Abstract
Background and Aims Studies in the carnivorous family Lentibulariaceae in the last years resulted in the discovery of the smallest plant genomes and an unusual pattern of genomic GC content evolution. However, scarcity of genomic data in other carnivorous clades still prevents a generalization of the observed patterns. Here the aim was to fill this gap by mapping genome evolution in the second largest carnivorous family, Droseraceae, where this evolution may be affected by chromosomal holokinetism in Drosera.
Methods The genome size and genomic GC content of 71 Droseraceae species were measured by flow cytometry. A dated phylogeny was constructed, and the evolution of both genomic parameters and their relationship to species climatic niches were tested using phylogeny-based statistics.
Key Results The 2C genome size of Droseraceae varied between 488 and 10 927 Mbp, and the GC content ranged between 37·1 and 44·7 %. The genome sizes and genomic GC content of carnivorous and holocentric species did not differ from those of their non-carnivorous and monocentric relatives. The genomic GC content positively correlated with genome size and annual temperature fluctuations. The genome size and chromosome numbers were inversely correlated in the Australian clade of Drosera.
Conclusions Our results indicate that neither carnivory (nutrient scarcity) nor the holokinetism have a prominent effect on size and DNA base composition of Droseraceae genomes. However, the holokinetic drive seems to affect karyotype evolution in one of the major clades of Drosera. Our survey confirmed that the evolution of GC content is tightly connected with the evolution of genome size and also with environmental conditions.
Keywords: DNA content, Droseraceae, carnivorous plants, flow cytometry, genome size evolution, GC content, DNA base composition, holocentric chromosomes, holokinetic chromosomes
INTRODUCTION
Droseraceae consists of three carnivorous genera, two of which are monotypic and equipped with highly specialized snap-traps: Dionaea muscipula from the wetlands of North and South Carolina (USA); and Aldrovanda vesiculosa, an aquatic species with scattered distribution in Africa, Australia and Eurasia. The third genus, Drosera (sundews), includes approx. 250 sticky-leaved species distributed across all the continents except for Antarctica (McPherson, 2010; Gonella et al., 2015). Sundews generally grow in wetlands, but some are adapted to seasonal droughts, especially the species from Australia (McPherson, 2008, 2010).
Flowering plants (Angiosperms) exhibit an extremely broad divergence in genome size compared with other Eukaryotes (Bennett, 1972). For instance, the difference between the largest and smallest angiosperm genome is > 2500-fold (Bennett and Leitch, 2012). This variation is considered to be the result of different selective pressures (ecological, physiological, morphological, etc.) on the outcomes of molecular processes (retrotransposon amplification, polyploidy), which vary in their degree across various angiosperm clades (Wendel et al., 2013). The smallest angiosperm genomes are known from the carnivorous family Lentibulariaceae (Greilhuber et al., 2006; Fleischmann et al., 2014; Veleba et al., 2014), making these miniature carnivorous species excellent candidates for whole-genome sequencing. Indeed, complete genomic sequences have already been published for Utricularia gibba (Ibarra-Laclette et al., 2013), Genlisea aurea (Leushkin et al., 2013), G. nigrocaulis and G. hispidula (Vu et al., 2015). Unlike Lentibulariaceae, the other prominent group of carnivorous plants, Droseraceae, has been analysed only sporadically, and the genome size is known for only nine of approx. 250 existing Droseraceae species (Rothfels and Heimburg, 1968; Veselý et al., 2012; Jensen et al., 2015). The reported genome sizes (2C = 587 Mbp in Drosera capensis to 2C = 5912 Mbp in Dionaea muscipula) seem to be generally larger than in Lentibulariaceae (2C = 126 Mbp in Genlisea aurea to 2C = 3020 Mbp in Genlisea hispidula; Greilhuber et al., 2006) but still relatively small compared with genome sizes known in other angiosperms (Bennett and Leitch, 2012). Given the small number of analysed species and other characteristics noted below, it cannot be excluded that this family may still hide species with similarly miniaturized genomes as in the carnivorous family Lentibulariaceae.
It has been hypothesized that selection for small genome sizes may be promoted by nutrient limitation, namely by phosphorus and nitrogen (Leitch and Leitch, 2008), because both are abundant components of nucleic acids (Sterner and Elser, 2002). Carnivory is considered an adaptation to nutrient-poor habitats (Givnish et al., 1984), and carnivorous plants could, therefore, act as suitable models to test this hypothesis by comparing the genome sizes of carnivorous species and closely related non-carnivorous clades. Indeed, the predicted decrease in genome size has been observed together with the evolution/appearance of carnivory in Lentibulariaceae (Veleba et al., 2014); however, studies on other carnivorous clades are necessary to generalize this trend.
Possibly, the major peculiarity of Droseraceae compared with other carnivorous lineages (including Lentibulariaceae) is its holokinetic chromosomes, which are typical for Drosera species (Rothfels and Heimburg, 1968; Sheikh et al., 1995; Kondo and Nontachaiyapoom, 2008; Shirakawa et al., 2011a, b; Zedek et al., 2016) with a possible exception of D. regia (Shirakawa et al., 2011b). In contrast to monocentric chromosomes, whose kinetochore formation is restricted to the small areas of the centromeres, holokinetic chromosomes lack primary constrictions and their kinetochores are formed along their poleward surfaces (Bureš et al., 2013; Cuacos et al., 2015). Holokinetic chromosomes, therefore, tolerate chromosomal fissions or fusions and do not allow more than two crossovers in meiosis (reviewed in Bureš et al., 2013; Heckmann and Houben, 2013) which may substantially affect genome and karyotype evolution of their bearers (Escudero et al., 2012; Bureš et al., 2013; Bureš and Zedek, 2014; Lukhtanov et al., 2015; Šíchová et al., 2016). One such effect may be a negative correlation between genome size and chromosome number in holokinetic lineages (Nishikawa et al., 1984; Roalson et al., 2007; Záveská Drábková and Vlček, 2010; Bureš et al., 2013; Lipnerová et al., 2013; Bureš and Zedek, 2014). Based on the comparison of four holokinetic clades (cyperids, Drosera, Chionographis and Myristica) with their close monocentric relatives, Bureš et al. (2013) suggested that holokinetism might be associated with genome size decrease. This association was later confirmed for the cyperid clade with a larger data set and phylogeneticaly corrected analyses by Šmarda et al. (2014) who also found a decreased overall genomic percentage of guanine and cytosine (GC content) in this clade. However, the extent to which these trends are general outcomes of holokinetism remains unclear because relevant comparisons of these genomic parameters are lacking in other holokinetic clades.
Thus far, the GC content is known only in two Droseraceae species (D. menziesii, 41·3 %; and D. peltata, 44·2 %; Veselý et al., 2012). In general, the GC content is extremely variable, particularly in bacteria, where it is known to relate to the ecology of particular taxa and lineages (correlated with the thermal optimum and thermal tolerance range; Nishio et al., 2003; Foerstener et al., 2005; Musto et al., 2006; Mann and Phoebe-Chen, 2010). Although the variation in GC content is much narrower in flowering plants (Šmarda and Bureš, 2012), its ecological impact has also been found in monocots, in which a higher GC content was found to be correlated with cold and drought tolerance (Šmarda et al., 2014). Droseraceae may serve as a good model for testing some of these predictions on a finer phylogenetic scale, particularly due to the contrasting ecology of Droseraceae species.
In this study, we aim (1) to analyse trends in the genome size and GC content evolution in the family Droseraceae and its close relatives and (2) to test whether the holokinetism in Droseraceae is associated with the predicted effects and patterns in the genome and karyotype evolution, namely (2a) genome downsizing, (2b) decreased GC content and (2c) the existence of a negative correlation between DNA content and chromosome number. Finally, we aim (3) to test the relationship between climatic parameters and GC content on a narrower phylogenetic scale than in our previous analysis across whole monocots (Šmarda et al., 2014).
MATERIALS AND METHODS
Most of the samples of Droseraceae were collected from the private collection of Adam Veleba; several samples originated from collections of other carnivorous plant enthusiasts. The related non-carnivorous plants were obtained from the Botanical Garden of the Faculty of Science, Masaryk University in Brno, or collected in the wild. The genomic data of 17 species were taken from the C-value database (Bennett and Leitch, 2012) and several other sources (for a detailed list, see Supplementary Data Table S1).
The samples for flow cytometry were prepared according to the protocol of Šmarda et al. (2008) and measured on two CyFlow flow cytometers (Partec GmbH, Münster, Germany; recently Sysmex) with internal standards whose genome size was derived from comparison with the completely sequenced Oryza sativa subsp. japonica ‘Nipponbare’ (International Rice Genome Sequencing Project, 2005; Supplementary Data Table S2). Each sample was processed with two fluorochromes: PI (propidium iodide) and DAPI (4',6-diamidino-2-phenylindole). The intercalating, base-unspecific PI was used to determine the absolute genome size, and the AT-selective DAPI, together with the results from measurements with PI, were used to calculate the genomic GC content. The procedure is detailed in Šmarda et al. (2008, 2014); for further details, see the Supplementary Data Methods.
The phylogenetic relationships of the analysed species (listed in Supplementary Data Table S1) were reconstructed based on a concatenated alignment of chloroplast (rbcL and matK) and nuclear (ITS) markers (Supplementary Data Methods). The resulting maximum likelihood phylogenetic tree was calibrated using available fossil records and published age estimates (Supplementary Data Methods). Both non-dated and dated phylogenetic trees in Newick format are supplied in Supplementary Data Fig. S1).
The GIS layer of geographic distribution was prepared for each species based on the distribution data of Droseraceae species in the World Checklist of Selected Plant Families, Kew Databases (Govaerts and Cheek, 2014), using the digitized layers of ‘TDWG areas of level 3’ (sensu Brummitt et al., 2001). The species concept was revised according to the current literature. For each species, the geographical distribution was transformed to the statistical distributions across each of the 19 bioclimatic variables (19 histograms) from the WorldClim database (Hijmans et al., 2005), i e. for each species and a given bioclimatic parameter a histogram was constructed in which the height of each column was given by the area of intersection of the respective bioclimatic GIS (sub-)layer (= sub-range of a given bioclimatic variable) with the GIS layer of geographic distribution of the respective species. Subsequently, the minimum, median and maximum values of the calculated bioclimatic variables were calculated (Supplementary Data Table S3). The precipitation variables were log-transformed prior to all statistical analyses; the temperature variables were used as raw values.
Recent polyploidy events were identified based on a comparison of chromosome numbers taken from the published literature and the measured genome sizes between closely related species (Supplementary Data Table S1). The analyses of genome size evolution were conducted with monoploid genome size (Cx; i.e. total 2C genome size divided by the ploidy level; Greilhuber et al., 2005) instead of the raw measures of DNA content. The monoploid genome size was log10 transformed prior to all statistical analyses; the GC contents and the chromosome numbers were used as raw values.
The statistical tests of the relationships between monoploid genome size, GC content and chromosome numbers were performed using the phylogenetic generalized least-squares method (function ‘pgls’) using the ‘caper’ package (v. 0.5.2; Orme et al., 2012) in R (v. 3·3; R Core Team, 2013) with λ (branch length transformation) determined by maximum likelihood.
The ancestral states of the monoploid genome size and GC content were reconstructed using the residual maximum likelihood method under the Brownian Motion model (function ace in the R package ape v. 3·5; Paradis et al., 2004) and visualized on the phylogenetic tree using the function ‘contMap’ in the R package ‘phytools’ v. 0·5-20 (Revell, 2012). Significant changes of the monoploid genome size or GC content in particular nodes were detected by the random tip-value reshuffling algorithm in R (this procedure compares actual node values with values obtained from random reshuffling of the tip values; Šmarda et al., 2014) based on 4999 randomizations.
The difference between the monoploid genome size of carnivorous and non-carnivorous species and between the monoploid genome size and the GC content of holokinetic and monocentric species was tested by phylogenetic analysis of variance (ANOVA; function ‘aov.phylo’, package ‘geiger’ v. 2.0.6; Harmon et al., 2015).
The relationships between the genomic GC content and climatic variables were analysed with a multiple phylogenetic regression approach using the ‘pgls’ function (package ‘caper’ in R) and λ (branch length transformation) determined by maximum likelihood. In this analysis, the climatic variables were handled as explanatory variables and were manually forward selected into the final explanatory model of GC content based on the amount of explained variation (in each step, the significant variable with the highest explained variation was included in the model). The α-level for this analysis was 8·33E-4, as the Bonferroni correction was applied to avoid false-positive results.
With respect to particular analysed parameters, analyses were performed with the respective sub-sets of data (Datasets 1–6 in Supplementary Table S1).
RESULTS
Variation of genomic parameters in Droseraceae and related clades
The genomes of the 71 analysed Droseraceae species (Table 1; 66 newly reported here) were relatively small, with medians of 1252 Mbp for 2C and 509 Mbp for Cx. The smallest genome was found in Drosera hamiltonii (2C = 488 Mbp, Cx = 244 Mbp), while the absolute largest was detected in the tetraploid D. ordensis (2C = 10 927 Mbp, Cx = 2732 Mbp) and the largest monoploid genome size in D. micrantha (2C = 7489 Mbp, Cx = 3745 Mbp). The genomes of 42 species (Table 1; 17 newly reported here) of related families (Drosophyllaceae, Nepenthaceae, Ancistrocladaceae, Dioncophyllaceae, Plumbag-inaceae, Polygonaceae and Tamaricaceae) varied from the smallest, 2C = 669 Mbp, Cx = 335 Mbp in Plumbago auriculata (Plumbaginaceae), to 2C = 20 833, Cx = 10 416 Mbp in carnivorous Drosophyllum lusitanicum (Drosophyllaceae).
Table 1.
Results of genome size and genomic DNA base composition (GC content) measurements
| Species | 2C (Mbp) | GC (%) | Ploidy level* | Cx (Mbp) |
|---|---|---|---|---|
| Droseraceae | ||||
| Aldrovanda vesiculosa | 938 | 42·8 | 2 | 469 |
| Dionaea muscipula | 5705 | 43·9 | 2 | 2853 |
| Drosera aberrans | 987 | 41·9 | 2 | 494 |
| D. adelae | 594 | 37·6 | 2 | 297 |
| D. admirabilis | 792 | 39·7 | – | – |
| D. afra | 613 | 39·6 | – | – |
| D. aliciae | 1949 | 40·0 | 8 | 244 |
| D. allantostigma | 2858 | 43·5 | 2 | 1429 |
| D. anglica | 4715 | 44·2 | 4 | 1179 |
| D. arcturi | 1050 | 39·7 | 2 | 525 |
| D. auriculata | 846 | 42·3 | 2 | 423 |
| D. barbigera | 4215 | 43·2 | 2 | 2108 |
| D. binata | 1465 | 41·5 | 2 | 733 |
| D. binata var. multifida | 1519 | 41·4 | – | – |
| D. burmannii | 504 | 38·7 | 2 | 252 |
| D. capensis | 789 | 39·0 | 4 | 197 |
| D. cistiflora | 671 | 41·8 | 4 | 168 |
| D. collinsiae | 905 | 40·1 | 4 | 226 |
| D. cuneifolia | 702 | 40·3 | 4 | 176 |
| D. dilatatopetiolaris | 4868 | 42·8 | 2 | 2434 |
| D. erythrorhiza | 1687 | 42·7 | – | – |
| D. falconeri | 5253 | 43·0 | 2 | 2627 |
| D. filiformis | 4877 | 43·5 | 2 | 2439 |
| D. filiformis var. tracyi | 5930 | 42·8 | – | – |
| D. gigantea | 1060 | 40·2 | 2 | 530 |
| D. grantsaui | 1069 | 39·9 | – | – |
| D. graomogolensis | 1629 | 40·4 | 4 | 407 |
| D. hamiltonii | 488 | 40·1 | 2 | 244 |
| D. helodes | 3586 | 43·0 | 2 | 1793 |
| D. hilaris | 738 | 40·9 | 4 | 185 |
| D. indica | 1307 | 40·9 | 2 | 654 |
| D. intermedia | 2516 | 42·5 | 2 | 1258 |
| D. kaieteurensis | 2695 | 41·7 | – | – |
| D. lanata | 854 | 39·0 | 2 | 427 |
| D. latifolia | 1102 | 40·9 | 4 | 276 |
| D. leucoblasta | 4121 | 42·1 | 2 | 2061 |
| D. menziesii | 967 | 40·8 | 2 | 484 |
| D. meristocaulis | 2969 | 38·9 | 2 | 1485 |
| D. micrantha | 7489 | 44·4 | 2 | 3745 |
| D. modesta | 1158 | 40·7 | – | – |
| D. monantha | 776 | 40·3 | – | – |
| D. natalensis | 1040 | 40·8 | 4 | 260 |
| D. neocaledonica | 1136 | 38·1 | 4 | 284 |
| D. nidiformis | 1027 | 40·8 | – | – |
| D. oblanceolata | 1933 | 40·1 | – | – |
| D. omissa | 2170 | 42·2 | 2 | 1085 |
| D. ordensis | 10 927 | 44·2 | 4 | 2732 |
| D. oreopodion | 3292 | 44·7 | – | – |
| D. peltata | 829 | 43·0 | 2 | 415 |
| D. petiolaris | 4707 | 42·2 | 2 | 2354 |
| D. prolifera | 502 | 37·1 | 2 | 251 |
| D. pulchella | 1862 | 43·4 | 2 | 931 |
| D. pygmaea | 1252 | 41·6 | 2 | 626 |
| D. ramentacea | 1361 | 40·8 | – | – |
| D. regia | 835 | 40·2 | 2 | 418 |
| D. roraimae | 2683 | 41·9 | – | – |
| D. roseana | 3513 | 43·3 | 2 | 1757 |
| D. rotundifolia | 2331 | 44·5 | 2 | 1166 |
| D. sessilifolia | 497 | 38·4 | 2 | 249 |
| D. sewelliae | 3863 | 40·4 | 2 | 1932 |
| D. schizandra | 1186 | 40·1 | 2 | 593 |
| D. spatulata | 586 | 38·5 | 2 | 293 |
| D. spiralis | 1259 | 40·4 | 4 | 315 |
| D. tomentosa | 1105 | 40·0 | 4 | 276 |
| D. trinervia | 573 | 40·1 | 4 | 143 |
| D. ultramafica | 2325 | 40·6 | – | – |
| D. venusta | 1054 | 39·8 | – | – |
| D. verrucata | 4653 | 43·5 | 2 | 2327 |
| D. viridis | 2316 | 42·9 | – | – |
| D. whittakeri | 946 | 41·2 | – | – |
| D. zonaria | 889 | 41·5 | – | – |
| Dioncophyllaceae | ||||
| Triphyophyllum peltatum | 1167 | 40·2 | 2 | 584 |
| Drosophyllaceae | ||||
| Drosophyllum lusitanicum | 20 833 | 41·0 | 2 | 10 417 |
| Plumbaginaceae | ||||
| Armeria alpina | 7600 | 41·0 | 2 | 3800 |
| A. vulgaris | 8663 | 42·7 | 2 | 4332 |
| Ceratostigma plumbaginoides | 743 | 39·7 | 2 | 372 |
| Plumbago auriculata | 669 | 38·8 | 2 | 335 |
| Polygonaceae | ||||
| Bistorta major | 5354 | 42·2 | 4 | 1339 |
| Fallopia dumetorum | 1324 | 40·5 | 2 | 662 |
| Muehlenbeckia complexa | 1414 | 39·9 | 2 | 707 |
| Oxyria digyna | 1909 | 41·3 | 2 | 955 |
| Persicaria amphibia | 2732 | 39·7 | 4 | 683 |
| P. hydropiper | 1300 | 41·0 | 2 | 650 |
| P. lapathifolia | 1458 | 43·8 | 2 | 729 |
| P. maculosa | 3015 | 40·8 | 4 | 754 |
| P. mitis | 3071 | 40·3 | 4 | 768 |
| Polygonum arenastrum | 1445 | 44·9 | 4 | 361 |
| Reynoutria japonica | 8279 | 40·5 | 8 | 1035 |
| Rumex acetosa | 6104 | 45·1 | 2 | 3052 |
| R. alpinus | 868 | 44·0 | 2 | 434 |
| R. arifolius | 5912 | 44·1 | 2 | 2956 |
| R. conglomeratus | 1370 | 44·5 | 2 | 685 |
| R. crispus | 3948 | 40·8 | 4 | 987 |
| R. maritimus | 1962 | 40·3 | 4 | 491 |
| R. patientia | 4305 | 41·2 | 6 | 718 |
| Tamaricaceae | ||||
| Myricaria germanica | 2872 | 40·8 | 2 | 1436 |
| Tamarix tetrandra | 2823 | 37·0 | 2 | 1412 |
For sources of chromosome number data, see Supplementary Data Table S1.
The GC content variation in Droseraceae (Table 1) was 7·6 %, with the lowest value found in Drosera prolifera (37·1 %) and the highest in D. oreopodion (44·7 %). The values of species of related clades varied between 36·3 % (Nepenthes pervillei) and 45·1 % (Rumex acetosa).
Phylogeny of Droseraceae and related clades
The Caryophyllales diversified at the turn of the lower and upper Cretaceous (Supplementary Data Fig. S2). The carnivorous Caryophyllales (families Droseraceae, Nepenthaceae, Dioncophyllaceae, Ancistrocladaceae and Drosophyllaceae) form a monophyletic clade in which Ancistrocladaceae and Dioncophyllaceae were ancestrally carnivorous (Heubl et al., 2006). They diverged from the Frankeniaceae + Tamaricaceae clade 93·31 Mya. The Polygonaceae + Plumbaginaceae clade diverged from the carnivorous Caryo phy llales + (Frankeniaceae + Tamaricaceae) clade 98·69 Mya.
Within the carnivorous Caryophyllales, the crown node is 74·48 Ma old, marking the minimum age of carnivory in the Caryophyllales. The individual carnivorous genera evolved during the Palaeogene. The estimated crown age of the Droseraceae is 54·67 Ma (Fig. 1; Supplementary Data Fig. S2). Within Droseraceae, the two genera of snap-traps (Aldrovanda and Dionaea) split at least 45·09 Mya. There are two basal species of Drosera, i.e. D. arcturi and D. regia, which diverged from the rest of the genus 54·11 and 52·21 Mya, respectively. The remaining species of Drosera form two main clades that split 46·47 Mya. The first clade comprises the subgenera Stelogyne, Theocalyx and Drosera (D. sessilifolia–D. trinervia clade; hereafter referred to as the ‘Cosmopolitan clade’ because its members occur on all the continents except for Antarctica), and the second clade includes the subgenera Bryastrum, Lasiocephala, Ergaleium and Phycopsis (D. binata–D. omissa clade; hereafter referred to as the ‘Australian clade’ because most of its members are restricted to Australia and adjacent areas).
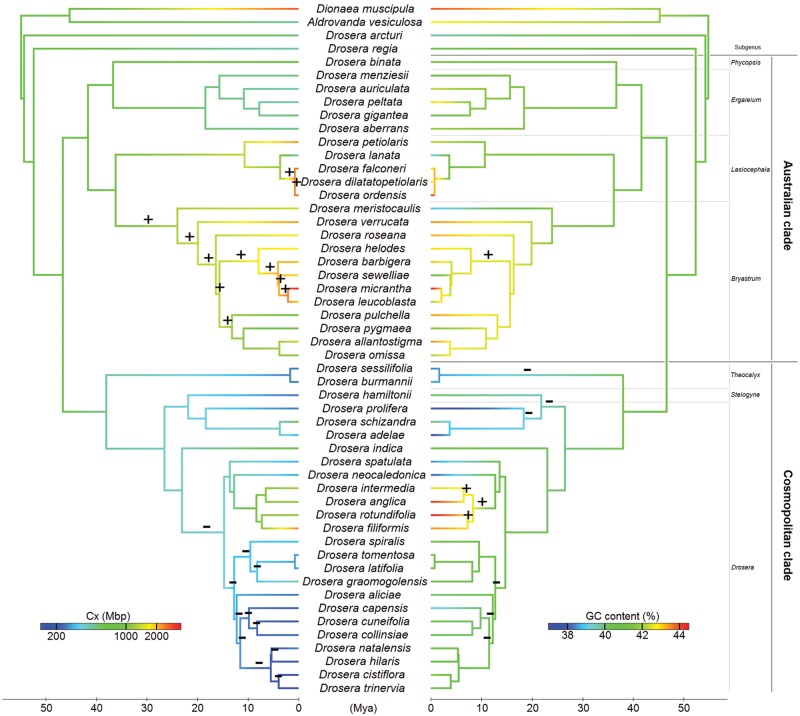
Fig. 1.
Ancestral state reconstruction of monoploid genome size (left) and GC content (right) in Droseraceae. Significant increases and decreases (P < 0·05) of monoploid genome size or GC content are marked with ‘+’ and ‘–’ signs above the branches leading to particular nodes.
Genome size evolution in Droseraceae
The reconstructed evolution of the monoploid genome size shows opposite trends in the two main clades of the genus Drosera (Fig. 1). The genomes of the species from the Cosmopolitan clade show a reduction tendency, and multiple significant downsizing events have been detected in several nodes of this clade (Fig. 1). In contrast, the genomes of species from the Australian clade (particularly in the subgenera Bryastrum and Lasiocephala) exhibit a tendency for genome growth with multiple significant upsizing events detected (Fig. 1). The genome size in the rest of the Australian clade (i.e. the subgenera Ergaleium and Phycopsis) is relatively stable.
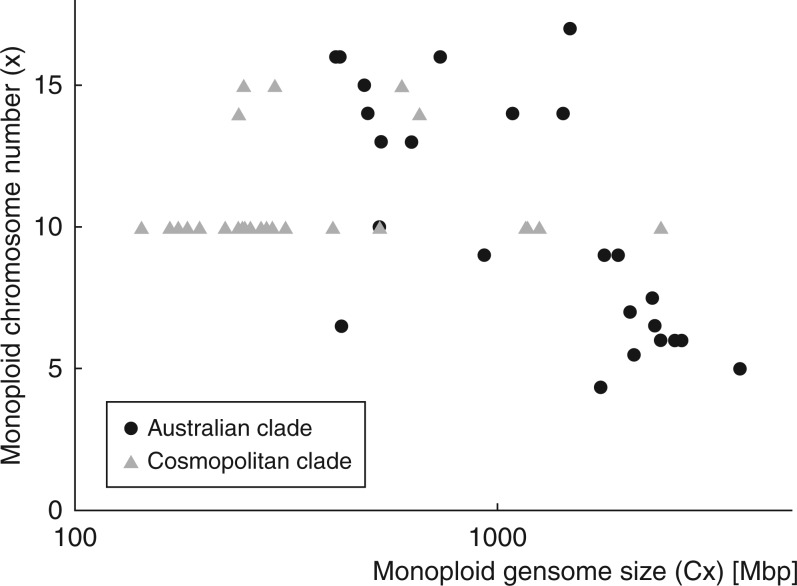
No difference was detected in a phylogeny-based comparison between monoploid genome sizes of carnivorous and non-carnivorous species (P = 0·680; Supplementary Table S1: Dataset 1) or between holokinetic and monocentric species (P = 0·600; Supplementary Table S1: Dataset 1). Within the holokinetic species of Drosera, a weak negative correlation was observed between the Cx genome size and the monoploid chromosome number (‘pgls’ λ = 1, P = 0·08; Fig. 2; Supplementary Table S1: Dataset 4). This negative correlation was apparent in the Australian clade (‘pgls’ λ = 0, P < 0·001; Supplementary Table S1: Dataset 5), while it was absent in the Cosmopolitan clade (‘pgls’ λ = 1, P = 0·813; Supplementary Table S1: Dataset 6) when both clades were analysed separately (Fig. 2).
Fig. 2.
Relationship between monoploid genome sizes (Cx) and basic (monoploid) chromosome numbers (x) in holokinetic species of Drosera. Note a negative correlation between both parameters in the Australian clade (P < 0·001), which probably resulted from the holokinetic drive.
Genomic GC content evolution in Droseraceae
Several reductions in the GC content were observed in the Cosmopolitan clade, with the exception of four temperate species (Drosera anglica, D. filiformis, D. intermedia and D. rotundifolia), where the GC content increased (Fig. 1). A single GC increase was also detected in the Australian clade, subgenera Bryastrum (Fig. 1). No difference was found in the GC content between the holokinetic Drosera species and the closely related monocentric species (P = 0·975; Supplementary Table S1: Dataset 2).
The GC content variation of Droseraceae in the summary explanatory model is best explained by the genome size (log-transformed 2C), which is positively correlated with the GC content (P = 4·06E-8; explained residual variation = 45·57 %; Supplementary Table S1: Dataset 3). Removing the effect of genome size in the model, the GC content further increases with an increasing annual range of temperature (median temperature annual range Bioclim variable; P = 4·41E-5, explained residual variation = 21·5 %; Supplementary Table S1: Dataset 2). After removing the effect of genome size and median annual temperature range, no other variable was able to explain the significant portion of the remaining residual variation in CG contents.
DISCUSSION
The genomes of the carnivorous species of the Caryophyllales have a ‘standard’ size which is comparable with its non-carnivorous relatives. Indeed, they are far from being truly miniature as in the carnivorous family Lentibulariaceae, whose genomes are strikingly smaller than the genomes of their non-carnivorous relatives (Veleba et al., 2014). The family Lentibulariaceae represents a unique lineage with unusually structured genomes (Ibarra-Laclette et al., 2013; Leushkin et al., 2013) and overall morphology (absent roots and leaves in Utricularia and Genlisea), while the morphological constitution of carnivorous Caryophyllales species is similar to a typical plant body. This questions whether genome downsizing in Lentibulariaceae is a direct consequence of carnivory and eventual nutrient starvation, or rather associated with some peculiar molecular properties of Lentibulariaceae (Jobson and Albert, 2002; Ibarra-Laclette et al., 2011a, b), or connected with their extreme body reduction.
Holokinetism has been suggested to be associated with genome size and GC content decrease (Bureš et al., 2013; Šmarda et al., 2014). In the present study, we have not confirmed lower genome size previously reported in Drosera (Bureš et al., 2013). This is most probably because we tested it phylogenetically this time. Similarly, we have not detected a decrease in the GC content associated with the evolution of holokinetism in Drosera. This suggests that genome downsizing and GC content decrease need not to be a direct consequence of holokinetism.
Aside from positive or no correlation between genome size and chromosome number (Zedek et al., 2010; Chung et al., 2012; Escudero et al., 2015), a negative correlation is commonly detected in holokinetic lineages (Nishikawa et al., 1984; Roalson et al., 2007; Bureš et al., 2013; Lipnerová et al., 2013; Záveská Drábková and Vlček, 2010; Bureš and Zedek, 2014). It has been hypothesized that this negative correlation is promoted by the holokinetic drive, which is based on a size-dependent competition between homologous chromosomes in asymmetric meiosis (Bureš and Zedek, 2014). Indeed, we have observed such a negative correlation in the Australian clade of Drosera, where the holokinetic drive seems therefore to have shaped the karyotype evolution. There are species with a few large chromosomes (e.g. Drosera micrantha, 2C = 7489 Mbp, 2n = 10, mean chromosome size, 2C/2n = 749 Mbp) as well as species with many small chromosomes (e.g. Drosera peltata, 2C = 829 Mbp, 2n = 32, mean chromosome size, 2C/2n = 26 Mbp), which results in the above-mentioned negative correlation (Fig. 2). The presence of holokinetic chromosomes does not automatically indicate the presence of the holokinetic drive (Bureš and Zedek, 2014). Likewise, the relatively stable chromosome counts and small differences in genome size among species in the Cosmopolitan clade indicate that the holokinetic drive plays no or only a negligible role there. Alternatively, it is possible that the holokinetic drive and the carnivory-driven selection for small genomes have opposite effects on genome size in Droseraceae. If the holokinetic drive prefers larger chromosomes, which may indeed be the case in the Australian clade of Drosera (Table 1; Fig. 1), the carnivory-driven selection for small genomes may be counteracted by genome size enlargement due to the holokinetic drive. Such opposition of the two evolutionary forces may have obscured any differences in genome size between carnivorous and non-carnivorous as well as holokinetic and monocentric species.
It should be noted that a recent study doubted the occurrence of holokinetic chromosomes in Drosera aliciae, D. binata and D. rotundifolia based on the chromosomal staining by a supposedly universal mitotic centromere marker H2AThr120ph (Demidov et al., 2014). However, in D. rotundifolia, chromosomal fragments induced by gamma irradiation are regularly inherited by daughter cells during mitosis (Shirakawa et al., 2011a) which is strong evidence for chromosomal holokinetism; D. aliciae and D. binata have not been studied this way. It is therefore possible that H2AThr120ph is not a universal mitotic centromere marker or at least not able definitely to distinguish between monocentric and holokinetic chromosomes. On the other hand, there is a hypothetical possibility that some species may be monocentric in mitosis but holokinetic in meiosis (Zedek and Bureš, 2016) which might be the case for D. aliciae and D. binata.
Both the genome size and GC content are perhaps often driven by the same process, such as the proliferation or removal of GC-rich or GC-poor transposable elements (Šmarda and Bureš, 2012), causing a commonly detected positive correlation of GC content with genome size in genera with relatively small genomes (Bureš et al., 2007). However, the GC content also seems to have an adaptive role (Šmarda et al., 2014), reflecting differences in the physical properties of GC and AT base pairs, such as the higher stacking interaction in GC base pairs and consequently a higher thermal stability of GC-rich DNA (Biro, 2008; Šmarda and Bureš, 2012). This trend has also been confirmed in monocots where higher GC contents are favoured in cold and dry climates (Šmarda et al., 2014).
A similar pattern has also been found in Droseraceae, where species with higher GC content are mostly found in areas with stronger annual temperature fluctuations, typical of temperate and Mediterranean regions. As an example may serve northern temperate sundews (Drosera anglica, D. rotundifolia, D. intermedia and D. filiformis) or Drosera subgenera Bryastrum from the Mediterranean climate of West Australia (McPherson, 2008, 2010), all possessing relatively high GC contents (Table 1; Fig. 1). In contrast, low GC contents can be expected in areas with low temperature fluctuations, typically in the tropical regions. Examples include the species of the ‘rainforest sundews’ (Drosera adelae, D. prolifera and D. schizandra) from northern Queensland, in Australia (McPherson, 2008, 2010), or Drosera meristocaulis from the Neblina massif on the Brazilian–Venezuelan border (Rivadavia et al., 2012).
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.com and consist of the following. Supplementary Methods: detailed description of flow cytometry measurements, sequencing and phylogenetic tree construction. Figure S1: the phylogenetic tree with posterior values. Figure S2: the phylogenetic tree with node ages. Table S1: detailed information about accession numbers used for phylogenetic tree construction and genomic parameters of all analysed species. Table S2: results of flow cytometry measurements. Table S3: genomic and BioClim variables.
ACKNOWLEDGEMENTS
The authors thank Patrik Hudec, Ondřej Knápek, Michal Kouba, Jakub Štěpán and the Botanical Garden of the Faculty of Science, Masaryk University (Brno, Czech Republic), for providing fresh plant material of several species, the Centre of Plant Structural and Functional Genomics (Olomouc, Czech Republic) for providing seeds of reference DNA standards, Pavel Veselý with Ondřej Hájek for the calculation of the bioclimatic variables and graphical assistance, and Ivana Lipnerová for R consultations. The study was supported by the Czech Science Foundation (projects No. 13-29362S and 14-30313S).
LITERATURE CITED
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences 181: 109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. 2012. Angiosperm DNA C-values database (release 8·0, December 2012). http://data.kew.org/cvalues/ (last accessed 17 January 2016).
- Biro JC. 2008. Correlation between nucleotide composition and folding energy of coding sequences with special attention to wobble bases. Theoretical Biology and Medical Modelling 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummitt RK. 2001. World geographical scheme for recording plant distributions, 2nd edm Pittsburgh: Hunt Institute for Botanical Documentation, Carnegie-Mellon University. [Google Scholar]
- Bureš P, Zedek F. 2014. Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution 68: 2412–2420. [DOI] [PubMed] [Google Scholar]
- Bureš P, Šmarda P, Hralová I, et al. 2007. Correlation between GC content and genome size in plants. Cytometry A 71A: 764. [Google Scholar]
- Bureš P, Zedek F, Marková M. 2013. Holocentric chromosomes In: Wendel J, Greilhuber J, Doležel J, Leitch IJ, eds. Plant genome diversity. Vol. 2. Physical structure of plant genomes. Heidelberg: Springer, 187–208. [Google Scholar]
- Chung KS, Hipp AL, Roalson EH. 2012. Chromosome number evolves independently of genome size in a clade with nonlocalized centromeres (Carex: Cyperaceae). Evolution 66: 2708–2722. [DOI] [PubMed] [Google Scholar]
- Cuacos M, H Franklin FC, Heckmann S. 2015. Atypical centromeres in plants – what they can tell us. Frontiers in Plant Sciences 6: 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D, Schubert V, Kumke K, et al. 2014. Anti-phosphorylated histone H2AThr120: a universal microscopic marker for centromeric chromatin of mono- and holocentric plant species. Cytogenetics and Genome Research 143: 150–156. [DOI] [PubMed] [Google Scholar]
- Escudero M, Hipp AL., Hansen TF, Voje KL., Luceño M. 2012. Selection and inertia in the evolution of holocentric chromosomes in sedges (Carex, Cyperaceae). New Phytologist 195: 237–247. [DOI] [PubMed] [Google Scholar]
- Escudero M, Maguilla E, Loureiro J, Castro M, Castro S, Luceño M. 2015. Genome size stability despite high chromosome number variation in Carex gr. laevigata. American Journal of Botany 102: 233–238. [DOI] [PubMed] [Google Scholar]
- Fleischmann A, Michael TP, Rivadavia F, et al. 2014. Evolution of genome size and chromosome numbers in the carnivorous plant genus Genlisea (Lentibulariaceae). Annals of Botany 114: 1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerstner KU, von Mering C, Hooper SD, Bork P. 2005. Environments shape the nucleotide composition of genomes. EMBO Reports 6: 1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Burkhardt EL, Happel RE, Weintraub JE. 1984. Carnivory in the bromeliad Brocchinia reducta, with a cost/benefit model for the general restriction of carnivorous plants to sunny, moist, nutrient-poor habitats. American Naturalist 124: 479–497. [Google Scholar]
- Gonella PM, Rivadavia F, Fleischmann A. 2015. Drosera magnifica (Droseraceae): the largest New World sundew, discovered on Facebook. Phytotaxa 220: 257–267. [Google Scholar]
- Govaerts R, Cheek M. 2014. World Checklist of Droseraceae Facilitated by the Royal Botanic Gardens, Kew. http://apps.kew.org/wcsp/ (last accessed 20 May 2015).
- Greilhuber J, Borsch T, Müller K, Worberg A, Porembski S, Barthlott W. 2006. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology 8: 770–777. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennet MD. 2005. The origin, evolution and proposed stabilization of the terms ‘Genome Size’ and ‘C-Value’ to describe nuclear DNA contents. Annals of Botany 95: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon L, Weir J, Brock C, et al. 2015. Geiger:analysis of evolutionary diversification. Version 2.0.6 https://cran.r-project.org/web/packages/geiger/geiger.pdf (last accessed 25 January 2016).
- Heckmann S, Houben A. 2013. Holokinetic centromeres In: Jiang J, Birchler JA, eds. Plant centromere biology. Oxford: Wiley-Blackwell, 83–94. [Google Scholar]
- Heubl G, Bringmann G, Meimberg H. 2006. Molecular phylogeny and character evolution of carnivorous plant families in Caryophyllales – revisited. Plant Biology 8: 821–830. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Ibarra-Laclette E, Albert VA, Herrera-Estrella A, Herrera-Estrella L. 2011a. Is GC bias in the nuclear genome of the carnivorous plant Utricularia driven by ROS-based mutation and biased gene conversion? Plant Signaling and Behavior 6: 1631–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Albert VA, Perez-Torres CA, et al. 2011b. Transcriptomics and molecular evolutionary rate analysis of the bladderwort (Utricularia), a carnivorous plant with a minimal genome. BMC Plant Biology 11: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Lyons E, Hernández-Guzmán G, et al. 2013. Architecture and evolution of a minute plant genome. Nature 498: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Vogt JK, Bressendorff S, et al. 2015. Transcriptome and genome size analysis of the venus flytrap. PLoS One 10: e0123887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Albert VA. 2002. Molecular rates parallel diversification contrasts between carnivorous plant sister lineages. Cladistics 18: 127–136. [DOI] [PubMed] [Google Scholar]
- Kondo K, Nontachaiyapoom S. 2008. An evidence on diffused centromeres in Drosera chromosomes provided by scanning electron microscopy. Chromosome Botany 3: 79–81. [Google Scholar]
- Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Leushkin EV, Sutormin RA, Nabieva ER, Penin AA, Kondrashov AS, Logacheva MD. 2013. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics 14: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipnerová I, Bureš P, Horová L, Šmarda P. 2013. Evolution of genome size in Carex (Cyperaceae) in relation to chromosome number and genomic base composition. Annals of Botany 111: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Shapoval NA, Anokhin BA, Saifitdinova AF, Kuznetsova VG. 2015. Homoploid hybrid speciation and genome evolution via chromosome sorting. Proceedings of the Royal Society B: Biological Sciences 282: 20150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S, Phoebe-Chen YP. 2010. Bacterial genomic G + C composition-eliciting environmental adaptation. Genomics 95: 7–15. [DOI] [PubMed] [Google Scholar]
- McPherson S. 2008. Glistening carnivores the sticky-leaved insect-eating plants. Poole, UK: Redfern Natural History Productions. [Google Scholar]
- McPherson S. 2010. Carnivorous plants and their habitats, vol. 2. Poole, UK: Redfern Natural History Productions. [Google Scholar]
- Musto H, Naya H, Zavala A, et al. 2006. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochemical and Biophysical Research Communications 347: 1–3. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Furuta Y, Ishitobi K. 1984. Chromosomal evolution in genus Carex as viewed from nuclear-DNA content, with special reference to its aneuploidy. Japanese Journal of Genetics 59: 465–472. [Google Scholar]
- Nishio Y, Nakamura Y, Kawarabayasi Y, et al. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Research 13: 1572–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, Freckleton R, Thomas G, et al. 2012. Caper: comparative analyses of phylogenetics and evolution in R. Version 0.5 http://cran.r-project.org/web/packages/caper/caper.pdf (last accessed 25 April 2013).
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2013. R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org/ (last accessed 25 April 2013).
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Rivadavia F, de Miranda VFO, Hoogenstrijd G, Pinheiro F, Heubl G, Fleichmann A. 2012. Is Drosera meristocaulis a pygmy sundew? Evidence of a long-distance dispersal between Western Australia and northern South America. Annals of Botany 110: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roalson E, McCubbin AG, Whitkus R. 2007. Chromosome evolution in Cyperales In: Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG, eds. Monocots: comparative biology and evolution of Poales. Claremont, CA: Allen, 62–71. [Google Scholar]
- Rothfels K, Heimburg M. 1968. Chromosome size and DNA values in sundews (Droseraceae). Chromosoma 25: 96–103. [Google Scholar]
- Sheikh SA, Kondo K, Hoshi Y. 1995. Study of diffused centromeric nature of Drosera chromosomes. Cytologia 60: 43–47. [Google Scholar]
- Shirakawa J, Hoshi Y, Kondo K. 2011a. Chromosome differentiation and genome organization in carnivorous plant family Droseraceae. Chromosome Botany 6: 111–119. [Google Scholar]
- Shirakawa J, Nagano K, Hoshi Y. 2011b. A chromosome study of two centromere differentiating Drosera species, D. arcturi and D. regia. Caryologia 64: 453–563. [Google Scholar]
- Šíchová J., Ohno M, Dincă V, Watanabe M, Sahara K, Marec F. 2016. Fissions, fusions, and translocations shaped the karyotype and multiple sex chromosome constitution of the northeast-Asian wood white butterfly, Leptidea amurensis. Biological Journal of the Linnean Society 118: 457–451. [Google Scholar]
- Šmarda P, Bureš P. 2012. The variation of base composition in plant genomes In: Wendel JF, Greilhuber J, Doležel J, Leitch IJ, eds. Plant genome diversity, vol. 1 Vienna: Springer, 209–235. [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. 2008. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany 101: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proceedings of the National Academy of Sciences, USA 111: E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner RW, Elser JJ. 2002. Ecological stoichiometry. The biology of elements from molecules to biosphere. Princeston and Oxford: Princeston University Press. [Google Scholar]
- Veleba A, Bureš P, Adamec L, Šmarda P, Lipnerová I, Horová I. 2014. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytologist 203: 22–28. [DOI] [PubMed] [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. 2012. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany 109: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu GTH, Schmutzer T, Bull F, et al. 2015. Comparative genome analysis reveals divergent genome size evolution in a carnivorous plant genus. Plant Genome 8: 3. doi:10.3835/plantgenome2015.04.0021. [DOI] [PubMed] [Google Scholar]
- Wendel J, Greilhuber J, Doležel J, Leitch IJ. 2013. Plant genome diversity. Vol. 2. Physical structure of plant genomes. Heidelberg: Springer. [Google Scholar]
- Záveská Drábková L, Vlček C. 2010. Molecular phylogeny of the genus Luzula DC. (Juncaceae, Monocotyledones) based on plastome and nuclear ribosomal regions: a case of incongruence, incomplete lineage sorting and hybridisation. Molecular Phylogenetics and Evolution 57: 536–551. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bureš P. 2016. Absence of positive selection on CenH3 in Luzula suggests that holokinetic chromosomes may suppress centromere drive. Annals of Botany (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Šmerda J, Šmarda P, Bureš P. 2010. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biology 10: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Veselý P, Horová L, Bureš P. 2016. Flow cytometry may allow microscope-independent detection of holocentric chromosomes in plants. Scientific Reports 6: 27161. doi: 10.1038/srep27161. [DOI] [PMC free article] [PubMed] [Google Scholar]