Abstract
Chronic hyperglycemia impairs intracellular redox homeostasis and contributes to impaired diabetic tissue regeneration. The Keap1/Nrf2 pathway is a critical regulator of the endogenous antioxidant response system, and its dysfunction has been implicated in numerous pathologies. Here we characterize the effect of chronic hyperglycemia on Nrf2 signaling within a diabetic cutaneous regeneration model. We characterized the effects of chronic hyperglycemia on the Keap1/Nrf2 pathway within models of diabetic cutaneous wound regeneration. We assessed reactive oxygen species (ROS) production and antioxidant gene expression following alterations in the Nrf2 suppressor Keap1 and the subsequent changes in Nrf2 signaling. We also developed a topical small interfering RNA (siRNA)–based therapy to restore redox homeostasis within diabetic wounds. Western blotting demonstrated that chronic hyperglycemia–associated oxidative stress inhibits nuclear translocation of Nrf2 and impairs activation of antioxidant genes, thus contributing to ROS accumulation. Keap1 inhibition increased Nrf2 nuclear translocation, increased antioxidant gene expression, and reduced ROS production to normoglycemic levels, both in vitro and in vivo. Topical siKeap1 therapy resulted in improved regenerative capacity of diabetic wounds and accelerated closure. We report that chronic hyperglycemia weakens the endogenous antioxidant response, and the consequences of this defect are manifested by intracellular redox dysregulation, which can be restored by Keap1 inhibition. Targeted siRNA-based therapy represents a novel, efficacious strategy to reestablish redox homeostasis and accelerate diabetic cutaneous tissue regeneration.
Introduction
Poor diabetic tissue repair is a significant economic and social burden within the U.S., with monetary costs close to $300 billion per annum; approximately 20% of that cost is spent specifically on diabetic ulcer care (1,2). Despite improvements in diagnosis, outreach, and glucose management, multiple large clinical trials suggest that significant diabetic complications persist (3).
In particular, chronic hyperglycemia is implicated in the “metabolic memory” phenomenon that underlies diabetes-related complications including retinopathy, nephropathy, and poor wound healing (4). A common theme that has evolved from research into the cellular mechanisms behind diabetic complications is the mishandling of oxidative stress within diabetic tissues (5). Oxidative stress is characterized not only by an accumulation of reactive oxygen species (ROS) but also by an imbalance between ROS-producing pathways and the endogenous ROS-scavenging programs within cells.
The signaling pathway involving the transcription factor nuclear factor, erythroid 2-like 2 (Nrf2) and its key cytoplasmic repressor, Kelch-like ECH-associated protein 1 (Keap1), represents a central cellular defense mechanism to maintain redox homeostasis. Nrf2, a basic-region leucine zipper transcription factor, is an important regulator that translocates into the nucleus and activates a host of stress-response genes (6–8). Nrf2 regulates the basal and inducible expression of phase II detoxification enzymes and antioxidant stress proteins in response to endogenous and exogenous oxidative or electrophilic stresses (9,10). Keap1, a Cul-E3 ligase, regulates Nrf2 turnover and the ability of Nrf2 to translocate into the nucleus and activate the cellular antioxidant response (11). Under “normal” physiologic conditions, Keap1, also an actin-binding cytoplasmic protein, interacts with Nrf2 and the cell’s actin cytoskeleton to sequester Nrf2 in the cytoplasm and promote its ubiquitination and subsequent degradation by the 26S proteasome (12,13). This “tethering” effect maintains basal gene expression of cytoprotective enzymes and proteins for redox homeostasis (14).
Several studies have indicated that the Keap1/Nrf2 pathway is altered in the context of chronic hyperglycemia, particularly in diabetic nephropathy (15). Secondary to the immediate response mechanisms by the mammalian body to protect against infection, wounds accumulate ROS. Under euglycemic conditions, cytoprotective mechanisms and pathways, such as the Keap1/Nrf2 axis, ensure the ROS do not overwhelm the healing tissues. Whether this pathway is dysfunctional in the context of nonhealing cutaneous wounds in chronic hyperglycemia is unknown. We hypothesize that Nrf2 dysfunction during diabetic cutaneous regeneration is a critical element contributing to diabetic wound pathology. As a corollary, we further hypothesize that Keap1 inhibition will allow for a return to redox homeostasis, thus alleviating dysfunctional regeneration within this environment.
Research Design and Methods
Fibroblast Cell Culture
Our in vitro studies used murine NIH 3T3 fibroblasts (CRL-1658; American Type Culture Collection, Manassas, VA) in a model system of chronic hyperglycemic (HG) conditions using 25 mmol/L d-glucose DMEM (Life Technologies, Grand Island, NY) versus normoglycemic (NG) conditions using 5 mmol/L d-glucose DMEM (Life Technologies) based on previously published models of hyperglycemia (16). Media was supplemented with 10% FBS (Life Technologies) and 1% penicillin/streptomycin. Cells were cultured in 37°C at 5% CO2.
In Vitro Transfection
The role of Nrf2 signaling in 3T3 cells was investigated by inhibiting Keap1 using small interfering RNA (siRNA; siRNA mouse silencer select Keap1 ID:s78526; Life Technologies). 3T3 fibroblasts at 70% confluence were plated in serum-starved conditions 6 h before transfection using 0.5% FBS in antibiotic-free media. siRNA–liposomal complexes were prepared per the manufacturer’s instructions. Briefly, 2.5 nmol/L siKeap1 or siScramble (AM4635; Life Technologies) were mixed with OptiMEM (Life Technologies). Separately, Lipofectamine 2000 (Life Technologies) was also mixed with OptiMEM. The siRNA and Lipofectamine dilutions were mixed at room temperature and incubated for 20 min. The siRNA–liposomal complexes were added to 3T3 cells and incubated for 24 h; 3 h following the addition of siRNA–liposomal complexes, the cell culture media was supplemented to 10% FBS. The media was changed 24 h later. For RNA and protein isolation, cells were collected 48 h after transfection.
Quantitative RT-PCR
Total RNA was harvested using the RNeasy Mini kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions, and 500 ng of total RNA was reverse transcribed using the High-Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, CA). mRNA quantification was determined by real-time quantitative RT-PCR using a SYBR Green detector (Life Technologies) and the ABI Prism 7900HT Sequence Detection System or QuantStudio 7 Flex (Applied Biosystems). Relative mRNA levels were calculated using a standard-curve method.
Preparation of Protein Lysates
To generate protein lysates, transfected cells were trypsinized and harvested 48 h after transfection. Separate nuclear and cytoplasmic protein extracts were prepared as follows. Cells were lysed in a buffer of 10 mmol/L HEPES (pH 7.9), 10 mmol/L KCl, 0.1 mmol/L EDTA, 10 mmol/L dithiothreitol, 1× protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), and 1× phosphatase inhibitor cocktail (Sigma-Aldrich). After vortexing for 15 s, the lysate was spun down at 15,000 rcf for 5 min for cells or 20 min for tissue to separate the cytoplasmic extract (supernatant) and nuclear extract (pellet). The cytoplasmic extract was removed into a prechilled tube and used directly or stored at −80°C. The nuclear extract pellet was resuspended in 20 mmol/L HEPES (pH 7.9), 0.4 mol/L NaCl, 1 mmol/L EDTA, 25% glycerol, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail and incubated for 10 min. Nuclear lysate was separated by centrifugation at 15,000 rcf for 5 min for cells or 20 min for tissue and used immediately or stored at −80°C. Protein concentration was measured using Pierce 660 nm reagent (ThermoFisher) on a NanoDrop 2000 spectrophotometer (ThermoFisher).
Western Blotting and Immunoprecipitation
Cytosolic or nuclear lysate (20 μg of either) was run on a 10% SDS-PAGE (Bio-Rad, Hercules, CA) and transferred to a polyvinylidene fluoride membrane (Bio-Rad). The membrane was blocked for 2 h using a solution of 5% milk in Tris-buffered saline with 0.1% Tween (Pierce, Rockford, IL) and probed with antibodies specific for Nrf2 (ab92946; Abcam, Cambridge, MA), Keap1 (60027–1-Ig; Proteintech, Chicago, IL), proliferating cell nuclear antigen (2586; Cell Signaling Technology, Danvers, MA), or β-actin (ab8229; Abcam). Immunoprecipitation of cytoplasmic lysates was performed using the Pierce Co-Immunoprecipitation Kit (26149; ThermoFisher), per the instructions, supplemented with protein A agarose beads (9863; Cell Signaling Technology). Secondary antibodies were species specific and conjugated in horseradish peroxidase (Cell Signaling Technology). Protein expression was detected on hyperfilm (Amersham Biosciences, Sunnyvale, CA) with ECL Plus reagent (ThermoFisher).
ROS Assay Using Chloromethyl-Dichlorodihydrofluorescein Diacetate
3T3 cells were trypsinized, centrifuged, and resuspended in 1× Hanks’ balanced salt solution. Chloromethyl-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) dissolved in DMSO was added to a final concentration of 5 µmol/L. Cells were incubated for 1 h in the dark at 37°C in 5% CO2. Incubation with 1 mmol/L diethyl malate was used as a positive control for high ROS. Following incubation, cells were placed on ice to temper further metabolism, and propidium iodide was added to a final concentration of 1 µg/mL. Live (propidium iodide–negative) cells were analyzed at 492- to 495-nm excitation and 517- to 527-nm emission on a FACSCalibur flow cytometry system (Becton Dickinson). Data were analyzed using FlowJo (www.flowjo.com/).
Measurement of 8-Hydroxy-2′-Deoxyguanosine
ROS levels were determined by measuring the levels of 8-hydroxy-2′-deoxyguanosine (8-OHdG), a ROS-mediated DNA damage by-product, with an oxidative DNA damage ELISA kit (OxiSelect Oxidative DNA Damage ELISA; Cell Biolabs, San Diego, CA). ELISA was performed according to the manufacturer’s instructions. Briefly, 100 μL of 8-OHdG–BSA conjugate in PBS was incubated overnight at 4°C in 96-well plates. After washing with distilled water, the plate was blocked with 200 μL of assay diluent for 1 h at room temperature. Samples and standards (50 μL) then were incubated for 10 min, followed by incubation for 1 h with 50 μL of anti–8-OHdG detection antibody. After washing, 100 μL of secondary antibody coupled with horseradish peroxidase was incubated for 1 h. After several washes, the peroxidase activity was detected by the addition of 100 μL of substrate solution and incubation for 5 min. The reaction was stopped with 100 μL of stop solution and measured at 450 nm on the Synergy H1 ELISA plate reader (BioTek, Winooski, VT). Data were standardized to total DNA concentration.
Animal Protocol
All animal protocols were approved by the New York University Medical Center Animal Care Committee (International Animal Care and Use Committee protocol 061104-01). Male diabetic (Leprdb/db) mice (db/db) and nondiabetic mice C57BL/6J (wild type [WT], +/+), aged 12 weeks, were obtained from The Jackson Laboratory (Bar Harbor, ME). Glucometers were used to confirm hyperglycemic status. All db/db mice used in the experiments had blood glucose concentrations >400 mg/dL on two random separate occasions. Mice were housed in a temperature-controlled, virus-free barrier animal facility with a 12-h light/12-h dark cycle and were maintained on chow diet/water ad libitum.
Preclinical Wounding Model
An established murine model of excisional wound healing was used (17). The animals were anesthetized with inhalational 2% isoflurane. Hair on the dorsal skin was removed using a hair trimmer and Nair (Church & Dwight, Princeton, NJ). Paired 10-mm full-thickness wounds extending through the panniculus carnosus were created on the dorsum of the mice using a punch biopsy tool. The wounds then were splinted open with silicone stents fashioned from a 0.5-mm-thick silicone sheet (Grace Bio-Labs, Bend, OR). Stents were fixed to the skin using an immediate-bonding adhesive (Krazy Glue; Elmer’s Products Inc., Columbus, OH), with the wound centered within the stent. This was followed by the placement of interrupted 5–0 nylon sutures (Ethicon, Inc., Somerville, NJ) to secure them in position. After the immediate-bonding adhesive dried, wounds were covered with an occlusive dressing (Tegaderm; 3M, St. Paul, MN). Standardized photographs were taken on days 1, 7, 14, 21, and 28 after wounding; they were digitally analyzed for the percent wound closure and calibrated against the internal diameter of the silicone stent to correct for magnification, perspective, or parallax effects. Time to wound closure (number of days for complete reepithelialization) and percent wound closure (1 − [{wound area}/{original wound area}]) were measured digitally (Adobe Systems, San Jose, CA). Area under the curve was calculated using the trapezoidal rule (GraphPad Prism software) to assess the regenerative potential of each wound.
Topical siRNA Treatment
A 12-kDa U.S. Food and Drug Administration–approved 0.4% (weight for volume) agarose colloid matrix was created for the siRNA–liposomal transfection complex (18). Experimental arms included a treatment group, 200 pmol Keap1 siRNA (n = 16), and a control group, 200 pmol scramble siRNA (n = 16). On the basis of prior experiments that outlined optimal dosing routines, Keap1 siRNA and scramble siRNA were applied weekly (on days 1, 7, 14, and 21 after wounding). To assess the presence of siRNA in the skin we used siGLO Red (ThermoFisher) in the siRNA–liposomal complex and applied it to intact skin.
Animal Tissue Harvest
Eight animals from each group were killed on day 10. Wounds were collected for RNA, DNA, protein, and histological analyses. Tissue samples undergoing RNA extraction were placed in a Buffer RLT/β-mercaptoethanol solution (RNeasy Fibrous Tissue Kit; Qiagen) and homogenized using a polytron tissue homogenizer (Kinematica, Bohemia, NY). Tissues undergoing DNA extraction for 8-OHdG analysis were processed with a Buffer ALT/proteinase K solution according to the instructions of the DNeasy Blood & Tissue Kit (Qiagen). Following extractions, total RNA and DNA were quantified with a NanoDrop 2000 spectrophotometer (ThermoFisher). For siGLO Red–treated tissues, skin was fixed in 4% paraformaldehyde overnight and embedded in optimal cutting temperature medium.
In Vivo ROS Imaging
All mice were anesthetized with inhalational 2% isoflurane. L-012 (Wako Chemicals, Richmond, VA) was prepared in PBS at 0.5 mg/100 μL and injected intraperitoneally into mice (5 mg/200 g body weight). Bioluminescence was measured at 5-min intervals, and images were captured using the Spectrum in vivo imaging system (PerkinElmer, Waltham, MA).
Histology
On day 10 after wounding, wounds were excised and fixed in 10% formalin. The samples subsequently underwent routine histologic processing and were embedded in paraffin or optimal cutting temperature medium. Deparaffinized skin-tissue sections were stained with hematoxylin and eosin. The images were digitized to ×400 magnification using Nikon NIS Elements software (Nikon, Melville, NY) and subsequently analyzed for epithelial gap and granulation tissue formation using a previously established protocol (17). For epithelial gap measurements, four serial photographs taken on a Zeiss Axioskop 40 at ×5 magnification were superimposed using Adobe Photoshop to generate an image panning the complete width of the wound.
Immunofluorescence
Paraffin tissue section slides were deparaffinized using subsequent washes in a gradient of xylenes, ethanol, and PBS. Antigens were retrieved using Tris–EDTA solution (1 mmol/L EDTA [pH 8.0], 10 mmol/L Tris [pH 8.0]) in a water bath microwaved for 6 min at 90% power. The tissue sections were blocked with PBS with 0.1% Tween-20 solution containing 2% BSA (Sigma-Aldrich) and 5% normal donkey serum (Jackson ImmunoResearch Laboratories). Additional blocking was performed using the Vector M.O.M. Immunodetection Kit (Vector Laboratories), according to the manufacturer’s instructions. The tissue was probed using anti-Nrf2 antibody (sc-722; Santa Cruz Biotechnology) and anti-Keap1 antibody (sc-15246; Santa Cruz Biotechnology). Secondary antibodies (Alexa Fluor 488 AffiniPure donkey anti-mouse IgG, Alexa Fluor 594 AffiniPure donkey anti-rabbit IgG) were purchased from Jackson ImmunoResearch Laboratories and used at a dilution of 1:500 each.
Frozen skin-tissue sections were stained with rat anti-mouse CD31 primary antibody (BD Biosciences, Franklin Lakes, NJ) and goat anti-rat IgG secondary antibody (Alexa Fluor 594; Invitrogen). Slides were mounted with DAPI (Vector Laboratories) and viewed on a Nikon Eclipse Ni-U. DAPI was used to determine the sample outline, whereas immunofluorescent CD31 was used to identify vascular structures. Dual-filter images were superimposed to illustrate wound architecture and vascular staining (Photoshop; Adobe Systems Inc.). Vessels per high-powered field (hpf) also were counted. All experiments were performed in triplicate.
Total Glutathione Assay Methods
Cells were washed with ice-cold PBS twice before analysis with the GSH/GSSG-Glo kit (Promega, Madison, WI). Briefly, cells were lysed with total glutathione lysis reagent or oxidized glutathione lysis reagent, and the lysates were used within 10 min. Standards were prepared with the total glutathione reagent. The plates were shaken for 5 min. Luciferin generation reagent was added (50 μL/well), and after brief mixing by shaking the plate was incubated for 30 min at room temperature. Then, 100 μL luciferin detection reagent was added, mixed, and incubated for 15 min at room temperature. Luminescence was measured at room temperature with an integration time of 1 s/well. The linear portions of the standard curves were used to fit equations of lines, and the concentrations of unknown samples were calculated. The reduced glutathione (GSH)–to–glutathione disulfide (GSSG) ratio was calculated by adjusting for every mole of GSSG being derived from 1 mol of GSH. Samples were assayed in triplicate.
Statistical Analysis
All data are expressed as mean ± SD of at least three separate trials, each with n = 3. Student t tests and one-way ANOVA were applied to assess for statistical significance. A P value less than 0.05 determined statistical significance.
Results
Nrf2 Nuclear Translocation Is Dysfunctional in the Context of Chronic Hyperglycemia
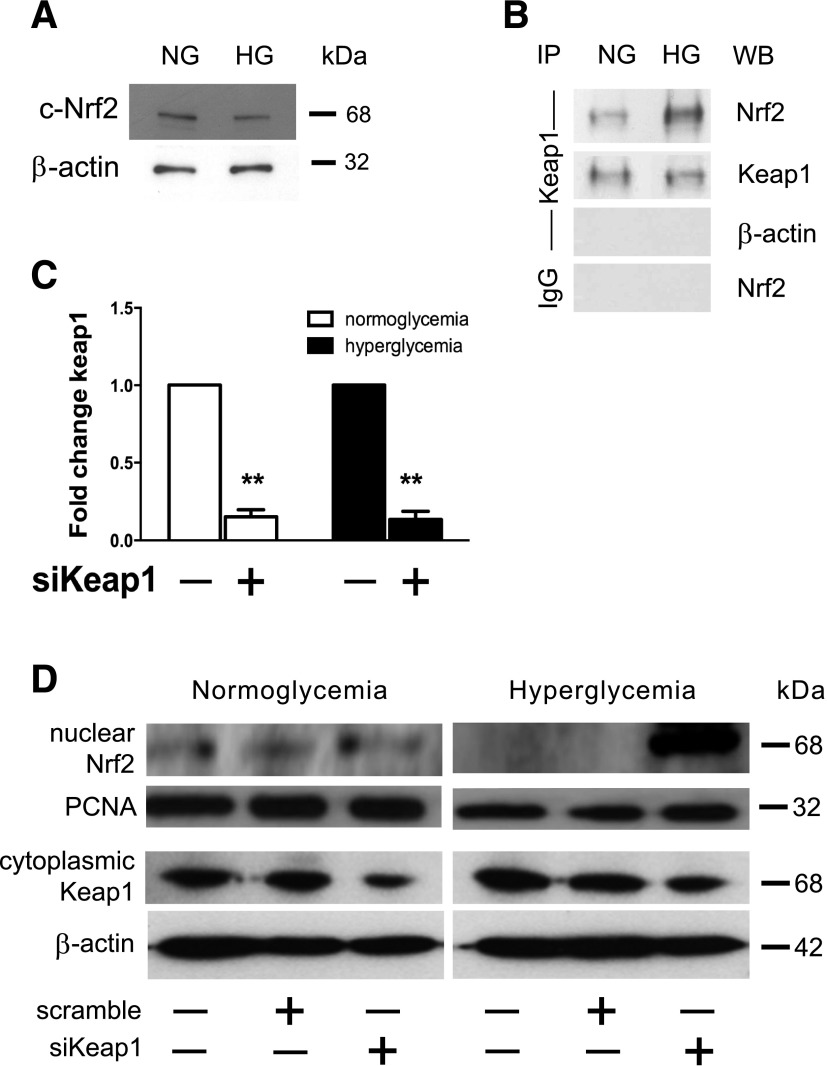
Our first goal was to determine the pattern of the Keap1/Nrf2 signaling pathway within diabetic cells. Using our in vitro model, we analyzed cytoplasmic protein lysates from NG- and HG-cultured 3T3 cells, and there were no striking differences in Nrf2 expression (Fig. 1A). However, coimmunoprecipitation showed a relative increase in Keap1-bound Nrf2 in the cytosol of HG fibroblasts (Fig. 1B). Hypothesizing that Keap1 was mediating defective Nrf2 translocation, we inhibited Keap1 using an siRNA-based strategy. We optimized a Lipofectamine-based method to inhibit Keap1 gene expression to <10% of baseline within 48 h of transfection in both NG and HG fibroblasts (Fig. 1C). While NG conditions induce a basal level of Nrf2 nuclear translocation, little Nrf2 migrated to the nucleus in HG-cultured fibroblasts (Fig. 1D). We observed maximal knockdown of Keap1 protein 2 days following transfection, resulting in a marked increase in the presence of nuclear Nrf2 in HG fibroblasts (Fig. 1D). Using an in vitro model, our data showed evidence of altered Nrf2 protein expression within fibroblasts undergoing hyperglycemia-related oxidative stress.
Figure 1.
Defective Nrf2 nuclear translocation in chronic hyperglycemia can be alleviated by Keap1 inhibition. Protein lysates of 3T3 fibroblasts cultured in either NG or chronic HG conditions were generated 48 h after introduction of siKeap1 or control scramble siRNA. A: Cytoplasmic protein lysate (10 μg) from NG and chronic HG samples reveal relatively equivalent amounts of Nrf2 within the cytoplasm. B: Immunoprecipitation of cytosolic Keap1 demonstrates that there are notable differences between the proportions of Nrf2 sequestered by Keap1 in the cytoplasm between NG and HG 3T3s. C: Quantitative RT-PCR demonstrates that Lipofectamine-based siKeap1 transfection effectively reduces Keap1 expression in cultured fibroblasts to <10% of baseline within 48 h compared with siScramble-transfected controls. D: Western blot of 3T3 fibroblast nuclear lysates from NG and chronic HG conditions demonstrates a nuclear signaling translocation defect that is restored 48 h after transfection with siKeap1. The same cytoplasmic lysate was used in A and D. **P < 0.01. IP, immunoprecipitation with antibody against indicated protein; PCNA, proliferating cell nuclear antigen; WB, immunoblot with indicated antibody.
Dysfunctional Nrf2 Signaling Creates Redox Imbalance That Can Be Rescued With Keap1 Inhibition
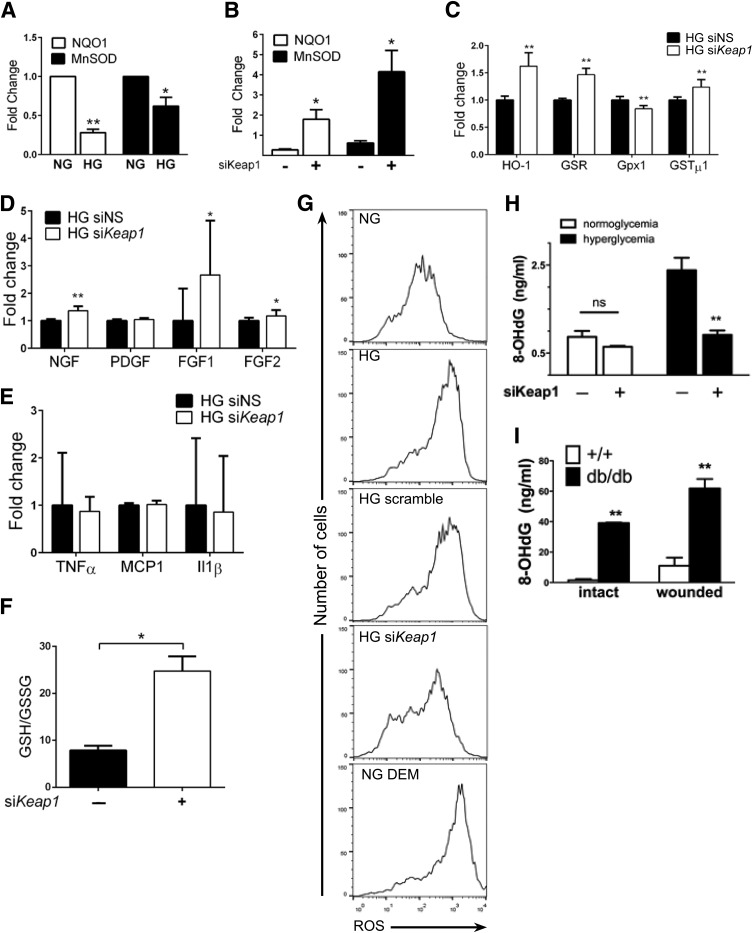
We further investigated the downstream consequences of the Nrf2 translocation defect in chronic hyperglycemia. Using the same in vitro model, total RNA was isolated from fibroblasts, and gene expression was analyzed by quantitative RT-PCR. Expression of NAD(P)H dehydrogenase, quinone 1 (NQO1) and manganese superoxide dismutase (MnSOD), essential antioxidant genes controlling ROS within the cellular environment, was decreased in HG compared with their NG counterparts (72 ± 7.7% decrease and 38 ± 19.3% decrease, respectively; P < 0.05) (Fig. 2A). Following treatment with Keap1 siRNA, the hyperglycemia-related reduction in antioxidant gene expression was rescued with 180% and 410% increases in gene expression of NQO1 and MnSOD, respectively (P < 0.05) (Fig. 2B). Keap1 silencing significantly altered the gene expression of Nrf2 downstream targets heme oxygenase-1 (HO-1), glutathione reductase (GSR), glutathione peroxidase 1 (Gpx1), and glutathione-S-transferase µ1 (GSTµ1). With the exception of Gpx1, whose expression is downregulated to 84 ± 6%, gene expression of HO-1, GSR, and GSTµ1 increased to 162 ± 34%, 147 ± 12%, and 124 ± 14%, respectively (Fig. 2C). Several growth factors associated with wound healing are also affected by Keap1 silencing: nerve growth factor (NGF; 136 ± 15%), fibroblast growth factor (FGF) 1 (267 ± 190%), and FGF2 (117 ± 20%) (Fig. 2D). Platelet-derived growth factor expression is not affected by siKeap1. We also evaluated the expression of inflammatory markers such as tumor necrosis factor (TNF)-α, monocyte chemoattractant protein 1 (MCP1), and interleukin (IL)-1β, but did not find any remarkable change (Fig. 2E).
Figure 2.
Keap1 inhibition improves ROS handling within fibroblasts by enhancing activation of the endogenous antioxidant program. Gene expression and ROS handling were assessed in vitro following Keap1 silencing. A: Chronic hyperglycemia impairs activation of NQO1 and MnSOD by 72% and 38%, respectively, compared with NG controls. B: siRNA-mediated reduction of Keap1 expression rescues MnSOD and NQO1 expression to 180% and 410% of controls, respectively. C–E: Gene expression analysis of Nrf2 target genes (C), growth factors (D), and inflammatory markers (E) with and without Keap1 silencing. F: Assessment of GSH/GSSG content with siKeap1. G: CM-H2DCFDA–based fluorescent assessment of ROS production reveals that real-time ROS production in HG fibroblasts can be reduced to that of NG cells with siKeap1. H: Keap1 inhibition within HG fibroblasts reduces the ROS by-product 8-OHdG by 62% compared with control. 8-OHdG in NG cells is nonsignificantly decreased by 25%. I: db/db Mice have 5.6-fold greater accumulation of ROS in wounded skin compared with WT controls; intact db/db skin accumulates 3.5-fold more ROS than wounded WT skin. *P < 0.05, **P < 0.01. DEM, diethyl malate; ns, not significant; PDGF, platelet-derived growth factor; siNS, nonsense siRNA.
To correlate these changes in gene expression with the cellular redox state, we characterized the oxidative state of fibroblasts using several measures. Because glutathione is abundant in cells and the ratio of GSH to GSSG is considered an indicator of redox environment, we assayed total glutathione following siKeap1 treatment. We found the GSH-to-GSSG ratio increased to 24.77 ± 3.089 in siKeap1-treated fibroblasts in comparison with 7.85 ± 0.98 in scramble-treated ones, indicating a decrease in the oxidized GSSG form and a corresponding increase in the GSH form (Fig. 2F). To investigate ROS stress, fibroblasts were incubated with the intracellular ROS indicator CM-H2DCFDA. Using flow cytometry to assess the level of fluorescence of the ROS-induced adduct, we found that Keap1-silenced HG fibroblasts demonstrated significantly reduced intracellular oxidative status, consistent with an enhanced ROS-scavenging potential, compared with NG and HG scramble controls (Fig. 2G). Similarly, an ELISA-based determination of 8-OHdG levels allowed assessment of longer-term ROS accumulation. We found that HG fibroblasts demonstrated an increased baseline value that is 290% of NG 8-OHdG (Fig. 2H). However, silencing Keap1 reduced 8-OHdG in HG fibroblasts by 62% (Fig. 2H). Inhibition of Keap1 in NG fibroblasts reduced 8-OHdG accumulation by only 25% compared with the NG control (Fig. 2H). 8-OHdG quantification and comparison between uninjured db/db skin and WT skin demonstrated that diabetic skin accumulates 25-fold more 8-OHdG (Fig. 2I). Tissue samples from the wound bed also demonstrated sixfold more 8-OHdG within db/db samples compared with WT samples (Fig. 2I).
Topical siKeap1 Therapy Improves the Redox Profile in Diabetic Mice and Accelerates Their Regenerative Capacity
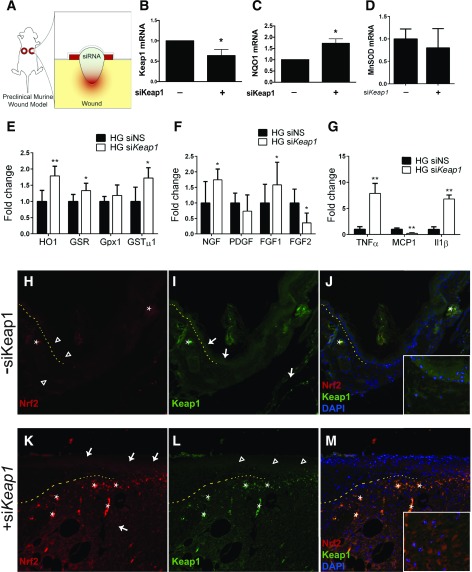
To transition to an in vivo assessment of Nrf2 signaling on redox homeostasis and regeneration, we used a validated stented-wound model of diabetic wound healing (Fig. 3A). We adapted our in vitro use of Keap1 siRNA to an in vivo preclinical therapy by topical weekly application of 200 pmol siRNA suspended in an agarose matrix to 10-mm diameter wounds weekly (Fig. 3A). Quantitative RT-PCR analysis of mRNA extracted from wounds 10 days into healing demonstrated that siKeap1 therapy reduced Keap1 expression within diabetic wounds by 36% (Fig. 3B). Expression of antioxidant gene NQO1 increased by 73% in diabetic wounds treated with siKeap1 compared with control therapy at 10 days (Fig. 3C). Expression of MnSOD, however, was not significantly altered following siKeap1 therapy in vivo (Fig. 3D). Following Keap1 silencing in the wound bed, Nrf2 target gene HO-1 expression increased by 179 ± 30%, GSR by 133 ± 23%, and GSTµ1 by 172 ± 32% (Fig. 3E). Growth factors NGF and FGF1 also increased with silenced Keap1 to 174 ± 35% and 157 ± 73%, respectively (Fig. 3F). FGF2 expression significantly decreased in the wound bed with siKeap1 therapy (Fig. 3F). In contrast to in vitro Keap1 silencing, inflammatory markers TNF-α, MCP1, and IL-1β were all significantly affected with Keap1 inhibition in vivo. TNF-α expression increased by 788 ± 193% and IL-1β expression increased by 680 ± 77% (Fig. 3G). MCP1 expression decreased to 24 ± 12% of that in scramble-treated diabetic wounds (Fig. 3G).
Figure 3.
Keap1 silencing in vivo upregulates Nrf2-mediated antioxidant mechanisms. Wound healing in db/db mice was analyzed using a validated stented-wound model, and the effect of weekly topical siKeap1 therapy was evaluated on macroscopic and molecular levels. A: Schematic of topical siKeap1 therapy. B: mRNA from siKeap1-treated wounds shows a 36% reduction in Keap1 expression 10 days into treatment. C: Ten days into treatment, topical siKeap1 therapy increases NQO1 expression in wounds by 73% compared with scramble siRNA. D: MnSOD expression in 10-day treated diabetic wounds. E–G: Gene expression of Nrf2 target genes (E), growth factors (F), and inflammatory factors (G) in wound beds following siKeap1 topical therapy. H–M: Immunofluorescence of Nrf2 and Keap1 in tissue sections of wounded diabetic tissue, with indicated topical siRNA therapy. All images are ×10 magnification. Insets in J and M are ×20 magnification. Open arrowheads show low expression in the indicated region; white arrows show upregulated expression in the indicated region; asterisks note autofluorescence. The dotted line demarcates the epidermis (above) and dermis (below) at the wound edge. PDGF, platelet-derived growth factor; siNS, nonsense siRNA. *P < 0.05. **P < 0.01.
Next we investigated the effect of Keap1 therapy on Nrf2 and Keap1 protein expression in skin-tissue sections of 10-day wounds harvested from db/db mice. In the absence of siKeap1 there is lack of Nrf2 expression in the wound bed and bordering skin (Fig. 3H and J). Upon application of siKeap1, Nrf2 expression was upregulated in epidermis, dermis, and wound granulation tissue at the wound edge (Fig. 3K and M). Specifically, more nuclear expression of Nrf2 is obvious in the granulation tissue of wounds that received topical siKeap1 (Fig. 3M, inset). In a corresponding manner, Keap1 immunoreactivity in epidermis and dermis decreased following silencing (Fig. 3I, L, and M). Our results demonstrate that topical siKeap1 is sufficient to upregulate Nrf2 expression and its downstream effects.
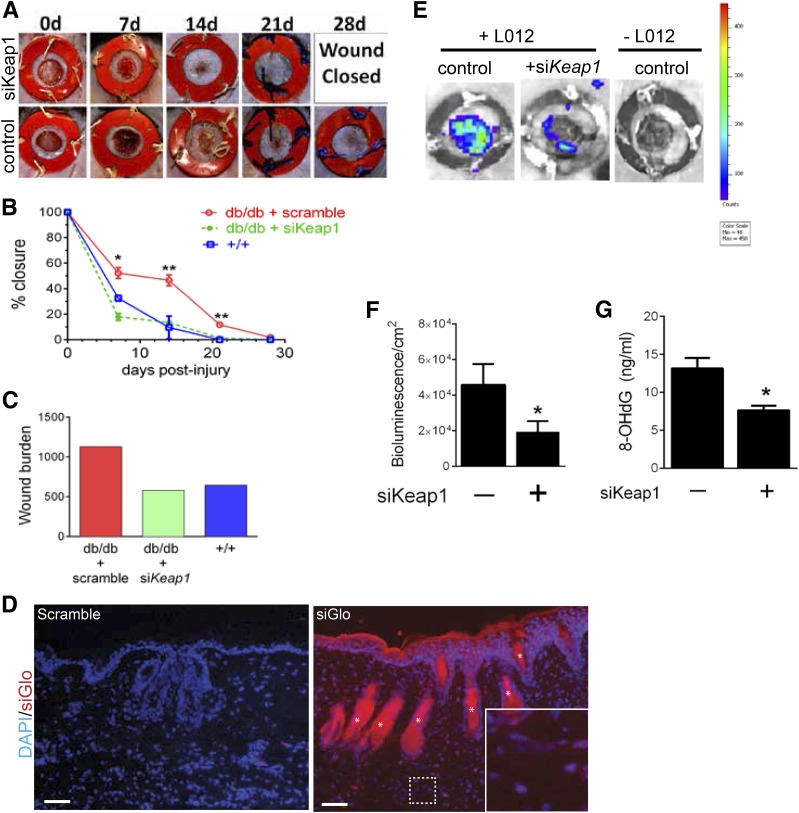
The clinical relevance of this altered metabolic state is demonstrated in Fig. 4. The full-thickness wounds on the dorsal skin of db/db mice that were treated with scramble or siKeap1 were monitored until the wound healed completely. Topical siKeap1 therapy reduced time to wound closure to 21 days, compared with the 30 days required by scramble-treated wounds (Fig. 4A). The wound closure rates between diabetic and WT mice are noticeably different as early as 7 days after excision (52% vs. 18% closure; P < 0.05) (Fig. 4B). Topical siRNA therapy targeted against Keap1 accelerated wound closure in db/db mice when compared with scramble siRNA–treated wounds (P < 0.01 at all time points observed), closely mimicking the WT closure pattern (Fig. 4B). Area under the curve analysis revealed that siKeap1 treatment corrected the regenerative delay and wound burden by 49% (Fig. 4C).
Figure 4.
Topical siKeap1 gene therapy accelerates diabetic wound closure. A: Photographs of stented wounds demonstrating accelerated wound closure with weekly topical siKeap1 therapy. B: Topical siKeap1 therapy accelerates diabetic wound closure by 7 days compared with nontreated controls. C: Wound burden analysis demonstrates that topical siKeap1 therapy enhances diabetic wound healing by 49% compared with scramble control. D: Dermal penetration and accumulation of siRNA in cryosections of diabetic mouse skin treated with siGLO Red–liposomal complex. At 10 days after application, siGLO Red is present up to the panniculus carnosus. Scale bar, 100 μm. Inset is magnification of area in dashed box. E: In vivo imaging system imaging of 7-day-old wounds using systemically delivered L-012 reagent demonstrates that topical siKeap1 treatment reduces real-time ROS accumulation compared with control diabetic wounds. F: Quantification of L-012 bioluminescence demonstrating a 58% decrease in ROS levels within diabetic wounds treated with topical siKeap1 therapy. G: The downstream ROS by-product 8-OHdG is also reduced by 42% with topical siKeap1 therapy compared with scramble-treated wounds. d, day. *P < 0.05. **P < 0.01.
We next examined the extent of penetration of the topical siRNA therapy in the mouse skin using a fluorophore-tagged siRNA, siGLO Red, in the siRNA–liposomal complex. We found accumulated siGLO Red in the skin down to the panniculus carnosus (Fig. 4D). To investigate whether the accumulation of siKeap1 in mouse skin, changes in redox gene expression, and accelerated wound closure correspond with ROS handling, we assessed real-time in vivo ROS concentrations within diabetic wounds by measuring L-012 bioluminescence. Seven days after topical siKeap1 application, diabetic wounds demonstrated improved redox homeostasis (Fig. 4E). Compared with scramble siRNA–treated wounds showing 45,775 ± 11,649 ROS bioluminescence units/cm2, siKeap1-treated wounds showed a remarkable 58% decrease to 19,405 ± 5,939 ROS bioluminescence units/cm2 (P = 0.04; Fig. 4F). Similarly, 8-OHdG ELISA analysis revealed that topical siKeap1 therapy reduced downstream by-products of ROS accumulation by 42%: from 13.16 ± 2.4 ng/mL in scramble control wounds to 7.64 ± 1 ng/mL in siKeap1 wounds (P < 0.05; Fig. 4G).
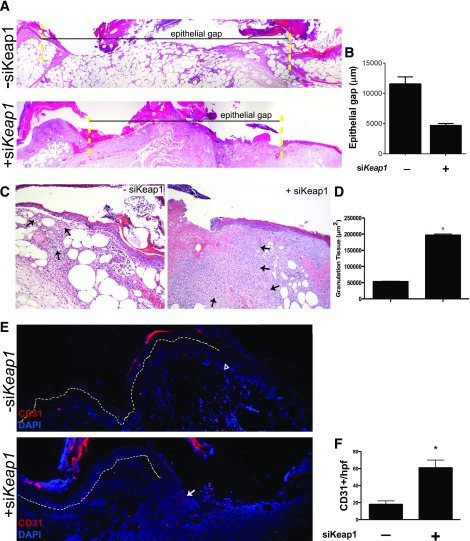
When correlating other measures of regeneration, we observed that topical siKeap1 treatment advantageously altered the histologic wound profile. The epithelial gap, or the distance between the healing edges of a wound, significantly decreased from 11,561 ± 1,175 μm in siScramble-treated wounds to 4,728 ± 302.4 µm in siKeap1-treated ones (Fig. 5A and B). Supporting this observation, a significant increase in wound granulation tissue was appreciated with topical siKeap1 treatment (197,148 µm2 in siKeap1-treated wounds vs. 53,165 µm2 in controls; P < 0.0001; Fig. 5C and D). Immunofluorescent staining showed that Keap1 inhibition increased the area of expression of CD31, a marker of neovascularization (Fig. 5E). We found 61 CD31+ cells/hpf in siKeap1-treated wounds versus 18 CD31+ cells/hpf in controls (P = 0.0016; Fig. 5F). Our data strongly suggest that silencing Keap1 in diabetic cutaneous wounds improves their redox status and promotes the generation of features necessary for successful neovascularization and reepithelialization.
Figure 5.
Topical siKeap1 gene therapy improves the histologic profile of diabetic wounds. Ten-day-old diabetic wounds were analyzed histologically to study the effect of topical siKeap1 therapy on reepithelialization and neovascularization. A: Hematoxylin and eosin–stained sections demonstrate accelerated reepithelialization in siKeap1-treated wounds. The yellow dotted lines indicate wound boundaries. B: Quantification of the epithelial gap. C: siKeap1-treated diabetic wounds demonstrate increased granulation tissue area. The black arrows indicate granulation tissue. D: Quantification of granulation tissue. siKeap1 therapy increases granulation tissue production by >3.5-fold compared with scramble-treated controls (*P < 0.01). E: CD31 immunofluorescence on tissue sections demonstrates increased neovascularization within the wound bed in siKeap1-treated wounds. The yellow dotted line demarcates the epidermis (above) and dermis (below). The wound bed is to the right of the yellow dotted line. The open arrowhead indicates low expression; the white arrow indicates high expression. F: Quantification of CD31+ cells in granulation tissue (*P < 0.01).
Discussion
Our study has several important findings. First, we demonstrate a critical Nrf2 signaling defect in chronic hyperglycemia that impairs downstream activation of the antioxidant response program. This defect is manifested on a cellular level through mishandling of intracellular ROS and, on a tissue level, impaired wound regeneration. Moreover, we describe a novel therapeutic strategy using local repression of Keap1, the endogenous Nrf2 inhibitor, to reverse the HG signaling defect, to restore cellular redox homeostasis, and to reestablish regenerative potential in a preclinical model of diabetic tissue injury.
There is growing recognition of the significant role redox homeostasis plays in the pathogenesis of diabetic complications (5). In particular, the Keap1/Nrf2 signaling pathway regulates the activity of over 50 antioxidant genes (19). Alterations in this pathway are already implicated in several diabetes-related complications including cardiomyopathy (9,20), kidney disease (10,12,13), hepatic steatosis (14,21), cerebral dementia, and metabolic syndrome (10,21,22). The common underlying pathologic mechanism involves the loss of redox homeostasis, which in turn contributes to impaired physiologic function on the tissue level. Nrf2-deficient mice are more susceptible to acetaminophen-related liver damage and are characterized by accelerated inflammation and renal injury in diabetic models (23). Conversely, Keap1 deficiency in mice has a protective role in a cholestatic liver injury model that attenuates liver injury through activation of Nrf2-related detoxifying enzymes and stress genes (24–26).
Problems of ROS handling within the heart, liver, and kidneys in the diabetic state have been well documented. However, few studies examine the role of ROS mishandling within cutaneous tissue, despite the overwhelming burden diabetic ulcers pose in the health care system (27). We sought to understand the role, if any, that Nrf2 signaling has in the pathogenesis of poor healing in the regenerative niche of a diabetic wound.
Our first step was to identify Nrf2 signaling in the context of a diabetic wound. Using an in vitro model of chronic hyperglycemia, we observed that cytoplasmic Nrf2 levels were relatively equivalent to NG conditions, whereas nuclear Nrf2 was detected in smaller amounts when compared with those in normoglycemia. Consistent with this finding, we observed increased amounts of Keap1-bound Nrf2 sequestered in the cytosol. This signaling defect, however, could be forcibly reversed through RNA interference (RNAi) of its repressor, Keap1. This is the first time that the endogenous repressor Keap1 has been targeted to increase Nrf2 signaling in fibroblasts and proposes a new therapeutic strategy in combating ROS mishandling within diabetic wounds.
Prior work has investigated mechanisms affecting Nrf2 localization and activity. Work by Bitar and Al-Mulla (27) similarly look at Nrf2 signaling differences in diabetic and nondiabetic fibroblasts, demonstrating that Nrf2 degradation occurs at a higher rate in diabetic fibroblasts than in WT ones. They correlate hyperactivity of glycogen synthase kinase 3β to poor nuclear translocation of Nrf2, possibly through phosphorylation of Nrf2 residues. Their use of lithium and thiadiazolidinone to inhibit glycogen synthase kinase 3β partially rescued the observed Nrf2 translocation defect. In addition to the ability of Nrf2 to translocate, other factors may affect its ability to initiate the endogenous antioxidant program. Kawai et al. (28) report that Nrf2 transcriptional activity may also be influenced by intranuclear acetylation. While our study is the first to target Keap1 to rescue the nuclear translocation defect, there may still be non-Keap1–related factors that explain persistent dysfunction despite rescue of the Nrf2 translocation defect.
We directly observed the immediate downstream effects of the Nrf2 signaling defect through decreased expression of the workhorse antioxidant genes NQO1 and MnSOD. The consequences of their relative inactivation, in conjunction with other redox metabolism–associated genes, correlated with both real-time ROS accumulation, as demonstrated by detection of the ROS stress indicator CM-H2DCFDA, and downstream ROS-related DNA adduct accumulation, as shown by 8-OHdG ELISA. Prior work by our lab (29,30) and others (5,31–34) identified such features of ROS accumulation as key characteristics of diabetic pathogenesis, and this study demonstrates that disruption of the Keap1/Nrf2 pathway is a central player in this process.
As a testament to the importance of the Keap1/Nrf2 pathway in the pathogenesis of diabetic complications, our in vivo model demonstrated that diabetic cutaneous regeneration can be enhanced through targeted reduction of Keap1 expression. Consistent with our in vitro results, diabetic wounds treated with topical siKeap1 demonstrated improved redox homeostasis, as shown by both real-time measurement via L-012 bioluminescence as well as accumulation of 8-OHdG. We found noticeable accumulation of siRNA from the topical therapy, contributing to the improved redox status of diabetic wounds. NQO1, widely recognized as one of the most important antioxidants, is upregulated in the siKeap1-treated wound bed. MnSOD, also a crucial antioxidant, however, is not affected by siKeap1 silencing. The Nrf2 target genes HO-1, GSR, and GSTµ1 are well-known antioxidants (35–37), and our results suggest their contribution in the reduction of wound-tissue ROS following Keap1 silencing. The increases in GSR and GSTµ1 also correlate with the increased GSH-to-GSSG ratio following Keap1 knockdown; these reducing enzymes are key in maintaining the balance between the GSH and oxidized GSSG. The presence of oxidized glutathione leads to reduced GSH-to-GSSG ratios and characterizes oxidative stress (38). The increased GSH-to-GSSG ratio is a testament to the restored redox balance with Keap1 knockdown. Gpx1 is also typically involved in antioxidant activity. However, the unremarkable change in Gpx1 expression in our study may reflect a redundant role or imply that Gpx1 is not involved in cutaneous cell redox homeostasis. The slight discrepancy between antioxidant gene expression between in vitro and in vivo studies may be explained by the inability of cell culture to capture a chronic inflammatory status. Nonetheless, the combined effect of Nrf2-downstream antioxidant gene products successfully restored redox handling in both our in vitro and in vivo models.
In conjunction with normalization of redox homeostasis, diabetic wound closure was accelerated and correlated with features of enhanced tissue repair and regeneration, including granulation tissue formation and CD31+ neovascularization. Well-vascularized, healthy granulation is essential to reepithelialization (39,40). We found increased expression of NGF, FGF1, and FGF2 in the wound bed following Keap1 silencing as well. Not surprisingly, Peplow and Baxter (41) found downregulation of these genes in diabetic wounds with delayed healing. Consistent with its role in angiogenesis and tissue repair, FGF1 has been shown to enhance wound healing in an alternative diabetic mouse model NONcNZO10/LtJ, as well (42). Though there are recognized limitations to the db/db model, here we recapture a similar upregulation of tissue repair–associated growth factors (43). FGF2 is similarly known to be involved in successful wound healing (44). Though the molecular and cellular targets of NGF upregulation during wound healing in skin have not been well explored, NGF promotes wound closure in mice and humans (45,46). Our data show a clear association between the repair of the Keap1/Nrf2 axis in the diabetic model and restoration of the regenerative niche.
In studies of vascularization after tissue injury, Nrf2 expression has been identified as an important promoter in physiological, noninflammatory neovascularization (47). In the context of neoplasia, however, Nrf2 overexpression is a known mechanism of tumor angiogenesis (48). Interestingly, HO-1 acts as an anti-inflammatory molecule in addition to its antioxidant role. In this study we found that activated Nrf2 resulted in upregulation of the traditionally proinflammatory cytokines TNF-α and IL-1β in 10-day-old siKeap1-treated diabetic wounds. Contrary to expectations of promoting inflammation, these cytokines are known to promote cutaneous wound healing, particularly in the proliferative phases (49). We show that even in a diabetic skin model, TNF-α and IL-1β reprise their prohealing roles. By contrast, Nrf2-mediated activity downregulated MCP1, a known chemokine for neutrophils and macrophages. Our results agree with the published literature in which forced expression of Nrf2 mitigated the MCP1 induced by ROS stress in human epithelial cells (50,51). Our data demonstrate that normalizing the Keap1/Nrf2 pathway in diabetic skin reinstates a multitude of healing mechanisms that are characteristic of nondiabetic tissue.
Our cutaneous gene therapy strategy to restore redox homeostasis via Keap1 suppression is of particular interest. RNAi-based gene therapy, using siRNAs or microRNAs, is a technology that has largely been limited to investigational uses. This therapy is limited by its ability to be effectively targeted to specific areas, requiring high dosages for systemic delivery, with untoward effects (52). However, the use of topically delivered siRNAs may obviate the traditional barriers to gene therapy because it is limited to local applications, is temporary, and exerts its function extrachromosomally. In this particular case, topical therapy for diabetic wounds would be an ideal application. Since significant Keap1 knockdown on both the RNA and protein levels occurs within 48 h of transfection, Keap1/Nrf2 turnover lends itself to effective application for an RNAi-based strategy, unlike other targets that may require an extended period of therapy to achieve target protein turnover. Moreover, in prior studies using this technology (18,53), concerns about off-target effects have been limited to the area of cutaneous application. This is of particular importance because constitutive and unregulated Nrf2 activation has been linked to carcinogenesis and the promotion of malignant neoplastic phenotypes (54).
There are several implications of this study. First, chronic oxidative stress on fibroblasts impairs Nrf2-mediated maintenance of cellular redox homeostasis, which can be restored through targeted inhibition of Keap1. Second, targeted therapeutic strategies to reduce Keap1 expression can accelerate diabetic tissue regeneration to near-normal levels, improving redox homeostasis and augmenting neovascularization. Finally, a targeted topical siRNA against Keap1 represents an ideal, rapidly translatable therapeutic strategy to address difficulties in treating cutaneous defects in diabetes.
Article Information
Acknowledgments. The authors would like to thank the Flow Cytometry, Genome Technology Center, Histopathology, and Small Animal Imaging cores at New York University (NYU) Langone Medical Center.
Funding. This study was supported in part by grant UL1 TR00038 from the National Center for Advancing Translational Sciences, National Institutes of Health (NIH) (to D.J.C.). The Flow Cytometry, Genome Technology Center, Histopathology, and Small Animal Imaging cores at NYU Langone Medical Center were also partially supported by this grant, as well as the NYU Cancer Center Support Grant, P30CA016087. The study was supported by the NIH National Cancer Institute training grant T32CA160002 to M.A.S.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.A.S. researched data and wrote the manuscript. O.D.C. researched data and reviewed the manuscript. Y.C.L. and R.A.S. researched data. T.E., U.A., and L.A. performed experiments, analyzed data, and reviewed the manuscript. J.B.C. reviewed the manuscript. P.B.S. reviewed and edited the manuscript. P.S.R. performed experiments, analyzed data, and wrote, reviewed, and edited the manuscript D.J.C. researched data and reviewed and edited the manuscript. D.J.C. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Driver VR, Fabbi M, Lavery LA, Gibbons G. The costs of diabetic foot: the economic case for the limb salvage team. J Vasc Surg 2010;52(Suppl.):17S–22S [DOI] [PubMed] [Google Scholar]
- 2.Reiber GE, Lipsky BA, Gibbons GW. The burden of diabetic foot ulcers. Am J Surg 1998;176(2A Suppl.):5S–10S. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi C, Del Prato S. Metabolic memory and individual treatment aims in type 2 diabetes–outcome-lessons learned from large clinical trials. Rev Diabet Stud 2011;8:432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi C, Miccoli R, Del Prato S. Hyperglycemia and vascular metabolic memory: truth or fiction? Curr Diab Rep 2013;13:403–410 [DOI] [PubMed] [Google Scholar]
- 5.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res 2010;107:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem 2006;387:1311–1320 [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul 2006;46:113–140 [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi A, Kang MI, Watai Y, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 2006;26:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, Cui W, Xin Y, et al. Prevention by sulforaphane of diabetic cardiomyopathy is associated with up-regulation of Nrf2 expression and transcription activation. J Mol Cell Cardiol 2013;57:82–95 [DOI] [PubMed] [Google Scholar]
- 10.Nakai K, Fujii H, Kono K, et al. Vitamin D activates the Nrf2-Keap1 antioxidant pathway and ameliorates nephropathy in diabetic rats. Am J Hypertens 2014;27:586–595 [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA 2004;101:2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi BH, Kang KS, Kwak MK. Effect of redox modulating NRF2 activators on chronic kidney disease. Molecules 2014;19:12727–12759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoja C, Benigni A, Remuzzi G. The Nrf2 pathway in the progression of renal disease. Nephrol Dial Transplant 2014;29(Suppl. 1):i19–i24 [DOI] [PubMed] [Google Scholar]
- 14.Meakin PJ, Chowdhry S, Sharma RS, et al. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol 2014;34:3305–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Whitman SA, Wu W, et al. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes 2011;60:3055–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Souza DR, Salib MM, Bennett J, et al. Hyperglycemia regulates RUNX2 activation and cellular wound healing through the aldose reductase polyol pathway. J Biol Chem 2009;284:17947–17955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galiano RD, Michaels J 5th, Dobryansky M, Levine JP, Gurtner GC. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 2004;12:485–492 [DOI] [PubMed] [Google Scholar]
- 18.Thanik VD, Greives MR, Lerman OZ, et al. Topical matrix-based siRNA silences local gene expression in a murine wound model. Gene Ther 2007;14:1305–1308 [DOI] [PubMed] [Google Scholar]
- 19.Surh YJ, Kundu JK, Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med 2008;74:1526–1539 [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Zhang Z, Cai L. Diabetic cardiomyopathy and its prevention by nrf2: current status. Diabetes Metab J 2014;38:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Kulkarni SR, Donepudi AC, More VR, Slitt AL. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes 2012;61:3208–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue P, Hou Y, Chen Y, et al. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 2013;62:845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enomoto A, Itoh K, Nagayoshi E, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci 2001;59:169–177 [DOI] [PubMed] [Google Scholar]
- 24.Okada K, Warabi E, Sugimoto H, et al. Nrf2 inhibits hepatic iron accumulation and counteracts oxidative stress-induced liver injury in nutritional steatohepatitis. J Gastroenterol 2012;47:924–935 [DOI] [PubMed] [Google Scholar]
- 25.Okada K, Warabi E, Sugimoto H, et al. Deletion of Nrf2 leads to rapid progression of steatohepatitis in mice fed atherogenic plus high-fat diet. J Gastroenterol 2013;48:620–632 [DOI] [PubMed] [Google Scholar]
- 26.Okada K, Shoda J, Taguchi K, et al. Nrf2 counteracts cholestatic liver injury via stimulation of hepatic defense systems. Biochem Biophys Res Commun 2009;389:431–436 [DOI] [PubMed] [Google Scholar]
- 27.Bitar MS, Al-Mulla F. A defect in Nrf2 signaling constitutes a mechanism for cellular stress hypersensitivity in a genetic rat model of type 2 diabetes. Am J Physiol Endocrinol Metab 2011;301:E1119–E1129 [DOI] [PubMed] [Google Scholar]
- 28.Kawai Y, Garduño L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem 2011;286:7629–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceradini DJ, Yao D, Grogan RH, et al. Decreasing intracellular superoxide corrects defective ischemia-induced new vessel formation in diabetic mice. J Biol Chem 2008;283:10930–10938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Low YC, Ham MJ, Lotfi P, et al. Keap1 silencing improves diabetes-specific defects in mesenchymal stem cell function (Abstract 171). Plast Reconstr Surg 2013;131(5S):127 [Google Scholar]
- 31.Yan SF, Ramasamy R, Schmidt AM. The RAGE axis: a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res 2010;106:842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coughlan MT, Thorburn DR, Penfold SA, et al. RAGE-induced cytosolic ROS promote mitochondrial superoxide generation in diabetes. J Am Soc Nephrol 2009;20:742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aktunc E, Ozacmak VH, Ozacmak HS, et al. N-acetyl cysteine promotes angiogenesis and clearance of free oxygen radicals, thus improving wound healing in an alloxan-induced diabetic mouse model of incisional wound. Clin Exp Dermatol 2010;35:902–909 [DOI] [PubMed] [Google Scholar]
- 34.Araki E, Nishikawa T. Oxidative stress: a cause and therapeutic target of diabetic complications. J Diabetes Investig 2010;1:90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 1999;274:26071–26078 [DOI] [PubMed] [Google Scholar]
- 36.McGrath-Morrow S, Lauer T, Yee M, et al. Nrf2 increases survival and attenuates alveolar growth inhibition in neonatal mice exposed to hyperoxia. Am J Physiol Lung Cell Mol Physiol 2009;296:L565–L573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 2002;62:5196–5203 [PubMed] [Google Scholar]
- 38.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 2001;30:1191–1212 [DOI] [PubMed] [Google Scholar]
- 39.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med 1999;341:738–746 [DOI] [PubMed] [Google Scholar]
- 40.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature 2008;453:314–321 [DOI] [PubMed] [Google Scholar]
- 41.Peplow PV, Baxter GD. Gene expression and release of growth factors during delayed wound healing: a review of studies in diabetic animals and possible combined laser phototherapy and growth factor treatment to enhance healing. Photomed Laser Surg 2012;30:617–636 [DOI] [PubMed] [Google Scholar]
- 42.Blaber SI, Diaz J, Blaber M. Accelerated healing in NONcNZO10/LtJ type 2 diabetic mice by FGF-1. Wound Repair Regen 2015;23:538–549 [DOI] [PubMed] [Google Scholar]
- 43.Fang RC, Kryger ZB, Buck DW 2nd, De la Garza M, Galiano RD, Mustoe TA. Limitations of the db/db mouse in translational wound healing research: Is the NONcNZO10 polygenic mouse model superior? Wound Repair Regen 2010;18:605–613 [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Alberico S, Saidu E, et al. Organic light emitting diode improves diabetic cutaneous wound healing in rats. Wound Repair Regen 2015;23:104–114 [DOI] [PubMed] [Google Scholar]
- 45.Aloe L. Nerve growth factor, human skin ulcers and vascularization. Our experience. Prog Brain Res 2004;146:515–522 [DOI] [PubMed] [Google Scholar]
- 46.Matsuda H, Koyama H, Sato H, et al. Role of nerve growth factor in cutaneous wound healing: accelerating effects in normal and healing-impaired diabetic mice. J Exp Med 1998;187:297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Florczyk U, Jazwa A, Maleszewska M, et al. Nrf2 regulates angiogenesis: effect on endothelial cells, bone marrow-derived proangiogenic cells and hind limb ischemia. Antioxid Redox Signal 2014;20:1693–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theodore M, Kawai Y, Yang J, et al. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J Biol Chem 2008;283:8984–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hübner G, Brauchle M, Smola H, Madlener M, Fässler R, Werner S. Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine 1996;8:548–556 [DOI] [PubMed] [Google Scholar]
- 50.Starrett W, Blake DJ. Sulforaphane inhibits de novo synthesis of IL-8 and MCP-1 in human epithelial cells generated by cigarette smoke extract. J Immunotoxicol 2011;8:150–158 [DOI] [PubMed] [Google Scholar]
- 51.Reddy NM, Potteti HR, Mariani TJ, Biswal S, Reddy SP. Conditional deletion of Nrf2 in airway epithelium exacerbates acute lung injury and impairs the resolution of inflammation. Am J Respir Cell Mol Biol 2011;45:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng Y, Wang CC, Choy KW, et al. Therapeutic potentials of gene silencing by RNA interference: principles, challenges, and new strategies. Gene 2014;538:217–227 [DOI] [PubMed] [Google Scholar]
- 53.Layliev J, Wilson S, Warren SM, Saadeh PB. Improving wound healing with topical gene therapy. Adv Wound Care (New Rochelle) 2012;1:218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geismann C, Arlt A, Sebens S, Schäfer H. Cytoprotection “gone astray”: Nrf2 and its role in cancer. Onco Targets Ther 2014;7:1497–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]