Abstract
Due to its long half-life compared to messenger RNA, bacterial transfer RNA is known as stable RNA. Here, we show that tRNAs become highly unstable as part of Escherichia coli's response to amino acid starvation. Degradation of the majority of cellular tRNA occurs within twenty minutes of the onset of starvation for each of several amino acids. Both the non-cognate and cognate tRNA for the amino acid that the cell is starving for are degraded, and both charged and uncharged tRNA species are affected. The alarmone ppGpp orchestrates the stringent response to amino acid starvation. However, tRNA degradation occurs in a ppGpp-independent manner, as it occurs with similar kinetics in a relaxed mutant. Further, we also observe rapid tRNA degradation in response to rifampicin treatment, which does not induce the stringent response. We propose a unifying model for these observations, in which the surplus tRNA is degraded whenever the demand for protein synthesis is reduced. Thus, the tRNA pool is a highly regulated, dynamic entity. We propose that degradation of surplus tRNA could function to reduce mistranslation in the stressed cell, because it would reduce competition between cognate and near-cognate charged tRNAs at the ribosomal A-site.
INTRODUCTION
Transfer RNAs (tRNA), the adaptor molecules that translate the codons in mRNA to amino acids in proteins, are a fundamental component of the translation machinery and is typically referred to as abundant ‘house-keeping’ RNA. Accordingly, our knowledge about the regulation of tRNA transcription in Escherichia coli is by and large limited to the growth-rate control of RNA polymerase partitioning, which ensures that the RNAs of the translation machinery are produced in amounts that match the cell's need for protein synthesis (1–3). In agreement with the house-keeping role, bacterial tRNA is considered highly stable RNA, which is generally only degraded when compromised in quality (4–6). In contrast to this conservative image of tRNA synthesis and turnover, the availability of individual tRNAs has been shown to vary with differences in growth rate (7). Intriguingly, although the variations in tRNA levels are modest, a clear correlation between the dynamic changes in tRNA levels and the frequency of usage of the corresponding codons in the transcriptome at different growth rates has been noted in E. coli (7,8). Moreover, a large body of recent experiments with eukaryotic cells suggest that, in fact, the tRNA pool of these cells should be considered a highly dynamic and tightly regulated entity (recently reviewed in (9,10)). For example, measurements of the tRNA pool of hundreds of human cell samples showed that the tRNA content of proliferating cells differs greatly from that of differentiated cells, and that each tRNA repertoire matches the codon usage signature of proliferation-related or differentiation-related mRNAs, respectively (11). In addition to global changes in the tRNA pool, it also appears that individual tRNAs may be either stabilized, destabilized or functionally altered depending on the cellular state (12). Also, different cytotoxic chemicals have been shown in yeast to induce specific changes in tRNA modifications, which in several cases appear to be an important part of the cellular stress response, since mutants that lack the corresponding modification enzymes show increased sensitivity to the toxicant (13). Along the same line, it has been observed in E. coli that amino acid limitation results in highly uneven charging levels of the different tRNA isoacceptors for the growth-limiting amino acid (14), and the codon usage of the corresponding amino acid biosynthetic genes appear to have evolved to match the changed availability of charged isoacceptors during stress. Specifically, codons corresponding to tRNA isoacceptors that remain highly charged during amino acid limitation predominate the coding sequences of the genes, whereas codons corresponding to the tRNA isoacceptors that are most sensitive to amino acid limitation are present in the regulatory leader sequences, which are controlled by ribosome-mediated transcriptional attenuation (15). Finally, a recent report showed that tRNA halflife in E. coli is strongly affected during oxidative stress (16). Together, these and other studies support a general view of dynamic control of both the production, modification, aminoacylation, and degradation of tRNAs, which serves to align the supply of aminoacylated tRNAs with the changing demands posed by the transcriptome characteristic of different cellular states or growth conditions.
Imbalance between the supply and demand for particular aminoacylated tRNAs is not only energetically suboptimal, it also results in increased mistranslation, which can be detrimental for the cell. Specifically, starvation for a single amino acid results in reduced levels of the corresponding aminoacylated tRNA, which in turn results in increased incorporation of incorrect amino acids supplied by near-cognate tRNAs at the corresponding ‘hungry’ codons on the mRNA (17,18). In E. coli, this increased error rate upon amino acid starvation is 10-fold less dramatic in wildtype cells than in mutants defective for the stringent response (19,20). The stringent response orchestrates large-scale changes in cellular gene expression upon decreases in nutrient availability. The major transcriptional effects of the stringent response are decreased expression of genes encoding the translational machinery (including tRNA and rRNA), as well as a general decrease in mRNA synthesis, except for mRNAs encoding some amino acid biosynthetic enzymes and stress response factors (reviewed in (21,22)). The reduced mRNA synthesis rate makes mRNA the limiting component for translation during the stringent response (23,24), and has been found to be essential for alleviating misreading at hungry codons, presumably because it reduces the demand for the cognate aminoacylated tRNA, thereby allowing it to remain at a level where it can outcompete near-cognate tRNAs at the hungry codons (18,23). Another means to decrease competition between cognate and near-cognate charged tRNAs at the hungry codons would be to reduce the levels of the near-cognate tRNAs. The reduced transcription of tRNA genes imposed by the stringent response upon starvation may to some degree accomplish this goal, but if the existing tRNA pool is long-lived it is unlikely that termination of tRNA synthesis upon amino acid starvation has noteworthy effects on the pools of near-cognate aminoacylated tRNAs in the short term.
In this work, we investigated the hypothesis that tRNA half-life may be actively decreased under conditions such as amino acid starvation where a reduction of near-cognate tRNA pools would be expected to improve translation accuracy.
Interestingly, it has been reported that different kinds of long-term nutrient limitation eventually results in degradation of the otherwise highly stable rRNA (25–30). Degradation of rRNA could serve the purpose of recycling the nucleotides as nutrients for the starved cell, and the degradation appears to aid recovery from long-term starvation (31). On the shorter time-scale of a few hours, 24% of ribosome-associated RNA is degraded after four hours of threonine starvation (32) and data from the Deutscher laboratory showed degradation of an appreciable amount of rRNA after 3 hours of carbon starvation (30). These experimental results show that different types of starvation eventually result in degradation of rRNA, and possibly all stable RNA. However, only very recently a mechanism for the rRNA degradation has been described (33), while no degradation mechanism has been identified for tRNA, and no one has investigated the short-term kinetics of tRNA decay. In this work, we show that more than half of the cellular tRNA content is degraded within twenty minutes after inhibition of translation.
MATERIALS AND METHODS
E. coli strains, growth media, and conditions for growth and starvation
The E. coli strains used are all K12 strains. Supplementary Table S2 lists the strains used in this study. The relA+/relA2 isogenic strain set NF915 and NF916 was chosen here because they have been used in a vast number of reports about the Rel+/Rel− relationship in growth physiology (see (18) for references).
Cultures were grown exponentially at 37.0°C in morpholinepropanesulfonic acid (MOPS) minimal medium (34) supplemented as described for each experiment. All cultures had been growing exponentially for at least ten generations before being exposed to amino acid starvation or antibiotic treatment. Amino acid starvation for auxotrophic strains was introduced by filtration of the culture and resuspension in medium lacking the amino acid indicated as previously described (18) or by addition of 400 μg/ml valine to the culture to introduce an isoleucine starvation (35).
Growth conditions for Sulfolobus solfataricus (internal standard)
The following final concentrations were used per 500 ml of 2× liquid medium stock (modified from (36)): 1.3 g (NH4)2SO4, 70 mg CaCl2•2H2O, 2.25 mg Na2B4O7•10H2O, 2.8 mg FeSO4•7H2O, 15 μg NaMoO4•2H2O, 5 μg CoCl2•7H2O, 250 mg MgSO4•7H2O, 900 μg MnCl2•4H2O, 110 μg ZnSO4•7H2O, 25 μg CuCl2•2H2O, 15 μg VOSO4•2H2O. The pH was adjusted to 3.0–3.5 with a solution of sulphuric acid and autoclaved. Prior to use, the 2× stock was supplemented with 2 g sucrose, 280 mg KH2PO4 and autoclaved water to 1 l. The pH was adjusted to 3.0–3.5 once again. This minimal medium supported a generation time of ∼16 h at 80°C in a shaking water bath.
tRNA preparation, blotting and hybridization
Each sample was harvested by transferring 4 ml of culture into 4 ml of 10% (w/v) trichloroacetic acid (TCA) at 37°C. This procedure stops enzymatic activities immediately, which is especially important here to avoid alterations of the tRNA charging levels after sample collection. After mixing, the 8 ml 5% TCA samples were stored at 0°C until all samples had been collected. At this time, 10% S. solfataricus cells (measured as OD436) were added as 0°C TCA-stopped culture to each sample before the first centrifugation. From this mixture, tRNA was purified by vigorous vortexing with acidic phenol followed by electrophoresis and blotting essentially as described by Varshney et al. (37) with a few modifications: each sample of tRNA was redissolved in 20 μl of 10 mM sodium acetate (pH 4.5), 1 mM EDTA. 4 μl of such a sample were used for each lane in the gels. Deacylated tRNA aliqouts were obtained by treatment with 0.1 M Tris–HCl (pH 9.0) at 37°C for at least 1 h in a total volume of 50 μl. The tRNA was neutralized, precipitated, and then dissolved in 4 μl of 10 mM sodium acetate (pH 4.5) and 1 mM EDTA. Untreated or deacylated tRNA (4 μl) was mixed with 6 μl of loading buffer (0.1 M sodium acetate (pH 5.0), 8 M urea) plus 0.05% (w/v) bromophenol blue and 0.05% (w/v) xylene cyanol, and separated by electrophoresis through a 6% (19:1 (w/w) acrylamide/bisacrylamide) gel containing 0.1 M sodium succinate (pH 5.0), 8 M urea. The gels were 50 or 60 cm long and electrophoresis was carried out at 10 V/cm at 4°C until the bromophenol blue reached the bottom of the gel (∼20 h).
Next, 20 cm of the gel between the two dyes was electroblotted onto a Hybond N+ membrane (GE Healthcare) at 20 V for 90 min with 40 mM Tris-acetate (pH 8.1), using 2 mM EDTA as transfer buffer. The tRNA was crosslinked to the membrane by 0.12 J of UV light in a Stratalinker 1800. The membranes were pre-hybridized at 42°C for 1 h in 6 ml of hybridization solution (0.9 M NaCl, 0.05 M NaH2PO4 (pH 7.7), 5 mM EDTA, 5× Denhardt's solution (38), 0.5% (w/v) SDS and 100 mg/ml of sheared, denatured herring sperm DNA). Hybridization was at 42°C overnight in the same solution with 30 pmol radioactive oligo-DNA probe 5′-end-labeled with 32P by the use of polynucleotide kinase and gamma-32P-ATP. Membranes were washed several times in 0.3 M NaCl, 30 mM sodium citrate, 0.1% SDS at room temperature. In addition, membranes used for detection of tRNAThrV were washed for 30 min at 60°C to remove unspecific cross-hybridization. The radioactivity present in specific bands was measured using a phosphorimager scanner. Before reprobing a blot, repetitive washes were performed by pouring >100 ml of boiling ‘stripping’ buffer (0.015 M NaCl, 1.5mM sodium citrate 0.1% SDS) over the blot, until no more radioactivity could be detected on the blot by a Geiger-Müller tube. The probe sequences are listed in Supplementary Table S3. None of the probes we have used have shown any cross reactivity with RNA from the other species.
Throughout this paper, tRNA species are named according to the name(s) of the gene(s) that encode them.
Normalization of tRNA levels
Each blot was probed for several tRNAs as well as the S. solfataricus 5S RNA. Therefore, we could use the signal from the S. solfataricus 5S RNA as an internal standard for gel loading and recovery of total RNA in each sample. The counts found in tRNA bands were divided by the counts found in S. solfataricus 5S RNA in the same lane and this ratio was normalized to the ratio found in the sample that was harvested before starvation.
Precision of tRNA level measurements in northern blots: In >90% of all the cases where we harvested several samples from the same culture at the same time point (e.g. the pre-starvation time point of Supplementary Figure S4 and data not shown), all values are within ±20% of the average. Charging levels of tRNA were determined as described (18).
Deep RNA sequencing
The RNA used for data in Supplementary Table S1 was extracted from a MG1655 derivative containing a plasmid carrying an IPTG-inducible birA gene (pBirA), but grown in the absence of IPTG. The strain is also modified to contain a biotinylation-tag at the C-terminal of the chromosomally encoded Hfq protein. We confirmed that the Hfq-tag did not influence the growth rate of the strain. Prior to harvesting the RNA, the cells were grown in minimal MOPS medium supplemented with 0.2% glucose and 5 μg/ml biotin. Isoleucine starvation was introduced by addition of 400 μg/ml valine (39). Cells were harvested right before and 10 min after valine addition. Large volumes of cultures were harvested by cooling to 0°C in ice-water, centrifugation and wash in 0°C biotin-free medium, centrifugation and resuspension in 2 ml of lysis buffer, followed by repeated rounds of sonic treatment. At this point, an aliquot of each sample was phenol extracted and stored as total RNA at −80°C. Before sending the RNA samples off for sequencing, we saw that the 23S RNA band was twice as intense as the 16S RNA band on an argarose gel. This indicated that the samples had been purified without significant RNase activity during preparation. The treatment of the samples and the Illumina sequencing was performed by GATC BIOTECH (http://www.gatc-biotech.com) on total RNA samples without prior rRNA depletion. The order of treatments were: (i) fragmentation of RNA, (ii) first strand synthesis with random primers, (iii) second strand synthesis with random primers, (iv) end repair and A tailing, (v) adapter ligation, (vi) size selection and quantification, and lastly, (vii) sequencing in reads of 50 bp. More than 2 billion base pairs were sequenced from each sample. We used the free software program Artemis release 16.0.0 (40) from the Sanger Institute for our analysis. Under the assumption that the frequency of reads mapping to a specific sequence actually represents the frequency of that sequence in the total RNA population, and that the 23S RNA concentration does not change considerably during 10 min of starvation, we used the number of reads mapped to rrlA (23S gene of ribosomal operon A) in each sample to normalize the amounts of RNA in each sample. This resulted in multiplication of the actual number of reads in the sample ‘10 min starvation’ by 0.74 to give the numbers in Supplementary Table S1.
RESULTS
The majority of tRNA is rapidly degraded upon amino acid starvation
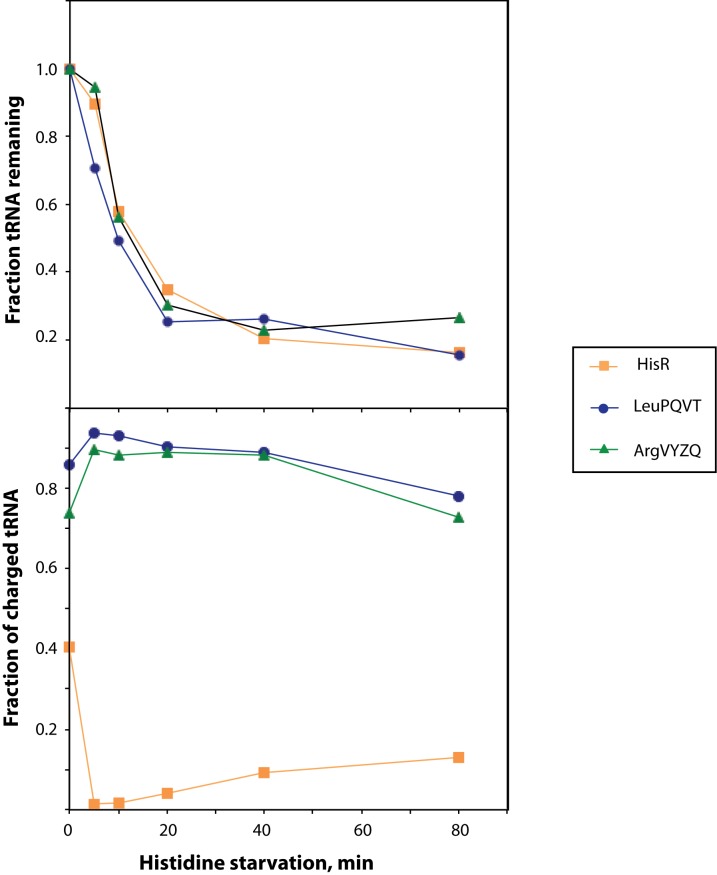
To determine tRNA half-life upon amino acid starvation, the histidine auxotroph E. coli strain NF915 (thr leu his argH thi mtl supE44 relA+ spoT+) was grown in minimal medium containing 5 μg/ml histidine, and then starved for histidine as described previously (18). Aliquots of the cultures were harvested for tRNA preparation immediately before and at 5–80 min after starvation, and the levels of selected tRNAs were examined by northern blot. Since one of the major effects of amino acid starvation is changes in the rates of synthesis, and perhaps degradation, of mRNA, tRNA and rRNA, there are no cellular RNA species that could serve as an appropriate loading control in these experiments. Similarly, simply normalizing to the total RNA harvested from each sample is ambiguous, given that we are precisely performing the experiment to measure the changes in RNA content per cell upon amino acid starvation. To circumvent these issues, the cell culture aliquots were instead spiked with equal volumes of a culture of Sulfolobus solfataricus prior to RNA preparation, and the northern blots were probed for S. solfataricus 5S RNA with a probe that does not cross-hybridize with any E. coli RNA. Such an experiment is shown in Figure 1A, and the results are quantified in Figure 2A.
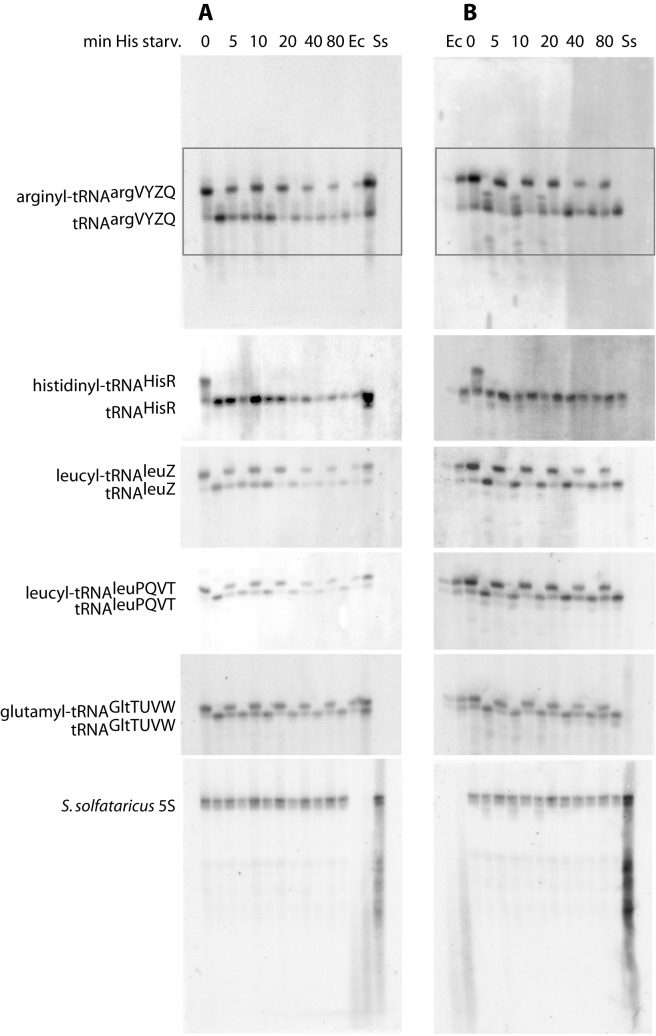
Figure 1.
Both cognate and non-cognate tRNA levels decrease rapidly upon histidine starvation. (A) Phosphorimager scan of a Northern blot membrane containing RNA harvested from NF915 (Rel+) and probed for the indicated tRNA species from E. coli (top) or S. solfataricus 5S RNA (bottom). Starvation was initiated at time zero and the samples were harvested at the time indicated above each lane (minutes). There are two lanes for each timepoint. The first lane contains the untreated sample, whereas the second lane contains chemically deacylated tRNA from the same sample. The same membrane was successively stripped and reprobed for the tRNAs indicated on the left. The boxed area in the upper blot indicates the part of the blot, which is shown for the additional tRNAs. Lanes marked Ec and Ss are control lanes containing RNA from E. coli or S. solfataricus, respectively. We note that due to the starvation for histidine, the charged form of tRNAHisR (histidinyl-tRNA) is only present in the sample collected right before starvation (time zero). (B) Same as (A), but RNA was harvested from the otherwise isogenic relA2 mutant NF916. The growth medium was MOPS 0.4% glycerol, 50 μg/ml Thr, Leu, Arg and 5 μg/ml His. The generation time prior to amino acid starvation was 80 min for both strains.
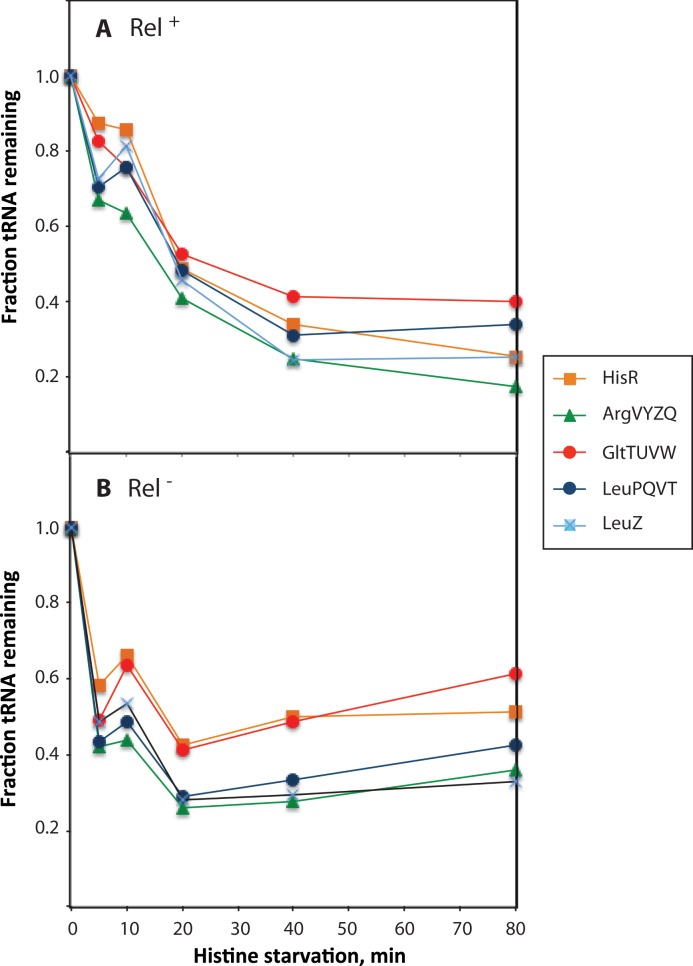
Figure 2.
Quantification of tRNA abundance during histidine starvation. The intensity of the bands found on the Northern blots shown in Figure 1 was quantified using ImageQuant software, and counts in total tRNA (independent of charging state) was normalized by dividing with the counts in S. solfataricus 5S RNA from the same sample. The ratio found at time zero was set to 1 and the ratios found in the following lanes are shown relative to this value. Panel A contains data for strain NF915 and panel B shows data for strain NF916.
Figure 2A shows that the majority of tRNA was degraded upon starvation for histidine. Importantly, both the cognate tRNAhisR (orange squares), as well as tRNAargVYZQ, tRNAleuZ, tRNAleuPQVT and tRNAgltTUVW, decreased in levels within 5 min after the initiation of histidine starvation, and reached a new steady state level of 20–40% of their level in unstarved cells within ∼40 min of starvation. Aminoacylated tRNA can be distinguished from uncharged tRNA on the northern blot due to the difference in size of the two species. To illustrate this difference, a chemically deacylated aliquot of each sample was loaded on the gel in the lane next to the untreated aliquot from the same sample (Figure 1A). We found that degradation of tRNA occurs independently of the charging status of the tRNA, that is, both charged and uncharged tRNA was rapidly degraded upon amino acid starvation (Figure 1A). Notably, no particular tRNA degradation products (fragments of tRNAs) were detected in any of the northern blot experiments.
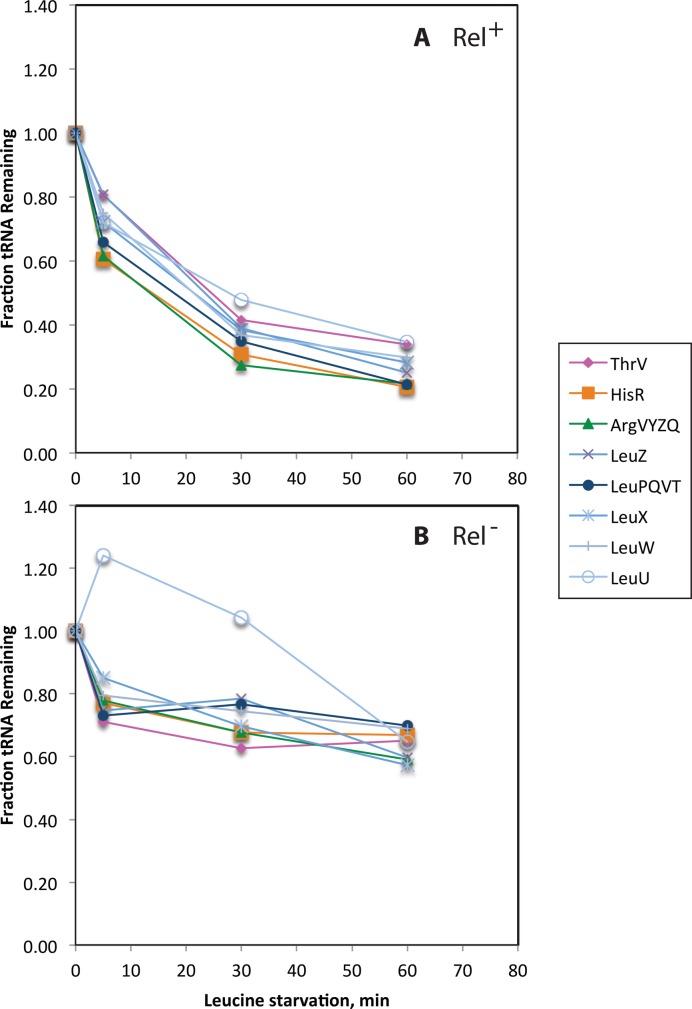
To examine whether this response was specific to histidine starvation, we next tested the stability of a battery of tRNAs in NF915 upon starvation for leucine. As shown in Figure 3A, the levels of tRNAargVYZQ, tRNAleuZ, tRNAleuX tRNAleuW, tRNAleuU, tRNAleuPQVT, tRNAhisR and tRNAthrV decreased rapidly upon removal of leucine. The leucine starvation procedure completely blocked growth as shown in Supplementary Figure S1A. We made sure that the observed loss of tRNA upon starvation was not due to an unintended consequence of the starvation procedure, such as cell lysis, by confirming with colony counts that the number of live cells remained constant for the duration of the starvation period (Supplementary Figure S1).
Figure 3.
Both cognate and non-cognate tRNA levels also decrease rapidly upon leucine starvation. (A) Levels of different tRNA species in strain NF915 immediately before and 5, 30 and 60 min after starvation for leucine. Cells were grown in MOPS glycerol containing 50 μg/ml threonine, leucine, and arginine, and 5 μg/ml histidine with a generation time of 82 min prior to starvation. RNA was harvested at the indicated time points and subjected to northern blot analysis and quantification, as in Figures 1 and 2 respectively. (B) same as A), but RNA was harvested from the otherwise isogenic relA2 mutant NF916.
To ensure that the results were independent of the choice of strain background, we also carried out a similar set of experiments with E. coli MG1655 under valine-induced starvation for isoleucine (35,41). Similar to the previous experiments, degradation of tRNAargVYZQ, tRNAgltTUVW and tRNAhisR was observed upon starvation for isoleucine (Supplementary Figure S2A, closed symbols). tRNA degradation was specific to the starvation condition, as all the tRNAs accumulated again upon addition of isoleucine to the cultures after 80 min of starvation (Supplementary Figure S2A, closed symbols). In the extraction method used for the experiments described above, cells are not lysed before phenol extraction, because the method has been optimised to preserve tRNA charging levels (see Materials and Methods and references therein). To ensure that our results are not an artefact of the RNA extraction procedure, we also extracted RNA using a different method (phenol extraction of total RNA after sonic treatment of cells in lysis buffer, see Materials and Methods), and used the total RNA from this experiment to perform Illumina Deep Sequencing of the transcriptome of E. coli MG1655. The average number of reads (after data normalization) that map to tRNAs before and 10 min after starvation for isoleucine are shown in Supplementary Table S1. It is evident that all tRNA species with an expression level detectable above background decreased in abundance upon starvation (Supplementary Table S1). We note that because reverse transcriptase does not faithfully replicate the modified bases in tRNAs, RNAseq data does not provide the full estimates of tRNA levels (42,43) but we assume that the modification pattern of tRNAs are stable during 10 min of starvation and therefore underrepresentation of a given tRNA sequence due to modified bases should be the same for both time points. Hence, the data support, at least in a qualitative way, our main conclusion that tRNAs become highly unstable during amino acid starvation.
Together, this body of data demonstrates that a hitherto undescribed part of the cellular response to the onset of amino acid starvation is a rapid degradation of the majority of cellular tRNA species, including both the non-cognate and the cognate tRNA for the amino acid that the cell is starving for, and including both aminoacylated and non-aminoacylated tRNA species.
Starvation induced tRNA degradation is independent of the (p)ppGpp-mediated stringent response
The cellular response to amino acid starvation is orchestrated by the stringent response, specifically by RelA-mediated (p)ppGpp production in response to the presence of a deacylated tRNA at the ribosomal A-site (44,45). Therefore, we tested whether tRNA degradation is part of the stringent response triggered by ppGpp, by measuring tRNA levels in a mutant defective for RelA.
Figures 1B and 2B show the results of a histidine starvation experiment equivalent to that shown in Figures 1A and 2A, but carried out with strain NF916, an otherwise isogenic relA2 mutant of E. coli strain NF915. Similarly, Figure 3B shows a leucine starvation experiment of strain NF916 carried out alongside that of NF915 shown in Figure 3A. It is evident that tRNA degradation initially occurs with similar kinetics in the Rel+ and Rel− strains, hence activation of the stringent response by RelA cannot be the trigger for activation of tRNA degradation. Upon amino acid starvation, Rel− strains initiate the so-called relaxed response, which includes a drop in the (p)ppGpp pool and an increase in the total transcription rate, in particular of the rRNA and tRNA genes (1,46). Therefore, the kinetics of degradation of tRNA we observe in the Rel− strain in Figures 2B and 3B is actually the sum-curve of increased production and degradation of tRNA upon amino acid starvation. Presumably, the increased tRNA production in the Rel− strain accounts for the difference in tRNA pools between the two strains observed at later time points: All tRNA levels show an upwards trend in the Rel− strain at later time points after starvation, whereas they appear to reach a new steady state in the Rel+ strain (Figures 2 and 3). The altered transcriptional response of the starved Rel− strain may also explain why a single tRNA, tRNAleuU, remains present at high levels up to 30 mins after leucine starvation in the relaxed mutant, but not in wild type (Figure 3B). The leuU gene is atypically located downstream of secG, encoding a component of the SecYEG translocase. We assume that regulation of transcription of this unique mRNA-tRNA operon is atypical compared to the regulation of the other tRNA- and rRNA-operon promoters. leuU transcription might therefore be differently increased after amino acid starvation in the relaxed mutant, which lacks the ability to down-regulate σ70-directed transcription of highly expressed genes, due to reduced levels of (p)ppGpp (46). This hypothesis is supported by microarray data of secG mRNA, which was found to be down regulated nearly 6-fold in Rel+ cells upon outgrowth in isoleucine-limited medium, but unaltered in the relaxed mutant (47). Finally, we note that it is possible that a quantitative difference exists in the late levels of tRNAs in the starved Rel− strain depending on the kind of amino acid starvation (compare Figures 2B with 3B). We may assume (20) that starvation for histidine is not as severe as starvation for leucine because histidine is a rare amino acid (∼2% of total protein) while leucine is a very common amino acid (∼8% of total protein) (48). The relaxed response may very well differ quantitatively and cause different transcription rates of stringently regulated promoters during starvation for the two different amino acids and therefore cause different late-starvation levels of tRNAs.
To further investigate the involvement of (p)ppGpp in tRNA degradation, we also measured degradation of two tRNAs, tRNAargVYZQ and tRNAthrV, upon starvation for arginine in a strain carrying the spoT1 allele (49), which has increased basal ppGpp levels due to reduced ppGpp-hydrolase activity of the bifunctional (p)ppGpp-hydrolase/synthase SpoT (50) (Supplementary Figure S3). The pattern of degradation of the tested tRNAs in the SpoT1 strain is similar to that in wildtype and relA2 backgrounds, supporting that tRNA degradation upon amino acid starvation is independent of (p)ppGpp.
One potential consequence of amino acid starvation in E. coli is inactivation of the antitoxin part of toxin-antitoxin modules in a fraction of the cell population, resulting in active toxins, many of which encode RNA endonucleases (51–53). Thus, we speculated that tRNA degradation could be a consequence of activation of an RNA endonuclease from a toxin-antitoxin module. However, tRNA degradation occurs according to the wildtype pattern in a strain that lacks the 10 type II toxin-antitoxin modules of E. coli (54), showing that tRNA degradation occurs independently of these 10 RNA endonucleases (Supplementary Figure S2, open symbols).
Inhibition of mRNA synthesis also causes tRNA degradation
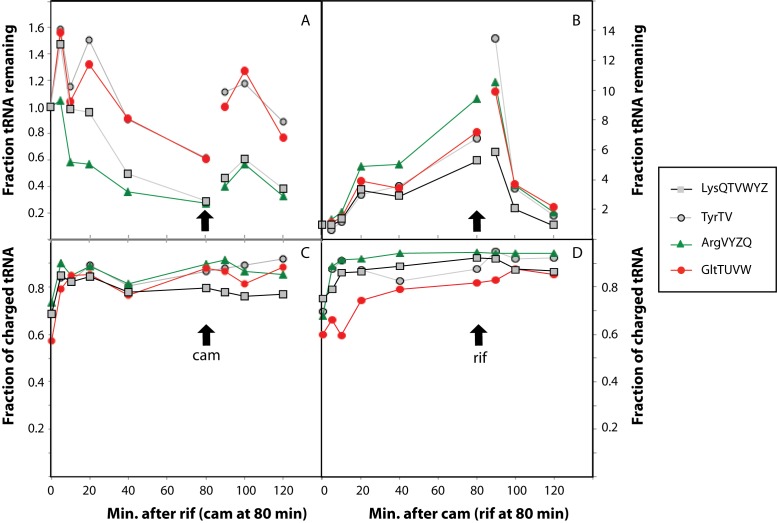
Since tRNA degradation is not dependent on the stringent response, we speculated that the trigger of tRNA degradation might not specifically be amino acid starvation, but rather, tRNA degradation might be triggered by any condition that results in a decreased demand for the tRNAs in protein synthesis. Accumulation of unemployed tRNAs would directly result from the decreased protein synthesis rates of an amino-acid starved cell, where ribosomes are expected to stall at the ‘hungry’ codons, independent of the (p)ppGpp. If so, other treatments that result in disengaged tRNAs may also lead to rapid tRNA degradation. To test this hypothesis, we measured tRNA levels upon addition of rifampicin, which blocks transcription initiation by binding to RNA polymerase, and therefore quickly leads to reduced protein synthesis due to a lack of mRNA substrate (55,56). Figure 4A shows that, indeed, blockage of transcription initiation with rifampicin results in degradation of tRNAtyrTV, tRNAgltTUVW, tRNAargVYZQ and tRNAlysQTVWYZ. Unexpectedly, we noted that rifampicin treatment reproducibly caused a brief rise in the levels of three of the four measured tRNAs before the degradation became evident (Figure 4A, 10–30 min time points). Since rifampicin inhibits transcription initiation, we do not consider it likely that tRNA synthesis was increased in the rifampicin-treated cells. Moreover, addition of the translation inhibitor chloramphenicol 80 min after rifampicin addition also appeared to cause a transient rise in tRNA levels. It is known that chloramphenicol treatment can induce transcription of tRNA (57,58), but in this particular case RNA polymerase is inhibited by the rifampicin in the culture, which should prevent tRNA transcription (Figure 4A, 100 min time point). To address whether the unexpected temporary effect of adding these antibiotics were specific to the RNA extraction protocol or the particular E. coli strain we used, we repeated the experiment using TriReagent® (Sigma-Aldrich) for RNA extraction of both E. coli strain MAS827 (as in Figure 4) and strain MG1655. These experiments are presented in Supplementary Figure S4A and S4B respectively. It is evident that the rifampicin- and chloramphenicol-induced transient increases in tRNA levels are reproduced under these conditions, and thus cannot be regarded as extraction-specific or strain-specific peculiarities. At the moment, we have no clear hypothesis to explain this phenomenon.
Figure 4.
tRNA is degraded when ribosomes are unoccupied. Cellular levels of tRNAs were quantified by northern blot as in Figures 1 and 2. (A) 300 μg/ml rifampicin was added to exponentially growing cells at time zero to block transcription initiation. At time 80 min, the translation inhibitor chloramphenicol (100 μg/ml) was added to the culture. (B) 100 μg/ml chloramphenicol was added to exponentially growing cells at time zero to block translation and induce tRNA transcription. At time 80 min, 300 μg/ml rifampicin was added to the culture to block transcription initiation. The samples were harvested at the indicated times. The strain was MAS827 growing in MOPS 0.2% glucose, 5 μg/ml histidine and 10 μg/ml uracil. The generation time was 52 min prior to the addition of antibiotics. (C and D) The fraction of tRNA that is detected in its aminoacylated form on the northern blots used to produce panels A and B, respectively.
Upon addition of rifampicin, the fraction of tRNAs that are aminoacylated increases (Figure 4C) as we would expect when tRNA deacylation on the translating ribosomes comes to a standstill due to the rifampicin-induced lack of mRNA substrate. We note that charging levels do not reach 100%, suggesting either some residual protein synthesis after addition of rifampicin, or that the charging reactions catalyzed by the aminoacyl tRNA synthases do not proceed to completion.
To examine the cellular capacity for tRNA degradation, we took advantage of the fact that the cellular tRNA (and rRNA) transcription rates can be increased several fold by addition of chloramphenicol (57,58). First, we induced tRNA transcription with chloramphenicol for 80 min, and then we added rifampicin to arrest transcription. Figure 4B shows that, as expected, tRNA levels rose upon addition of chloramphenicol to the culture, and reached up to ∼10-fold the normal level after 80 minutes. Also, the fraction of tRNAs that are aminoacylated increased, consistent with the lack of protein synthesis in the chloramphenicol-treated cells (Figure 4D). However, upon subsequent addition of rifampicin, tRNA levels rapidly plummetted, indicating that the effector(s) of tRNA degradation have a large capacity for tRNA turnover. Importantly, the effector(s) of tRNA degradation must already be present in the cells prior to the first addition of antibiotic in these experiments since synthesis of new proteins is inhibited by chloramphenicol, and de novo synthesis of RNA is inhibited by rifampicin.
In conclusion, we here present evidence that amino acid starvation results in a reduction of the tRNA level to between 20% and 40% of the pre-starvation level within 60–80 min of the onset of starvation. This conclusion is reached on the basis of a large number of experiments in which we have varied the identity of the amino acid starved for, the strain of E. coli used, the RNA extraction method, and the tRNA quantification method. While the levels of residual tRNA during starvation certainly varies between different experimental set-ups, we conclude based on the combined data that tRNA degradation is a general part of E. coli’s response to amino acid starvation.
tRNA charging levels during amino acid starvation are consistent with a reduced pool of total tRNA
In many of our experiments, we have also measured the changes in tRNA charging levels during amino acid starvation (see for example Supplementary Figures S2 and S3). Naturally, the charging levels of tRNAs carrying the amino acid starved for drop rapidly upon initiation of starvation. The charging levels of tRNAs accepting other amino acids, on the other hand, increase slightly during amino acid starvation. Notably, they do not reach fully charged levels as one would predict if the tRNA pools remained unchanged while translation was reduced to negligible levels (15). However, the modest increases in charging levels can be explained by our observation that the total tRNA pool is reduced upon starvation, because decharging of the remaining small pool of tRNAs by residual translation activity could be sufficient to keep the charging levels <100%. We return to this point in the discussion.
To illustrate how the reduction of the total tRNA pool can affect the charging levels of starved and unstarved tRNAs, we carried out a partial histidine starvation by using an E. coli strain that has a leaky His− phenotype, that is, the strain retains the ability to synthesize a small amount of histidine. The histidine limitation resulted in an increase in generation time from 52 min before starvation to 300 min upon removal of histidine from the growth medium. Upon removal of histidine, the charging level of tRNAHisR immediately dropped, and the majority of total tRNA was degraded with the expected kinetics (Figure 5A). During the starvation period, the charging level of tRNAHisR then began to increase (Figure 5B), which is possible because the diminished mRNA pool that results from activation of the stringent response becomes the limiting factor for translation (23,24). In addition, the stringent response induces synthesis of histidine biosynthetic enzymes (reviewed in (59)). Due to the ability of the leaky His− strain to increase histidine biosynthesis above zero, the translational activity would recover slightly from the initial drop caused by removal of histidine during the starvation period. The opposite charging pattern was observed for tRNAs charged with amino acids that are not in shortage: initially, the charging level of tRNAArgVYZQ and tRNALeuPQVT increased, as expected due to the lack of translation, but during the histidine starvation period the charging levels of tRNAArgVYZQ and tRNALeuPQVT returned to levels similar to their pre-starvation levels. This relatively strong reduction in charging levels resulting from a small increase in translation activity can best be explained by the fact that the concentration of total tRNAArgVYZQ and tRNALeuPQVT was about five fold lower than before the starvation, due to tRNA degradation (Figure 5A). The end result is a reduction of the difference in concentration between the charged His-tRNAHis and the tRNAs charged with other amino acids, which should lower the competition between His-tRNAHis and near-cognate aa-tRNAs at starved His codons during the mild amino acid starvation, and therefore most probably reduce the error rate in translation (17,23,60).
Figure 5.
Charging levels of unstarved tRNAs are reflected in their concentration. (A) Levels of tRNAArgVYZQ, tRNALeuPQVT and tRNAHisR in strain MAS827 immediately before and after starvation for histidine. Cells were grown in MOPS glucose containing 5 μg/ml histidine with a generation time of 52 min prior to starvation and 300 min after removal of histidine. RNA was harvested at the indicated time points and subjected to Northern blot analysis and quantification, as in Figures 1 and 2, respectively. (B) Charging levels of the tRNAs shown above.
DISCUSSION
Escherichia coli bacteria carefully match the capacity of their translation apparatus to the demand for protein synthesis posed by their growth rate, which can vary greatly depending on the availability of nutrients. This relation was first noted almost 60 years ago (61), and depends largely on RNA polymerase partitioning between mRNA synthesis and so-called stable RNA synthesis (tRNA and rRNA). For example, tRNA and rRNA synthesis amounts to 30% of total RNA synthesis at a growth rate of 0.5 doublings per hour, whereas it amounts to 80% of total RNA synthesis at a growth rate of 2.5 doublings per hour in E. coli B/r, and independent of the exponential growth rate, this fraction drops to 24% within 15 min of amino acid starvation due to the (p)ppGpp-mediated stringent response (1). Here, we report that tRNA is not, in fact, stable. tRNA is rapidly degraded in response to at least two conditions where the demand for tRNAs in protein synthesis is reduced, namely upon amino acid starvation and after blockage of transcription initiation with rifampicin. The degradation of tRNA seems to be independent of the amino acid starved for (Figures 1–3, Supplementary Figures S2, and S3) as well as the strain background (Figures 1, 2, 5, Supplementary Figures S2 and S3). Furthermore, all the types of tRNA we have tested by northern blot analysis showed similar fast kinetics of degradation, and the transcriptome analysis included in Supplementary Table S1 indicates that all detectable tRNAs are, in fact, unstable during amino acid starvation. In summary, we report here that the match between levels of components of the translational machinery and the demand for protein synthesis is not limited to alterations in the rates of synthesis of said components, but includes also their active degradation, at least in the case of tRNA. Thus, similar to the situation in eukaryotic cells, both the rate of synthesis and the rate of degradation of tRNA are subject to substantial regulation in E. coli.
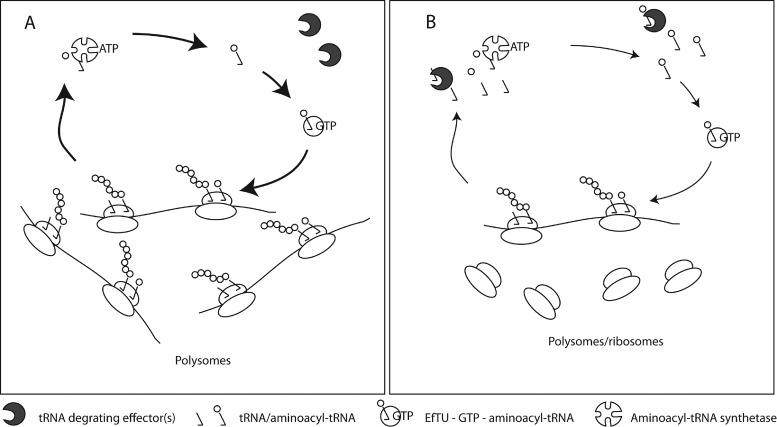
The degradation of tRNA is measurable within minutes of amino acid starvation (Figures 2A, 3A, 5A, Supplementary Figures S2 and S3), appears to be independent of the (p)ppGpp levels (Figures 2B, 3B and Supplementary Figure S3), and can occur when both transcription and translation is inhibited by antibiotics (Figure 4), strongly suggesting that neither the signal(s) nor the effector(s) required for tRNA degradation are synthesized de novo upon starvation. Instead, we favor a passive model, in which tRNA is rapidly degraded when it is not associated with components of the translational apparatus, such as the ribosome, elongation factor Tu or aminoacyl tRNA synthetases (Figure 6). In this model, there is no signal per se for tRNA degradation, and the effector(s) is likely to be any of a number of ribonucleases already present in the cell prior to starvation, which would have a lower general affinity for the tRNA than that of the specialized binding partners mentioned above. Especially for EF-Tu, whose level is expected to match the number of tRNAs, already in the unstarved cells, we have to assume that the affinity for aminoacylated tRNA is sufficiently low to permit degradation of transiently unbound tRNAs. Further verification of this model and identification of the effector(s) is ongoing work in our laboratory. In essence, this passive model for degradation of tRNA is equivalent to the model suggested for rRNA degradation upon starvation (30), albeit operating at the faster time scale of minutes rather than hours. Interestingly, a recent report showed that E. coli responds to oxidative stress (0.5 mM H2O2) by rapidly degrading its tRNA and strongly decelerating the translation elongation rate (16). Oxidative stress results in oxidation of cellular RNA and subsequent stalling of ribosomes at 8-oxoG residues in the mRNA (62). Therefore, we suggest that application of oxidative stress may resemble the addition of rifampicin reported here, in the sense that both treatments are expected to block translation due to deprivation of functional mRNA substrate. The regulatory pathway that results in tRNA degradation upon oxidative stress has not been identified, but like the case reported here, it was found that tRNA degradation occurred so rapidly that it is unlikely to be carried out by an effector synthetized de novo in response to the stress condition (16). On this basis, we argue that tRNA degradation upon oxidative stress may be included as another example of the mechanism described in this work, whereby tRNA is degraded as a result of blockage of translation, in agreement with the passive model for degradation described above.
Figure 6.
Model for passive tRNA degradation upon translational arrest. (A) During undisturbed growth, tRNA is occupied by binding to ribosomes, aminoacyl-tRNA-synthetases and elongation factor Tu. (B) Any disturbance that reduces the activity of ribosomes (e.g. amino acid starvation) reduces the demand for tRNA in translation, thus producing free tRNAs, which are subject to degradation by an as yet unidentified pathway (indicated by packman-shaped factor). The only constraint in this model is that the degradation effector must have a slightly lower affinity for tRNA than the translational apparatus.
Starvation for a single amino acid results in dramatically reduced charging levels of tRNAs accepting that particular amino acid, whereas models suggest that the charging levels of tRNAs accepting the remaining 19 amino acids should increase to ∼100%, because aminoacylated tRNAs would accumulate in the cells when the rate of tRNA deacylation on the ribosomes is reduced as a consequence of decreased translation activity (15). In our experiments, we clearly detected a rapid decrease in charging levels of the tRNA accepting the amino acid subject to starvation (see e.g. tRNAargVYZQ in Supplementary Figure S3), but we only detected a slight increase in tRNA charging levels of the remaining amino acids upon starvation (see Figure 5 and Supplementary Figures S2 and S3). We propose that this apparent discrepancy is neatly resolved by consideration of the passive model for tRNA degradation presented in Figure 6. According to this model, only those tRNAs engaged in translation are protected and excess tRNA, whether acylated or not, is degraded. Thus, when translation activity is decreased upon starvation, the tRNA pool is decreased to match the residual translation activity of the starving cell. The tRNA charging levels then remain unaltered because the remaining tRNAs are engaged in the decharging-recharging cycle of translation at nearly the same rate as during unrestricted growth. In further support of this model, we note that addition of the lacking amino acid to the starved culture caused a significant drop in the charging level of tRNAs (see Supplementary Figure S3), suggesting the tRNA concentrations during the early nutritional shift-up may be limiting or close to limiting for protein production.
At first glance, degradation of a fully functional and abundant macromolecular class such as the tRNAs in response to amino acid starvation may seem contradictory to the needs of the starving cell, as energy would then need to be spent on replenishing the tRNA pool for continued protein synthesis once the amino acid supply had been restored. We speculate that this energetic cost is outweighted by a beneficial effect of tRNA degradation on the accuracy of translation of the proteins that are synthesized during starvation conditions. In this context, it is important to recall that the stringent response to amino acid starvation involves upregulation of amino acid biosynthesis and stress response genes (reviewed in (22)). As described in the introduction, proteins that are synthesized during amino acid starvation are prone to errors because of competition from near-cognate tRNAs at the codons that code for the growth-limiting amino acid. Most of our amino acid starvation experiments were carried out with E. coli strains that couldn't upregulate biosynthesis of the growth-limiting amino acid, and therefore couldn't resolve the starvation. However, in the example where we starved for histidine using a leaky histidine auxothroph strain (Figure 5) it became more evident that the fold difference in charging level between the starved and unstarved tRNAs was reduced as the starvation progressed (Figure 5B). Thus, we hypothesize that the error rate during amino acid starvation would be greater if E. coli lacked the ability to degrade surplus tRNA. In addition, the presence of excess tRNA can adversely affect co-translational protein folding (63), and therefore protein function, and has been shown to increase aggregation of some E. coli proteins (64). In conclusion, we propose that degradation of tRNA upon amino acid starvation serves to rapidly adjust the pool of cellular tRNA to match the reduced demand, which would be expected to improve the quality of the residual protein synthesis during starvation.
ACCESSION NUMBER
RNA sequencing data used for results shown in Supplementary Table S1 are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5221.
Acknowledgments
The authors thank Marit Warrer for excellent technical assistance and Shiraz Shah for help with the bioinformatic analysis of data in Supplementary Table S1. We are grateful to K. Gerdes and S. Pedersen for critical reading of the manuscript. S.L.S. and M.A.S. wrote the manuscript; S.L.S., M.A.S. and M.K. designed experiments; M.A.S., M.K., T.S. and A.G. performed experiments; M.A.S. conceived and designed research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Council for Independent Research|Natural Sciences [1323-00343B to S.L.S.]; Lundbeck Foundation [R108-A10583 to M.A.S.]; Danish National Research Foundation [DNRF120 to S.L.S. and M.A.S.]. Funding for open access charge: The Danish National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ryals J., Little R., Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 1982;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez V.J., Bremer H. Characterization of RNA and DNA—synthesis in Escherichia coli strains devoid of Ppgpp. J. Biol. Chem. 1993;268:10851–10862. [PubMed] [Google Scholar]
- 3.Maaløe O., Kjeldgaard N.O. In: Control of Macromolecular Synthesis:A Study of DNA, RNA, and Protein Synthesis in Bacteria. Benjamin WA, editor. NY; 1966. p. 284. [Google Scholar]
- 4.Li Z., Reimers S., Pandit S., Deutscher M.P. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis B.D., Luger S.M., Tai P.C. Role of ribosome degradation in the death of starved Escherichia coli cells. J. Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohanty B.K., Maples V.F., Kushner S.R. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res. 2012;40:4589–4603. doi: 10.1093/nar/gks006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong H., Nilsson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 8.Wohlgemuth S.E., Gorochowski T.E., Roubos J.A. Translational sensitivity of the Escherichia coli genome to fluctuating tRNA availability. Nucleic Acids Res. 2013;41:8021–8033. doi: 10.1093/nar/gkt602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz J.E. Controlling translation via modulation of tRNA levels. Wiley interdiscipl. Rev. RNA. 2015;6:453–470. doi: 10.1002/wrna.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 11.Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sorensen K.D., et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C.T., Dyavaiah M., DeMott M.S., Taghizadeh K., Dedon P.C., Begley T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmar K.A., Sorensen M.A., Elf J., Ehrenberg M., Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elf J., Nilsson D., Tenson T., Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science (New York, N.Y.) 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 16.Zhong J., Xiao C., Gu W., Du G., Sun X., He Q.Y., Zhang G. Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet. 2015;11:e1005302. doi: 10.1371/journal.pgen.1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker J., Johnston T.C., Borgia P.T. Mistranslation in cells infected with the bacteriophage MS2: direct evidence of Lys for Asn substitution. Mol. Gen. Genet. 1980;180:275–281. doi: 10.1007/BF00425839. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen M.A. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(-) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 2001;307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 19.Hall B., Gallant J. Defective translation in Rc- cells. Nat.-N. Biol. 1972;237:131. doi: 10.1038/newbio237131a0. [DOI] [PubMed] [Google Scholar]
- 20.O'Farrell P.H. The suppression of defective translation by ppGpp and its role in the stringent response. Cell. 1978;14:545–557. doi: 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 21.Potrykus K., Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 22.Magnusson L.U., Farewell A., Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen M.A., Jensen K.F., Pedersen S. High-concentrations of Ppgpp decrease the RNA chain growth-rate—implications for protein-synthesis and translational fidelity during amino-acid starvation in Escherichia-coli. J. Mol. Biol. 1994;236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 24.Tian C., Roghanian M., Jorgensen M.G., Sneppen K., Sorensen M.A., Gerdes K., Mitarai N. Rapid curtailing of the stringent response by toxin-antitoxin encoded mRNases. J. Bacteriol. 2016 doi: 10.1128/JB.00062-16. doi:10.1128/JB.00062-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson A., Gillespie D. Metabolic events occurring during recovery from prolonged glucose starvation in Escherichia coli. J. Bacteriol. 1968;95:1030–1039. doi: 10.1128/jb.95.3.1030-1039.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julien J., Rosset R., Monier R. [Metabolism of ribonucleic acids in Escherichia coli deficient in phosphate] Bull. Soc. Chim. Biol. 1967;49:131–145. [PubMed] [Google Scholar]
- 27.McCarthy B.J. The effects of magnesium starvation on the ribosome content of Escherichia coli. Biochim. Biophys. Acta (BBA) 1962;55:880–889. [Google Scholar]
- 28.Maruyama H., Mizuno D. Ribosome degradation and the degradation products in starved Escherichia coli. I. Comparison of the degradation rate and of the nucleotide pool between Escherichia coli B and Q-13 strains in phosphate deficiency. Biochim. Biophys. Acta. 1970;199:159–165. [PubMed] [Google Scholar]
- 29.Norris T.E., Koch A.L. Effect of growth rate on the relative rates of synthesis of messenger, ribosomal and transfer RNA in Escherichia coli. J. Mol. Biol. 1972;64:633–649. doi: 10.1016/0022-2836(72)90088-5. [DOI] [PubMed] [Google Scholar]
- 30.Zundel M.A., Basturea G.N., Deutscher M.P. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan R., Apirion D. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J. Biol. Chem. 1975;250:1854–1863. [PubMed] [Google Scholar]
- 32.Mandelstam J., Halvorson H. Turnover of protein and nucleic acid in soluble and ribosome fractions of non-growing Escherichia coli. Biochim. Biophys. Acta. 1960;40:43–49. doi: 10.1016/0006-3002(60)91313-5. [DOI] [PubMed] [Google Scholar]
- 33.Sulthana S., Basturea G.N., Deutscher M.P. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA. 2016;22:1163–1171. doi: 10.1261/rna.056275.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavitt R.I., Umbarger H.E. Isoleucine and valine metabolism in Escherichia coli. X. The enzymatic formation of acetohydroxybutyrate. J. Biol. Chem. 1961;236:2486–2491. [PubMed] [Google Scholar]
- 36.Brock T.D., Brock K.M., Belly R.T., Weiss R.L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 37.Varshney U., Lee C.-P., RajBhandary U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 38.Denhardt's solution (100X) Cold Spring Harbor Protoc. 2008 doi:10.1101/pdb.rec11538. [Google Scholar]
- 39.Leavitt R.I., Umbarger H.E. Isoleucine and valine metabolisn in Escherichia coli. XI. K-12: valine inhibition of the growth of Escherichia coli strain. J. Bacteriol. 1962;83:624–630. doi: 10.1128/jb.83.3.624-630.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 41.Bonner D. Further studies of mutant strains of Neurospora requiring isoleucine and valine. J. Biol. Chem. 1946;166:545–554. [PubMed] [Google Scholar]
- 42.Czech A., Fedyunin I., Zhang G., Ignatova Z. Silent mutations in sight: co-variations in tRNA abundance as a key to unravel consequences of silent mutations. Mol. BioSyst. 2010;6:1767–1772. doi: 10.1039/c004796c. [DOI] [PubMed] [Google Scholar]
- 43.Puri P., Wetzel C., Saffert P., Gaston K.W., Russell S.P., Cordero Varela J.A., van der Vlies P., Zhang G., Limbach P.A., Ignatova Z., et al. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol. 2014;93:944–956. doi: 10.1111/mmi.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haseltine W.A., Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agirrezabala X., Fernandez I.S., Kelley A.C., Carton D.G., Ramakrishnan V., Valle M. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagosky P.A., Chang F.N. Correlation between RNA synthesis and basal level guanosine 5′-diphosphate 3′-diphosphate in relaxed mutants of Escherichia coli. J. Biol. Chem. 1981;256:11651–11656. [PubMed] [Google Scholar]
- 47.Traxler M.F., Summers S.M., Nguyen H.T., Zacharia V.M., Hightower G.A., Smith J.T., Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waller J.P. The NH2-terminal residues of the protein from cell-free extracts of E. coli. J. Mol. Biol. 1963;7:483–496. doi: 10.1016/s0022-2836(63)80096-0. [DOI] [PubMed] [Google Scholar]
- 49.Laffler T., Gallant J. spoT, a new genetic locus involved in the stringent response in E. coli. Cell. 1:27–30. [Google Scholar]
- 50.Fiil N.P., Willumsen B.M., Friesen J.D., von Meyenburg K. Interaction of alleles of the relA, relC and spoT genes in Escherichia coli: analysis of the interconversion of GTP, ppGpp and pppGpp. Mol Gen Genet. 1977;150:87–101. doi: 10.1007/BF02425329. [DOI] [PubMed] [Google Scholar]
- 51.Germain E., Roghanian M., Gerdes K., Maisonneuve E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Maisonneuve E., Castro-Camargo M., Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 53.Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin–antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 54.Maisonneuve E., Shakespeare L.J., Jorgensen M.G., Gerdes K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Hartmann G., Honikel K.O., Knusel F., Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 56.Reid P., Speyer J. Rifampicin inhibition of ribonucleic acid and protein synthesis in normal and ethylenediaminetetraacetic acid-treated Escherichia coli. J. Bacteriol. 1970;104:376–389. doi: 10.1128/jb.104.1.376-389.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurland C.G., Maaloe O. Regulation of ribosomal and transfer RNA synthesis. J. Mol. Biol. 1962;4:193–210. doi: 10.1016/s0022-2836(62)80051-5. [DOI] [PubMed] [Google Scholar]
- 58.Dennis P.P. Effects of chloramphenicol on the transcriptional activities of ribosomal RNA and ribosomal protein genes in Escherichia coli. J. Mol. Biol. 1976;108:535–546. doi: 10.1016/s0022-2836(76)80135-0. [DOI] [PubMed] [Google Scholar]
- 59.Winkler M., Ramos-Montañez S. Biosynthesis of Histidine. EcoSal Plus. 2009 doi: 10.1128/ecosalplus.3.6.1.9. doi:10.1128/ecosalplus.3.6.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramer E.B., Farabaugh P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaechter M., Maaloe O., Kjeldgaard N.O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 62.Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang G., Hubalewska M., Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 64.Fedyunin I., Lehnhardt L., Böhmer N., Kaufmann P., Zhang G., Ignatova Z. tRNA concentration fine tunes protein solubility. FEBS Lett. 2012;586:3336–3340. doi: 10.1016/j.febslet.2012.07.012. [DOI] [PubMed] [Google Scholar]