Abstract
We present a conceptually new reversible nanosensor regulated by a DNA nanoswitch. This system is not only responsive to external stimuli (e.g. ATP) but also can be reversibly switched between ‘OFF’ and ‘ON’ states via toehold mediated strand displacement reactions. It functions like a molecular net woven by DNA to capture or release the target molecules. As a proof-of-principle experiment, ATP is here chosen as the model to demonstrate our new strategy, which holds great promise for applications such as switchable DNA nanomachines and nanocarriers for drug delivery.
INTRODUCTION
In the past decades, DNA nanosensors have attracted much attention for their wide applications in environmental monitoring (1) and biological diagnosis (2), including the specific and sensitive detections of metal ions (3,4), proteins (5,6), nucleic acids (7), small molecules (8,9), etc. Recently, several DNA nanodevices have been built upon DNA nanosensors such as DNA logic sensors (10), DNA sensing dendrimers (11) and DNA sensing nanomachines (12). It reveals an emerging interest in these fields of nucleic acids. Nevertheless, only a few of reported DNA nanosensors can be reset (13,14). There remains a challenge in the development of a reusable nanosensor. Hence, it is of great significance to design a reversible nanosensor which can be reset like a switch.
In general, a DNA nanoswitch undergoes a reversible structural change upon addition/removal of external stimuli such as metal ions (1,15), light (16,17), pH value (18,19), electrons (20,21), etc. Incorporating the properties of DNA switches into sensors, herein we devise a conceptually new reversible DNA nanosensor where capturing and releasing adenosine triphosphate (ATP) molecules are switched by toehold mediated strand displacement reactions. This is in contrast to previous studies (8,22,23) that typically utilize ATP as a target to trigger DNA strand displacement reactions. In our design, a fluorescent ligand for an ATP-binding aptamer (24), Thioflavin T (ThT), is utilized as an indicator for the binding and release of ATP, as it serves as a competitive reporter and reflects ATP binding by a fluorescence decrease.
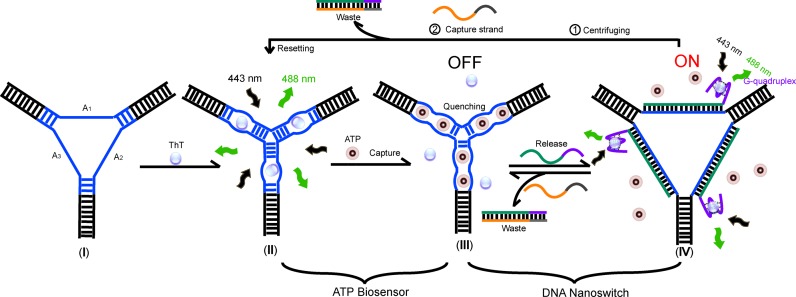
Scheme 1 briefly depicts this strategy. A functionalized contractile three-way structure was designed by incorporating three 27-mer ATP aptamer sequences (25) (in blue, Scheme 1, I) into single-stranded motifs in the center of the three-way junction. The resulting functional DNA nanostructure behaves as a specific ‘ATP-responsive three-way nanosensor’ (ARTWN) whose structural integrity was confirmed by native polyacrylamide gel electrophoresis (PAGE). Upon addition of ThT molecules, the embedded ATP aptamer sequences undergo an inter-loop folding instead of an intra-loop one, namely, each pair of ATP aptamers in two adjacent loops forms an active bimolecular structure with minor groove binding pockets where ATP binds (Scheme 1, II). As a result, the folded ARTWN strongly resembles a Watson–Crick base-paired three-arm junction rather than the six-arm, as verified by PAGE and fluorescence quenching (Figure 1). Meanwhile, the fluorescent ligand ThT is replaced by ATP and removed from ARTWN, accompanied by a sharp decrease in the fluorescent intensity of ThT (Scheme 1, III). By this means, ARTWN is able to sensitively probe ATP with high specificity.
Scheme 1.
Illustration of the working principle of the DNA nanoswitch-controlled reversible nanosensor. (I) The nanostructure of ARTWN is self-assembled from equivalent of three component strands A1, A2 and A3. (II) The initial state of nanosensor. ThT molecules intercalate into the two adjacent embedded ATP aptamers intermolecularly, giving rise to a fluorescence increase of ThT. (III) The ‘OFF’ state of the switch cycle. The bound ThT molecules are displaced by ATP, accompanied by a sharp fluorescence decrease. (IV) The ‘ON’ state of the switch cycle. Upon introduction of the release strand, the central ATP aptamers are fully hybridized thereby releasing the ATP molecules. The free ThT molecules bound to G-quaduplexes serves as a fluorescence indicator for the switch cycle.
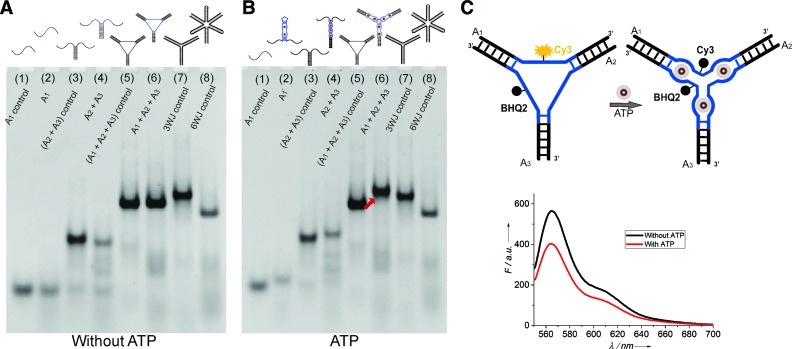
Figure 1.
Native PAGE electrophoretogram for analyzing the formation and structural transformations of ARTWN in the absence (A) and presence (B) of 300 μM ATP. Lanes 1–8: A1 control, A1, (A2+A3) control, A2+A3, (A1+A2+A3) control, A1+A2+A3, three-way junction (3WJ) control, six-way junction (6WJ) control. (C) Fluorescence quenching experiment using the Cy3-BHQ2-labeled ARTWN.
To drive ARTWN to operate repeatedly, two single-stranded DNAs were further used to release and capture ATP. The release strand consists of the complementary sequence of the ATP aptamer (Scheme 1, marked in green) and a DNA G-quaduplex PW17 (in purple). The G-quadruplex not only serves as a toehold to facilitate the strand displacement reactions, but also complexes with ThT (26) to indicate the repeated operations. The capture strand is fully complementary to the release strand. When all bound ThT molecules are replaced by ATP, ARTWN is in the ‘OFF’ state (Scheme 1, III) with low fluorescence intensity. Upon addition of the release strand, the central ATP aptamers are fully hybridized to form rigid duplexes and consequently release the ATP molecules (Scheme 1, IV). Simultaneously, the free ThT molecules bind to G-quaduplexes, giving rise to a fluorescence increase to indicate the ‘ON’ state of the switch cycle. By introducing the capture strand to remove the release strand, the system can return to the ‘OFF’ state. As the ATP molecule is released from the DNA structure in the ‘ON’ state (Scheme 1, IV), namely, it's free and unbound in the system, it can be easily separated from the DNA structure using centrifugal filter devices thereby removed from the working system. This will allow ARTWN to be reset to the initial state (Scheme 1, II) of the ATP biosensor upon addition of the capture strand.
Although a similar strategy using ATP and toehold strand displacement reactions has been reported previously (22), there are main differences between ours and the reported approach. The DNA structural integrity is here maintained throughout the whole conformational changing processes (illustrated in scheme 1), which guarantees a good reversibility of our designed DNA nanosensor. Furthermore, the reported approach (22) focused on the ‘hidden toehold’ on a metastable DNA bulge-loop structure, whereas our strategy highlights the ‘reversible nanosensor’ itself, which can be reset after the sensing processes and thus it holds great promise for applications such as switchable nanomachines, nanocarriers for drug delivery and other related DNA nanodevices. In a sense, the designed nanoswitch behaves like a molecular net woven by DNA to capture or release ATP molecules through toehold strand-displacements.
MATERIALS AND METHODS
Chemicals
All of purified DNAs used in this work were purchased from Sangon Biotechnology (Shanghai, China) and prepared in Tris-Ac (TA) buffer. Thioflavin T was purchased from Sigma-Aldrich (USA). Adenosine 5′-triphosphate (ATP), cytidine 5′-triphosphate (CTP), guanosine 5′-triphosphate (GTP) and uridine 5′-triphosphate (UTP) were purchased from Sangon Biotechnology (Shanghai, China).
Native page
The DNA nanosensors were assembled by incubating appropriate amounts of component strands at 90°C for 10 min, followed by slow cooling to room temperature. Then, they were subject to stimuli-induced structural changes under different conditions (for details, see Supplementary Data). Finally, the resulting DNA structures were analyzed by 6% native PAGE at room temperature. The gels were stained with GelRed and photographed under UV light using a Tanon Gel Imaging System (Shanghai, China).
Fluorescence spectroscopy
The responses of DNA nanosensors to external stimuli were performed as described above. The corresponding fluorescence spectra in the sensing and switching processes were all collected at room temperature using a F-4600 Fluorescence spectrometer (Hitachi, Japan). For more details, see Supplementary Data.
RESULTS
Assembly of the ‘ATP-responsive three-way nanosensor’
As a DNA scaffold, ARTWN was first formed by the self-assembly of component strands A1, A2, A3 under appropriate conditions, verified by native PAGE (Figure 1A, lane 6 versus lanes 2, 4). It demonstrates that the ARTWN is stable, as expected. This DNA structure has three loops consisting of ATP aptamer molecules and thus interacts with ATP. Since its ATP-binding sites are located on the G-rich minor groove pocket (27) formed by three loops, we replace all the guanine residues of three loops with thymine. The resulting structure is used as the control, which has no ability to bind ATP. By this means, we can show the interaction between ARTWN and ATP by an observable shift. Figure 1B depicts the structural transformations induced by ATP, a slower band of ARTWN (lane 6) is observed in the presence of ATP, as compared with the control structure (lane 5). The phenomenon is the same for A1 (lane 2), A2+A3 (lane 4). This suggests the formation of a more compact structure of ARTWN induced by ATP binding.
Binding mechanism
To demonstrate that the ARTWN undergoes a structural change to a three-way structure instead of a six-way structure in the presence of ATP, two control DNA junctions (three-way and six-way) without ATP aptamers were analyzed by PAGE together with ARTWN. The two control structures have the same number of bases as ARTWN, whereas they exhibit a big difference in the electrophoretic mobility. The three-way junction moves much slower than the six-way (Figure 1A, lane 7 versus lane 8), due to the increasing steric hindrance of lengthened arms. After binding ATP, ARTWN displays a lower mobility, which is close to that of the three-way junction rather than the six-way (Figure 1B, lane 6, compared with lanes 7, 8). It means that the folded ARTWN has a secondary structure resembling the three-way junction, as we expected.
The ATP-induced three-way structure of ARTWN is further confirmed by fluorescence quenching (Figure 1C), in which a fluorescent dye (Cy3) and a quencher (BHQ2) are labelled in the middle of A1 and A3. Without ATP, Cy3 displays a moderate fluorescence emission in the DNA structure. Upon addition of ATP, there is a sharp decrease in the fluorescence intensity of Cy3, indicating that in this case BHQ2 is close to Cy3 thus quenching it. This observation strongly suggests that ATP induces the ARTWN structure to adopt a three-arm form rather than six-arm form, because in the latter Cy3 should be separated from BHQ2 and give rise to an increase in the fluorescence intensity.
Detection of ATP molecules using the nanosensor
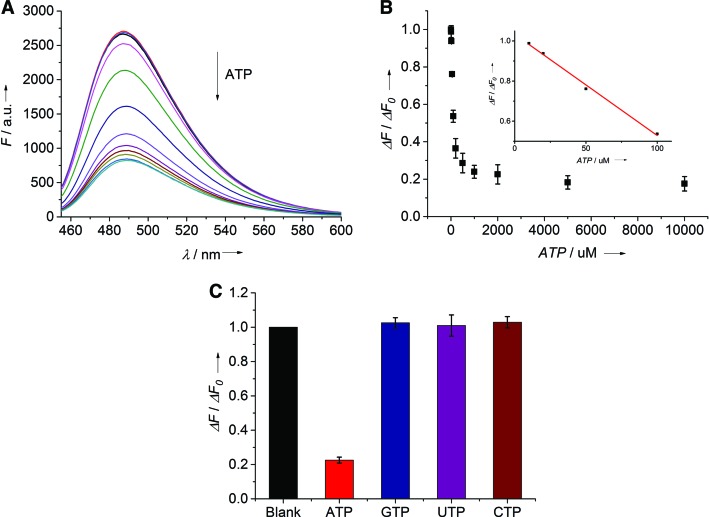
To test the ability of ARTWN for ATP analysis, the fluorescence emission spectra of ThT were recorded by varying the concentrations of ATP (Figure 2). As the concentration of ATP increases, the fluorescence intensity decreases (Figure 2A) due to the replacement effect of ThT by ATP on ARTWN. The plot of the fluorescence intensity at 488 nm (F) versus the concentration of ATP shows 10 μM of ATP can induce an observable change in the fluorescence intensity (Figure 2B, inset). Furthermore, the selectivity for ATP analysis was tested by using other analogues including GTP, CTP and UTP in place of ATP (Figure 2C). It shows that only ATP causes a remarkable decrease in fluorescence signal, indicating a good selectivity of ARTWN for ATP analysis.
Figure 2.
Fluorescent analysis of ATP molecules by using ThT as a reporter. (A) Fluorescence emission spectra of ThT with increasing concentrations of ATP (from top to bottom): 0, 10, 20, 50, 100, 200, 500, 1000, 2000, 5000, 10000 μM. (B) Plot of the relative fluorescnence intensity change at 488 nm (ΔF/ΔF0) versus the concentration of ATP. The inset displays a linear relationship (R2 = 0.994) in the concentration range from 10 to 100 μM. (C) Selectivity of ATP against other analogs including GTP, UTP, CTP at 2 mM.
Reversibility
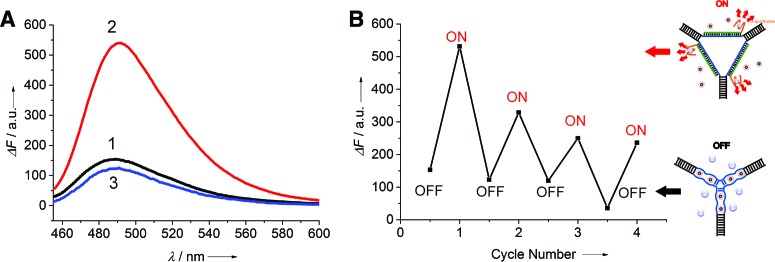
From the point of view of DNA devices, the above-mentioned ATP analysis in fact corresponds to capturing ATP by ARTWN. Our next step is to release ATP by adding the release stand, and then reset the system by using the capture strand (Scheme 1). Figure 3A shows the signal readout of such a switched system in the ‘OFF’ and ‘ON’ states, indicating that it can be operated repeatedly. The results of several switching cycles are shown in Figure 3B. We find the signal change gradually becomes weaker as the cycle number increases, possibly owing to the negative influence of the accumulation of double-stranded waste products. This problem might be overcome by using light-regulated strand-displacement reactions (28), in which light is used to drive DNA devices work thereby eliminating any waste product.
Figure 3.
Fluoresecent analysis for the switching cycles of ARTWN. (A) Fluorescence emission spectra of the nanoswitch corresponding to: (1) ‘OFF’ state (black). (2) ‘ON’ state (red), generated by introduction of the release strands. (3) ‘OFF’ state (blue), generated by adding capture strands to the ‘ON’ state. (B) Switching performance of the DNA nanoswitch between ‘OFF’ and ‘ON’ state by alternately adding the release strands and capture strands into the switching system.
Generality
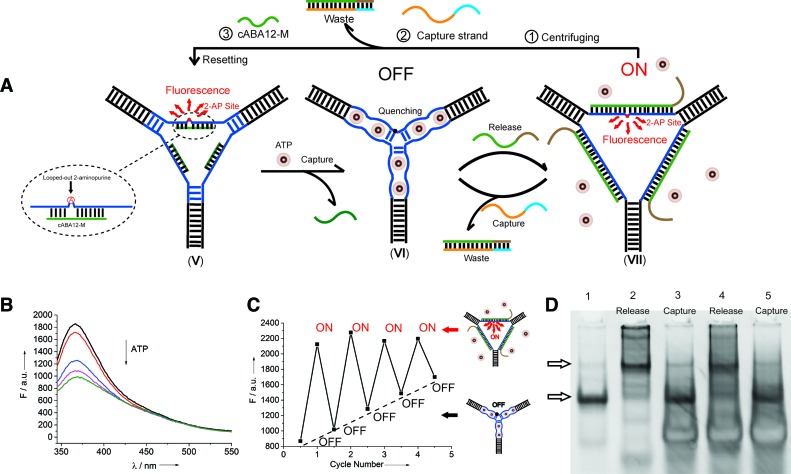
To further demonstrate the generality of our concept for the design of reversible nanosensors, we introduce another mode for the signal readout by inserting a looped-out (29) fluorescent 2-aminopurine into one of the ATP aptamers (Figure 4A) and introducing a new sequence partly complementary to the ATP aptamer (in green, Figure 4A, V). This modification has no influence on the assembly of our designed nanosensor (Supplementary Figure S5). When the two partly complementary DNA sequences are hybridized, the mismatched 2-aminopurine adopts a looped-out conformation and behaves as a bulged base out of the helix (red dot), which is proven to exhibit a high fluorescent emission owing to the 2-aminopurine not stacking with any other base (29). Furthermore, upon addition of ATP molecules, an obvious fluorescence decrease is observed (Figure 4B) due to the ATP-induced unwinding of DNA duplexes. The resulting structure holds the 2-aminopurine in a stacked position (black dot, Figure 4A, VI), and thus the fluorescence is significantly quenched due to the base-stacking interaction during this transition.
Figure 4.
Generality of DNA switch-controlled reversible nanosensors. (A) Illustration of the working principle of the modified system: (V) Initial state of the modified ARTWN with a fluorescent 2-aminopurine bulge. (VI) ATP molecules intercalate into the embedded ATP aptamers, unwinding the 2-aminopurine bulge duplex with an obvious fluorescence decrease and placing the switch in the ‘OFF’ state. (VII) Upon the introduction of the release strands, the central ATP aptamers fully hybridize with them to form three rigid central duplexes, consequently releasing the ATP molecules and allowing the 2-aminopurine bulge to reappear, giving rise to a fluorescence increase. The switch is then in the ‘ON’ state. (B) Fluorescence emission spectra of 2-aminopurine with increasing concentrations of ATP (from top to bottom): 0, 200, 500, 1000, 2000 μM. (C) Switching processes of the DNA nanoswitch between ‘OFF’ and ‘ON’ state characterized by fluorescence emission spectra. (D) Switching performance characterized by native PAGE.
Here two new release and capture strands were utilized as fuels to drive this system to switch repeatedly (Figure 4A, VII). The switching operations in several cycles are recorded by using fluorescence spectroscopy (Figure 4C) and native PAGE (Figure 4D), indicating a good reversibility of this switchable system.
DISCUSSION
In summary, we proposed a new principle to construct DNA nanoswitch-controlled reversible nanosensors. The designed nanosensor can be switched between the ‘OFF’ and ‘ON’ states via toehold strand displacement reactions. In a sense, it functions like a molecular net woven by DNA to capture or release ATP molecules. Removing free ATP molecules from DNA nanostructure, the final state of the switchable system will be returned to the initial state of the ATP biosensor. Such a novel reversible nanosensor holds great promise for the applications to switchable nanomachines (18,30), nanocarriers for drug delivery (11) and other related DNA nanodevices (31).
Acknowledgments
We thank Prof. Zachary J. Smith of the University of Science and Technology of China for reading and revising the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [21575133]; National Key Research and Development Program of China [2016YFA0201300]; Recruitment Program of Global Experts. Funding for open access charge: National Natural Science Foundation of China [21575133]; National Key Research and Development Program of China [2016YFA0201300]; Recruitment Program of Global Experts.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li T., Dong S., Wang E. A lead(II)-driven DNA molecular device for turn-on fluorescence detection of lead(II) ion with high selectivity and sensitivity. J. Am. Chem. Soc. 2010;132:13156–13157. doi: 10.1021/ja105849m. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J., Zhang L., Teng Y., Lou B., Jia X., Gu X., Wang E. G-quadruplex enhanced fluorescence of DNA-silver nanoclusters and their application in bioimaging. Nanoscale. 2015;7:13224–13229. doi: 10.1039/c5nr03092g. [DOI] [PubMed] [Google Scholar]
- 3.Li T., Dong S., Wang E. Label-free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated G-quadruplex-based DNAzymes. Anal. Chem. 2009;81:2144–2149. doi: 10.1021/ac900188y. [DOI] [PubMed] [Google Scholar]
- 4.Li T., Wang E., Dong S. G-Quadruplex-based DNAzyme as a sensing platform for ultrasensitive colorimetric potassium detection. Chem. Commun. 2009:580–582. doi: 10.1039/b815814b. [DOI] [PubMed] [Google Scholar]
- 5.Ranallo S., Rossetti M., Plaxco K.W., Vallee-Belisle A., Ricci F. A modular, DNA-based beacon for single-step fluorescence detection of antibodies and other proteins. Angew. Chem. Int. Ed. 2015;54:13214–13218. doi: 10.1002/anie.201505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Zhong X., Zhang H., Le X.C., Zhu J.J. Binding-induced fluorescence turn-on assay using aptamer-functionalized silver nanocluster DNA probes. Anal. Chem. 2012;84:5170–5174. doi: 10.1021/ac3006268. [DOI] [PubMed] [Google Scholar]
- 7.Li T., Dong S., Wang E. Enhanced catalytic DNAzyme for label-free colorimetric detection of DNA. Chem. Commun. 2007:4209–4211. doi: 10.1039/b712165b. [DOI] [PubMed] [Google Scholar]
- 8.Tang Z., Mallikaratchy P., Yang R., Kim Y., Zhu Z., Wang H., Tan W. Aptamer switch probe based on intramolecular displacement. J. Am. Chem. Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 9.Roncancio D., Yu H., Xu X., Wu S., Liu R., Debord J., Lou X., Xiao Y. A label-free aptamer-fluorophore assembly for rapid and specific detection of cocaine in biofluids. Anal. Chem. 2014;86:11100–11106. doi: 10.1021/ac503360n. [DOI] [PubMed] [Google Scholar]
- 10.Pei H., Liang L., Yao G., Li J., Huang Q., Fan C. Reconfigurable three‐dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem. Int. Ed. 2012;124:9154–9158. doi: 10.1002/anie.201202356. [DOI] [PubMed] [Google Scholar]
- 11.Meng H.M., Zhang X., Lv Y., Zhao Z., Wang N.N., Fu T., Fan H., Liang H., Qiu L., Zhu G., et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8:6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Tang Y., Mason S.D., Chen J., Li F. Enzyme-powered three-dimensional DNA nanomachine for DNA walking, payload release, and biosensing. ACS Nano. 2016;10:2324–2330. doi: 10.1021/acsnano.5b07102. [DOI] [PubMed] [Google Scholar]
- 13.Li T., Famulok M. I-motif-programmed functionalization of DNA nanocircles. J. Am. Chem. Soc. 2013;135:1593–1599. doi: 10.1021/ja3118224. [DOI] [PubMed] [Google Scholar]
- 14.Li T., Lohmann F., Famulok M. Interlocked DNA nanostructures controlled by a reversible logic circuit. Nat. Commun. 2014;5:4940. doi: 10.1038/ncomms5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sannohe Y., Endo M., Katsuda Y., Hidaka K., Sugiyama H. Visualization of dynamic conformational switching of the G-quadruplex in a DNA nanostructure. J. Am. Chem. Soc. 2010;132:16311–16313. doi: 10.1021/ja1058907. [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Huang J., Zhou Y., Yan S., Weng X., Wu X., Deng M., Zhou X. Conformational switching of G-quadruplex DNA by photoregulation. Angew. Chem. Int. Ed. 2010;49:5305–5309. doi: 10.1002/anie.201002290. [DOI] [PubMed] [Google Scholar]
- 17.Endo M., Yang Y., Suzuki Y., Hidaka K., Sugiyama H. Single-molecule visualization of the hybridization and dissociation of photoresponsive oligonucleotides and their reversible switching behavior in a DNA nanostructure. Angew. Chem. Int. Ed. 2012;51:10518–10522. doi: 10.1002/anie.201205247. [DOI] [PubMed] [Google Scholar]
- 18.Liu D., Balasubramanian S. A Proton-Fuelled DNA Nanomachine. Angew. Chem. Int. Ed. 2003;115:5912–5914. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 19.Wang C., Huang Z., Lin Y., Ren J., Qu X. Artificial DNA nano-spring powered by protons. Adv. Mater. 2010;22:2792–2798. doi: 10.1002/adma.201000445. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y., Liu G., Liu H., Li D., Fan C., Liu D. An electrochemically actuated reversible DNA switch. Nano Lett. 2010;10:1393–1397. doi: 10.1021/nl100169p. [DOI] [PubMed] [Google Scholar]
- 21.Ranallo S., Amodio A., Idili A., Porchetta A., Ricci F. Electronic control of DNA-based nanoswitches and nanodevices. Chem. Sci. 2016;7:66–71. doi: 10.1039/c5sc03694a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing Y., Yang Z., Liu D. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. Int. Ed. 2011;50:11934–11936. doi: 10.1002/anie.201105923. [DOI] [PubMed] [Google Scholar]
- 23.Kong L., Xu J., Xu Y., Xiang Y., Yuan R., Chai Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on SYBR Green I dye. Biosens. Bioelectron. 2013;42:193–197. doi: 10.1016/j.bios.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Peng P., Liu S., Li T. Thioflavin T behaves as an efficient fluorescent ligand for label-free ATP aptasensor. Anal. Bioanal. Chem. 2016;408:7927–7934. doi: 10.1007/s00216-016-9926-9. [DOI] [PubMed] [Google Scholar]
- 25.Huizenga D.E., Szostak J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 26.Mohanty J., Barooah N., Dhamodharan V., Harikrishna S., Pradeepkumar P.I., Bhasikuttan A.C. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2013;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- 27.Lin C.H., Patei D.J. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997;4:817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann F., Ackermann D., Famulok M. Reversible light switch for macrocycle mobility in a DNA rotaxane. J. Am. Chem. Soc. 2012;134:11884–11887. doi: 10.1021/ja3042096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao Y., Stringfellow S., Yu H. Distinguishing ‘looped-out’ and ‘stacked-in’ DNA bulge conformation using fluorescent 2-aminopurine replacing a purine base. J. Biomol Str. Dyn. 2002;19:929–934. doi: 10.1080/07391102.2002.10506795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubrich D., Lin J., Yan J. A contractile DNA machine. Angew. Chem. Int. Ed. 2008;47:7026–7028. doi: 10.1002/anie.200800476. [DOI] [PubMed] [Google Scholar]
- 31.Zhu G., Hu R., Zhao Z., Chen Z., Zhang X., Tan W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]