Abstract
Human mitochondrial DNA is transcribed by POLRMT with the help of two initiation factors, TFAM and TFB2M. The current model postulates that the role of TFAM is to recruit POLRMT and TFB2M to melt the promoter. However, we show that TFAM has ‘post-recruitment’ roles in promoter melting and RNA synthesis, which were revealed by studying the pre-initiation steps of promoter binding, bending and melting, and abortive RNA synthesis. Our 2-aminopurine mapping studies show that the LSP (Light Strand Promoter) is melted from −4 to +1 in the open complex with all three proteins and from −4 to +3 with addition of ATP. Our equilibrium binding studies show that POLRMT forms stable complexes with TFB2M or TFAM on LSP with low-nanomolar Kd values, but these two-component complexes lack the mechanism to efficiently melt the promoter. This indicates that POLRMT needs both TFB2M and TFAM to melt the promoter. Additionally, POLRMT+TFB2M makes 2-mer abortives on LSP, but longer RNAs are observed only with TFAM. These results are explained by TFAM playing a role in promoter melting and/or stabilization of the open complex on LSP. Based on our results, we propose a refined model of transcription initiation by the human mitochondrial transcription machinery.
INTRODUCTION
Mitochondria are the only autonomous organelle in the eukaryotic cell with their own replication and transcription machineries. Understanding the basic transcription machinery that leads to mitochondrial gene expression is fundamental in dissecting the role of mitochondria in human health and disease (1,2). The human mitochondrial DNA is transcribed by POLRMT, which is homologous to single subunit RNA polymerases from phage T7 and yeast mitochondria (3). T7 RNA polymerase can initiate transcription on its own without any transcription factors (4), the Saccharomyces cerevisiae Rpo41 requires Mtf1 to initiate transcription (5–7), and the human POLRMT requires two initiation factors, TFAM (Transcription Factor A of the Mitochondria) and TFB2M (Transcription Factor B2 of the Mitochondria) for transcript synthesis (8,9).
The initiation factor TFB2M is structurally homologous to bacterial RNA methyltransferase ErmC' (8,10); however, it has lost most of its RNA methyltransferase activity (11) and serves mainly as the transcription initiation factor. Crosslinking studies have shown that TFB2M interacts with POLRMT, the promoter initiation region, and the initiating ATPs (12,13). TFAM is a small protein (∼29 kDa) that serves both as a mitochondrial DNA packaging protein and transcription initiation factor. TFAM binds to double-stranded DNA of any sequence and also site-specifically to the upstream −39 to −12 regions of the light strand and heavy strand promoters (LSP and HSP, respectively), inducing U-turn bends in the DNA (14–18). Additionally, TFAM interacts with POLRMT on the promoter DNA (13,19,20). Interestingly, transcription from HSP and LSP are differentially regulated by the level of TFAM (21,22), and TFAM levels in the cells are reported from 35–50 molecules per mitochondrial DNA to 2000 TFAM molecules per mitochondrial DNA (23–27).
There is consensus that TFAM and TFB2M are necessary to make RNA transcripts on both the LSP and HSP, and presently it is believed that the two factors have distinct roles (12,19,28,29). The role of TFAM is to recruit POLRMT to the promoter initiation site and TFB2M comes in later to melt the promoter (19,28). However, there is little mechanistic understanding of this multistep initiation pathway of the human mitochondrial DNA transcription, including the pre-initiation steps of promoter binding, bending, melting and the initiation steps of ATP binding and abortive synthesis. Therefore, we do not fully understand the roles of TFAM and TFB2M in each of the aforesaid steps. For instance it is thought that POLRMT+TFB2M do not form a complex on the promoter (9,19,28), but the thermodynamic Kd values of the various protein–DNA complexes possible with POLRMT, TFAM and TFB2M on the promoter have not been determined. Similarly, methods to directly assay the closed to open complex transition on the human mitochondrial promoters and the kinetics of de novo RNA synthesis with and without TFAM or TFB2M are not characterized. Hence, the roles of TFAM and TFB2M in promoter binding, melting and initiation of RNA synthesis are not fully understood.
To fill these gaps in the understanding of the mechanism of transcription initiation, we have used fluorescence anisotropy methods to measure the thermodynamic Kd values of the protein–DNA complexes with the individual protein components (POLRMT, TFAM and TFB2M) and their various combinations. We have used Förster Resonance Energy Transfer (FRET) as a molecular ruler to measure the DNA bending conformational changes and 2-aminopurine (2-AP) fluorescence changes to study promoter melting in these various protein–DNA complexes and to determine the size of the initiation bubble in the open complex. These methods have provided new insights into the roles of POLRMT, TFAM and TFB2M in closed to open complex transition on the LSP and will be applicable to the studies of the mechanism of initiation at the HSP. We show that the size of the initiation bubble is conserved from phage T7 to human mitochondrial RNA polymerases, but the mechanism of promoter opening is not. We provide evidence that the role of TFAM is not only in the ‘recruitment’, but that TFAM has ‘post-recruitment’ roles in promoter melting and RNA elongation. Based on our studies, we propose a refined model of the transcription initiation pathway at the LSP in the human mitochondrial system.
MATERIALS AND METHODS
Cloning, expression and purification of POLRMT, TFAM and TFB2M proteins
POLRMT (43–1230 with N-terminal 6x His-tag) and TFAM (43–245, N-terminal 6x His-tag) were cloned into the bacterial expression vector pPROEXHTb. The TFB2M (21–396) expression vector pT7TEV-HMBP4 expresses a fusion protein of TFB2M with N-terminal His-tagged maltose binding protein (MBP) that is cleavable by Tobacco Etch Virus (TEV) protease and was a kind gift from Professor Miguel Garcia-Diaz at the Stony Brook University School of Medicine.
POLRMT was expressed in Escherichia coli BL21 codon plus (RIL) at 18°C after IPTG induction. The cells were lysed by lysozyme treatment and sonicated in lysis buffer (40 mM Tris–Cl, pH 7.9, 300 mM NaCl, 0.1% Tween 20, 1 mM PMSF, 15% glycerol, 1 mM EDTA, 0.05 mM DTT and protease inhibitors). The lysate was subjected to polyethyleneimine treatment, ammonium sulfate precipitation, and the protein was purified through two column chromatographic steps: Ni Sepharose 6 HP followed by Heparin Sepharose (GE Lifesciences).
TFAM was expressed in BL21(DE3)-RIL cells at 37°C after IPTG induction. The cells were lysed using lysozyme and sonication in lysis buffer (50 mM Na-phosphate (pH 7.8), 500 mM NaCl, 10% glycerol, 0.1% Tween and protease inhibitors). The clarified lysate containing 20 mM imidazole was subjected to two column chromatography steps: Ni Sepharose 6 HP followed by Heparin Sepharose.
TFB2M was purified as previously reported (20). Briefly, TFB2M plasmid was expressed in Arctic Express (DE3) cells (Stratagene) and induced overnight at 14°C. The cells were lysed with lysozyme and sonicated in lysis buffer (20 mM HEPES–KOH pH 8, 1 M KCl, 5% glycerol, 10 mM imidazole and protease inhibitors). The clarified lysate was subjected to Ni Sepharose 6 HP column chromatography, followed by overnight TEV protease cleavage, Heparin Sepharose, and finally gel filtration chromatography.
All the proteins were concentrated using Amicon Ultra-10K and stored at −80 °C (Supplementary Figure S1B). Molar concentrations of purified proteins were determined in 6M guanidium-HCl buffer from light absorbance at 280 nm and the calculated respective molar extinction coefficients.
Oligonucleotides
All the DNA oligonucleotides were custom synthesized and HPLC purified from IDT (Integrated DNA Technologies). The molar concentration of purified DNA was determined from their absorbance at 260 nm and the respective extinction coefficients. For transcription assays, the double-stranded DNA was prepared by annealing complementary non-template and template strand in 1.05 to 1 ratio. Oligonucleotides for FRET assays were TAMRA (NHS Ester) labeled at the −12 position of the non- template LSP sequence (−41 to +10) and fluorescein dT (C6 spacer) attached either at the 5′ or 3′ end of the template strand. For FRET, dual labeled DNA was prepared by annealing complementary non-template and template strands in 1:1 ratio. Donor alone control for FRET and DNA for fluorescence anisotropy assays were prepared using 5′ or 3′ end fluorescein template strand annealed to its complementary non-template strand in 1:1 ratio.
Steady state transcription assays
Transcription was measured using the gel-based RNA synthesis assays at 25°C (30). Briefly, a complex of 1 μM POLRMT, 1 μM TFAM and 1 μM TFB2M along with 1 μM human LSP promoter duplex DNA (−42 to +21) was incubated at 25°C in reaction buffer (50 mM Tris acetate, pH 7.5, 50 mM Na-glutamate, 10 mM magnesium acetate, 1 mM DTT and 0.05% Tween-20). The transcription reaction was initiated by adding a mixture of 250 μM of ATP, UTP, GTP, 3′dCTP or without 3′dCTP (wherever indicated) and spiked with [γ-32P]ATP. After 10 min, reactions were stopped using 400 mM EDTA and formamide dye mixture (98% formamide, 0.025% bromophenol blue, 10 mM EDTA). For Km studies, the transcription reaction was initiated with different concentrations of ATP spiked with [γ-32P]ATP and let to proceed for 15 min. Samples were heated to 95°C for 2 min, and the RNA products were resolved on 24% polyacrylamide/4M urea sequencing gels. The gels were exposed to a phosphor screen and scanned on a Typhoon 9410 PhosphorImager instrument (Amersham Biosciences). The free ATP and RNA bands were quantified using ImageQuant and molar amounts of RNA synthesized were calculated according to Equation (1).
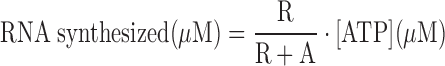 |
(1) |
where R and A are the band intensities of RNA products and free ATP respectively, and [ATP] is the molar concentration of ATP added to the reaction.
The rate of RNA synthesis (μM/s) divided by protein–DNA complex concentration was plotted against the ATP concentrations and the Km and kcat were determined from fit to Equation (2).
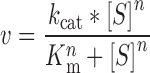 |
(2) |
where v is the rate of RNA synthesis divided by enzyme–DNA concentration, [S] is the substrate concentration and n is the Hill coefficient.
Steady state fluorescence anisotropy assays
Fluorescence anisotropy measurements were carried out on Fluoromax-4 spectrofluorometer (Jobin Yvon-Spex Instruments S.A., Inc.) equipped with an autopolarizer and a thermoelectrically controlled cell holder for constant temperature. Fluorescein labeled −41 to +10 LSP DNA (1 nM) was titrated with increasing concentrations of POLRMT, TFAM, TFB2M or equimolar amounts of combinations of proteins in transcription reaction buffer. Anisotropy values (robs) were recorded after excitation at 490 nm (4-nm bandwidth) and emission at 514 nm (8-nm bandwidth) and plotted against protein concentrations [P]. The Kd values of the protein–DNA complexes were determined from fit to Equation (3).
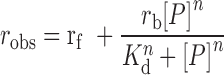 |
(3) |
where rf and rb are fluorescence anisotropy values of free DNA and bound DNA complex and n is the Hill coefficient.
Promoter DNA melting by 2-AP fluorescence analysis
Steady-state fluorescence intensity measurements were measured on a Fluoromax-4 spectrofluorometer at 25°C. Fluorescence spectra from 350 to 420 nm (6-nm bandwidth) were collected after excitation at 315 nm (2-nm bandwidth) in samples containing 100 nM 2-AP incorporated double-stranded DNA promoter, 150 nM each of TFAM, POLRMT and TFB2M and reaction buffer (50 mM Tris-acetate, pH 7.5, 50 mM sodium glutamate, 10 mM magnesium acetate, 1 mM DTT). Addition of the components was carried out in the following order: DNA followed by TFAM followed by POLRMT and then TFB2M or DNA followed by POLRMT, TFB2M and then TFAM. When ATP was used, its concentration was 500 μM. A parallel experiment was carried out with unmodified LSP DNA to correct for background fluorescence from buffer alone and proteins with unmodified DNA. The 2-AP fluorescence intensities between 360 and 380 nm were integrated and were divided by the fluorescence intensity of free DNA to obtain the fold-increase.
Steady state förster resonance energy transfer (FRET) assays
FRET assays were performed at 25°C on Fluoromax-4 spectrofluorometer using 10 nM donor–acceptor (D–A) or donor (D)-labeled LSP DNA and increasing protein concentrations (TFAM alone or equimolar mixture of proteins) in reaction buffer (50 mM Tris-acetate, pH 7.5, 50 mM sodium glutamate, 10 mM magnesium acetate and 1 mM DTT). The fluorescence intensity in the 500–630 nm range was measured after excitation at 494 nm. The sensitized acceptor fluorescence was determined in the 555–630 nm range after correcting for donor-only fluorescence. The FRET efficiency was calculated using the (ratio)A method (16,31,32), which provided D–A distances and DNA bend angles, as detailed in the Supplementary Methods.
RESULTS
Runoff and abortive RNA synthesis on the LSP DNA
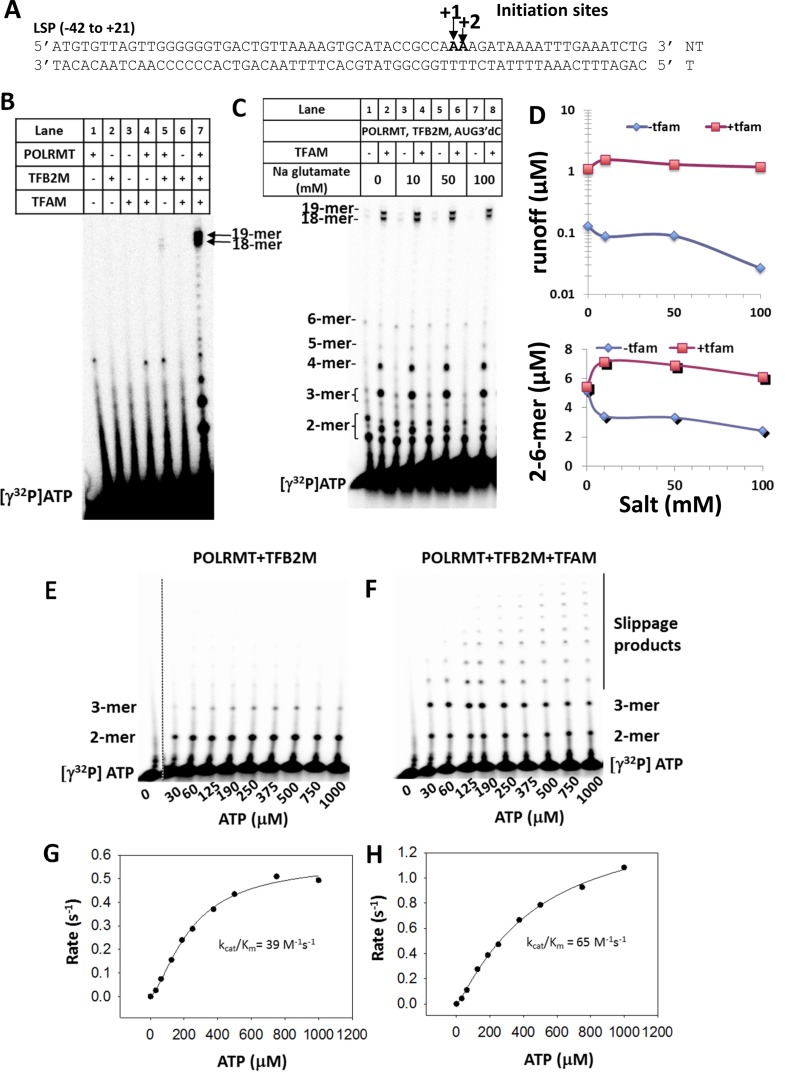
The Light Strand Promoter (LSP) DNA fragment from −42 to +21 (Figure 1A) was chemically synthesized and POLRMT, TFB2M and TFAM proteins were bacterially expressed and purified (Supplementary Figure S1B). To ensure that the proteins were active in transcription initiation, we tested their RNA synthesis activity on the LSP DNA fragment that makes 18 and 19 mer run-off products in the presence of ATP, GTP, UTP, 3′dCTP and [γ32P]ATP. In our transcription buffers, we have used the more physiological glutamate anion instead of commonly used chloride anion that has been demonstrated (33) to compete with the DNA phosphate backbone. The high-resolution (24%) polyacrylamide/urea sequencing gel resolves the free ATP from RNA products ranging from 2-mer to the run-off, and shows that the run-off products are made in presence of all three proteins (Figure 1B, lane 7). In contrast, POLRMT (lane 1) and POLRMT+TFAM (lane 4) are inactive in abortive 2–6 mer and run-off RNA synthesis. However, POLRMT+TFB2M (lane 5) make abortives and the correct length run-off products (18 and 19-mer), although with a 50-fold lesser efficiency than the three proteins together (lane 5 versus lane 7). This indicates that POLRMT+TFB2M can form the correct promoter-specific transcription initiation complex on the LSP and make run-off RNAs, but not as efficiently as in the presence of TFAM. This is in accordance with published literature (12,29).
Figure 1.
Transcription initiation activity of POLRMT, TFB2M and TFAM on the LSP DNA. (A) Sequence of the LSP DNA (−42 to +21) is shown. (B) The 24% polyacrylamide–4M urea gel image shows the abortives (2 to 6-mer) and the run off products (18 and 19-mer) after 10 min of transcription reaction at 25°C on the LSP DNA (1 μM) with individual and various combinations of POLRMT, TFB2M and TFAM (1 μM) and NTPs (ATP, GTP and UTP, 3′dCTP at 250 μM each) + [γ32P]ATP. (C) Salt concentration dependence of LSP DNA transcription by POLRMT+TFB2M in the presence and absence of TFAM. The transcription reactions were carried out as described in (B) under the indicated sodium glutamate salt concentrations. The abortives and run off products are shown. (D) The quantitation of total run-off (y-axis shown in log scale) and 2–6 mer abortives as a function of salt concentration. The experiment was performed once. (E) Transcription reactions were carried out as in (B) but with different concentrations of ATP and [γ32P]ATP for 15 minutes in presence and absence of TFAM. The 24% polyacrylamide–4M urea gel image shows the 2 mer, 3 mer and the slippage products. The catalytic efficiency (kcat/Km) was calculated by plotting the rate of RNA synthesis/enzyme concentration against the ATP concentration and fitting the curve to Hill Equation (Equation 2 in Materials and Methods). The experiment was performed twice. The average kcat/Km with the standard deviation in the presence of all three proteins is 64 ± 0.8 M−1 s−1 while in the absence of TFAM is 30 ± 13 M−1 s−1.
When the Na-glutamate salt was removed from the reactions, the run-off products with POLRMT+TFB2M increased by 3–4-fold, but then steadily decreased at higher salt concentrations (Figure 1C and 1D). This salt effect has been observed in previous studies (9,12,22,29) and the higher activity of POLRMT+TFB2M is attributed to ease of DNA melting at lower salt. Interestingly, these earlier studies did not resolve the 2–3 mer abortives; hence, did not report that the amount of 2-mer synthesis by POLRMT+TFB2M remains high even at high salt concentrations (Figure 1D, bottom graph). This indicates that even at high salt, the POLRMT+TFB2M can efficiently initiate RNA synthesis and make 2-mers but is unable to extend it efficiently to form run-off products. However, with TFAM the 2-mers are elongated to longer RNA products.
To explore the roles of TFAM and TFB2M in initial abortive RNA synthesis, we performed transcription reactions in the presence of only ATP where we expect 2-mer and 3-mer synthesis on the LSP. Synthesis of 2-mer was observed with and without TFAM (Figure 1E and 1F), but efficient elongation of 2-mer to 3-mer was observed only in the presence of TFAM (Figure 1F). Similarly, the presence of TFAM promoted poly(A) formation from slippage synthesis. The total RNA was quantified as a function of ATP concentration to determine the initiating ATP Km and kcat values, which provided the catalytic efficiency (kcat/Km) of initiation (Figure 1G and 1H). The catalytic efficiency of initiation is 65 M−1 s−1 with all three proteins and 39 M−1 s−1 in the presence of POLRMT+TFB2M. These results indicate that POLRMT+TFB2M is active in catalyzing the first phosphodiester bond formation reaction between ATPs to make the 2-mer RNA, but it needs TFAM for RNA elongation. Thus, TFAM could be directly involved in stabilizing the 2-mer on the active site, or TFAM could indirectly perform this function by stabilizing the open initiation complex. Furthermore, this is the first report of the initiation rate measurement on the human mitochondrial LSP, which when compared to the yeast counterpart shows that transcription initiation at the human LSP occurs with ∼10-fold lower kcat/Km in comparison to the observed activity at the most active 15s yeast mitochondrial promoter (34).
Measurement of the binding affinities of POLRMT, TFAM, and TFB2M complexes on LSP supports independent rather than sequential assembly of proteins
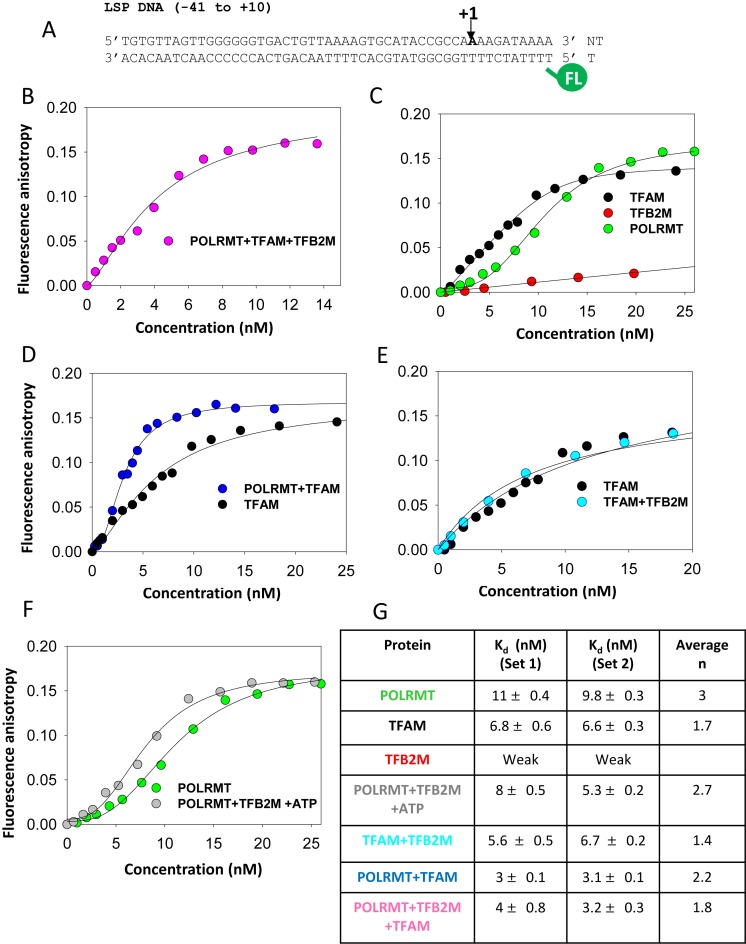
To obtain a better understanding of the roles of TFAM in initiation, we took a systematic approach to quantitatively analyze the pre-initiation steps of promoter binding, bending and melting in all possible protein–DNA complexes with the three protein components. We developed a fluorescence anisotropy based titration method to measure the affinities of the protein–DNA complexes with the individual components and various combinations to determine how the three protein components of the transcriptional initiation machinery assemble on the LSP DNA. This method allows Kd measurements under equilibrium conditions, hence provides the relative thermodynamic affinities of the various protein–DNA complexes formed during initiation of transcription.
The LSP DNA fragment from −41 to +10 was labeled with fluorescein at the 5′-end of the template DNA (Figure 2A) and titrated with increasing protein concentrations. First, we titrated the LSP DNA with an equimolar mixture of POLRMT+TFAM+TFB2M (Figure 2B). The binding curve did not fit to the 1:1 binding model, which indicates that the three protein components do not form a pre-complex before binding to the LSP DNA. The binding data fit well to the Hill equation (Equation 3), which provided an apparent Kd of 4 nM for the three-component protein–DNA complex.
Figure 2.
Interactions of the LSP DNA with TFAM, POLRMT and TFB2M measured by fluorescence anisotropy titrations. (A) The 5′-end fluorescein labeled LSP DNA (−41 to +10) was used in the fluorescence anisotropy titrations. (B) Binding of TFAM+POLRMT+TFB2M to the LSP DNA. Fluorescein-labeled LSP DNA (1 nM) was titrated with increasing concentration of equimolar mixture of TFAM+POLRMT+TFB2M and fluorescence anisotropy was recorded after excitation at 490 nm and emission at 514 nm. The plots show the fluorescence anisotropy change corrected for free DNA (anisotropy of 0.16). The binding data were fit to Equation (3). (C) Binding of the individual proteins to the LSP DNA. (D) Comparison of LSP DNA binding to TFAM alone and TFAM+POLRMT. (E) Comparison of LSP DNA binding to TFAM alone and TFAM+TFB2M. (F) Comparison of LSP DNA binding to POLRMT and POLRMT+TFB2M+ATP. Fluorescein labeled LSP DNA (1 nM) and 1 mM ATP was titrated with increasing concentration of an equimolar mixture of POLRMT+TFB2M. (G) The table summarizes the apparent Kd values and n (Hill coefficient) values from fits to the Hill equation. The Kd values shown are from two sets of independent experiments, n values are shown as an average. Associated error is the standard error of fitting.
Next, we studied the binding of the individual protein components to the LSP DNA. It is known that TFAM forms a tight complex with the −40 to −16 or −39 to −12 upstream LSP region DNA fragment (16,17), but the binding parameters of TFAM to the entire LSP from −41 to +10 have not been measured. Titration of the long fluorescein labeled LSP DNA (−41 to +10) with increasing TFAM shows a steady increase in fluorescence anisotropy reaching a plateau at saturating TFAM (Figure 2C, black). Interestingly, the saturating anisotropy value, which is proportional to the size of the DNA-protein complex, is greater than expected for the relatively small sized TFAM-DNA complex, indicating that the 51 bp LSP fragment binds multiple TFAM molecules. This was confirmed by mobility shift assays, which show a single complex on the shorter −41 to −12 LSP fragment (Supplementary Figure S2A), but multiple complexes on the longer −41 to +10 promoter fragment (Supplementary Figure S2B). The anisotropy binding data were fit to the Hill equation, which provided an apparent Kd of 7 nM for the TFAM-LSP complex (Figure 2G table). This value is similar to the reported Kd values of TFAM, ∼5 nM bound to the shorter LSP fragment (−40 to −16 or −39 to −12) (16,17). Thus, our results show that the −41 to +10 LSP DNA binds to multiple TFAM molecules to form a complex with nanomolar Kd value.
Interestingly, POLRMT alone also forms a tight complex with the LSP DNA with an apparent Kd of 10–11 nM (Figure 2C, green and Figure 2G table). The sigmoidal binding of POLRMT suggests binding of more than one POLRMT molecule to the LSP DNA fragment. On the other hand, TFB2M shows very weak binding to the LSP DNA (Figure 2C, red). Thus, overall our binding data indicate that TFAM and POLRMT on their own bind on the LSP with high affinities, most likely occupying both the upstream and downstream promoter regions of the LSP.
Next, we studied the binding of the LSP DNA fragment to an equimolar mixture of two protein components—POLRMT+TFAM, POLRMT+TFB2M and TFAM+TFB2M. The POLRMT+TFAM mixture binds to the LSP DNA with a higher affinity (3 nM Kd) than TFAM alone (7 nM Kd) or POLRMT alone (10–11 nM Kd) (Figure 2D) and similar affinity as the three-protein complex (3–4 nM Kd). On the other hand, TFAM+TFB2M binds to LSP with the same affinity as TFAM alone (6–7 nM Kd) (Figure 2E). POLRMT+TFB2M mixture in presence or absence (data not shown) of ATP binds to the LSP DNA with a higher affinity (5–8 nM Kd) than POLRMT alone (10–11 nM Kd) and TFB2M alone (Figure 2F). Additionally, sigmoidal binding of POLRMT+TFB2M indicates that multiple molecules of POLRMT+TFB2M bind on the LSP fragment. We confirmed that POLRMT and POLRMT+TFB2M bind non-specifically to the upstream −41 to −12 LSP fragment with Kd values of 8 and 5 nM, respectively (Supplementary Figure S3A and B).
To summarize, the binding data show that the individual protein components, except for TFB2M, and various combinations of protein components bind to the LSP DNA with low nanomolar Kd values. The very similar Kd values of the various protein–DNA complexes suggest that the proteins can come together on the LSP DNA independently and not necessarily in a stepwise or sequential manner. Additionally, TFAM, POLRMT and POLRMT+TFB2M can form non-specific complexes on the DNA.
The LSP DNA is bent both upstream and downstream of the promoter initiation region by TFAM and POLRMT, respectively
It is known that TFAM induces a distinctive U-turn in the DNA containing the upstream LSP region or short non-specific DNA (15–18,35), but the corresponding DNA conformational change by TFAM on the entire LSP containing both the upstream and downstream DNA region has not been studied.
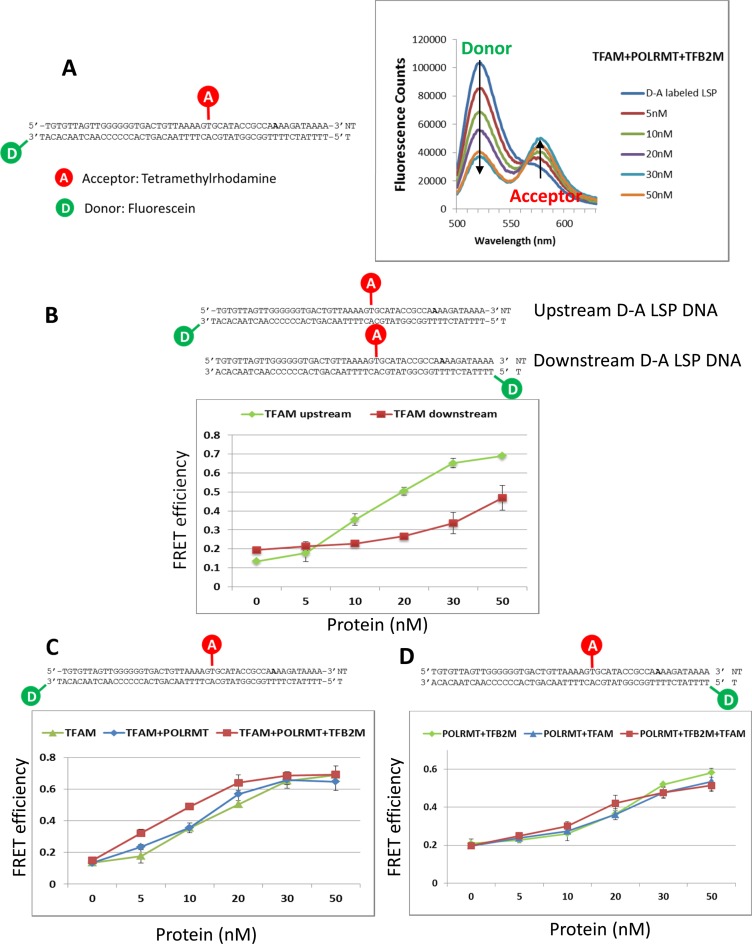
Here, we have used FRET to study the bent state of both the upstream TFAM binding site and downstream initiation site on the 51 bp LSP DNA fragment from −41 to +10. To study upstream DNA bending, fluorescein and TAMRA donor–acceptor (D–A) pair was introduced at −41 and −12 positions, and to study downstream promoter bending, the probes were introduced at −12 and +10 positions (Figure 3B). We show a representative set of fluorescence spectra that illustrates FRET upon addition of TFAM+POLRMT+TFB2M to the upstream D–A LSP DNA. The data shows a dose dependent decrease in donor fluorescence and corresponding increase in acceptor fluorescence (Figure 3A). The FRET efficiency was calculated from the sensitized acceptor fluorescence using the steady state (ratio)A method (31,36) (Materials and Methods and Supplementary Methods). Protein induced fluorescence quenching of these fluorophores was found to be <10%.
Figure 3.
FRET studies to investigate protein induced bending of the LSP DNA. (A) Representative fluorescence emission spectra of 10 nM LSP DNA (−41 to +10) with D–A pair in the upstream region show the dose dependent decreasing donor fluorescence intensity and increasing acceptor fluorescence intensity with addition of TFAM+POLRMT+TFB2M. The FRET efficiency was calculated using sensitized acceptor fluorescence calculated by the (ratio)A method (Materials and Methods and Supplementary Methods). (B) Comparison of upstream and downstream LSP DNA bending by TFAM. The LSP DNA with D–A pair in the upstream region and downstream region was titrated with TFAM. The y-axis shows the FRET efficiency. (C) Comparison of upstream LSP DNA bending by TFAM, TFAM+POLRMT and TFAM+POLRMT+TFB2M. (D) Comparison of downstream LSP DNA bending by POLRMT+TFB2M, POLRMT+TFAM and POLRMT+TFAM+TFB2M. The errors were calculated from two independent experiments.
Titration of the upstream D–A LSP DNA with TFAM resulted in a dose dependent increase in FRET efficiency (Figure 3B, green line), which is indicative of TFAM binding and inducing DNA bending in the upstream LSP region. At saturating TFAM concentrations, the FRET efficiency reached a value of 0.7, which corresponds to a D–A distance of ∼50 Å (Supplementary Figure S4A) and a change of 25 Å relative to free DNA. This D–A distance is consistent with the U-turn in the crystal structure of TFAM-DNA complex (PDB 3TQ6) with an end-to-end distance of 55 Å (35). Thus, our data indicates that TFAM causes a U-turn bend in the upstream region of the transcriptionally active LSP DNA. Similar FRET experiments were carried out with the downstream D–A LSP DNA, which shows that TFAM also induces DNA bending in the downstream initiation region, but the average bending of the downstream DNA is lower in comparison to the bending observed in the upstream DNA region (Figure 3B, red). These results indicate that the transcriptionally active LSP DNA can bind multiple TFAM molecules, which is consistent with our data from fluorescence anisotropy titration, the gel shift assays, and the very close Kd values for specific and non-specific DNAs (16,17). Additionally, TFAM bound to the downstream promoter region does not seem to affect the bend angle of the upstream promoter region.
Next, we determined the DNA bent state of the upstream LSP region in the POLRMT+TFAM and POLRMT+TFB2M+TFAM protein–DNA complexes. Interestingly, POLRMT+TFAM showed the same DNA bend in the upstream LSP region as the TFAM complex (Figure 3C, compare blue and green lines). These results indicate that POLRMT alone has little effect on upstream DNA bending by TFAM. On the other hand, upstream DNA bending by TFAM was more efficient in the presence of POLRMT+TFB2M (Figure 3C, red line). Comparison of FRET efficiencies of TFAM+POLRMT (blue line) and all three protein complex (red line) shows DNA bending starting at lower protein concentrations in titrations with all three proteins. The POLRMT+TFAM and POLRMT+TFAM+TFB2M complexes have similar Kd values; hence, these results indicate that POLRMT+TFB2M somehow stabilizes the TFAM-induced upstream DNA bend. Curiously, at higher protein concentrations, the FRET efficiency reached the same value in all the TFAM complexes. The exact reason is not known, but it is possible that TFAM oligomers at higher concentrations stabilize the upstream DNA bend. TFB2M has no effect on upstream DNA bending by TFAM (data not shown).
DNA bending in the downstream transcription initiation region was investigated in POLRMT complexes - POLRMT+TFAM (Figure 3D, blue line), POLRMT+TFB2M (Figure 3D, green line), and POLRMT+TFB2M+TFAM (Figure 3D, red line). All protein–DNA complexes showed a similar protein concentration dependent increase in FRET efficiency that reached a maximum value of 0.55. The FRET efficiency of 0.55 corresponds to a D–A distance of 54 Å (Supplementary Figure S4B) and a change of 15 Å from the free DNA. Using the single kink model of DNA bending (Supplementary Methods), we calculated that bend angle in the promoter initiation region is ∼80o. Interestingly, the degree of DNA bending in the initiation region is conserved and close to 90o in both human and yeast and also in the related phage T7 RNA polymerase complex (37–39).
Overall, our results show that TFAM bends the upstream LSP region more severely than the downstream LSP region. Similarly, the upstream promoter region of LSP is bent by the TFAM in all its complexes whereas the downstream initiation region is bent by POLRMT in all its complexes.
The LSP DNA is melted from −4 to +1 in the open complex and from −4 to +3 in the initiation complex
The 2-AP fluorescence method has been successfully used to map the initiation bubbles of several RNA polymerases (40–44). Human mitochondrial promoters present an extra challenge because the DNA region upstream of the transcription start-sites is GC rich, unlike the T7 and yeast mitochondrial promoters. Fortunately, the fluorescent base 2-AP pairs with both thymine and cytosine bases (Figure 4B) (45,46) and hence can be used on human mitochondrial promoters. The desirable property of 2-AP is that its fluorescence is quenched in duplex DNA and enhanced when the 2-AP base pair is melted and unstacked.
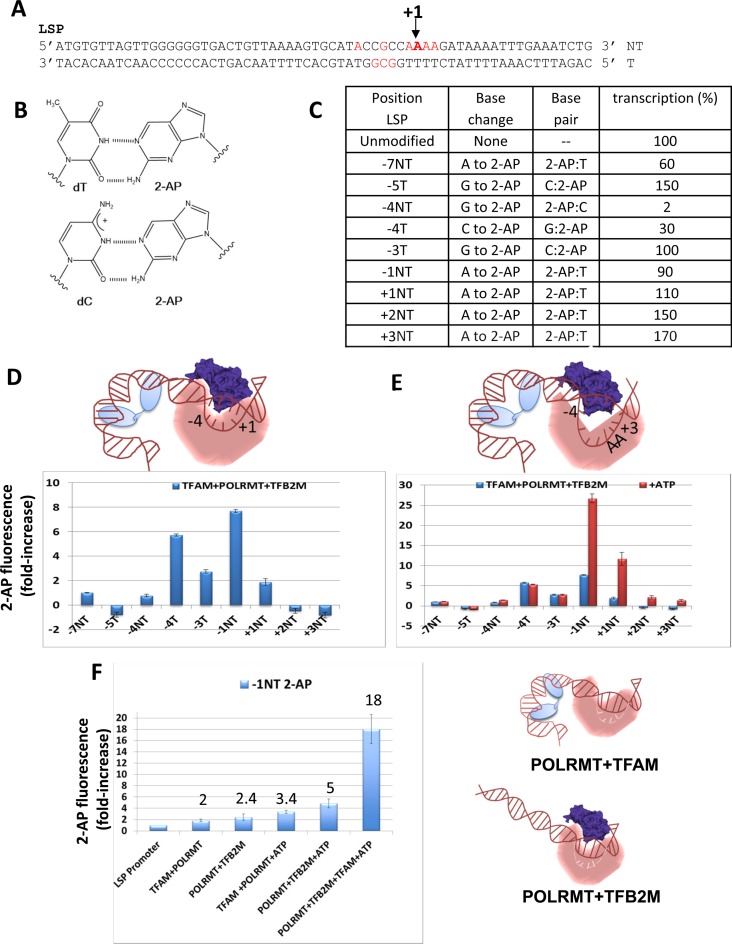
Figure 4.
2-AP fluorescence mapping to determine the melted DNA region in the LSP promoter and the roles of TFAM, TFB2M,and POLRMT in open complex formation. (A) Sequence of the LSP promoter fragment (−42 to +21) is shown with the positions of 2-AP substitutions in red and the numbering is relative to the transcription initiation site indicated as +1. (B) Structures of 2-AP:dT and 2-AP:dC base pairs (45,46). (C) The table indicates the individual base changes to 2-AP, the resulting base pairs, and the transcriptional activity of the singly 2-AP labeled LSP promoters. (D) The bar chart shows the fold increase in 2-AP fluorescence (excitation at 315 nm and emission at 370 nm) in the LSP promoters (100 nM) substituted with 2-AP relative to free 2-AP labeled LSP promoter in the presence of TFAM+POLRMT+TFB2M (equimolar 150 nM). The cartoon summarizes the FRET and 2-AP results showing the open complex generated with all three proteins (TFAM, light blue; TFB2M, dark blue; POLRMT, orange). (E) ATP (0.5 mM) was added after addition of equimolar TFAM+POLRMT+TFB2M to each of the 2-AP labeled LSP promoter DNAs. The cartoon summarizes the FRET and 2-AP results showing the initiation complex with all three proteins and ATP. (F) The bar chart shows the fold increase in 2-AP fluorescence at the -1NT position with POLRMT+TFAM and POLRMT+TFB2M with and without ATP in comparison to all three proteins and ATP. The numbers over the bars are the fold increase in fluorescence intensity over the free DNA fluorescence. The cartoon shows the two possible semi-open complexes of POLRMT+TFAM and POLRMT+TFB2M on LSP. The error bars in the above experiments are from two independent experiments.
To identify the base pairs that are melted in the open complex of the LSP, the 2-AP base was introduced at nine individual base positions in the −7 to +3 region of the LSP fragment (−42 to +21) (Figure 4A and 4C). All the 2-AP substitutions in the LSP DNA maintained base pairing, except the −4T (−4 template) position where 2-AP substitution resulted in a G:2-AP mismatch (Figure 4A and 4C). All the 2-AP substituted promoters were active in transcription in presence of TFAM, TFB2M and POLRMT, except 2-AP substitution at −4NT (−4 non-template base), which showed a decrease in activity to 2% (Figure 4C and Supplementary Figure S1C). The complementary −4T substitution showed 30% activity, which indicates that the −4NT G-residue on the LSP DNA is critical for transcription initiation. 2-AP substitutions at some positions, such as −5T, +2NT and +3NT, have higher activities, but the reasons are not known.
To map the DNA bubble in the open complex, the 2-AP fluorescence intensity was measured after adding equimolar amounts of TFAM, POLRMT and TFB2M to each of the 2-AP substituted LSP DNAs. A parallel experiment was carried out with unmodified LSP DNA to correct for background fluorescence enhancement. The corrected 2-AP fluorescence in the presence of protein was divided by the fluorescence of 2-AP labeled LSP DNA in the absence of protein to calculate the fold-increase in 2-AP fluorescence. Only positions from −4 to +1 showed a reproducible 2–8-fold increase in 2-AP fluorescence (Figure 4D). Bases upstream and downstream of −4 to +1 position did not show fluorescence increases and some showed a slight negative change, but the negative change was too small to make any conclusions. The −4NT showed no fluorescence increase, which is consistent with its low transcription activity. The transcriptionally active −4T on the other hand showed a large 6-fold increase in fluorescence, which indicates that the −4T is significantly unstacked in the open complex. The upstream −5T and −7NT showed no change or a slight negative change in 2-AP fluorescence. Together, these results indicate that the upstream edge of the DNA bubble in the open complex is most likely at the −4 position. The downstream positions +2 and +3 showed no increase in fluorescence; hence, the downstream edge of the DNA bubble in the open complex is +1. These results indicate that 5 base pairs from −4 to +1 are melted in the open complex with TFAM, POLRMT, and TFB2M.
ATP is the initiating nucleotide on the LSP DNA; hence, ATP addition converts the open complex to the initiation complex. Addition of ATP to the open complex with TFAM+POLRMT+TFB2M increased the DNA bubble size from 5 to 7 bp from −4 to +2/+3 (Figure 4E). Interestingly, the greatest effect of ATP was observed on the −1 and +1 positions, which showed a 26-fold and 12-fold increase in fluorescence, respectively. This indicates that ATP binding results in substantial unstacking of both −1 and +1 bases. There was no effect of ATP binding at the other positions (Figure 4E); thus, the conformational change due to ATP binding is localized within the transcription initiation bubble from −1 to +2/+3. Overall, the 2-AP fluorescence mapping studies show that TFAM, POLRMT and TFB2M melt the LSP promoter from −4 to +1 and ATP binding unstacks the −1 and +1 bases as well as expands the transcription initiation bubble from −4 to +2/+3.
Both TFB2M and TFAM are necessary for efficient melting and/or stabilization of the DNA bubble of the LSP
Having developed an assay to measure promoter melting in the LSP DNA, we used the most sensitive −1NT 2-AP substituted position to investigate the roles of TFAM and TFB2M in promoter melting. Interestingly, POLRMT+TFB2M and POLRMT+TFAM showed a reproducible 2–3-fold increase in −1NT 2-AP fluorescence upon binding to the LSP DNA. Addition of ATP to the complex increased the fluorescence further to about 3-fold in the POLRMT+TFAM+ATP mixture and to about 5-fold in the POLRMT+TFB2M+ATP mixture. When both TFAM and TFB2M are present with POLRMT and ATP, there is 18–26-fold increase in −1NT 2-AP fluorescence. This indicates that TFAM and TFB2M on their own lack the mechanism to efficiently melt the promoter initiation region. The low efficiency of promoter melting is not due to lack of protein–DNA complex formation on the LSP DNA, because the Kd values of the two component protein–DNA complexes are in the 3–8 nM range and the 2-AP experiments were carried out at 100 nM concentrations. Thus, our data suggests that TFAM and TFB2M work synergistically to catalyze open complex formation, which indicates that TFAM has additional roles beyond POLRMT recruitment. One of these ‘post-recruitment’ roles is in promoter melting and/or stabilization of the melted DNA in the initiation complex, which is consistent with our transcription results showing that POLRMT+TFB2M can catalyze the synthesis of 2-mer RNA but requires TFAM for the synthesis of longer run-off RNA products (Figure 1E, 1F).
DISCUSSION
Transcription initiation by POLRMT at the human mitochondrial light strand and heavy strand promoters (LSP and HSP) requires two transcription initiation factors, TFAM and TFB2M. To understand the mechanism of initiation complex formation at the LSP and the roles of TFAM and TFB2M in this process, we have quantified the pre-initiation steps of promoter binding, bending and melting and the initiation steps of abortive RNA synthesis with ATP. The current model of initiation complex formation on the mitochondrial DNA posits that POLRMT, TFAM, and TFB2M are assembled on the promoter in a stepwise manner (13,19,20,28). According to the stepwise model, TFAM bound to the upstream promoter region of the mitochondrial DNA recruits POLRMT to the downstream promoter initiation region forming a pre-initiation complex, in which POLRMT interacts with the C-tail of TFAM and the far upstream regions in the promoter. Finally, TFB2M binds to POLRMT and catalyzes the melting of the promoter initiation region that allows POLRMT to interact with the transcription start site.
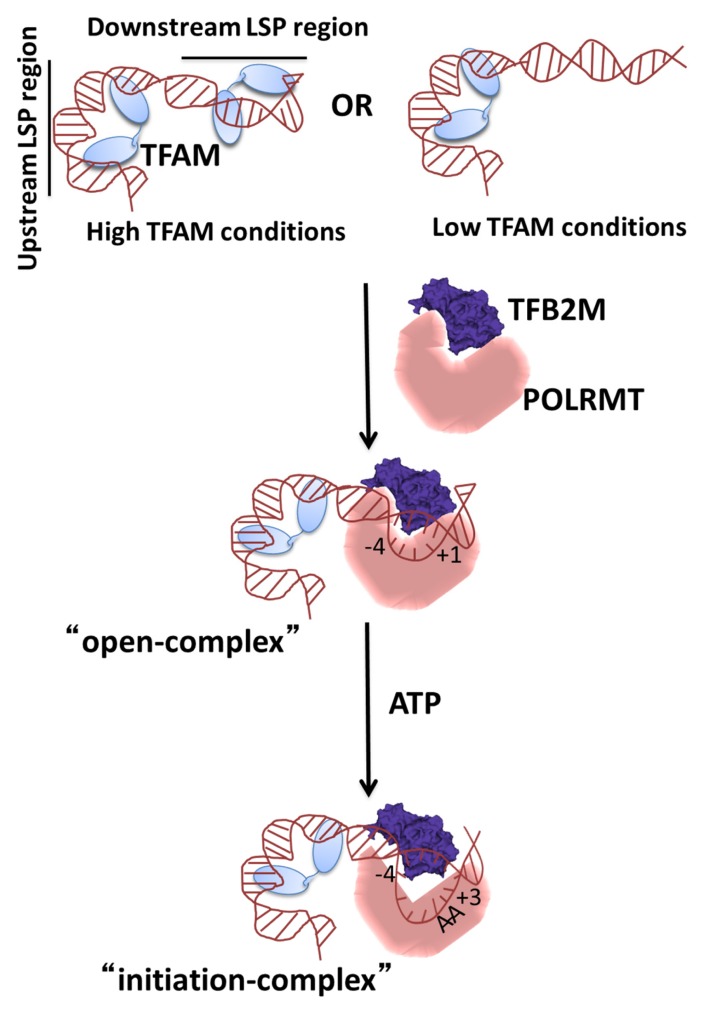
The in vivo scenario for transcription initiation at the mitochondrial promoters could be different. There is a range of in vivo TFAM concentrations reported in the literature, from 35–50 to 2000 molecules per mtDNA (23–27). Therefore, TFAM either sparsely covers the mitochondrial DNA or the entire mitochondrial DNA is covered with TFAM. Thus, under high TFAM conditions in vivo the upstream and downstream LSP regions are occupied with TFAM and under low TFAM conditions perhaps only the upstream site on LSP is covered with TFAM (Figure 5). We propose that POLRMT+TFB2M complex, owing to its similar Kd as the TFAM complex on the DNA, displaces the TFAM molecule bound at the promoter or directly binds to the promoter site to initiate transcription. There are many arguments for POLRMT+TFB2M directly binding to the DNA. First, there is no in vivo evidence for the POLRMT+TFAM intermediate on the mitochondrial DNA; second, the POLRMT and TFB2M concentrations in the cell are very similar (24); third, the two proteins have been shown to form a tight pre-complex with a Kd of 100 nM (28); and fourth, POLRMT+TFB2M has a higher affinity for the DNA than POLRMT alone (Figure 2F).
Figure 5.
A minimal model of transcription initiation on LSP in human mitochondria. The cartoon shows that under high TFAM conditions, the LSP DNA region is bound to two TFAM molecules, one on the upstream and the other on the downstream LSP region. Similarly, under low TFAM conditions, one TFAM molecule is bound to the upstream site. As shown by our FRET studies and those in the literature (16), the TFAM bound upstream DNA is more severely bent than the TFAM bound downstream region. Transcription initiates when the POLRMT+TFB2M displaces the TFAM molecule bound to the downstream initiation region or it directly binds to the protein-free initiation region. The resulting POLRMT+TFAM+TFB2M promoter DNA complex contains a U-turn upstream DNA bend and an ∼80o downstream DNA bend that brings all three proteins in close vicinity to one another to form a compact open complex in which the DNA is melted from −4 to +1. The binding of the initiating ATP converts the open complex to the initiation complex in which the DNA bubble is expanded from −4 to +3.
The stepwise model in literature is based on the in vitro observation of a stable POLRMT+TFAM complex on the promoter and the suggestion that POLRMT and POLRMT+TFB2M cannot form a stable complex on the DNA. Specifically, crosslinking studies noted an efficient crosslink between POLRMT and TFB2M in the presence of TFAM, and a lower amount of crosslinked complex in the absence of TFAM (13,19). Similarly, DnaseI footprinting studies did not detect a specific footprint of POLRMT+TFB2M on the promoter DNA until TFAM was added (9,28). In contrast, our equilibrium fluorescence anisotropy titrations show that both POLRMT+TFAM and POLRMT+TFB2M form high affinity complexes on LSP with nanomolar Kd values (Figure 2G). Similarly, POLRMT+TFB2M complex on the DNA was observed by gel-filtration (20). Based on these studies, we conclude that the lower amount of crosslinked POLRMT+TFB2M complex in the absence of TFAM might be due to the different conformational states of POLRMT+TFB2M and POLRMT+TFB2M+TFAM on the promoter. Similarly, the lack of a specific footprint by POLRMT+TFB2M on the promoter is explained by the tendency of POLRMT and POLRMT+TFB2M to form non-specific complexes on the DNA in the absence of TFAM (Supplementary Figure S3A and B). There is also the possibility that the POLRMT+TFB2M complex is more dynamic on the DNA (higher kon and koff) and chased away by DnaseI, which can act as a trap and compete with the rebinding of POLRMT+TFB2M. The low nanomolar Kd values of all protein–DNA complexes, except for the TFB2M–DNA complex, indicates that at least in vitro, these proteins can assemble on the LSP independently. We propose that the ‘recruitment’ role of TFAM is to ensure specific binding of POLRMT+TFB2M to the promoter initiation site.
Our studies of promoter bending, melting and abortive synthesis, revealed previously unknown post-recruitment functions of TFAM in promoter melting and RNA elongation. First, we show that both the upstream and downstream DNA regions of the LSP are bent in the open complex with POLRMT+TFAM+TFB2M (Figure 5). The upstream DNA is bent mainly by TFAM, which is not surprising, but we show that the downstream DNA is also bent in all the POLRMT complexes. The 2-AP fluorescence mapping studies show that the size of the DNA bubble in the open complex is conserved in single subunit RNA polymerases of phage T7, yeast mitochondria, and human mitochondria. However, the mechanism of promoter opening is not conserved. The LSP DNA is melted from −4 to +1 in the open complex when all three proteins are present and from −4 to +3 when ATP is added to generate the initiation complex. Thus, the three single subunit RNA polymerases melt the promoter region from −4 to +2/+3, but using different mechanisms. T7 RNA polymerase does not require additional protein factors to melt the promoter, but relies on the initiating nucleotides, especially the +2 NTP, to efficiently melt the promoter (47). The yeast mitochondrial Rpo41 relies on Mtf1, which is sufficient for promoter melting (5,30,48), although ATP stabilizes the open complex (49). In case of human mitochondria, POLRMT forms a stable complex with TFAM or TFB2M on the LSP DNA, but these complexes with the individual factors lack the mechanism to efficiently open the promoter and/or keep the open promoter stably in the melted state. Thus, POLRMT requires both TFAM and TFB2M to melt the promoter.
The inefficient melting of the promoter by POLRMT+TFAM and POLRMT+TFB2M is consistent with their low transcription activities. Under our reaction conditions, we find that the POLRMT+TFB2M complex has a 50-fold lower transcription activity to make runoff RNA relative to the activity with all three proteins. Other studies have shown that POLRMT+TFB2M and POLRMT+TFAM complexes are active with the ApA primer on the LSP and HSP DNAs (22,29,50). Our studies of abortive synthesis with ATP alone show that POLRMT+TFB2M makes 2-mer RNA with a catalytic efficiency (kcat/Km) only two-fold lower than that of the POLRMT+TFB2M+TFAM initiation complex. Interestingly, POLRMT+TFB2M cannot efficiently elongate the 2-mer to 3-mer or synthesize poly(A) slippage products without TFAM. This indicates that TFAM plays a role in the stabilization of the DNA bubble and/or the stabilization of the nascent 2-mer on the active site. Thus, both the direct promoter melting studies and the abortive RNA synthesis studies are indicating that TFAM is necessary to stabilize the open and initiation complexes.
We propose here a working model of promoter melting based on our findings and those in the literature as well as similarities to the yeast system. Our FRET studies show that the upstream promoter region of LSP is bent by TFAM and the downstream initiation region is bent by ∼80o by the POLRMT. These severe bending conformational changes bring all three proteins in close vicinity to one another to form a compact complex, as observed in the electron microscopy studies (20). Crosslinking studies indicate that TFB2M is in close proximity to the initiation region and the +1 /+2 ATP when TFAM is part of the complex with POLRMT (12). We propose that this catalytically active conformational state is achieved when TFAM interacts with the N-terminal extension domain of POLRMT and relieves the ‘autoinhibition’ exhibited by the N-terminal extension (28), allowing TFB2M to interact with the melted template (12) and perhaps the non-template strand (51) of the DNA bubble and position the beta hairpin and the specificity loop of the POLRMT to stabilize the melted template strand. We propose that the role of TFAM in promoter melting is 3-fold: (i) TFAM enables specific binding of POLRMT+TFB2M to the promoter DNA, (ii) TFAM induces DNA bending to promote interactions between its C-tail and the N-terminal extension of POLRMT and (iii) TFAM stabilizes a conformational state of POLRMT that aids in the melting and/or stabilization of the open complex by TFB2M. These proposed roles of TFAM in promoter melting and stabilization of the nascent RNA might be allosteric and/or direct. A molecular level understanding of the roles of TFAM will require further studies including high resolution structures of the initiation complexes.
Thus, contrary to common belief that TFAM is required only to recruit POLRMT, our studies indicate that at least on the LSP, TFAM is not just a ‘recruitment’ factor, but has ‘post-recruitment’ roles: it facilitates the conversion of closed complex to open complex and helps stabilize the initiation complex. In this regard, TFAM is similar to the class II transcription activator, the Catabolite Activator Protein or CAP at the gal promoter of the E. coli, which facilitates both RNA polymerase binding and the conversion of the closed complex to the catalytically active open complex (52,53). It is suggested that there may be differential regulation by TFAM and TFB2M factors at the LSP and HSP (21,54). Thus, it will be interesting to determine whether the HSP promoters show a similar degree of dependency on both transcription factors and ATP for promoter melting.
Acknowledgments
We are thankful to Professor Miguel Garcia-Diaz at the Stony Brook University School of Medicine for his kind gift of the TFB2M expression vector and Dr Elena Yakubovskaya for her invaluable advice for TFB2M protein purification. We thank Swapnil Devarkar for critical reading of the manuscript and the Patel lab members for their critical insights throughout these studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences (NIGMS) [R35GM118086 to S.S.P.]; American Heart Association (AHA) [16PRE30400001 to U.B.]. Funding for open access charge: NIGMS [R35GM118086 grant to S.S.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Shadel G.S. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am. J. Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shutt T.E., Shadel G.S. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cermakian N., Ikeda T.M., Miramontes P., Lang B.F., Gray M.W., Cedergren R. On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 4.Davanloo P., Rosenberg A.H., Dunn J.J., Studier F.W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshpande A.P., Patel S.S. Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase. Biochim. Biophys. Acta. 2012;1819:930–938. doi: 10.1016/j.bbagrm.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang S.H., Jaehning J.A. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- 7.Mangus D.A., Jaehning J.A. Transcription in vitro with Saccharomyces cerevisiae mitochondrial RNA-polymerase. Methods Enzymol. 1996;264:57–66. doi: 10.1016/s0076-6879(96)64009-9. [DOI] [PubMed] [Google Scholar]
- 8.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 9.Gaspari M., Falkenberg M., Larsson N.G., Gustafsson C.M. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubot F.D., Chen C.J., Rose J.P., Dailey T.A., Dailey H.A., Wang B.C. Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 2001;10:1980–1988. doi: 10.1110/ps.11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotney J., Shadel G.S. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 12.Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morozov Y.I., Parshin A.V., Agaronyan K., Cheung A.C., Anikin M., Cramer P., Temiakov D. A model for transcription initiation in human mitochondria. Nucleic Acids Res. 2015;43:3726–3735. doi: 10.1093/nar/gkv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dairaghi D.J., Shadel G.S., Clayton D.A. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 15.Fisher R.P., Lisowsky T., Parisi M.A., Clayton D.A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 16.Malarkey C.S., Bestwick M., Kuhlwilm J.E., Shadel G.S., Churchill M.E. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngo H.B., Lovely G.A., Phillips R., Chan D.C. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngo H.B., Kaiser J.T., Chan D.C. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morozov Y.I., Agaronyan K., Cheung A.C., Anikin M., Cramer P., Temiakov D. A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 2014;42:3884–3893. doi: 10.1093/nar/gkt1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yakubovskaya E., Guja K.E., Eng E.T., Choi W.S., Mejia E., Beglov D., Lukin M., Kozakov D., Garcia-Diaz M. Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 2014;42:4100–4112. doi: 10.1093/nar/gkt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodeiro M.F., Uchida A., Bestwick M., Moustafa I.M., Arnold J.J., Shadel G.S., Cameron C.E. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6513–6518. doi: 10.1073/pnas.1118710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shutt T.E., Bestwick M., Shadel G.S. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription. 2011;2:55–59. doi: 10.4161/trns.2.2.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alam T.I., Kanki T., Muta T., Ukaji K., Abe Y., Nakayama H., Takio K., Hamasaki N., Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotney J., Wang Z., Shadel G.S. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukat C., Wurm C.A., Spahr H., Falkenberg M., Larsson N.G., Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniura-Weber K., Goffart S., Garstka H.L., Montoya J., Wiesner R.J. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posse V., Hoberg E., Dierckx A., Shahzad S., Koolmeister C., Larsson N.G., Wilhelmsson L.M., Hallberg B.M., Gustafsson C.M. The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Res. 2014;42:3638–3647. doi: 10.1093/nar/gkt1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y., Dierckx A., Wanrooij P.H., Wanrooij S., Larsson N.G., Wilhelmsson L.M., Falkenberg M., Gustafsson C.M. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paratkar S., Deshpande A.P., Tang G.Q., Patel S.S. The N-terminal domain of the yeast mitochondrial RNA polymerase regulates multiple steps of transcription. J. Biol. Chem. 2011;286:16109–16120. doi: 10.1074/jbc.M111.228023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clegg R.M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 32.Tang G.Q., Patel S.S. Rapid binding of T7 RNA polymerase is followed by simultaneous bending and opening of the promoter DNA. Biochemistry. 2006;45:4947–4956. doi: 10.1021/bi052292s. [DOI] [PubMed] [Google Scholar]
- 33.Ha J.H., Capp M.W., Hohenwalter M.D., Baskerville M., Record M.T., Jr Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the “glutamate effect" on protein–DNA interactions. J. Mol. Biol. 1992;228:252–264. doi: 10.1016/0022-2836(92)90504-d. [DOI] [PubMed] [Google Scholar]
- 34.Deshpande A.P., Patel S.S. Interactions of the yeast mitochondrial RNA polymerase with the +1 and +2 promoter bases dictate transcription initiation efficiency. Nucleic Acids Res. 2014;42:11721–11732. doi: 10.1093/nar/gku868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubio-Cosials A., Sidow J.F., Jimenez-Menendez N., Fernandez-Millan P., Montoya J., Jacobs H.T., Coll M., Bernado P., Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 36.Tang G.Q., Patel S.S. T7 RNA polymerase-induced bending of promoter DNA is coupled to DNA opening. Biochemistry. 2006;45:4936–4946. doi: 10.1021/bi0522910. [DOI] [PubMed] [Google Scholar]
- 37.Cheetham G.M., Jeruzalmi D., Steitz T.A. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 38.Cheetham G.M., Steitz T.A. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 39.Tang G.Q., Deshpande A.P., Patel S.S. Transcription factor-dependent DNA bending governs promoter recognition by the mitochondrial RNA polymerase. J. Biol. Chem. 2011;286:38805–38813. doi: 10.1074/jbc.M111.261966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandwar R.P., Patel S.S. Peculiar 2-aminopurine fluorescence monitors the dynamics of open complex formation by bacteriophage T7 RNA polymerase. J. Biol. Chem. 2001;276:14075–14082. doi: 10.1074/jbc.M011289200. [DOI] [PubMed] [Google Scholar]
- 41.Liu C., Martin C.T. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 42.Liu C., Martin C.T. Promoter clearance by T7 RNA polymerase. Initial bubble collapse and transcript dissociation monitored by base analog fluorescence. J. Biol. Chem. 2002;277:2725–2731. doi: 10.1074/jbc.M108856200. [DOI] [PubMed] [Google Scholar]
- 43.Tang G.Q., Paratkar S., Patel S.S. Fluorescence mapping of the open complex of yeast mitochondrial RNA polymerase. J. Biol. Chem. 2009;284:5514–5522. doi: 10.1074/jbc.M807880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deshpande A.P., Sultana S., Patel S.S. Fluorescent methods to study transcription initiation and transition into elongation. EXS. 2014;105:105–130. doi: 10.1007/978-3-0348-0856-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reha-Krantz L.J., Hariharan C., Subuddhi U., Xia S., Zhao C., Beckman J., Christian T., Konigsberg W. Structure of the 2-aminopurine-cytosine base pair formed in the polymerase active site of the RB69 Y567A-DNA polymerase. Biochemistry. 2011;50:10136–10149. doi: 10.1021/bi2014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkins M.E. Topics in Fluorescence Spectroscopy, Volume 7: DNA Technology. Vol. 7. Springer; 2003. pp. 151–175. [Google Scholar]
- 47.Stano N.M., Levin M.K., Patel S.S. The +2 NTP binding drives open complex formation in T7 RNA polymerase. J. Biol. Chem. 2002;277:37292–37300. doi: 10.1074/jbc.M201600200. [DOI] [PubMed] [Google Scholar]
- 48.Savkina M., Temiakov D., McAllister W.T., Anikin M. Multiple functions of yeast mitochondrial transcription factor Mtf1p during initiation. J. Biol. Chem. 2010;285:3957–3964. doi: 10.1074/jbc.M109.051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H., Tang G.Q., Patel S.S., Ha T. Opening-closing dynamics of the mitochondrial transcription pre-initiation complex. Nucleic Acids Res. 2012;40:371–380. doi: 10.1093/nar/gkr736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C.M., Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paratkar S., Patel S.S. Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem. 2010;285:3949–3956. doi: 10.1074/jbc.M109.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busby S., Ebright R.H. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 53.Feng Y., Zhang Y., Ebright R.H. Structural basis of transcription activation. Science. 2016;352:1330–1333. doi: 10.1126/science.aaf4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shutt T.E., Lodeiro M.F., Cotney J., Cameron C.E., Shadel G.S. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]