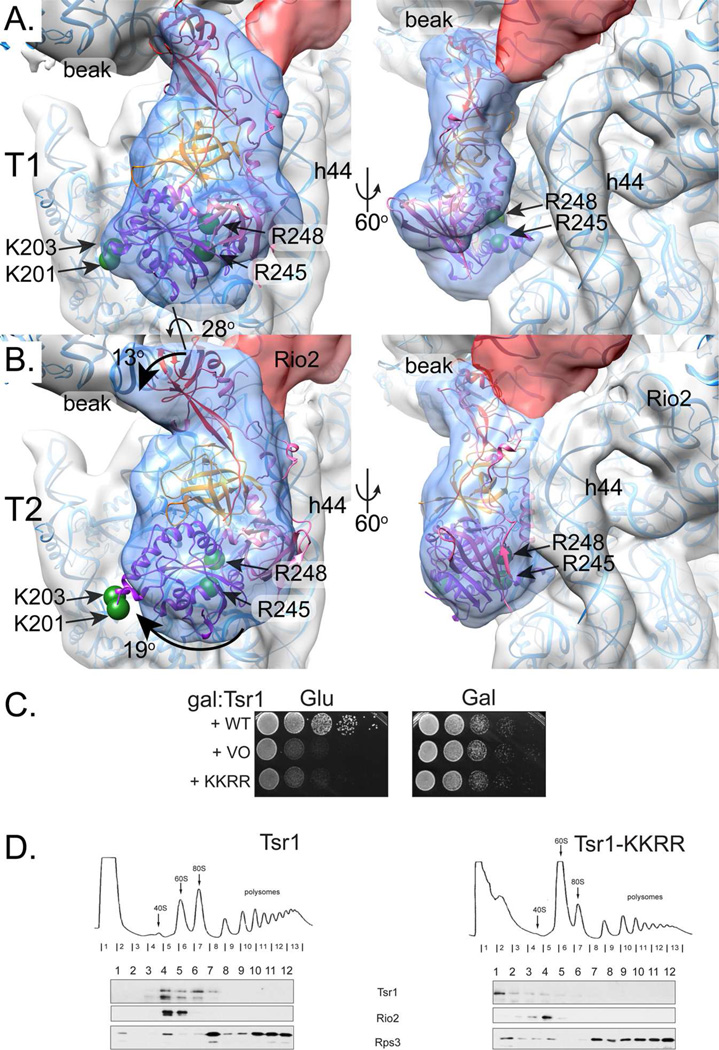
Fig. 2. Tsr1 adopts two discrete conformations.
A. Local 3D classification revealed density shaped like Tsr1 (transparent blue) rotated away from h44 (subclass T1). Tsr1’s X-ray crystal structure (PDB 5IW7,(McCaughan et al., 2016) fit as a rigid body, positioning the four amino acids that are essential for its pre-40S binding and in vivo function on the ribosome-binding face of Tsr1 (two green spheres represent Cα atom position of R245 and R248) and at the edge of the molecule, far from h44 (two green spheres represent Cα atom position of K201 and K203). Tsr1 domains are colored per McCaughan et al., 2016: Tsr1I is purple, Tsr1II is pink, Tsr1III is orange, and Tsr1IV is red. MDFF-relaxed rRNA and 40S ribosomal proteins are shown as blue ribbons to clarify the interpretation of the pre-40S structural features. See also Figure S2.
B. A second subclass of Tsr1 density, T2, shows Tsr1 rotated toward h44 by about 28° but the shape of the density did not correspond as well to the crystal structure. Rotation of Tsr1I, Tsr1II, and Tsr1IV relative to Tsr1III, colored as in A, would be needed to adjust Tsr1 from its T1-form. Arrows demark these suggested movements and the relevant amino acids are represented as in A. Angles were estimated by independently positioning each domain into the density corresponding to that feature and then calculating the angle relative to the rigid body fit with the “measure rotation” function in Chimera (Pettersen et al., 2004).
C. Growth of galactose-inducible/glucose-repressible Tsr1 (GAL::Tsr1) yeast cells carrying an empty vector (VO), or vectors coding wild-type (WT) or Tsr1_KKRR, is compared on glucose and galactose-containing plates.
D. Sucrose gradients of cell extracts from GAL::Tsr1 cells transformed with vectors carrying WT-Tsr1 or Tsr1_KKRR, grown in glucose for 16 h. Shown are absorbance profiles at 254 nm and Western blots for Tsr1, Rio2 and Rps3.