Abstract
Homologous recombination (HR) is a crucial pathway for double-stranded DNA break (DSB) repair. During the early stages of HR, the newly generated DSB ends are processed to yield long single-stranded DNA (ssDNA) overhangs, which are quickly bound by replication protein A (RPA). RPA is then replaced by the DNA recombinase Rad51, which forms extended helical filaments on the ssDNA. The resulting nucleoprotein filament, known as the presynaptic complex, is responsible for pairing the ssDNA with homologous double-stranded DNA (dsDNA), which serves as the template to guide DSB repair. Here, we use single-molecule imaging to visualize the interplay between human RPA (hRPA) and human RAD51 during presynaptic complex assembly and disassembly. We demonstrate that ssDNA-bound hRPA can undergo facilitated exchange, enabling hRPA to undergo rapid exchange between free and ssDNA-bound states only when free hRPA is present in solution. Our results also indicate that the presence of free hRPA inhibits RAD51 filament nucleation, but has a lesser impact upon filament elongation. This finding suggests that hRPA exerts important regulatory influence over RAD51 and may in turn affect the properties of the assembled RAD51 filament. These experiments provide an important basis for further investigations into the regulation of human presynaptic complex assembly.
INTRODUCTION
Homologous recombination (HR) is a highly conserved pathway that enables the exchange of genetic information between DNA molecules and is a driving force in evolution. HR is indispensable for the repair of DNA double-strand breaks (DSBs), which are cytotoxic lesions that can lead to genomic rearrangements and chromosomal abnormalities. HR plays essential roles in the rescue of stalled or collapsed replication forks (1,2), chromosomal rearrangements (1), horizontal gene transfer (3,4) and meiosis (5,6). Defects in HR have been directly linked with hereditary breast cancer, and the cancer prone diseases Fanconi anemia and Bloom syndrome (7,8).
During DSB repair by HR, the DNA break ends are processed to yield long 3′ single-stranded DNA (ssDNA) overhangs. In eukaryotes, these ssDNA overhangs are rapidly coated by the abundant ssDNA binding protein RPA (replication protein A) and the RPA-coated ssDNA recruits DNA damage checkpoint kinases to trigger the DNA damage response (9–13). RPA participates in numerous aspects of eukaryotic nucleic acid metabolism, including DNA replication and repair (7,14–17). RPA is essential for survival and the maintenance of genome integrity (9). The importance of RPA is further underscored by the observations that loss of any of RPA subunits is lethal (18,19), haploinsufficiency in mice is associated with lymphoid tumors and a decrease in lifespan (20,21), and a reduction in protein level results in defects in the DNA damage response (13,22,23). RPA binds very tightly to ssDNA, exhibiting a sub-nanomolar dissociation constant 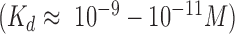 , consistent with its role in minimizing ssDNA secondary structure and preventing ssDNA degradation by cellular nucleases (9,14). Yet, emerging evidence suggests that the binding of RPA is highly dynamic, commensurate with the requirement that it be readily displaced from ssDNA by proteins that function in DNA repair or replication (9,24–28). RPA is a heterotrimeric complex composed of 70-, 32- and 14-kDa subunits, with a total of six OB-folds (oligosaccharide- or oligonucleotide-binding), and four ssDNA-binding domains (DBD): three DBDs on the 70 subunit linked by flexible polypeptide chains, and one on the 30 subunit (9,14,24). This multidomain architecture is thought to confer conformational flexibility that allows RPA to both bind ssDNA very tightly and yet be readily displaced by downstream DNA repair or replication factors (9,24).
, consistent with its role in minimizing ssDNA secondary structure and preventing ssDNA degradation by cellular nucleases (9,14). Yet, emerging evidence suggests that the binding of RPA is highly dynamic, commensurate with the requirement that it be readily displaced from ssDNA by proteins that function in DNA repair or replication (9,24–28). RPA is a heterotrimeric complex composed of 70-, 32- and 14-kDa subunits, with a total of six OB-folds (oligosaccharide- or oligonucleotide-binding), and four ssDNA-binding domains (DBD): three DBDs on the 70 subunit linked by flexible polypeptide chains, and one on the 30 subunit (9,14,24). This multidomain architecture is thought to confer conformational flexibility that allows RPA to both bind ssDNA very tightly and yet be readily displaced by downstream DNA repair or replication factors (9,24).
In the initial stage of HR, RPA is replaced by the adenosine triphosphate (ATP)-dependent recombinase RAD51, which forms extended helical filaments on the ssDNA (1,7,29–31). This filament assembly process begins with a nucleation event, in which a small complex, comprised of 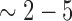 RAD51 monomers, binds to the ssDNA (32–34). Then, additional Rad51 subunits are added to the ends of the growing filaments to form a long protein polymer (30). The resulting Rad51-ssDNA filaments are referred to as the presynaptic complex, and this complex is responsible for locating a homologous DNA sequence in the genome and then catalyzing strand invasion, which pairs the ssDNA overhang with the homologous duplex (30,31). The 3′ end of the invading strand is then used to prime DNA synthesis and the resulting intermediates can be resolved through a number of mechanistically distinct pathways, culminating with the repair of the DSB (1,7,35).
RAD51 monomers, binds to the ssDNA (32–34). Then, additional Rad51 subunits are added to the ends of the growing filaments to form a long protein polymer (30). The resulting Rad51-ssDNA filaments are referred to as the presynaptic complex, and this complex is responsible for locating a homologous DNA sequence in the genome and then catalyzing strand invasion, which pairs the ssDNA overhang with the homologous duplex (30,31). The 3′ end of the invading strand is then used to prime DNA synthesis and the resulting intermediates can be resolved through a number of mechanistically distinct pathways, culminating with the repair of the DSB (1,7,35).
Here, we use ssDNA curtain assays to directly visualize the interplay between human RPA (hRPA) and human RAD51 during presynaptic complex assembly. These experiments reveal that hRPA binds tightly to ssDNA with a half-life >2 h, but is also able to rapidly exchange with free hRPA present in solution through a mechanism known as facilitated exchange (36,37). These findings are strikingly similar to results we have previously reported for Saccharomyces cerevisiae RPA (25–27), indicating that the facilitated exchange mechanism is conserved between yeast and humans. We also demonstrate that hRPA inhibits RAD51 assembly by restricting new filament nucleation events, but appears to have a lesser impact upon filament elongation. As a consequence, the RAD51 filaments are visibly longer when free RPA is present, allowing us to directly observe bidirectional RAD51 filament elongation. RAD51 filament assembly requires calcium, and our work reveals that pre-assembled RAD51 filaments remain intact upon removal of free ATP so long as Ca2+ remains present in the buffer, highlighting the dominating role of ATP hydrolysis in RAD51 filament stability. Together, our findings provide new insights into the assembly and disassembly of the presynaptic RAD51 filament on RPA-coated ssDNA.
MATERIALS AND METHODS
Protein expression and purification
6xHis-tagged hRPA-eGFP was expressed in Escherichia coli pLysS cells. A single colony was inoculated into 6L of LB containing 50 μg/ml carbenicillin and induced at 0.9 OD600 using 0.5 mM IPTG. Cells were grown overnight at 16°C and then harvested by centrifugation at 4000 × g for 25 min at 4°C. The cell pellet was resuspended in lysis buffer (500 mM NaCl, 20 mM Tris [pH 7.5], 2 mM β-ME, 5 mM imidazole, 10% glycerol, and 0.5 mM PMSF) and then lysed by sonication. The lysate was clarified by centrifugation at 25 000 rpm for 30 min at 4°C, and the clarified lysate was bound to Ni-NTA resin (Qiagen) equilibrated in R-buffer (50 mM KCl, 20 mM Tris–HCl [pH 7.4], 1 mM dithiothreitol (DTT), 0. 5 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol) for 1 h in batch at 4°C on a rotator. The Ni-NTA resin was then washed with R-buffer plus 50 mM imidazole and hRPA-eGFP was eluted using R-buffer with 200 mM imidazole. The pooled fractions were dialyzed into R buffer and then fractionated on a heparin column. Protein was eluted from the heparin column using a linear gradient 50–1000 mM KCl in R buffer. Pooled fractions were dialyzed into R150 buffer (150 mM KCl, 20 mM Tris [pH7.4], 1 mM DTT, 0.5 mM EDTA, 50% glycerol) overnight at 4°C using 10 000 MWCO Snakeskin™ dialysis tubing (Thermo-Fisher Scientific). Fractions were then flash frozen in liquid nitrogen and stored at −80°C. Protein concentrations were measured using a Nanodrop instrument and an extinction coefficient of ε488 nm = 55 000 cm−1M−1 for eGFP. Purity of the hRPA was assessed using a 4–20% gradient gel and Coomassie staining.
6xHis-tagged hRPA-mCherry and wild-type RPA (unlabeled) were expressed and purified as above with the following changes. After heparin column purification, pooled fractions were resolved on a Superdex 200 gel filtration column equilibrated in R-buffer and subsequently a Q-column (also eluted using R-buffer) before undergoing dialysis into R-buffer containing 50% glycerol. We did not observe a significant difference in purity with these additional steps and therefore did not perform them for RPA-eGFP. E. coli Single-Stranded Binding protein (SSB) and RAD51 were purified as previously described (26).
Single-strand DNA curtains
Flowcells were prepared as previously described (26,38). Biotinylated ssDNA was made by rolling circle replication using a circular M13 ssDNA template, a biotinylated primer and phi29 DNA polymerase, as described (38). For the extension and visualization of the ssDNA curtain, hRPA-eGFP (1 nM) was injected into the sample chamber at 1 ml/min in bovine serum albumin (BSA) buffer (40 mM Tris [pH 8.0], 2 mM MgCl2, 1 mM DTT and 0.2 mg/ml BSA). After 2 min, 150 μl of 7 M urea was flushed through the sample chamber to help remove protein aggregates and any remaining ssDNA secondary structure, and flow with 1 nM hRPA-eGFP was continued for another 15 min to ensure that both ends of the ssDNA were anchored to the flowcell surface.
RPA stability and facilitated dissociation measurements
All hRPA and RAD51 experiments were performed at 37°C. Experiments involving only hRPA were conducted in RPA buffer (30 mM Tris-Acetate [pH 7.5], 20 mM Mg-Acetate, 50 mM KCl, 1 mM DTT, 0.2 mg/ml BSA). For hRPA stability measurements in the absence of free protein, hRPA-eGFP (100 nM) was injected into the sample chamber and allowed to bind to the ssDNA. The flowcell chamber was then flushed with HR buffer at 1 ml/min for a minimum of 2 min to remove free hRPA, flow was terminated and the hRPA-eGFP signal was observed over time as previously described for S. cerevisiae RPA (26).
Facilitated dissociation was examined by chasing the hRPA-eGFP–ssDNA complexes with HR buffer containing unlabeled hRPA (100 nM). hRPA was injected at a flowrate of 0.2 ml/min, flow was then terminated and images (100 ms integration) were acquired at set intervals (from 2 to 20 s) for the duration of the experimental measurements. For analysis, kymographs of individual ssDNA molecules were generated using Fiji (ImageJ 1.48b, Wayne Rasband, National Institutes of Health, USA). Fluorescence intensity of the resulting kymographs was adjusted based upon the average background pixel intensity. Summing the intensities from across all the pixels for each ssDNA molecule and dividing by the number of pixels gave the adjusted pixel intensity for each molecule. For each time point, the overall adjusted pixel intensity was then averaged over N > 10 ssDNA molecules and the average intensities were then normalized against the first frame of the kymographs and plotted against time. The resulting exchange curves were fitted to a double exponential because the overall loss of signal intensity reflects contributions of protein exchange and photobleaching.
RAD51 filament assembly and disassembly
RAD51 filaments were assembled using HR buffer (30 mM Tris [pH 7.4], 1 mM MgCl2, 5 mM CaCl2, 100 mM KCl, 0.2 mg/ml BSA, 1 mM DTT and 2 mM ATP), unless otherwise indicated. After ssDNA-hRPA-eGFP curtain was formed using BSA buffer, we flushed out excess hRPA with HR buffer at 1 ml/min for a min of 2 min before injecting RAD51 plus or minus free hRPA-eGFP through a 50 μl sample loop. The adjusted intensity over time was calculated and normalized as for hRPA exchange. The intensity curves were fitted to a simple exponential decay function: 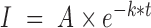 , where I is the normalized intensity for the filament, k = kexchange + kphotobleaching. We used the 0 nM RAD51 case to estimate kphotobleaching in order to calculate kexchange, which was plotted against the concentration of RAD51. This was subsequently fitted to the Hill equation: rate
, where I is the normalized intensity for the filament, k = kexchange + kphotobleaching. We used the 0 nM RAD51 case to estimate kphotobleaching in order to calculate kexchange, which was plotted against the concentration of RAD51. This was subsequently fitted to the Hill equation: rate 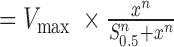 , where n is the Hill coefficient, Vmax is the maximum assembly rate, x is the RAD51 concentration, and S0.5 is the constant that represents the RAD51 concentration at half-maximal velocity. This equation was adapted for competitive inhibition by RPA to:
, where n is the Hill coefficient, Vmax is the maximum assembly rate, x is the RAD51 concentration, and S0.5 is the constant that represents the RAD51 concentration at half-maximal velocity. This equation was adapted for competitive inhibition by RPA to: 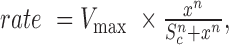 where
where 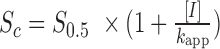 , I is the free hRPA concentration and kapp is the dissociation constant for hRPA. RAD51 binding also results in extension of the tethered ssDNA (27,39), and in these double-tethered ssDNA curtain assays this extension results in movement of the ssDNA further away from the flowcell surface. The experimentally observed decrease in hRPA-eGFP fluorescence intensity reflects a combination of hRPA-eGFP displacement from the ssDNA and movement of the protein-bound ssDNA further away from the slide surface.
, I is the free hRPA concentration and kapp is the dissociation constant for hRPA. RAD51 binding also results in extension of the tethered ssDNA (27,39), and in these double-tethered ssDNA curtain assays this extension results in movement of the ssDNA further away from the flowcell surface. The experimentally observed decrease in hRPA-eGFP fluorescence intensity reflects a combination of hRPA-eGFP displacement from the ssDNA and movement of the protein-bound ssDNA further away from the slide surface.
For disassembly reactions, presynaptic complexes were formed using 1 μM RAD51 as described above. After 30-min incubation, buffer flow was resumed using HR buffer containing 1 nM hRPA-eGFP and the indicated amounts of calcium and ATP. The rebinding of hRPA-eGFP to the ssDNA was used to measure the rate and extent of RAD51 dissociation, and the resulting kymographs were analyzed as above.
RAD51 nucleation, growth and disassembly
Nucleation positions were identified by first determining the time point (frame) in which the hRPA-eGFP signal had decreased to 40% of its original level following the injection of RAD51, as described above. The hRPA-eGFP signal intensity for each pixel along the selected ssDNA molecule was then plotted against position along the ssDNA. The resulting plot was analyzed to identify local minima, and these local minima were used as proxies to approximate the positions of RAD51 filament nucleation events. The local minima only serve as an approximation because we are unable to observe the initial nucleation events as our readout of hRPA-eGFP signal and a small nucleus of RAD51 initially might not result in macroscopic dissociation of RPA and it is also impossible to detect a single GFP loss in our system. This analysis was only performed on data obtained from experiments conducted in the presence of ≥50 nM free hRPA, because these were the only conditions under which we readily observed separate RAD51 filaments on the individual ssDNA molecules. Standard deviations for position distribution measurements were obtained from bootstrapping analysis, as previously described (39). The RAD51 filament elongation rates were estimated from experiments conducted in the presence of 750 nM RAD51 and 50 nM hRPA. The resulting kymographs were analyzed by visual inspection to identify readily resolved RAD51 that initiated from well-defined nucleation points, and the rate of filament elongation was estimated based on the rate at which these filaments spread along the ssDNA.
Filament disassembly rates were measured from experiments in which disassembly was initiated by the removal of calcium from the reaction buffer, and the rate of filament disassembly was estimated based on the rate at which the newly bound hRPA-eGFP spread along the ssDNA. The concentration of free hRPA-eGFP used (10 nM) is in vast excess to the amount of ssDNA in the flowcells. The positions of the individual filament ends were identified from kymographs by first selecting the time point (frame) in which the fluorescent signal arising from the re-binding of hRPA-eGFP had reached 15% of its maximal values. The hRPA-eGFP signal intensity for each pixel along the selected ssDNA molecule was then plotted against position along the ssDNA, and the resulting data analyzed to define local maxima. These local maxima were then used as proxies for the approximate the positi ons of the initial hRPA-eGFP binding events following the initiation of RAD51 filament disassembly.
ATP hydrolysis assays
ATP hydrolysis reactions were carried out at 37°C in HR buffer containing 23 μM nucleotide of M13 ssDNA, 2 mM MgCl2, 1 mM ATP, 5 μCi [γ32-P]ATP and 2 μM RAD51. Aliquots (3 μl) were removed at the indicated time intervals and reactions terminated by mixing with an equal volume of 100 mM EDTA. Each sample (1 μl) was spotted on a PEI-cellulose TLC plate (Sigma Aldrich) and resolved in running buffer containing 0.5 M LiCl and 1 M formic acid. The dried TLC plates were exposed to a phosphor screen and quantified using a GE Life Sciences Typhoon imager. The extent of ATP hydrolysis was then determined by calculating the fraction of radioactivity from free phosphate relative to the total radioactivity present in the sample.
Monte Carlo simulation of RAD51 disassembly
Monte Carlo simulations were performed essentially as described (33,40). Within the simulations, a 16 000 element 1-D array was used to emulate the RAD51 bound ssDNA, with each element representing a triple nucleotide that can be bound by one Rad51 monomer. Starting at one end of the array, the entire ssDNA was populated with RAD51 filaments of a pre-determined length with 30 nucleotides placed between adjacent filaments. Simulations were performed using RAD51 filaments that varied from 20 to 1220 monomers in length. Filament disassembly was modeled as a two-stage process starting with ATP hydrolysis and following with end-dependent protein monomer dissociation. Where indicated, the model was modified to reflect the possibility of internal RAD51 dissociation. The ATP hydrolysis rate was fixed at 0.009 sec−1 based upon our experimental measure of RAD51 ATP hydrolysis in the absence of Ca2+. A time step of 1 second was used with in the Monte Carlo simulation, and for each time step, a value was extracted from a random distribution of values ranging between 0 and 1. If the selected value was smaller than the rate for that step, then that monomer underwent this change. The resulting simulation data were evaluated using a least-squares method for comparing the simulated curve to the experimental data for RAD51 dissociation.
RESULTS
Visualizing human RPA with ssDNA curtains
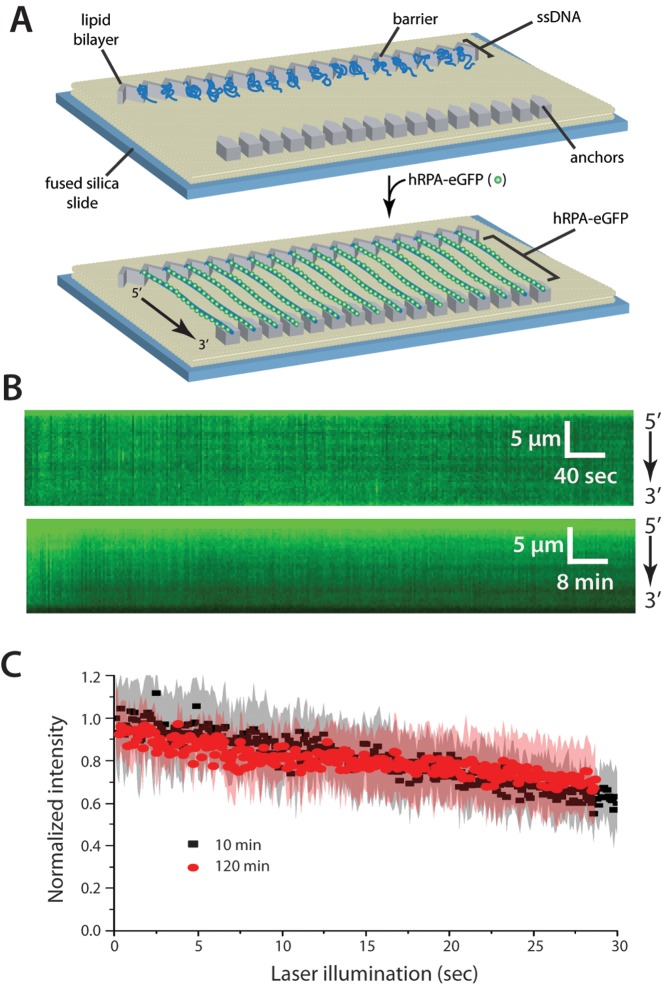
We have established an ssDNA curtain assay that allows us to observe large numbers of ssDNA molecules and their binding partners in real-time by total internal reflection fluorescence microscopy (27,38,39,41). Using this technique, we have previously shown that S. cerevisiae RPA (ScRPA) binds very tightly to ssDNA, with an observed half-life exceeding 2 h (26). Therefore, as an initial assessment of hRPA ssDNA-binding stability, we prepared ssDNA curtains bound by hRPA-eGFP (Figure 1A and B). After removing free hRPA-eGFP, we monitored the stability of the hRPA-eGFP–ssDNA complexes for either 10 min or 2 h while keeping the total illumination (laser exposure) time constant (Figure 1C). We observed a slight decrease in fluorescent signal intensity in both cases, but this decrease in signal was the same for the 10-min and 2-h observations, indicating that it was due to photobleaching, and did not reflect dissociation of hRPA-eGFP from ssDNA. Based on these results, we conclude that the half-life of the RPA-eGFP–ssDNA complex is much longer than 2 h.
Figure 1.
Human RPA can bind tightly to ssDNA. (A) Schematic of the double-tethered hRPA-ssDNA curtain, showing the nanofabricated patterns on the surface of a fused silica microscope slide. The ssDNA molecules are anchored to the lipid bilayer in defined a 5′→3′ orientation through a biotin-streptavidin-biotin linkage and aligned at the zig-zag shaped chromium (Cr) barriers. The ssDNA is labeled and extended by injection of hRPA-eGFP and downstream ends are anchored through non-specific adsorption of the RPA-ssDNA to the exposed Cr pedestals. (B) Kymographs showing single ssDNA molecules bound by hRPA-eGFP in the absence of free RPA. Images (100-msec exposure) where collected at 2-s intervals for 10 min (upper panel) or at 24-s intervals for 2 h (lower panel), as described (26). (C) Loss of RPA-eGFP signal over time is due to photobleaching. Collecting images with longer shutter time (24 s) led to the same rate of signal decrease when corrected for total illumination time. Each curve represents normalized averages over time calculated from at least 14 individual ssDNA molecules, and the shaded regions represent standard deviation.
Human RPA undergoes facilitated exchange
Although ScRPA binds very tightly to ssDNA, it also turns over rapidly when free ScRPA is present (26). This concentration-dependent protein turnover is referred to as facilitated exchange (36,37), and this phenomenon is thought to reflect the existence of microscopically dissociated states, which only undergo macroscopic dissociation when free RPA is available to occupy ssDNA that is exposed during the microscopic dissociation events (42–44).
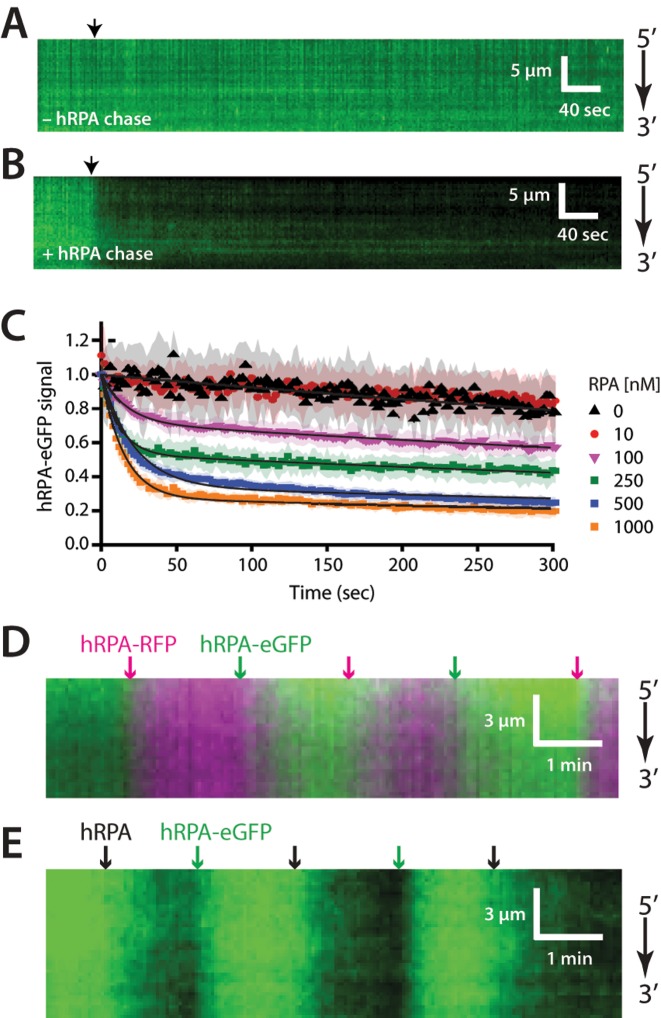
To determine whether human RPA also undergoes facilitated exchange in vitro, we pre-bound hRPA-eGFP to the ssDNA curtains, and then chased these complexes with buffer containing varying concentrations of free untagged hRPA. As expected, hRPA-eGFP remained tightly bound to the ssDNA in mock reactions lacking free hRPA (Figure 2A). In striking contrast, hRPA-eGFP rapidly dissociated from the ssDNA curtains when chased with buffer containing 1 μM free hRPA (Figure 2B) and both the rate and extent of hRPA-eGFP dissociation were dependent upon the concentration of the free hRPA (Figure 2C).
Figure 2.
Facilitated exchange of ssDNA-bound hRPA. (A) Representative kymograph showing hRPA stably bound to ssDNA in the absence of free RPA. Arrowhead indicates injection of HR buffer alone. (B) Kymograph showing dissociation of ssDNA-bound hRPA-eGFP upon injection of unlabeled wild-type hRPA (1 μM), as shown by the black arrowhead. (C) Normalized RPA-eGFP signal intensity versus time after injection of different concentrations of unlabeled, wild-type hRPA. Each curve represents normalized averages over time calculated from at least 18 different ssDNA molecules, and shaded regions represent standard deviation. Loss of RPA-eGFP signal over time is due to combination of photobleaching (e.g. at 0 nM and 10 nM free hRPA) and hRPA-eGFP turnover in the presence of 100–1000 nM free hRPA. Accordingly, the curves were fit to double exponential decays (solid black lines), with one of the rates corresponding to photobleaching. (D) Two-color kymograph showing facilitated exchange between successive injections of hRPA-eGFP (green) and hRPA-RFP (magenta) and the corresponding color-coded arrowheads indicate injection time each different hRPA protein. (D) Kymograph showing facilitated exchange between successive injections of hRPA-eGFP (green) and (E) wild-type hRPA (dark), and the corresponding arrowheads indicate the injection time for each protein.
We next conducted experiments involving iterative exchanges between hRPA-eGFP and hRPA-RFP (Figure 2D), and between hRPA-eGFP and untagged hRPA (Figure 2E). If hRPA is able to undergo facilitated exchange, then these iterative changes in hRPA identity should coincide with changes in the fluorescence color or signal intensity observed on the individual ssDNA molecules. This behavior has been previously observed for S. cerevisiae RPA (45). As anticipated, when ssDNA-bound hRPA-eGFP was chased with buffer containing hRPA-RFP (100 nM) the fluorescence signal on the ssDNA molecules changed rapidly from green to red, and vice versa (Figure 2D). Similarly, when ssDNA-bound hRPA-eGFP was chased with buffer containing wild-type hRPA (100 nM) the fluorescence signal on the ssDNA molecules oscillated between green and dark, and vice versa (Figure 2E). This last experiment demonstrates that untagged hRPA also undergoes facilitated exchange when chased with the eGFP-tagged protein, confirming that the observed behavior is not due to the presence of the eGFP (or RFP) tag. We conclude that, like its yeast counterpart, hRPA is able to under concentration-dependent facilitated exchange between free and bound states.
Human RPA exchange with heterotypic binding proteins
Saccharomyces cerevisiae RPA (ScRPA) can undergo facilitated exchange in vitro when chased with either ScRPA or the E. coli ssDNA-binding protein SSB, and vice versa (26). This result provides strong evidence that the facilitated exchange involving ScRPA is not reliant upon species-specific protein-protein contacts. Similarly, experiments using hRPA and either E. coli SSB or ScRPA revealed that the former is readily displaced from ssDNA by either of the heterologous proteins (Supplementary Figure S1). These findings reveal that hRPA displacement from ssDNA by another ssDNA-binding protein does not require any specific interactions with the competing protein.
Cooperative assembly of RAD51 on hRPA-coated ssDNA
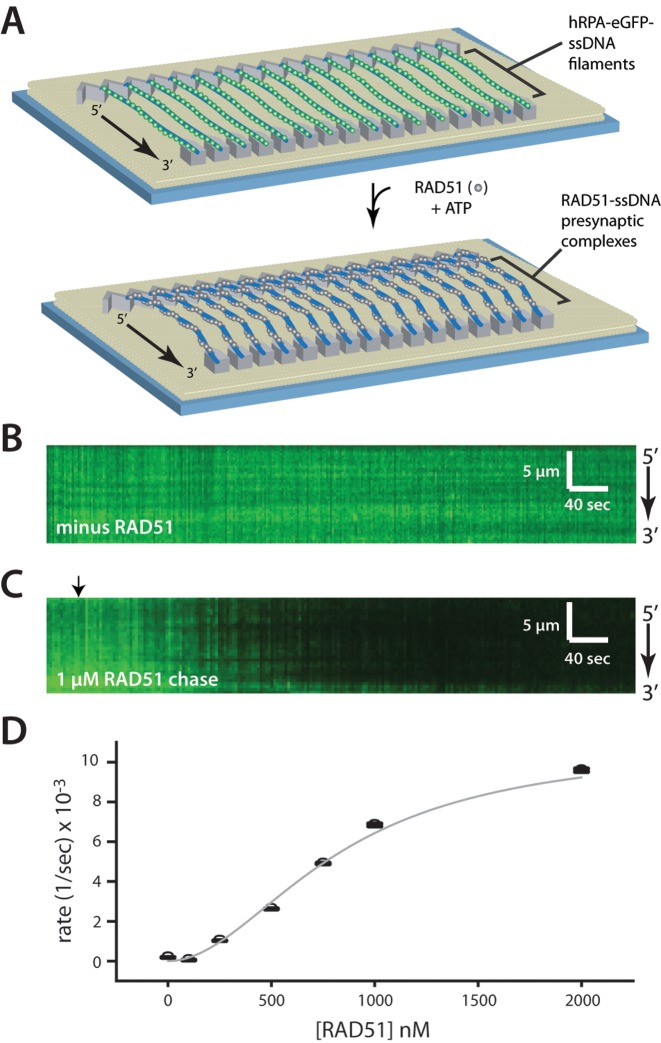
The RPA–ssDNA complex is the physiologically relevant substrate for presynaptic complex assembly, and although many single-molecule studies have focused on RAD51 assembly on either naked ssDNA or naked dsDNA, relatively little is known about how RAD51 assembles onto hRPA-coated ssDNA molecules. Therefore, we next examined the assembly of human RAD51 filaments on the hRPA-eGFP-coated ssDNA curtains (Figure 3A). The addition of RAD51 led to the rapid loss of fluorescent signal as hRPA-eGFP was displaced from the ssDNA (Figure 3B and C). Reactions performed at different RAD51 concentrations confirmed that RAD51 filament assembly was cooperative (Figure 3D), consistent with previous reports of RAD51 ssDNA binding behavior (32–34). The experimentally observed assembly rates were fit to the Hill equation, yielding a Hill coefficient of n = 1.95 ± 0.32 (Figure 3D), which is also consistent with previous reports (32–34).
Figure 3.
Assembly of the RAD51 presynaptic complex. (A) Schematic showing the ssDNA curtain experiment for visualizing assembly of wild-type (dark) RAD51 using ssDNA-bound by hRPA-eGFP as a starting substrate. (B) Kymograph showing hRPA-eGFP stably bound to ssDNA in the absence of RAD51. (C) Kymograph showing displacement of hRPA-eGFP from the ssDNA upon injection of unlabeled RAD51 (indicated by black arrowhead) in the presence of 2 mM ATP, 1 mM Mg2+ and 5 mM Ca2+. The decrease in signal intensity is the result of hRPA-eGFP photobleaching, dissociation of hRPA-eGFP by RAD51 and the movement of molecules away from the TIRF field due to extension of the ssDNA. (D) The observed dissociation rates (see Supplementary Figure S3A), corrected to remove the contribution of photobleaching, are plotted against the RAD51 concentration. The constants are subsequently fitted to the hill equation to extract the Hill coefficient of 1.95 ± 0.322, Vmax = 0.011 ± 0.0042 s−1 and km = 828 ± 319 nM.
Free RPA inhibits presynaptic filament assembly
Although the RPA–ssDNA complex is the physiologically relevant target for presynaptic filament assembly, RPA can interfere with the filament assembly process by preventing Rad51 binding. For instance, if RPA is added prior to or concurrently with Rad51 in bulk biochemical reactions, then RPA out competes Rad51 for available ssDNA binding sites (46). This inhibitory effect of RPA can be overcome by including recombination mediator proteins such as Rad52 (in yeast) or BRCA2 (in humans) (47,48). In vivo, Rad52 and BRCA2 help load Rad51 onto ssDNA to overcome the inhibitory effects of RPA and these mediators are clearly needed for the timely assembly of RAD51–ssDNA complexes at DNA lesions (1,7,29). Interestingly, RAD51 filament formation in our ssDNA curtain assays observed in the absence of mediator, and we see no evidence that the ssDNA-bound RPA prevents Rad51 loading for either S. cerevisiae (26,27) or human proteins (see above). However, our assays are performed within microfluidic chambers allowing for the removal of free RPA prior to RAD51 addition. This situation differs considerably from either bulk biochemical or the in vivo situation where free RPA is always present. Therefore, we next asked whether RAD51 ssDNA-binding activity was affected by the presence of free hRPA.
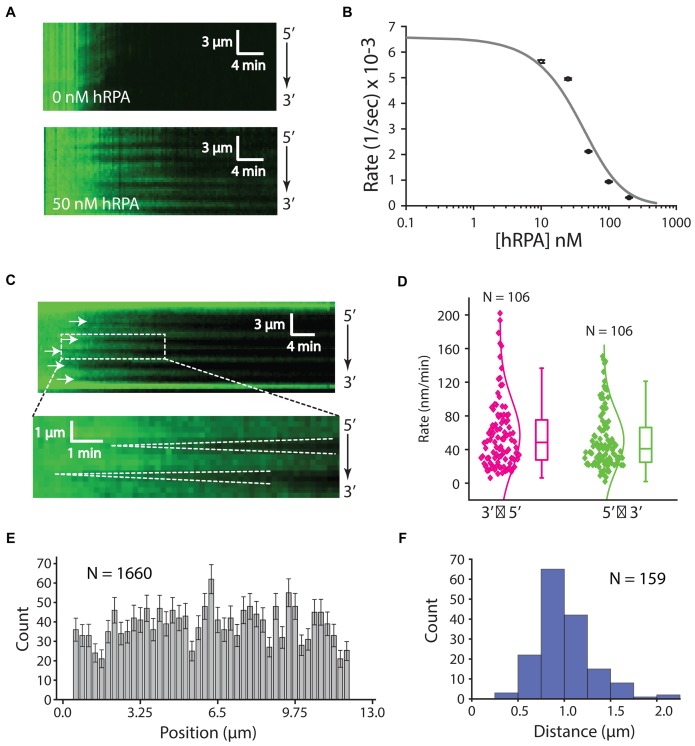
To determine the effect of free hRPA on RAD51 filament assembly, we concurrently injected a fixed concentration of RAD51 (750 nM) along with varying concentrations of free hRPA-eGFP (0–200 nM) into sample chambers containing hRPA-eGFP-coated ssDNA molecules (Figure 4A). Low concentrations of free hRPA (≤25 nM) had no appreciable effect on the rate of RAD51 filament assembly (Figure 4A and B). However, higher concentrations of hRPA (≥50 nM) significantly reduced the rate of RAD51 filament assembly (Figure 4A and B). It should be noted that each RPA heterotrimeric complex is capable of associating with up to 30 nucleotides, whereas each RAD51 monomer engages 3 nucleotides of ssDNA meaning that 10× more RAD51 than RPA is necessary to occupy the same amount of ssDNA. Given that free hRPA is likely to compete with RAD51 for the same ssDNA sites, we fitted the resulting assembly rate data to a competitive inhibition model (Figure 4B), yielding an apparent inhibition constant (Kapp) of 35.49 ± 9.33 nM. This number is an estimate as the binding site size for is much larger compared to RAD51, but we are able to conclude that free RPA restricts RAD51 filament assembly through a mechanism involving simple competitive inhibition.
Figure 4.
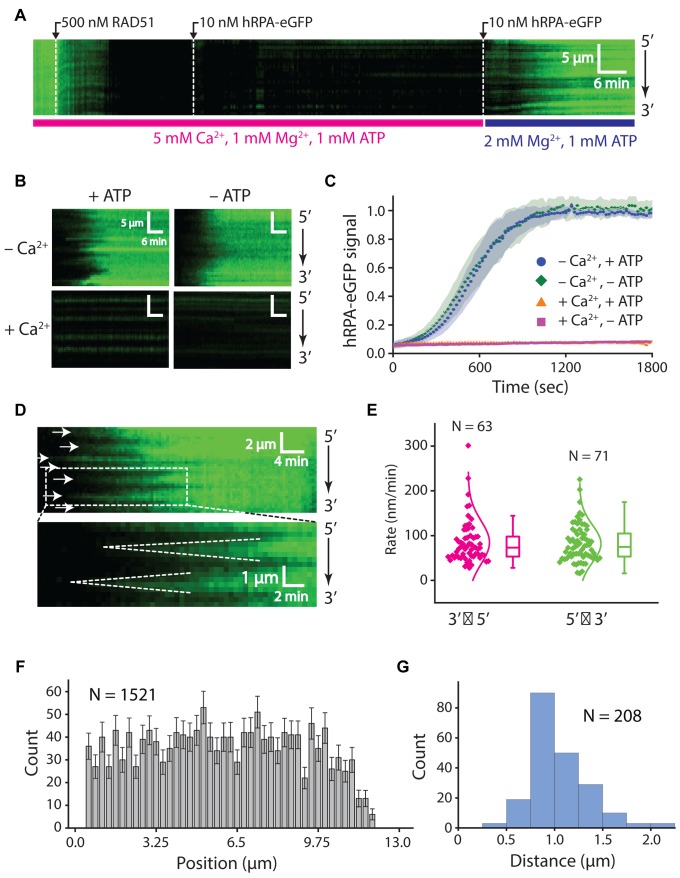
Influence of hRPA on RAD51 presynaptic complex assembly. (A) Assembly of the RAD51 filament on ssDNA-hRPA in the presence of free hRPA-eGFP. Representative kymograph showing dark RAD51 (750 nM) binding to hRPA-eGFP coated ssDNA in the presence or absence of 50 nM free hRPA-eGFP, as indicated. (B) Plot of hRPA-eGFP dissociation rates after injection of 750 nM RAD51 with different concentrations of free hRPA-eGFP in buffer containing 2 mM ATP, 1 mM Mg2+ and 5 mM Ca2+. Each rate was calculated from at least 42 different ssDNA molecules (see Supplementary Figure S3B). The resulting data were fitted to the adjusted hill equation for competitive inhibition by hRPA. (C) Kymographs highlighting individual nucleation events (white arrowheads) and the bi-directional RAD51 filament growth. (D) Plot showing filament elongation rates for 5′→3′ and 3′→5′ growth. (E) Position distribution histogram showing the locations of different RAD51 nucleation events along the length of the ssDNA substrate; error bars correspond to std. dev. obtained from bootstrapping. (F) Size distribution histogram reporting the lengths of RAD51 filaments based on the distances between adjacent nucleation events, as highlighted by the white arrowheads in (C) for 750 nM RAD51 with 50 nM RPA.
Interestingly, for concentrations of hRPA that inhibited assembly we also noticed a qualitative difference in the pattern of RAD51 assembly (Figure 4A and C). In the absence of hRPA, nucleation of the RAD51 filaments occurred rapidly and relatively uniformly along the length of the ssDNA (Figures 3C and 4A). In contrast, with increasing concentrations of hRPA, visual inspection of the resulting kymographs revealed clear evidence of independent RAD51 nucleation events followed by progressive elongation of the unlabeled RAD51 filaments in both directions along the ssDNA (Figure 4A and C). In reactions with 750 nM RAD51 and 50 nM hRPA, the apparent rates of assembly for 3′→5′ and 5′→3′ growth were 57 ± 42 and 51 ± 35 nm/min, respectively (Figure 4D). Our resolution of 1 pixel per ∼750 nucleotide of RAD51-ssDNA precludes us from being able to directly observing the initial nucleation events as there may be only a few RAD51 molecules involved in these events and our fluorescence signal depends on RPA dissociation rather than RAD51 binding. The nucleation sites appeared to be randomly distributed along the length of the ssDNA molecules (Figure 4C and E) and the mean distance between visibly resolved nucleation events was 0.88 ± 0.30 μm (Figure 4F). We anticipate that this value reflects an upper bound on the actual lengths of the RAD51 filaments under these reaction conditions, as there are likely to be nucleation events too close to resolve (see below) (33,49,50). We conclude that the presence of as little as 50 nM free hRPA can significantly reduce the rate of RAD51 filament assembly through inhibition of nucleation and also results in the formation of longer individual Rad51 filaments.
Disassembly of RAD51 filaments
Previous in vitro studies have demonstrated that calcium enhances RAD51-mediated DNA strand exchange by attenuating ATP hydrolysis and thus preventing the self-inactivation of RAD51 (51,52). Our DNA curtain assays recapitulate this effect, showing an increase in the rate of RAD51 filament assembly with Ca2+ present (Supplementary Figure S2). Indeed, RAD51 filament assembly on the hRPA-coated ssDNA was completely inhibited in the absence of Ca2+, highlighting the importance of preventing ATP hydrolysis in the assembly of the human presynaptic complex (Supplementary Figure S2). Moreover, RAD51 presynaptic complexes assembled in the presence of 5 mM Ca2+, 1 mM Mg2+ and 1 mM ATP remained stable even when free RAD51 was flushed from the sample chamber (Figure 5A). However, RAD51 dissociated from the ssDNA upon removal of Ca2+, even in the continued presence of 2 mM Mg2+ and 1 mM ATP (Figure 5A and B). This suggests that RAD51-ADP-ssDNA is not a stable complex and readily dissociates after ATP hydrolysis.
Figure 5.
Disassembly of the RAD51 presynaptic complex. (A) Kymograph showing the assembly of an RAD51 filament on hRPA-eGFP coated ssDNA in buffer containing 1 mM ATP, 1 mM Mg2+ and 5 mM Ca2+. Free RAD51 was removed by chasing with buffer containing 1 nM hRPA-eGFP, and filament disassembly was initiated by chasing with buffer lacking Ca2+. (B) Kymographs showing the behavior of pre-assembled the RAD51 filaments when chased with buffers with and without Ca2+, ATP, or both, as indicated. (C) hRPA-eGFP normalized signal intensity versus time after injection of free hRPA-eGFP as a readout of RAD51 dissociation. Signal increase is the result of binding of free hRPA-eGFP that follows the dissociation of RAD51 from ssDNA. Shaded regions on the curves represent standard deviation, at least 23 ssDNA molecules were measured for each condition. (D) Kymographs highlighting the bi-directional disassembly of RAD51 filaments as visualized by re-binding of fluorescent hRPA-eGFP. White arrows highlight the positions at which disassembly initiates, and dashed lines highlight examples of bi-directional filament disassembly. (E) Plot showing filament disassembly rates for 5′→3′ and 3′→5′ dissociation. (F) Position distribution histogram showing the locations at which RAD51 filament dissociation initiates along the ssDNA; error bars correspond to std. dev. obtained from bootstrapping. (G) Size distribution histogram reporting the lengths of RAD51 filaments based on the distances between adjacent positions at which disassembly initiates; as highlighted by the white arrowheads in (D).
We next asked whether RAD51 filaments prepared in the presence of Ca2+remained intact when chased with buffer lacking ATP. RAD51 dissociated from the DNA upon the removal of both ATP and Ca2+, at a similar rate to the removal of Ca2+ alone (Figure 5B and C). Surprisingly, the RAD51 filaments remained fully intact when Ca2+ was retained in the buffer even when ATP was flushed from the sample chamber (Figure 5B and C). These findings highlight the influence of Ca2+ on ATP hydrolysis and the stability of RAD51 filaments.
We examined RAD51 filament disassembly induced by Ca2+ removal by following the re-binding of hRPA-eGFP (Figure 5A, B and D). Interestingly, inspection of the resulting kymographs revealed that hRPA-eGFP binding began at defined positions along the ssDNA with subsequent spreading of the fluorescence signal from the initial binding sites in both the 5′→3′ and 3′→5′ directions at apparent rates of 85 ± 48 and 82 ± 41 nm/min, respectively (Figure 5D and E). The initial appearance of RPA-eGFP on the ssDNA likely signifies the initiation of RAD51 filament dissociation, with the bi-directional spreading of hRPA-eGFP reflecting the progressive, end-dependent disassembly of two adjacent RAD51 filaments. As with RAD51 filament nucleation (Figure 4), the positions at which RAD51 disassembly initiated appeared to be randomly distributed along the ssDNA, reflecting lack of sequence specificity for the initiation of filament disassembly within our resolution limits (Figure 4D and F). Assuming that the distance between sites of initial hRPA-eGFP nucleation represents the ends of adjacent RAD51 filaments, then the visually observed filaments exhibit a mean length of 0.92 ± 0.31 μm (Figure 5G). However, these length estimates should be taken with caution because there are likely numerous filaments too short to be spatially resolved in our measurements, therefore this value likely represents an upper limit on filament length under our reaction conditions (see ‘Discussion’ section) (33,49,50).
DISCUSSION
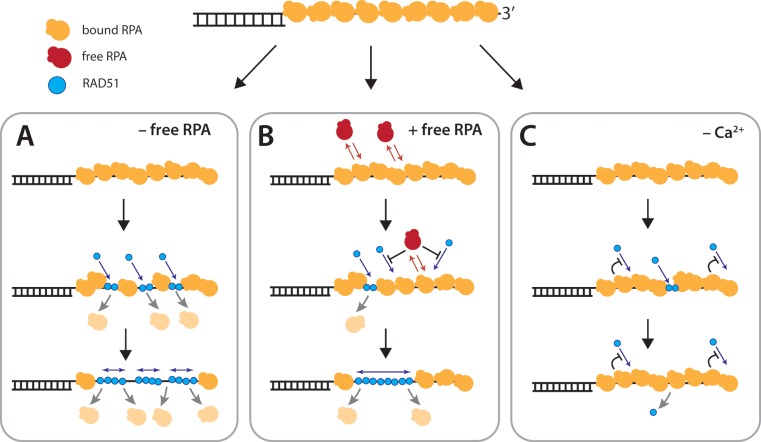
Single-stranded DNA curtains allow for the direct visualization of the dynamics of protein binding and dissociation on the ssDNA in real-time. Here, we have used these curtains to study the dynamic properties of human RPA and RAD51 during filament assembly and disassembly, to provide new insights into these processes. Together our data allow us to propose a model for the dynamic interplay between hRPA, RAD51 and ssDNA during the early stages of DSB processing and presynaptic complex assembly (Figure 6).
Figure 6.
Model for RAD51 filament assembly. (A) When free hRPA is absent from solution, the ssDNA-bound hRPA does not exchange between free and bound state and RAD51 nucleation is unrestricted, resulting in numerous, short filaments. (B) When free hRPA is present, hRPA is in dynamic equilibrium between free and bound states. Free hRPA also restricts RAD51 nucleation events, giving rise to fewer, but longer filaments. (C) In the absence of Ca2+ RAD51 either cannot nucleate on the DNA, or the nucleation events do not result in stable DNA-bound complexes that are capable of supporting filament elongation. Additional details are presented in the ‘Discussion’ section.
Mechanism and impact of hRPA turnover
Recent studies have begun to shed light on the highly dynamic behavior of RPA in its interactions with ssDNA (9), including the ability of hRPA to undergo rapid one-dimensional diffusion (53) and that of ScRPA to rapidly exchange between free and bound states (25–27). Our work shows that hRPA remains very stably bound to ssDNA in the absence of free protein (Figure 6A). However, hRPA undergoes rapid exchange between bound and unbound states when free hRPA is present in vitro (Figure 6B). These observations are most consistent with a facilitated exchange mechanism, wherein the four ssDNA-binding OB-folds interact with the ssDNA through multiple contacts to allow hRPA to undergo constant microscopic dissociation events without complete dissociation of the protein from the ssDNA (54–56). Previous studies have determined that these OB-folds have different affinities for ssDNA, resulting in two RPA binding modes that are differentiated by the number of OB-folds associated with ssDNA (a weak 8-nt mode with only 2-OB folds associated, and a stronger 24–30 nt binding mode with 3–4 OB folds associated with the ssDNA) (54,55). Macroscopic dissociation of RPA requires that all four OB-folds be disengaged from the ssDNA simultaneously and becomes possible only when free protein molecules are present to compete for the short patches of ssDNA that become exposed during each microscopic dissociation event. Importantly, we have previously reported a similar behavior for ScRPA and E. coli SSB (25–27) and bulk biochemical studies have also suggested that SSB undergoes facilitated dissociation (57). These findings suggest that the ability to undergo facilitated exchange in vitro is a broadly conserved property among ssDNA-binding proteins (45,56). We note that facilitated exchange has also been reported for various dsDNA-binding proteins in vitro, including Fis, HU, HMGB, NHP6A, EcoRI and the transcription factor CueR (37,43,44,58–60).
Interestingly, when chased with an excess of untagged RPA, 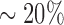 of hRPA-eGFP remained bound to ssDNA. We have previously reported a similar behavior for ScRPA (26), highlighting another similarity between the human and yeast proteins. It is not yet clear why a subpopulation of RPA remains resistant to facilitated exchange, although a recent report has also demonstrated the existence of two distinct hRPA populations with differential ssDNA dissociation kinetics (28). It is possible RPA has differential affinities for particular sequences within ssDNA, and bulk biochemical studies have shown that yeast and human RPA both display a moderate preference for polypyrimidine versus polypurine (14). Alternatively, it is possible that RPA can adopt a conformation that enhances its affinity for ssDNA or hRPA-hRPA contacts, as recently demonstrated for E. coli SSB (61), which may render individual RPA molecules resistant to facilitated exchange. Future studies will be necessary to help distinguish between these possibilities.
of hRPA-eGFP remained bound to ssDNA. We have previously reported a similar behavior for ScRPA (26), highlighting another similarity between the human and yeast proteins. It is not yet clear why a subpopulation of RPA remains resistant to facilitated exchange, although a recent report has also demonstrated the existence of two distinct hRPA populations with differential ssDNA dissociation kinetics (28). It is possible RPA has differential affinities for particular sequences within ssDNA, and bulk biochemical studies have shown that yeast and human RPA both display a moderate preference for polypyrimidine versus polypurine (14). Alternatively, it is possible that RPA can adopt a conformation that enhances its affinity for ssDNA or hRPA-hRPA contacts, as recently demonstrated for E. coli SSB (61), which may render individual RPA molecules resistant to facilitated exchange. Future studies will be necessary to help distinguish between these possibilities.
The ability of RPA to undergo facilitated exchange has important implications for understanding the role and behavior of RPA during DSB repair. RPA is highly abundant, and eGFP-RPA localizes to DSB sites rapidly (62–65). Our work predicts that DSB-bound RPA can undergo facilitated exchange as a feasible mechanism to hand ssDNA off to downstream proteins in the DSB repair pathway.
Effects of free hRPA on RAD51 presynaptic filament assembly
The role of RPA during the early stages of recombination is complex, as it both inhibits and facilitates ssDNA engagement by Rad51 (14,30). RPA can compete with Rad51 for ssDNA binding, but is also necessary for removing secondary structure from ssDNA to enable efficient Rad51 presynaptic complex formation. In vivo, the inhibitory effect of RPA is overcome by mediator proteins such as Rad52 or BRCA2, which promote the nucleation of Rad51 on RPA-coated ssDNA (7,29). Our ssDNA curtain studies with human and yeast Rad51 have shown that ssDNA-bound RPA does not prevent Rad51 filament formation in the absence of a mediator protein (Figure 6A). This result was somewhat surprising given the requirement for mediator proteins during recombination in cells and in numerous in vitro studies. However, this apparent discrepancy is explained by the finding that in our experimental system we are able to control the concentration of free RPA, leading to the suggestion that free RPA restricts RAD51 nucleation, whereas ssDNA-bound RPA only does not (Figure 6A and B). It is likely that mediators can work together to help suppress this inhibition in vivo, and this will be a major focus for future studies.
Interestingly, RAD51 filament assembly appears qualitatively different when free hRPA is present. In the absence of free hRPA, RAD51 filament assembly is very rapid and appears to entail numerous nucleation events, followed by relatively limited elongation, leading to a nearly uniform disappearance of hRPA-eGFP from the ssDNA (Figure 6A). These observations are consistent with previous studies suggesting that RAD51 filament formation occurs through many nucleation events, giving rise to relatively short filaments consisting of a few tens of RAD51 monomers each (33,49,50). In contrast, when free hRPA is present, RAD51 filament assembly becomes much slower and a significant number of the resulting RAD51 filaments could be segregated into visually identifiable nucleation events followed by bidirectional filament elongation. Taken together, these observations are consistent with a model where free hRPA inhibits RAD51 nucleation, but has a lesser impact on filament growth. As a consequence, the fewer RAD51 nucleation events that take place in the presence of free hRPA result in the formation of longer filaments (Figure 6B). This can be compared to how RecA is unable to nucleate and assemble on a SSB-bound ssDNA but can grow by elongation on the same substrate provided there are pre-existing RecA nuclei (66). Although we were able to assemble RAD51 filaments at high enough RAD51 concentration on RPA-bound ssDNA, this would suggest nucleation as a point of conservation in the regulation of filament assembly and the part of assembly that can be heavily influenced by mediators.
We note that free hRPA likely affects the characteristics of the resulting RAD51 filaments as well. Specifically, RAD51 nucleation events in the absence of free RPA may result in short RAD51 filaments (33,49,50) that are out of register with one another, resulting in potential discontinuities within the protein filament (33,49,50). In contrast, the infrequent RAD51 nucleation events that occur when free hRPA is present would yield longer contiguous RAD51 filaments that may possess a different catalytic potential. Future work will determine whether discontinuities within the RAD51 filament indeed affect its ability to promote the homology search and DNA strand exchange.
Our observations also raise the question of how RAD51 displaces hRPA from ssDNA. One possibility is that there exist protein–protein interactions between RAD51 and hRPA that somehow promote the association of RAD51 with the ssDNA while simultaneously enabling the removal of hRPA. However, even though hRPA and RAD51 can interact directly (56) and RPA may be able to localize RAD51 to ssDNA, the generally inhibitory nature of hRPA on the ssDNA-binding activity of RAD51 would suggest that this interaction only makes a small contribution to RAD51 binding activity. In addition, human RAD51 and bacterial RecA can both assemble on ssDNA bound by S. cerevisiae RPA, further diminishing the importance of protein–protein interactions for RAD51 binding (39,41). Another possibility is that RAD51 takes advantage of the inherent propensity of hRPA to undergo facilitated turnover, and simply nucleates at ssDNA sites that are exposed during the hRPA microscopic dissociation events, possibly at moments when human RPA shifts to a weaker binding mode with the smaller 8-nt footprint (55). In agreement with this hypothesis, we can detect assembly of human RAD51 and ScRad51 filaments on ssDNA bound by the heterologous RPA. We can even detect efficient assembly of E. coli RecA filaments on ssDNA bound by either human or S. cerevisiae RPA (26,39,41). This hypothesis also leads to a competitive inhibition model where free hRPA can compete with hRAD51 for binding to ssDNA that is exposed during microscopic dissociation of bound hRPA to inhibit nucleation and thereby slowing down filament assembly (Figure 6B). Based on these results, in our system, the most probable model for RAD51 binding relies mostly on RAD51 displacing ssDNA-bound hRPA by taking advantage of its inherent propensity to undergo facilitated exchange and this displacement is prevented by free hRPA through competitive inhibition (Supplementary Figure S4). However, although the interaction of RPA and RAD51 is not a major contributor in vitro, in cells, it may play a more significant role when other recombination mediators are present to counteract the inhibitory aspect of RPA binding.
Stability and disassembly of the human Rad51 presynaptic complex
Calcium strongly stimulates DNA strand exchange by human RAD51 (51,52), and it acts by inhibiting ATP hydrolysis thereby keeping the presynaptic filament in the ATP-bound state (51,52). Indeed, Ca2+ is critical to filament assembly in our assays, and in reactions containing 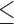 2 μM RAD51, we are unable to detect stable ssDNA binding or filament assembly in the absence of Ca2+. Interestingly, previous studies have reported RAD51 filament assembly on ssDNA in the absence of ATPase inhibition by Ca2+, but these studies did not included ssDNA pre-bound by RPA (32,33,51). Our finding that RAD51 cannot assemble on RPA-coated ssDNA suggests the possibility that the presence of RPA on the ssDNA may impose more stringent requirements for filament assembly. For instance, RAD51 may be unable to nucleate on RPA-coated ssDNA, or nucleation may give rise to highly unstable complexes that rapidly dissociate in the absence of an agent that prevents ATP hydrolysis (Figure 6C). In our experiments, we inhibited ATPase activity with calcium, but in vivo it is more likely to be one or a combination of recombination mediators in the nucleus as cellular calcium levels are much lower than those used in our experiments (67). Importantly, we do not observe RAD51 mediated hRPA dissociation in the absence of Ca2+; if transient nucleation can take place under these conditions then it does not result in any detectable displacement of the bound hRPA. Transient nucleation with few RAD51 molecules may not be sufficient to promote complete dissociation of all four RPA OD-binding domains. Remarkably, when ATP hydrolysis is inhibited with Ca2+, RAD51 filament assembly was rapid, complete, and stable even following removal of free ATP, provided Ca2+ was present. Upon removal of Ca2+, the release of inhibition on ATP hydrolysis causes the rapid dissociation of RAD51 from the ssDNA, even if ATP is present in the buffer. These findings agree with previous studies showing that ATP hydrolysis promotes RAD51 filament disassembly on dsDNA (40,52), and further suggest that inhibition of ATPase activity locks RAD51 in a conformation of high stability.
2 μM RAD51, we are unable to detect stable ssDNA binding or filament assembly in the absence of Ca2+. Interestingly, previous studies have reported RAD51 filament assembly on ssDNA in the absence of ATPase inhibition by Ca2+, but these studies did not included ssDNA pre-bound by RPA (32,33,51). Our finding that RAD51 cannot assemble on RPA-coated ssDNA suggests the possibility that the presence of RPA on the ssDNA may impose more stringent requirements for filament assembly. For instance, RAD51 may be unable to nucleate on RPA-coated ssDNA, or nucleation may give rise to highly unstable complexes that rapidly dissociate in the absence of an agent that prevents ATP hydrolysis (Figure 6C). In our experiments, we inhibited ATPase activity with calcium, but in vivo it is more likely to be one or a combination of recombination mediators in the nucleus as cellular calcium levels are much lower than those used in our experiments (67). Importantly, we do not observe RAD51 mediated hRPA dissociation in the absence of Ca2+; if transient nucleation can take place under these conditions then it does not result in any detectable displacement of the bound hRPA. Transient nucleation with few RAD51 molecules may not be sufficient to promote complete dissociation of all four RPA OD-binding domains. Remarkably, when ATP hydrolysis is inhibited with Ca2+, RAD51 filament assembly was rapid, complete, and stable even following removal of free ATP, provided Ca2+ was present. Upon removal of Ca2+, the release of inhibition on ATP hydrolysis causes the rapid dissociation of RAD51 from the ssDNA, even if ATP is present in the buffer. These findings agree with previous studies showing that ATP hydrolysis promotes RAD51 filament disassembly on dsDNA (40,52), and further suggest that inhibition of ATPase activity locks RAD51 in a conformation of high stability.
Interestingly, the sigmoidal shape of the disassembly curves obtained upon the removal of Ca2+ suggests the existence of a rate limiting step that must take place prior to RAD51 dissociation from the ssDNA. Although we do not yet know the nature of this slow step, previous reports have suggested that RAD51 dissociation from dsDNA also involves a slow step, being attributed to either ATP hydrolysis or the exchange of Ca2+ for Mg2+ (32,40). To explore this process further, we used Monte Carlo simulations to model potential mechanisms of RAD51 filament disassembly and the simulation data were evaluated by comparison to the experimentally observed dissociation rates (Supplementary Figure S6). Within the simulations we assumed that disassembly initiated upon the exchange of Ca2+ for Mg2+, which was necessary to enable ATP hydrolysis and occurred rapidly after the buffer change, that RAD51 monomer dissociation proceeded exclusively from the filament ends and that this end-dependent dissociation step was rate limiting (Supplementary Figure S6A). We also assumed that the rate of ATP hydrolysis was equivalent to the steady state hydrolysis rate in the presence of Mg2+ only (Supplementary Figure S5), and we made no implicit assumptions regarding the lengths of individual RAD51 filaments bound to the ssDNA, but instead ran simulations for a distribution of filaments with a fixed lengths (Supplementary Figure S6B). Interestingly, the MC simulations suggest that the average lengths of the independent RAD51 filaments were on the order of 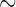 100–150 protein monomers (Supplementary Figure S6B and C), which is in good agreement with previous estimates of RAD51 filament lengths (33,49) and suggests that RAD51 filament assembly is dominated by nucleation. However, this simulation result seemingly disagrees with the visually observed lengths of the RAD51 filaments in our assays (Figures 4F and 5G). There are at least two plausible explanations for this discrepancy. First, our experimental analysis was restricted to well-defined filaments that could be identified by visual inspection and ascribed as individual filaments. Such readily defined filaments encompassed just
100–150 protein monomers (Supplementary Figure S6B and C), which is in good agreement with previous estimates of RAD51 filament lengths (33,49) and suggests that RAD51 filament assembly is dominated by nucleation. However, this simulation result seemingly disagrees with the visually observed lengths of the RAD51 filaments in our assays (Figures 4F and 5G). There are at least two plausible explanations for this discrepancy. First, our experimental analysis was restricted to well-defined filaments that could be identified by visual inspection and ascribed as individual filaments. Such readily defined filaments encompassed just 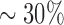 of the total RAD51-ssDNA length, so the resulting data likely represent an upper bound for the actual filament lengths. The second possibility is that our model for RAD51 filament dissociation used in the MC simulations may not accurately reflect the disassembly process. For instance, a model invoking even a small propensity of RAD51 dissociation internal positions within the filaments, rather than exclusively restricting dissociation to filament ends, can readily accommodate longer individual filaments (Supplementary Figure S6D–F). Future work will be necessary to more completely test the models for RAD51 dissociation, but together our findings provide a new basis for probing the properties of the human presynaptic complex.
of the total RAD51-ssDNA length, so the resulting data likely represent an upper bound for the actual filament lengths. The second possibility is that our model for RAD51 filament dissociation used in the MC simulations may not accurately reflect the disassembly process. For instance, a model invoking even a small propensity of RAD51 dissociation internal positions within the filaments, rather than exclusively restricting dissociation to filament ends, can readily accommodate longer individual filaments (Supplementary Figure S6D–F). Future work will be necessary to more completely test the models for RAD51 dissociation, but together our findings provide a new basis for probing the properties of the human presynaptic complex.
Mechanism of presynaptic complex assembly
In summary, from the results of our experiments we suggest a model where hRPA engages the ssDNA overhangs rapidly and then undergoes continuous turnover between free and bound states (54,55). This dynamic equilibrium is assured by free nuclear hRPA (64,65,68). Free hRPA restricts RAD51 nucleation, which may be important to keep RAD51 from inappropriate association with certain ssDNA intermediates, such as Okazaki fragments. Upon RAD51 nucleation we find that filament elongation is bidirectional, and the presence of free RPA may help ensure the formation of longer contiguous RAD51 filaments. Mediators, such as Rad52 in S. cerevisiae and BRCA2 in humans, would then function by both targeting RAD51 to the correct locations and possibly promoting nucleation of RAD51 filaments by increasing the local concentration ratio of RAD51 to RPA. This mechanism of mediator action would need to be verified in vivo but would allow cells to maintain high concentrations of RPA to quickly engage and protect ssDNA, and yet still enable proper presynaptic complex assembly at processed DSBs.
HR in eukaryotes involves the participation of at least 45 different proteins (1,7,29), many of which are also subject to regulation through post-translational modifications. This work has focused on only two primary protein participants, hRPA and RAD51, but the experimental approaches and results reported here provide the requisite foundation for further studies of human HR proteins, intermediates and mechanisms.
Acknowledgments
We thank Kyle Kaniecki, Corentin Moevus, Luisina de Tullio, Fabian Erdel, Ja Yil Lee and Justin Steinfeld for comments on the manuscript, and we thank Zhi Qi, Johannes Stigler and Sy Redding for assistance with data analysis and curve fitting.
Footnotes
Present address: Bryan Gibb, Department of Life Sciences, New York Institute of Technology, Old Westbury, NY, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R35GM118026 to E.C.G., R01ES015252, PO1CA92584 to P.S.]. Funding for open access charge: NIH [R35GM118026 to E.C.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Symington L.S., Rothstein R., Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C., Hanage W.P., Spratt B.G. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeling P.J., Palmer J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 5.Brown M.S., Bishop D.K. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 2014;7:a016659. doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neale M.J., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 8.Aguilera A., García-Muse T. Causes of genome instability. Annu. Rev. Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 9.Chen R., Wold M.S. Replication protein A: single-stranded DNA's first responder. Bioessays. 2014;36:1156–1161. doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Vaithiyalingam S., Shi Q., Chazin W.J., Zinkel S.S. BID binds to replication protein A and stimulates ATR function following replicative stress. Mol. Cell. Biol. 2011;31:4298–4309. doi: 10.1128/MCB.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X., Vaithiyalingam S., Glick G.G., Mordes D.A., Chazin W.J., Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 13.Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Wold M.S. REPLICATION PROTEIN A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA Metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 15.Oakley G.G., Patrick S.M. Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. (Landmark Ed) 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broderick S., Rehmet K., Concannon C., Nasheuer H.P. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Subcell. Biochem. 2010;50:143–163. doi: 10.1007/978-90-481-3471-7_8. [DOI] [PubMed] [Google Scholar]
- 17.Sugitani N., Chazin W.J. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog. Biophys. Mol. Biol. 2015;117:206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brill S.J., Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 19.Haring S.J., Mason A.C., Binz S.K., Wold M.S. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hass C.S., Gakhar L., Wold M.S. Functional characterization of a cancer causing mutation in human replication protein A. Mol. Cancer Res. 2010;8:1017–1026. doi: 10.1158/1541-7786.MCR-10-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Putnam C.D., Kane M.F., Zhang W., Edelmann L., Russell R., Carrion D.V., Chin L., Kucherlapati R., Kolodner R.D., et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 22.O'Driscoll M., Dobyns W.B., van Hagen J.M., Jeggo P.A. Cellular and clinical impact of haploinsufficiency for genes involved in ATR signaling. Am. J. Hum. Genet. 2007;81:77–86. doi: 10.1086/518696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Outwin E., Carpenter G., Bi W., Withers M.A., Lupski J.R., O'Driscoll M. Increased RPA1 gene dosage affects genomic stability potentially contributing to 17p13.3 duplication syndrome. PLoS Genet. 2011;7:e1002247. doi: 10.1371/journal.pgen.1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng S.K., Gibb B., de Almeida M.J., Greene E.C., Symington L.S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibb B., Ye L.F., Kwon Y., Niu H., Sung P., Greene E.C. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol. 2014;21:893–900. doi: 10.1038/nsmb.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R., Subramanyam S., Elcock A.H., Spies M., Wold M.S. Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 2016;44:5758–5772. doi: 10.1093/nar/gkw339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyer W.-D.D., Ehmsen K.T., Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianco P.R., Tracy R.B., Kowalczykowski S.C. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 31.Morrical S.W. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 2015;7:a016444. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilario J., Amitani I., Baskin R.J., Kowalczykowski S.C. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Heijden T, Seidel R., Modesti M., Kanaar R., Wyman C., Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candelli A., Holthausen J.T., Depken M., Brouwer I., Franker M.A.A., Marchetti M., Heller I., Bernard S., Garcin E.B., Modesti M., et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15090–15095. doi: 10.1073/pnas.1307824111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ha T. Single-molecule approaches embrace molecular cohorts. Cell. 2013;154:723–726. doi: 10.1016/j.cell.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadizadeh N., Johnson R.C., Marko J.F. Facilitated dissociation of a nucleoid protein from the bacterial chromosome. J. Bacteriol. 2016;198:1735–1742. doi: 10.1128/JB.00225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibb B., Silverstein T.D., Finkelstein I.J., Greene E.C. Single-stranded DNA curtains for real-time single-molecule visualization of protein-nucleic acid interactions. Anal. Chem. 2012;84:7607–7612. doi: 10.1021/ac302117z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Z., Redding S., Lee J.Y., Gibb B., Kwon Y., Niu H., Gaines W.A., Sung P., Greene E.C. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Mameren J., Modesti M., Kanaar R., Wyman C., Peterman E.J., Wuite G.J. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.Y., Terakawa T., Qi Z., Steinfeld J.B., Redding S., Kwon Y., Gaines W.A., Zhao W., Sung P., Greene E.C. DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015;349:977–981. doi: 10.1126/science.aab2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cocco S., Marko J.F., Monasson R. Stochastic ratchet mechanisms for replacement of proteins bound to DNA. Phys. Rev. Lett. 2014;112:238101. doi: 10.1103/PhysRevLett.112.238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham J.S., Johnson R.C., Marko J.F. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011;39:2249–2259. doi: 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sing C.E., Olvera de la Cruz M., Marko J.F. Multiple-binding-site mechanism explains concentration-dependent unbinding rates of DNA-binding proteins. Nucleic Acids Res. 2014;42:3783–3791. doi: 10.1093/nar/gkt1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sung P., Krejci L., Van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 47.Sung P., Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 48.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modesti M., Ristic D., van der Heijden T., Dekker C., van Mameren J., Peterman E.J., Wuite G.J., Kanaar R., Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure (London, England : 1993) 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Candelli A., Modesti M., Peterman E.J., Wuite G.J. Single-molecule views on homologous recombination. Q. Rev. Biophys. 2013;46:323–348. doi: 10.1017/S0033583513000073. [DOI] [PubMed] [Google Scholar]
- 51.Dmitry V.B., Alexander V.M. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ristic D., Modesti M., van der Heijden T., van Noort J., Dekker C., Kanaar R., Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen B., Sokoloski J., Galletto R., Elson E.L., Wold M.S., Lohman T.M. Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 2014;426:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan J., Pavletich N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brosey C.A., Yan C., Tsutakawa S.E., Heller W.T., Rambo R.P., Tainer J.A., Ivanov I., Chazin W.J. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 2013;41:2313–2327. doi: 10.1093/nar/gks1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stauffer M.E., Chazin W.J. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 2004;279:25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- 57.Kunzelmann S., Morris C., Chavda A.P., Eccleston J.F., Webb M.R. Mechanism of interaction between single-stranded DNA binding protein and DNA. Biochemistry. 2010;49:843–852. doi: 10.1021/bi901743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giuntoli R.D., Linzer N.B., Banigan E.J., Sing C.E., de la Cruz M.O., Graham J.S., Johnson R.C., Marko J.F. DNA-segment-facilitated dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J. Mol. Biol. 2015;427:3123–3136. doi: 10.1016/j.jmb.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joshi C.P., Panda D., Martell D.J., Andoy N.M., Chen T.Y., Gaballa A., Helmann J.D., Chen P. Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15121–15126. doi: 10.1073/pnas.1208508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sidorova N.Y., Scott T., Rau D.C. DNA concentration-dependent dissociation of EcoRI: direct transfer or reaction during hopping. Biophys. J. 2013;104:1296–1303. doi: 10.1016/j.bpj.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell J.C., Liu B., Kowalczykowski S.C. Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. Elife. 2015;4:e08646. doi: 10.7554/eLife.08646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 63.West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 64.Brenot-Bosc F., Gupta S., Margolis R.L., Fotedar R. Changes in the subcellular localization of replication initiation proteins and cell cycle proteins during G1- to S-phase transition in mammalian cells. Chromosoma. 1995;103:517–527. doi: 10.1007/BF00355316. [DOI] [PubMed] [Google Scholar]
- 65.Solomon D.A., Cardoso M.C., Knudsen E.S. Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell Biol. 2004;166:455–463. doi: 10.1083/jcb.200312048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joo C., McKinney S.A., Nakamura M., Rasnik I., Myong S., Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 67.Jensen R.B., Ozes A., Kim T., Estep A., Kowalczykowski S.C. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Rep. 2013;12:306–311. doi: 10.1016/j.dnarep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]