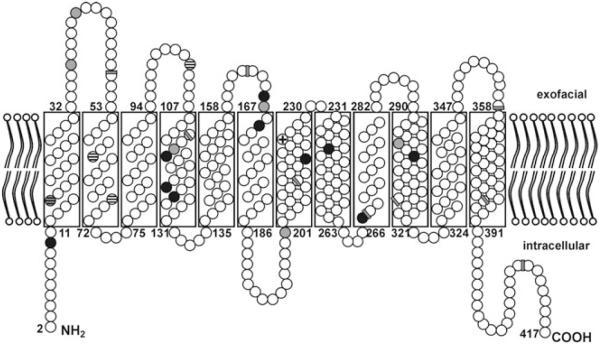
The mechanisms of diminished antigen expression may include conformational changes in the RhCE protein, missplicing of the RHCE gene transcript, and reduced translation. Similar to RhD, single-amino-acid substitutions were the most frequent molecular bases of diminished RhCE expression. While in RhD variants RHD Exons 5 to 7 are predominantly affected,33 we found many RhCE variants with single-nucleotide substitutions in RHCE Exons 1, 3, and 4 (Table 4). In 12 of 18 observed alleles, the single-amino-acid substitution were located in the transmembraneous sections of the RhCE protein (Table 4 and Fig. 2). Our samples with unresolved molecular bases exemplified that additional molecular mechanisms exist and can be addressed now, for example, using the samples identified in the current study.
Fig. 2.
Model of the RhCE protein topology in the RBC membrane. Amino acids are depicted as circles. Four amino acid positions differ between C and c antigens (horizontally striped circles) and one amino acid differs between E and e antigens (crossed circle). Previously known amino acid substitutions (gray circles) and novel variants identified in this study (black circles) are predominantly located within the N-terminal half of the protein. The 9 exon boundaries in the RHCE cDNA, as reflected in the amino acid sequence, are indicated (gray bars).
Three RhD and three RhCE variants harbor amino acid substitutions at positions 114 and 115, which are located in the transmembraneous section of Helix 4 (Fig. 2). The three RhD variants are weak D Type 17 (114W), weak D Type 47 (114G), and weak D Type 25 (114Q) with weakened D antigen expression. Both 114W and 114Q substitutions are known to be associated with JAL antigen expression,30-32 which has been documented so far in RhCE only. The L115R substitution introduces a basic residue into the putatively hydrophobic transmembraneous segment and the L115P substitution introduces proline, which is known to disturb the regular helix structure. Further examples observed by us for disruption of helical structures by proline are the substitutions S122P and L297P.
At the extracellular RhD vestibule several amino acid substitutions cause antigen loss and permit anti-D immunization.34 Similarly, the amino acid substitutions H166L, M167K, and L169Q in RhCE are predicted to lie at the extracellular RhCE protein vestibule and the corresponding RhCE variants likely represent “partial RhCE.” The partial D variant DFW (H166P) also carried an amino acid substitution at Position 166.35 We confirmed that the M167K amino acid substitution in cat EI (Ew) is associated with a weakened expression of antigen E but with a normal antigen c,13 which was also observed for the novel L169Q substitution.
Single-nucleotide substitutions in the promoter region or in proximity to splice sites can influence protein translation. First, RHce(5′-UTR-10C>T) may be explained by inhibiting ribosomal binding to the mRNA, which would decrease translation. The second mechanism may be exemplified by RHCe(IVS3-5G) rendering splicing less efficient, which reduces protein expression representing a quantitative effect, or causing missplicing of the transcript, which leads to an altered protein representing a qualitative effect. Although the nucleotide substitution in RHCe(M267K) lies at the splice site it is unlikely to affect the splicing process, because it represents a templated mutation, that is, deriving from the normal RHD gene in which it is not known to affect splicing.
The rare RhCE phenotypes observed in Southwestern Germany do not frequently permit a clinically relevant immunization in their carriers. While the humoral immune response in carriers of RhCE variants appears almost always to be left mute, the identification of rare RhCE variants enables to examine the possible cellular immune response in the carriers when they were transfused with blood harboring the regular RhCE protein.
Most of the RhD and RhCE protein is integrated into the RBC membrane and also most of the amino acid substitutions occur in this transmembrane part. Similar to weak D caused by substitutions in the transmembraneous section, the number of examples for such weak CE is increasing. Substitutions in the much smaller extracellular parts of these proteins are inherently rarer but make the carrier of the variant prone to immunization. Hence, the consequences with respect to immunization are expected to be the same for D and CE: as weak D, most weak CE will have no consequence for the carrier while carriers of partial D and partial CE can get immunized against the normal protein. Although the discrimination between weak and partial will never be sharp, the predominant amount of alleles confers to this scheme which is of great help to the practically oriented physician. A considerable amount of information has been collected with regard to the immunization of individuals with partial D; comparable information is now accumulating for CE.
ACKNOWLEDGMENTS
We thank Franz F. Wagner for analyzing samples in Ulm until 2003. We acknowledge the technical assistance of Gabi Rink in Mannheim, Kathrin Panter and Andrea Ernst in Baden-Baden, and Marianne Lotsch in Ulm.
This work was supported by intramural grants from the DRK-Blutspendedienst Baden-Württemberg–Hessen.
ABBREVIATIONS
- SSP
sequence-specific priming
- UTR
untranslated region
Footnotes
Authorship contribution: PB designed the study, performed molecular analysis, interpreted data, and wrote the first draft of the paper; EAS designed the study and collected and serologically characterized blood donor samples; CG collected and serologically characterized patient samples; IvZ serologically and molecularly characterized patient and blood donor samples and analyzed and interpreted data; and WAF designed the study, collected samples, interpreted data, and wrote the paper.
CONFLICT OF INTEREST
The authors declare no competing interests relevant to this article.
REFERENCES
- 1.Daniels GL, Castilho L, Flegel WA, Fletcher A, Garratty G, Levene C, Lomas-Francis C, Moulds JJ, Moulds JM, Olsson ML, Overbeeke MA, Poole J, Reid ME, Rouger P, van der Schoot CE, Scott M, Sistonen P, Smart E, Storry JR, Tani Y, Yu L-C, Wendel S, Westhoff CM, Yahalom V, Zelinski T. International Society of Blood Transfusion Committee on terminology for red cell surface antigens: Macao report. Vox Sang. 2009;96:153–6. doi: 10.1111/j.1423-0410.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 2.Wagner FF, Flegel WA. Review: the molecular basis of the Rh blood group phenotypes. Immunohematol. 2004;20:23–36. [PMC free article] [PubMed] [Google Scholar]
- 3.Ansart-Pirenne H. Stratégie d’identification des variants du gène RHCE au Centre national de référence pour les groupes sanguins: impact sur la sécurité transfusionelle [Identification strategy of RHCE gene variants at the National Blood Group Reference Laboratory: impact on transfusion safety] Transfus Clin Biol. 2006;13:13–8. doi: 10.1016/j.tracli.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Mouro I, Colin Y, Sistonen P, Le Pennec PY, Cartron JP, Le Van Kim C. Molecular basis of the RhCw (Rh8) and RhCx (Rh9) blood group specificities. Blood. 1995;86:1196–201. [PubMed] [Google Scholar]
- 5.Beckers EA, Faas BH, von dem Borne AE, Overbeeke MA, van Rhenen DJ, van der Schoot CE. The R0Har Rh:33 phenotype results from substitution of exon 5 of the RHCE gene by the corresponding exon of the RHD gene. Br J Haematol. 1996;92:751–7. doi: 10.1046/j.1365-2141.1996.382918.x. [DOI] [PubMed] [Google Scholar]
- 6.Daniels GL, Faas BH, Green CA, Smart E, Maaskant-van Wijk PA, Avent ND, Zondervan HA, von dem Borne AE, van der Schoot CE. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion. 1998;38:951–8. doi: 10.1046/j.1537-2995.1998.381098440860.x. [DOI] [PubMed] [Google Scholar]
- 7.Faas BH, Beckers EA, Wildoer P, Ligthart PC, Overbeeke MA, Zondervan HA, von dem Borne AE, van der Schoot CE. Molecular background of VS and weak C expression in blacks. Transfusion. 1997;37:38–44. doi: 10.1046/j.1537-2995.1997.37197176949.x. [DOI] [PubMed] [Google Scholar]
- 8.Noizat-Pirenne F, Mouro I, Le Pennec PY, Ansart-Pirenne H, Juszczak G, Patereau C, Verdier M, Babinet J, Roussel M, Rouger P, Cartron JP. Two new alleles of the RHCE gene in Black individuals: the RHce allele ceMO and the RHcE allele cEMI. Br J Haematol. 2001;113:672–9. doi: 10.1046/j.1365-2141.2001.02802.x. [DOI] [PubMed] [Google Scholar]
- 9.Noizat-Pirenne F, Lee K, Pennec PY, Simon P, Kazup P, Bachir D, Rouzaud AM, Roussel M, Juszczak G, Menanteau C, Rouger P, Kotb R, Cartron JP, Ansart-Pirenne H. Rare RHCE phenotypes in black individuals of Afro-Caribbean origin: identification and transfusion safety. Blood. 2002;100:4223–31. doi: 10.1182/blood-2002-01-0229. [DOI] [PubMed] [Google Scholar]
- 10.Rouillac C, Gane P, Cartron JP, Le Pennec PY, Colin Y. Molecular basis of the altered antigenic expression of RhD in weak D (Du) and RhC/e in RN phenotypes. Blood. 1996;87:4853–61. [PubMed] [Google Scholar]
- 11.Noizat-Pirenne F, Le Pennec PY, Mouro I, Rouzaud AM, Juszczak G, Roussel M, Lauroua P, Krause C, Rouger P, Cartron JP, Ansart-Pirenne H. Molecular background of D(C)(e) haplotypes within the white population. Transfusion. 2002;42:627–33. doi: 10.1046/j.1537-2995.2002.00097.x. [DOI] [PubMed] [Google Scholar]
- 12.Westhoff CM, Silberstein LE, Wylie DE, Skavdahl M, Reid ME. 16Cys encoded by the RHce gene is associated with altered expression of the e antigen and is frequent in the R0 haplotype. Br J Haematol. 2001;113:666–71. doi: 10.1046/j.1365-2141.2001.02803.x. [DOI] [PubMed] [Google Scholar]
- 13.Noizat-Pirenne F, Mouro I, Gane P, Okubo Y, Hori Y, Rouger P, Le Pennec PY, Cartron JP. Heterogeneity of blood group RhE variants revealed by serological analysis and molecular alteration of the RHCE gene and transcript. Br J Haematol. 1998;103:429–36. doi: 10.1046/j.1365-2141.1998.01004.x. [DOI] [PubMed] [Google Scholar]
- 14.Strobel E, Noizat-Pirenne F, Hofmann S, Cartron JP, Bauer MF. The molecular basis of the Rhesus antigen Ew. Transfusion. 2004;44:407–9. doi: 10.1111/j.1537-2995.2004.00655.x. [DOI] [PubMed] [Google Scholar]
- 15.Noizat-Pirenne F, Mouro I, Le Pennec PY, Roussel M, Boulard P, Rouger P, Cartron J. Molecular basis of category EIV variant phenotype [abstract] Transfusion. 1999;39:103S. [Google Scholar]
- 16.Flegel WA, Wagner FF, Chen Q, Schlanser G, Frame T, Westhoff CM, Moulds MK. The RHCE allele ceCF: the molecular basis of Crawford (RH43) Transfusion. 2006;46:1334–42. doi: 10.1111/j.1537-2995.2006.00901.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen Q, Hustinx H, Flegel WA. The RHCE allele ceSL: the second example for D antigen expression without D-specific amino acids. Transfusion. 2006;46:766–72. doi: 10.1111/j.1537-2995.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- 18.Vege S, Lomas-Francis C, Kosanke J, Westhoff CM. The RHCE*ceSL allele encodes a weak e antigen [abstract] Transfusion. 2007;47:160A. [Google Scholar]
- 19.Scharberg EA, Green C, Daniels G, Richter E, Klüter H, Bugert P. Molecular basis of the JAHK (RH53) antigen. Transfusion. 2005;45:1314–8. doi: 10.1111/j.1537-2995.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- 20.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion. 2003;43:1248–54. doi: 10.1046/j.1537-2995.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 21.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood. 1999;93:385–93. [PubMed] [Google Scholar]
- 22.Erdmann J, Großhennig A, Braund PS, König IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet D-A, Cambion F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O’Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schäfer A, März W, Renner W, Bugert P, Klüter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Italian Atherosclerosis, Thrombosis, and Vascular Biology Working Group; Myocardial Infarction Genetics Consortium; Wellcome Trust Case Control Consortium; Cardiogenics Consortium. Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H. Novel susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–2. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner FF, Kasulke D, Kerowgan M, Flegel WA. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in South-Western Germany. Infusionsther Transfusionsmed. 1995;22:285–90. doi: 10.1159/000223144. [DOI] [PubMed] [Google Scholar]
- 24.Doescher A, Vogt C, Bittner R, Gerdes I, Petershofen EK, Wagner FF. RHCE alleles detected in automated RH blood grouping in Northern Germany. Transfusion. doi: 10.1111/j.1537-2995.2009.02221.x. DOI: 10.1111/j.1537-2995.2009.02221.x. [DOI] [PubMed] [Google Scholar]
- 25.Murray J, Race RR, Taylor GL. Serological reactions caused by the rare human gene Rhz. Nature. 1945;155:112–3. [Google Scholar]
- 26.Race RR, Sanger R, Lawler SD. Rh genes allelomorphic to C. Nature. 1948;161:316. doi: 10.1038/161316a0. [DOI] [PubMed] [Google Scholar]
- 27.Race RR, Sanger R, Lawler SD. The Rh antigen called c-v: a revocation. Vox Sang. 1960;5:334–6. doi: 10.1111/j.1423-0410.1960.tb03747.x. [DOI] [PubMed] [Google Scholar]
- 28.von Zabern I, Flegel WA. IVS5-38del4 deletion in the RHD gene does not cause a DEL phenotype: relevance for RHD alleles including DFR-3. Transfusion. 2007;47:1552–5. doi: 10.1111/j.1537-2995.2007.01353.x. [DOI] [PubMed] [Google Scholar]
- 29.Wagner FF, Flegel WA. Polymorphism of the h allele and the population frequency of sporadic nonfunctional alleles. Transfusion. 1997;37:284–90. doi: 10.1046/j.1537-2995.1997.37397240210.x. [DOI] [PubMed] [Google Scholar]
- 30.Ong J, Walker PS, Schmulbach E, Storry JR, Vege S, Westhoff C, Lomas-Francis C, Reid ME. Alloanti-c in a c-positive, JAL-positive patient. Vox Sang. 2009;96:240–3. doi: 10.1111/j.1423-0410.2008.01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westhoff CM, Vege S, Wylie D, Nickle P, Lomas-Francis C, Hue-Roye K, Reid ME. The JAL antigen (RH48) is the result of a change in RHCE that encodes Arg114Trp. Transfusion. 2009;49:725–32. doi: 10.1111/j.1537-2995.2008.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hustinx H, Poole J, Bugert P, Gowland P, Still F, Fontana S, Scharberg EA, Tilley L, Daniels G, Niederhauser C. Molecular basis of the Rh antigen RH48 (JAL) Vox Sang. 2009;96:234–9. doi: 10.1111/j.1423-0410.2008.01142.x. [DOI] [PubMed] [Google Scholar]
- 33.Wagner FF. The RhesusBase. DRK-Blutspendedienst Baden-Württemberg—Hessen; Ulm: 1998-2009. [Google Scholar]
- 34.Flegel WA, von Zabern I, Doescher A, Wagner FF, Vytiskova J, Pisacka M. DCS-1, DCS-2 and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion. 2008;48:25–33. doi: 10.1111/j.1537-2995.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 35.Flegel WA, von Zabern I, Doescher A, Wagner FF, Strathmann KP, Geisen C, Palfi M, Pisacka M, Poole J, Polin H, Gabriel C, Avent ND. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion. 2009;49 doi: 10.1111/j.1537-2995.2009.02102.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]