Abstract
Borrelia burgdorferi, the bacterial pathogen responsible for Lyme disease, modulates its gene expression profile in response to the environments encountered throughout its tick-mammal infectious cycle. To begin to characterize the B. burgdorferi transcriptome during murine infection, we previously employed an in vivo expression technology-based approach (BbIVET). This identified 233 putative promoters, many of which mapped to un-annotated regions of the complex, segmented genome. Herein, we globally identify the 5′ end transcriptome of B. burgdorferi grown in culture as a means to validate non-ORF associated promoters discovered through BbIVET. We demonstrate that 119 BbIVET promoters are associated with transcription start sites (TSSs) and validate novel RNA transcripts using Northern blots and luciferase promoter fusions. Strikingly, 49% of BbIVET promoters were not found to associate with TSSs. This finding suggests that these sequences may be primarily active in the mammalian host. Furthermore, characterization of the 6042 B. burgdorferi TSSs reveals a variety of RNAs including numerous antisense and intragenic transcripts, leaderless RNAs, long untranslated regions and a unique nucleotide frequency for initiating intragenic transcription. Collectively, this is the first comprehensive map of TSSs in B. burgdorferi and characterization of previously un-annotated RNA transcripts expressed by the spirochete during murine infection.
INTRODUCTION
Borrelia burgdorferi or Borreliella burgdorferi (1) is the spirochetal agent of Lyme disease, the foremost vector-borne bacterial disease in the world (2). B. burgdorferi is naturally maintained in an enzootic cycle between an arthropod vector, Ixodes sp. ticks and diverse small vertebrate reservoir hosts, such as the white-footed mouse Peromyscus leucopus (3). Humans may become infected with B. burgdorferi through the bite of an infected tick, resulting in Lyme disease (4). The survival and pathogenesis of B. burgdorferi requires that the bacterium appropriately modifies its transcription profile in response to the distinct tick vector and mammalian host environments encountered throughout its infectious cycle (5–7). The paradigm example of this is the down-regulation of ospA, encoding outer surface protein A and up-regulation of ospC, encoding outer surface protein C, during B. burgdorferi transmission from the feeding tick to the mammalian host (8–12). Elucidation of the transcriptional activity of a pathogen during infection is critical for understanding the molecular mechanisms used by the pathogen to cause disease. Yet, despite the well characterized transcriptomes of other pathogens (13–17), B. burgdorferi's paucibacillary nature (18) has led to technical challenges toward global elucidation of the spirochete's transcription profile during mammalian infection.
Global transcriptome approaches performed using B. burgdorferi cultivated in modified growth conditions, used to simulate tick or mammalian environments, have allowed interrogation of the spirochete's gene expression profile in response to diverse stimuli. Temperature and pH shift of bacterial cultures in complex medium (19,20), co-incubation of spirochetes with host cells (21) and growth of B. burgdorferi in blood (22), or in dialysis membrane chambers in the peritoneal cavities of rats (7,23,24), have resulted in the discovery of a number of B. burgdorferi genes that are expressed under mammalian infection-like conditions and contribute to pathogenesis (5). However, these modeled conditions are unable to fully recapitulate the complex tissue environments that B. burgdorferi encounters during an active infection. Recently, an amplification-microarray approach was used to define the transcription profile of annotated open reading frames and pseudogenes in fed larval ticks, fed nymphal ticks and mammalian host adapted B. burgdorferi cultivated in dialysis membrane chambers (7). This analysis provided stage-specific mRNA expression levels and insights into the transcriptional network that the spirochete uses to sense and respond to the environmental cues it receives as it transitions between the tick vector and mammalian host (7). Microarray analysis of B. burgdorferi gene expression in a non-human primate model of neuroborreliosis revealed genes that are specifically induced in the central nervous system during infection (25). Targeted analyses of specific gene subsets have identified a number of B. burgdorferi genes that are upregulated during mouse infection (5,18,26). In addition, global mutagenesis approaches have identified a number of genes that contribute to murine infection (27,28). Nonetheless much remains to be discovered about the B. burgdorferi transcriptome and how it contributes to the spirochete's pathogenesis.
To overcome the challenges of identifying B. burgdorferi genes expressed during murine infection, our lab developed an in vivo expression technology (IVET)-based approach for B. burgdorferi (BbIVET) (29). IVET is a powerful and versatile promoter identification method and has been used with pathogenic bacteria and fungi in a wide variety of host environments, and led to the discovery of a number of previously uncharacterized virulence genes (30–34). This approach does not rely on the recovery of bacterial RNA from mouse tissues for transcriptional analysis and thus is not hindered by low bacterial numbers during infection. BbIVET was the first genetic screen of this type to identify B. burgdorferi putative promoter sequences that are transcriptionally active during a murine infection. Moreover, the experimental design of the BbIVET screen was such that the recovered infection-active promoters could be active both in culture and during infection or specifically active during infection alone. We previously reported the BbIVET-identified sequences that mapped upstream of annotated open reading frames (ORFs) (29). Unexpectedly, many sequences discovered through BbIVET were internal, antisense or intergenic to ORFs. This suggested the presence of novel transcripts in these genomic regions; however, due to the lack of B. burgdorferi RNA-seq studies that report transcripts other than annotated mRNAs, the significance of these findings were unknown.
Since their development, 5′ end RNA-seq protocols have been used to globally define transcription start sites (TSSs) and processed RNA 5′ ends across bacterial genomes. The initial use of this technique revealed the complexity of the Helicobacter pylori transcriptome through the discovery of global antisense transcription, numerous small regulatory RNAs (sRNAs) and previously unrecognized intragenic TSSs (35). Furthermore, 5′ end RNA-seq applications in other organisms have identified high instances of leaderless transcripts (36), novel regulatory RNA networks (37–39), strain-specific sRNAs (40) and a high prevalence of un-annotated small proteins (41), as well as provided valuable resources for future genetic applications in model organisms (42).
As a means to gain insight into the significance of the un-annotated sequences identified through BbIVET, as well as work to elucidate the BbIVET promoters that are principally active during infection alone, we have applied 5′ end RNA deep sequencing (5′RNA-seq) to B. burgdorferi grown in culture and overlapped this TSS map with the BbIVET-identified sequences. We provide substantial genome-wide data on the transcriptional start sites for B. burgdorferi and validate the expression of a subset of these both in culture and during an active murine infection. In total, this work contributes significant understanding to the complex transcriptome of B. burgdorferi and identifies novel B. burgdorferi sequences expressed during mammalian infection.
MATERIALS AND METHODS
Bacterial clones and growth conditions
Borrelia burgdorferi clones used in this study were derived from strain B31. Wild-type clone B31 A3 (43), which lacks cp9, was used for the cDNA library preparations. All B. burgdorferi genetic manipulations used infectious low-passage clone A3-68ΔBBE02, which lacks cp9, lp56 and gene bbe02 on lp25 (44). All shuttle vectors were created in DH5α Escherichia coli, grown in LB broth or on LB agar plates containing 300 μg/ml spectinomycin when appropriate, and transformed into B. burgdorferi as previously described (45). Spirochetes were cultivated in liquid Barbour–Stoenner–Kelly (BSK) II medium supplemented with gelatin and 6% rabbit serum (46) and plated in solid BSK medium as previously described (47). B. burgdorferi cultures were grown at 35°C supplemented with 2.5% CO2 in the presence of 50 μg/ml streptomycin and/or 200 μg/ml kanamycin when applicable.
RNA Isolation
In general, 45 ml of log phase (3–7 × 107 cells/ml) B. burgdorferi B31 A3 in BSK II medium, as determined using a Petroff–Hausser counting chamber under dark field microscopy, was pelleted at 3210 ×g for 15 min, supernatant decanted and snap frozen in liquid N2. Total RNA was extracted using a hot phenol protocol described previously (48). 50 μg total RNA was incubated with 10 U DNase I (Roche) and 80 U rRNasin (Promega) at 37°C for 15 min. The RNA samples were purified as described (48). RNA concentrations were measured and DNA removal and RNA integrity was verified by gel electrophoresis. For RNA preps used in deep-sequencing, RNA integrity number was determined using the Agilent 2100 Bioanalyzer. Samples with RNA integrity numbers over 9.0 were selected for RNA-seq library preparations.
cDNA library preparations
A simplified schematic diagram of the 5′RNA-seq protocol is provided in Supplementary Figure S1. Two biological replicates of B. burgdorferi B31 A3 were grown to a density of 3 × 107 cells/ml in BSK II medium, the presence of all circular and linear plasmids confirmed by PCR with a panel of primers (43), and RNA isolated and DNase treated, as above. Ribo-zeroTM rRNA Removal Kit for Gram-negative bacteria (Epicentre) was used to remove ribosomal RNA, per manufacturer's instructions. Elimination of rRNA was confirmed using an Agilent 2100 Bioanalyzer. A total of 1.5 μg samples of RNA from each of the two B. burgdorferi cultures were incubated at 37°C for 1 h with or without 45 U tobacco acid pyrophosphatase (TAP) (Epicentre), 1X TAP buffer and 40 U rRNasin (Promega). Samples were then loaded into Heavy Phase Lock Gel tubes (5 PRIME) with an equal volume of phenol stabilized:cholorform:isoamyl alcohol (25:24:1) (PCI), ethanol precipitated and reconstituted in 5 μl DEPC–H2O. A total of 30 μM of 5′ RNA adapter (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′), based on the Illumina adapter sequence, (Oligonucleotide sequences © 2007–2009 Illumina, Inc., all rights reserved) was ligated to the 5′ ends of the TAP+ and TAP− treated RNA samples using 20 U RNA Ligase 1 (NEB), 1X RNA Ligase 1 Buffer, 10% DMSO, 1 mM ATP (NEB) and 40 U rRNasin (Promega) in 20 μl DEPC- H2O and incubated at 20°C for 6 h. Reactions were then loaded into Heavy Phase Lock Gel tubes (5 PRIME) with equal volume PCI, ethanol precipitated and reconstituted in DEPC-H2O. Fragmentation of the 5′ ligated RNA samples was conducted using RNA fragmentation reagents (Ambion) following manufacturer's instructions with a fragmentation time of 4 min at 70°C. Reactions were ethanol precipitated, reconstituted in DEPC–H2O and analyzed with an Agilent 2100 Bioanalyzer. RNA libraries were then 3′ end dephosphorylated using 10 U T4 Polynucleotide Kinase (PNK) (NEB), minus ATP, with 1X PNK buffer and 20 U rRNasin (Promega) at 37°C for 3 h. A subsequent PCI and ethanol precipitation was performed to purify the libraries, which were then size selected on an 8% polyacrylamide 8 M urea gel. RNA gel segments from ∼150–300 nts were isolated and extracted overnight in RNA gel elution buffer (10 mM Tris pH 7.5, 0.1% SDS, 2 mM EDTA, 0.3 M NaOAC) at 4°C with agitation. Gel slurries were loaded onto 0.2 μm aqua Nanosep® MF columns (PALL), centrifuged at 14 000 ×g for 2 min and the eluates were ethanol precipitated. The 3′ RNA adapter, based on the Illumina multiplexing sequence and blocked on the 3′ end with an inverted dT (5′-(p)AGAUCGGAAGAGCACACGUCU(idT)-3′), was 5′ phosphorylated using T4 PNK (NEB) per manufacturer's instructions, and purified using illustra MicroSpin G-25 columns (GE). A total of 52 μM of phosphorylated 3′ RNA adapter was next ligated to the libraries using 25 U RNA Ligase 1 (NEB), 1X RNA Ligase 1 Buffer, 10% DMSO, 1 mM ATP (NEB) and 40 U rRNasin (Promega) in 25 μl DEPC–H2O and incubated at 20°C for 6 h. To remove unincorporated adapter, size selection (∼200–300 nts) on an 8% polyacrylamide 8 M urea gel was performed and RNA extracted from gel, as described above. Adapter conjugated RNA library preps were converted to cDNA with SuperScriptII reverse transcriptase (Invitrogen) and random nonamers, per manufacturer's instructions. A total of 2 U of RNaseH (Promega) was used to remove the remaining RNA from the libraries, and a subsequent 18 cycle PCR with Phusion High-fidelity DNA polymerase (NEB) was used to add appropriate barcodes using ScriptSeq Index Primers (Epicentre) (Supplementary Table S1). Products were analyzed on an Agilent 2100 Bioanalyzer and by KAPA qPCR (KAPABiosystems), and prepared for sequencing with Illumina technology on a HiSeq 1500. Pooled libraries (2 TAP+ and 2 TAP−) were clustered with TruSeq Rapid SR Cluster Kit (Illumina) and sequenced on a rapid run with single-end 50 base-pair reads at the analytical genomics core facility at Sanford Burnham Prebys Medical Discovery Institute (Orlando, FL, USA).
Deep-sequencing analyses
A schematic of the 5′RNA-seq analysis is presented in Supplementary Figure S2. Reads were mapped to the Borrelia (Borreliella) burgdorferi B31 genome (GenBank Ids: AE000783, AE001583, AE000793, AE001582, AE000785, AE000794, AE000786, AE000784, AE000789, AE000788, AE000787, AE000790, AE001584, AE000791, AE000792, AE001575, AE001576, AE001577, AE001578, AE001579, AE001580 and AE001581) using NextGenMap v0.4.12 (49) with default configuration but ensuring at least 90% sequence identity between the read and the B. burgdorferi B31 reference genome.
The B. burgdorferi genome harbors a number of regions comprised of highly repetitive sequence. This is particularly true for sequences residing on the cp32 replicons, lp56, lp21 and lp5 (50). The presence of extensive similarity between sequences found on multiple B. burgdorferi plasmids suggested that we may encounter challenges in our ability to definitively map a portion of the 50 bp sequence reads from 5′RNA-seq to their locations in the genome. A dot plot comparing the sequences of all B. burgdorferi B31 plasmids to each other confirmed regions of high sequence similarity, especially along the cp32s and lp56, as expected (Supplementary Figure S3A). The same analysis, however, revealed sub-sequences with reduced similarity on these plasmids, which provided evidence that a considerable fraction of the B. burgdorferi genome, including sequences on the cp32s and lp56, is sufficiently unique to be mapped with good confidence (Supplementary Figure S3B). For the main analysis we removed aligned reads with mapping quality values below 20, therefore, removing reads that cannot be aligned to a unique location in the genome with high probability. We then extracted one-dimensional, genome-wide signals from the mappable alignments by counting reads that have their 5′ nucleotide mapped to a particular genomic position. These signals were normalized by library size. The 5′ ends were called in coverage segments of the TAP+ and TAP− signals using an in house method. This procedure excluded peaks below a minimum threshold of 30, which corresponds roughly to a minimum of 30 reads covering a genomic position, and required a 5′ end to be sequenced in both biological replicates. For each sequenced 5′ end, read counts were averaged between biological replicates and the ratio of reads between the TAP+ and TAP− libraries was then calculated. We observed a strong correlation between the 5′ end read counts within the TAP+ and TAP− replicates (Supplementary Figure S4). A 5′ end was determined to be a TSS if the nucleotide reads were at least 2-fold higher in the averaged TAP+ compared to the averaged TAP− libraries. This is based on the notion that TAP hydrolyses phosphodiester bonds on RNA 5′ ends and that RNA ligase has specificity to only ligate the 5′ Illumina adapter to RNA sequences harboring a 5′ monophosphate, thereby enriching TSSs to TAP+ libraries. Ideally, 5′RNA-seq would unambiguously distinguish TSSs from processing events, thereby TAP− libraries would solely sequence processing events. However, practicality, 5′RNA-seq takes a snapshot of an actively growing bacterial population. Given that in routine RNA transcript turnover the 5′ triphosphate is removed, combined with unintentional phosphate loss during manipulation of RNA in library preparations, it is not surprising that some TSSs are captured in the TAP− library. Hence a 2-fold threshold was established, as has also been applied to other studies utilizing 5′RNA-seq (37,40). The 5′ nucleotides in the TAP+ libraries not identified as TSSs were defined as 5′ processed ends. The resulting TSSs were then associated with BbIVET and gene annotations present in the Schutzer annotation set (51) (including tRNA annotations from the UCSC database).
For the analysis that included the repetitive part of the genome, NextGenMap was configured to output the top 100 possible alignments that share the maximum alignment score for each read. These alignments were not filtered for mapping quality and therefore also contained a considerable number of multi-mapped reads with mapping quality zero (MQ0) that map to multiple positions in the reference sequence with equal probability. For such ambiguously mapped reads, the BAM file therefore contained multiple alignment entries. We then extracted a 5′ end from these alignments as above but this time each read contributed to this signal only with a weight 1/X0 to the signal where X0 is the number of optimal alignments for this read in the data set. In other words, a uniquely mapped read would count as before; whereas, a read that mapped to three different (repetitive) regions would contribute only 1/3 to the each of them, thereby equally dividing its contribution among all possible (optimal) mapping locations in the genome.
TSS classifications
A TSS was termed primary if it was located within 300 nucleotides upstream the annotated start codon of an ORF on the same DNA strand, and had the highest average TAP+ read count of all such TSSs associated with the same ORF. Secondary TSSs were those that fulfilled the above criteria with respect to location but were not the TSS with the highest average TAP+ read count. The delegation of antisense was assigned if the TSS was located within or just outside, extending 100 nucleotides (nts) upstream or downstream, of an ORF on the complementary DNA strand. An internal classification was assigned if the TSS was located on the same DNA strand at any position between the second nucleotide of the start codon and the last nucleotide of the stop codon of an ORF. A TSS that was not classified into any of the above categories was identified as an orphan. Each TSS was assigned to all of the categories for which it qualified (Figure 1). There were five cases for which a TSS was assigned the designation of primary for one annotated transcript and secondary for another, and three cases for which a TSS was secondary, internal and antisense for three different annotated transcripts (Supplementary Table S2). In each of these situations, the unusual pattern of TSS classification resulted from the close proximity of the TSS to a small ORF or tRNA transcript.
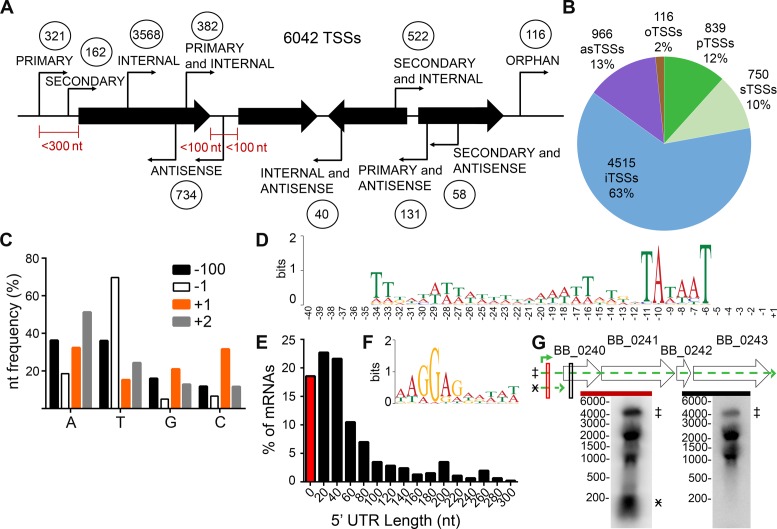
Figure 1.
Genome-wide identification and characterization of B. burgdorferi transcription start sites (TSSs). RNA was isolated from log phase B. burgdorferi clone B31 A3, and treated with and without TAP (tobacco acid phosphatase). (A) Schematic classification of the 6042 TSSs relative to the genome organization. Circled numbers indicate TSSs for each category. The maximum nucleotide distances from the 5′ and/or 3′ of annotated sequences, shown as black arrows, for a particular category of TSS are indicated in red. (B) Graphical representation of the TSS classifications. as, antisense; o, orphan; p, primary; s, secondary; i, internal. (C) Nucleotide frequency at the −100, −1, +1 (TSS) and −1 nucleotide positions. A, adenine, T, thymine, G, guanine, C, cytosine. (D) Consensus motif for promoter regions upstream of primary TSSs. The nucleotide sequences from −40 to +1 of the 321 identified primary TSSs were analyzed by MEME 4.11.2. (E) Distribution of 5′ untranslated regions (UTR) lengths among uniquely classified primary and secondary TSSs. The data are shown as the percent of mRNA sequences with a 5′ UTR of each bin length. 0–10 nucleotides (red bar), bin size of 20 nucleotides, where the number shown is the middle length in each bin (black bars). (F) Consensus ribosome binding motif for uniquely classified, primary 5′ UTRs. MEME 4.11.2 was used to analyze UTR sequences ranging from 10–293 nts in length for a conserved motif, which was found on average to begin 8 nts upstream of the annotated start codon. (G) Northern blot analyses validate a long 5′ UTR. Total RNA was extracted from mid-log phase spirochetes and separated by denaturing formaldehyde–agarose gel, blotted to nylon membranes and probed with 32P-labeled complementary probes, indicated by red and black boxes. Genomic context of ORFs BB_0240-BB_0243 (wide black arrows), the primary TSS (green bent arrow), putative transcripts and their position on the Northern blot (broken line, green arrows and marked with designated symbol) are indicated. Marker sizes in nucleotides are indicated to the left of each blot.
Bbive-TSS association
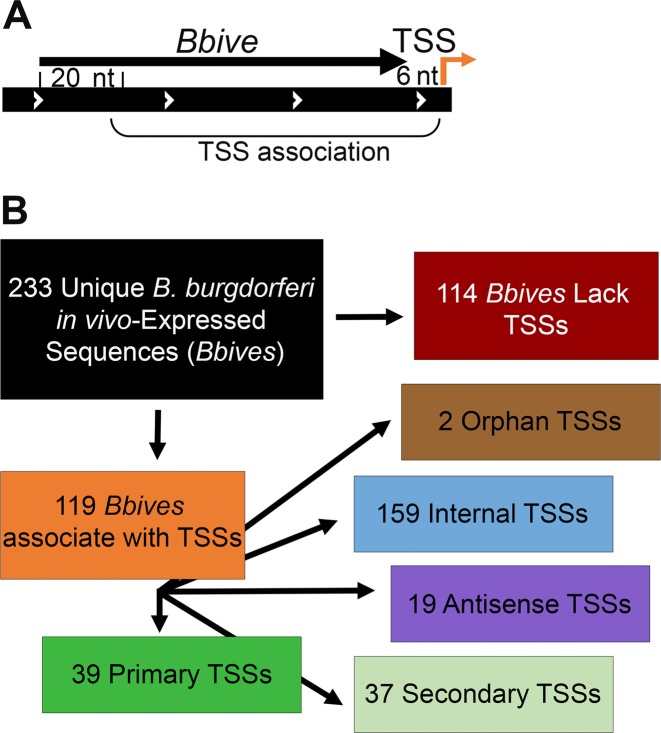
A conserved Pribnow box sequence was identified, on average, at the −6 nucleotide position relative to unique primary TSSs (Figure 1) and we reasoned that a minimum of 20 nts of a Bbive sequence may be sufficient to contain a functional promoter, due to the lack of strong conservation in the −35 region. According to this rationale, we established the criteria that if a TSS was located at a position 20 nts or greater downstream of the 5′ end of a Bbive sequence up to 6 nts beyond the 3′ end of the Bbive sequence then it was considered a BbIVET associated TSS (Bbive-TSS) (Figure 2).
Figure 2.
B. burgdorferi in vivo expression technology (BbIVET)-identified sequences with associated TSSs. (A) Schematic representation of BbIVET associations with 5′RNA-seq TSSs. Brackets designate the parameters for the association. Relative orientation of the genome region (wide black bar with white arrows), Bbive sequence (thin black arrow) and TSS (orange bent arrow) are indicated. The minimum and maximum Bbive-TSS association distances were defined as 20 nts from the 5′ end and 6 nts from the 3′ end of a Bbive sequence. (B) Categorization of Bbives that associate with 5′RNA-seq TSSs.
Northern blotting
Ten micrograms of DNase treated (Roche) total RNA was separated using a formaldehyde/MOPS agarose gel, modified from www.lonza.com/research and (52). Briefly, RNA was denatured in 1X MOPS (20 mM MOPS, 5 mM NaOAC, 1 mM EDTA, pH 7.0), 3.7% formaldehyde and 1X RNA loading dye (ThermoScientific) at 70°C for 10 min, and incubated on ice for 3 min prior to loading. Following separation on a 1% agarose, 1X MOPS, 2% formaldehyde gel at 150V at 4°C, RNA was transferred to Hybond XL membranes (Amersham Pharmacia Biotech) via capillary action overnight (52). Membranes were UV crosslinked with a UVC 500 crosslinker (Amersham Pharmacia Biotech) at 150 mJ, probed with 50 nM DNA oligonucleotide probes (Supplementary Table S1) and end-labeled with ATP (γ-32P) by T4 polynucleotide kinase (NEB), at 42°C overnight with ULTRAhyb oligo hybridization buffer (Ambion). Blots were washed twice with 2X SSC (3 M NaCl, 300 mM Na3C6H5O7), 0.5% SDS, exposed to a phosphor screen and imaged with a Typhoon Trio+ (GE).
Luciferase promoter fusion constructs
B. burgdorferi shuttle vectors pJSB161 and pJSB175 carrying a promoterless or flaBp-driven B. burgdorferi codon optimized Photinus pyralis firefly luciferease gene (luc), respectively, were generously provided by Dr Jon Blevins (53). Generation of the Bbive:luc fusion constructs was carried out as follows. Bbive putative promoter regions, (i) the −1 nucleotide, as determined from 5′RNAseq, to the 5′ end of the BbIVET sequence or (ii) the entire Bbive sequence, were PCR amplified using Phusion High-fidelity DNA polymerase (NEB) from B31 A3 genomic DNA with primer pairs that introduced a BlgII site at the 3′ and 5′ ends (Supplementary Table S1). For Bbive–TSSs, which were located less than 100 nts downstream the 5′ end of the BbIVET sequence or for the analysis of processed 5′ ends, ∼100 bp upstream the TSS/processed 5′ end were BlgII cloned into pJSB161. The PCR generated DNA fragments were digested with BlgII and ligated into BlgII digested pJSB161. The ospA p:luc and ospC p:luc fusion constructs were generated in the same manner, using primer pairs 1693, 1694 and 1845, 1846, respectively (Supplementary Table S1). All plasmid constructs were confirmed by PCR and sequence analysis.
Luciferase assays
B. burgdorferi clones carrying the luciferase fusion constructs were grown to logarithmic phase (4–7 × 107 spirochetes/ml) in BSK II medium and pelleted at 3210 ×g for 10 min. Cells were washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and resuspended to approximately 2–3 × 109 spirochetes/ml. One hundred microliters of each sample was used to measure the OD600 using a BioTek Synergy 4. A total of 92.5 μl of each sample was loaded into a black, solid bottom 96-well plate (Corning) and combined with 7.5 μl 10 mM D-luciferin (Regis) in PBS. The relative luciferase activity was determined by measuring photon emission in each well for 1 s, 10 times using the EnVision 2104 Multilabel Reader (PerkinElmer). Relative luciferase activity for each sample was averaged, subtracted from the PBS control and normalized to the OD600 reading for that clone, providing the relative luciferase units (RLU) for each promoter fusion. Experiments were conducted in biological triplicate and analyzed by two-tailed Student's t-test relative to pJSB161 using Prism Graphpad.
Ethics statement
The University of Central Florida is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the National Institutes of Health and were reviewed and approved by the University of Central Florida Institutional Animal Care and Use Committee.
Luciferase assays during murine infection
Prior to infection and imaging, anaesthetized 6- to 8-week-old female C3H/HeN mice (Envigo) were depilated ventrally by shaving and treatment with odorless hair removal creme (SallyHansen). One week prior to inoculation, and throughout the duration of the study, mice were treated with 5 mg/ml streptomycin and 1 mg/ml Equal® sweetener in their water to maintain selection for the luciferase plasmid in the B. burgdorferi clones. Groups of three mice were infected with 1 × 105 spirochetes intraperitoneally. To detect luciferase activity, mice were intraperitoneally injected with 150 mg/kg body weight sterile D-luciferin in PBS, 15 min prior to imaging. The IVISTM 50 Imaging System (Xenogen Imaging Technologies) was utilized to capture luminescence signals with a f/1 camera lens aperture, binning at 4 and 5 min exposure time. Mice were imaged at various time points between 1 and 24 days post inoculation. To determine the inoculum doses, spirochetes were counted using a Petroff–Hausser counting chamber and verified by the number of colony-forming units plated in solid BSK medium. All inocula were PCR verified to contain all expected B. burgdorferi plasmids as previously described (54). Immediately following the final in vivo imaging system (IVIS) reading time point, mice were sacrificed and ear and bladder tissues were cultured for spirochete reisolation as described (55), and all mice were confirmed positive for infection. Any background luciferase signals detected in the mice inoculated with B. burgdorferi carrying the promoterless luciferase gene were subtracted out of the readings.
RESULTS
5′ End Identification across the B. burgdorferi transcriptome
IVET-based approaches are powerful methods to identify transcriptionally active sequences, annotated as in vivo expressed (ive) sequences, in an environment of interest (34). Previously our lab developed IVET for BbIVET to identify putative promoters that are active during a murine infection (29). This approach exploited the use of a promoterless nicotinamidase, pncA, a gene required for B. burgdorferi survival in the mammalian host, and a randomized, fragmented B. burgdorferi genomic library (Supplementary Figure S1). The promoter trap system, after screening through a mouse model of B. burgdorferi infection, revealed 233 transcriptionally active sequences (Supplementary Table S2), herein referred to as B. burgdorferi in vivo expressed sequences (Bbives). These putative promoters were sufficient to drive expression of pncA thereby restoring infectivity to a B. burgdorferi mutant lacking the essential gene. The design of the BbIVET approach did not uniquely distinguish promoters that are specifically active in the mammalian host, and a subset of these are likely active both in culture and during infection. Further, this approach did not include assessment of B. burgdorferi transcripts that are produced during the tick phase of the enzootic cycle. To discover candidate genes that are expressed during mammalian infection, these sequences were mapped to annotated ORFs in the B. burgdorferi B31 genome under the parameter that a Bbive must be found within 300 nts upstream of an annotated start codon. A total of 91 Bbives fell into this category; whereas unexpectedly, the remaining 61% of the sequences discovered through BbIVET mapped to genomic locations internal, antisense or intergenic to ORFs (Supplementary Table S2). Furthermore, given the highly repetitive nature of regions of the B. burgdorferi genome, 34 Bbives exactly mapped to multiple genomic locations. To indicate these instances, decimal delegations were assigned to non-specific Bbives to indicate every possible genomic location that the sequence may have derived from (e.g. Bbive149.1 maps to lp25 and Bbive149.2 to lp36 but is the same nucleotide sequence; Supplementary Table S2).
To validate the non-ORF associated promoters, discovered through BbIVET and to gain a deeper overall understanding of the B. burgdorferi transcriptome, we identified genome-wide TSSs. Utilization of a 5′ end RNA-seq protocol (5′RNA-seq) defined the TSSs and 5′ processed ends of two biological replicates of B. burgdorferi clone B31 A3 grown to log phase in culture. Native RNA transcripts contain a triphosphate at the 5′ end, whereas those that are endogenously processed or degraded have a monophosphate or hydroxyl group at the 5′ end. These characteristics were exploited in our 5′RNA-seq protocol to discriminate between 5′ ends. Samples were either treated with or without TAP during the library preparation. RNA-seq libraries with TAP treatment captured all RNA 5′ ends; whereas, the TAP− libraries captured endogenously processed or degraded 5′ ends (Supplementary Figure S1).
Illumina sequencing resulted in 265 million, 50 bp single-end reads from across all four libraries, ranging from 60 to 70 million reads per library. This theoretically corresponds to a total genomic coverage of about 1900–2200X. Due to the highly repetitive nature of some regions of the B. burgdorferi genome, reads that were not uniquely mapped to a single genomic location were removed. This resulted in 23–42 million reads (749–1388X coverage) for the main analysis. The 5′ ends were then called across libraries and categorized based on the TAP+ enrichment of TSSs, which discovered 6042 potential TSSs and 30 716 processing and/or degradation events (Supplementary Tables S3 and S4). An online interactive interface is available to search the 5′RNA-seq data at www.ucf.edu/research/interactive-genomics/lyme-disease.
Examination of tRNA loci provided validation of our TSS and processed-end calling method. 5′ end processing is critical for tRNA maturation (56) and as expected both TSSs and 5′ processed ends were detected for tRNA transcripts (Supplementary Table S3 and S4). Furthermore, TSSs identified by 5′RNA-seq were compared to B. burgdorferi TSSs that were determined by 5′ RACE or primer extension. The 5′RNA-seq identified TSSs were found to be within three nucleotides of 70% of the previously described TSSs in B. burgdorferi strain B31 (Table 1). Furthermore, the 5′RNA-seq library preparation protocol used herein was also performed in E. coli. These data were compared to E. coli TSSs identified by Thomason et al., (2015), which used an independent enzyme, terminator 5′-phosphate-dependent exonuclease for TSS characterization by 5′ end RNA-seq (42). A majority of the TSSs called in our E. coli data set were consistent with the TSSs identified by Thomason et al. (manuscript in preparation, I.B., M.L. and R.S.). Together these data provide support for the robustness of our 5′RNA-seq method.
Table 1. Comparison of 5′ RACE/primer extension 5′ end determination and 5′RNA-seq identified TSSs in B. burgdorferi B31.
| Gene | ORF annotationa | 5′ RACE/primer extension 5′ end position | Reference | 5′ RNA-seq TSS position | Avg. TAP+ read countb | Avg. TAP- read countc | TAP+:TAP- Ratio | Nucleotide position difference (nts)d |
|---|---|---|---|---|---|---|---|---|
| ospAe | BB_A15 | 9421 | (94) | 9421 | 19 421 | 1407 | 13.8 | 0 |
| chbC | BB_B04 | 3849 | (95) | 3849 | 1693 | 28.5 | 59.4 | 0 |
| bb0360 operon | BB_0360-0364 | 368 456 | (71) | 368 456 | 3941.5 | 123.5 | 31.9 | 0 |
| antigen IpLA7 | BB_0365 | 374 265 | (96) | 374 190 | 1097.5 | 172.5 | 6.4 | 75 |
| cheYI operon | BB_0551 | 561 955 | (97) | 561 958 | 1433 | 55.5 | 25.8 | 3 |
| bbb22 | BB_B22 | 19 338 | (78) | 19 336 | 3020.5 | 160.5 | 18.8 | 2 |
| bbb23 | BB_B23 | 21 082 | (78) | 21 080 | 1609 | 59.5 | 27.0 | 2 |
| bpur | BB_0047 | 46 423 | (98) | 46 423 | 191 | 60 | 3.2 | 0 |
| bdrF2operon | BB_G29 | 24 469 | (99) | 24 347 | 222 | 11 | 20.2 | 122 |
| BB_G31 | 26 453 | Nonef | N/Ag | N/A | N/A | N/A | ||
| BB_G33 | 28 014 | 28 017 | 293.5 | 25.5 | 11.5 | 3 |
bAverage normalized read counts from two biological replicated 5′RNA-seq TAP+ libraries, for a given TSS.
cAverage normalized read counts from two biological replicated 5′RNA-seq TAP- libraries, for a given TSS.
dDifferences in TSS positions from 5′ RACE/primer extension compared to 5′RNA-seq.
eThe two previously published TSSs for the ospC gene were not included, as these data were acquired using a B. burgdorferi strain other than B31 (100–102). Nonetheless, the 5′RNA-seq defined TSS for ospC was found to be located one and four nucleotides away from the two previously defined sites of transcription initiation.
fNo TSS was identified within 300 nucleotides of BB_G31 ORF.
gNot applicable.
5′ End identification in repetitive genomic regions
Although all of the data discussed herein are derived from the uniquely mappable portions of the B31 genome, we acknowledge that there may be a biological interest in B. burgdorferi genomic regions that contain repetitive sequences. In addition, it is possible that the exclusion of such sequences from our analysis may have affected the identification and categorization of the TSSs and processed 5′ ends, both of which, as will be detailed in the following section, were highly dependent on the local genomic and transcriptional context. Therefore, we added for reference, an additional analysis of the 5′RNA-seq library that included the 5′ nucleotide reads that mapped to repetitive regions and therefore had a mapping quality score of zero (MQ0). We analyzed these data to determine TSSs as described above and provide the results in Supplementary Table S5.
TSS classification
TSSs were categorized relative to their orientation to annotated sequences through a systematic classification assignment scheme. Because annotated sequences can lie adjacent, divergent or convergent and often in close proximity to one another, our classification scheme did not uniquely sort TSSs into single categories, similar to previously applied approaches that used automated TSS classification algorithms (40,42). Therefore, a TSS could be assigned multiple classifications (Figure 1A). Accounting for overlaps among categories (i.e. counting each TSS for all categories to which it was assigned), 12% of the TSSs were classified as primary, 10% as secondary, 13% as antisense, 63% as internal and 2% as orphan (Figure 1B).
Unique initiating nucleotide usage for B. burgdorferi transcription
Purine nucleotides are typically the most common initiating nucleotide in bacteria (57,58). Surprisingly, in B. burgdorferi cytosine had a similar frequency as adenine at the +1 position for all TSSs identified in this study. The individual nucleotide frequencies were measured at 32.2% A, 15.2% T, 20.9% G and 31.6% for TSSs despite the A/T-rich (71.8%) nature of the genome (Figure 1C and Supplementary Figure S5). As a comparison, the nucleotide frequency at the −100 position was found to be 72.2% A + T (Figure 1C). This is consistent with the average nucleotide frequency of 70.5% for all positions 100 nucleotides upstream and downstream of the TSSs (Supplementary Figure S5A and B). TSSs were found most commonly flanked by a thymine at the −1 position (69.7%) and an adenine at the +2 position (51.2%) (Figure 1C). Internal TSSs constitute the majority of the TSSs identified and analysis among different TSS categories separately demonstrated that only the internal TSSs have the higher G + C nucleotide frequency at the +1 nucleotide. The primary, secondary, antisense and orphan +1 nucleotide frequencies were similar to the genome-wide average (Supplementary Figure S5C–G).
Identification of σ70 motifs in TSS promoter regions
As both a means of validation and Pribnow box discovery, sequences containing the +1 nucleotide and corresponding 40 nucleotides upstream of all of the 321 unique primary TSSs (Figure 1A) were analyzed for conserved sequence motifs using MEME 4.11.2 (59). Due to the abundance of A/T nucleotides in the B. burgdorferi genome, a training set of 100 000 random 41 nucleotide subsequences from the B31 reference genome was selected for a background control. Not surprisingly, given that the 5′RNAseq library was generated from logarithmic phase bacteria, which were expected to be expressing predominately sigma 70-dependent transcripts, we detected a 29 bp motif containing a canonical Pribnow box (beginning on average at the −6 position), in its consensus sequence, upstream of 320 of the 321 primary TSSs (Figure 1D). However, these sequences lacked strong conservation at the −35 region, which is consistent with observations made in other bacteria (35,42,57). A similar conserved Pribnow sequence was identified upstream of 88 of the 162 secondary TSSs (54.3%) and 656 of the 734 antisense TSSs (89.4%) (data not shown); these sequences also lacked a conserved −35 region. In contrast, no conserved sequence motifs were detected upstream of the 3568 unique internal TSSs. However, further analysis of internal TSSs with high TAP+ read counts (greater than 500) revealed a similar conserved Pribnow box in 109 out of 266 (41.0%) of these sequences (data not shown).
Leaderless and long UTRs
Depending on their size and secondary structure, 5′ untranslated regions (UTRs) may contain important RNA regulatory elements (60–62). Mechanisms of cis-regulatory RNAs have yet to be explored in B. burgdorferi. To identify ORFs that might be controlled by cis-regulatory RNAs we characterized the lengths of the 5′ UTRs associated with uniquely classified primary and secondary TSSs that were only assigned to a single annotated transcript (on occasion, due to small ORFs, a primary or secondary TSS was assigned to multiple ORFs and these were excluded from the analysis) (Supplementary Table S6). Our data indicated that the median 5′ UTR length in B. burgdorferi is ∼36 nts (Figure 1E), which is consistent with what has been determined for other bacterial species (35,36,57,63,64). Approximately 19% of the 5′ UTRs analyzed were less than 10 nts in length and a 29% subset of these short sequences represented leaderless transcripts with a 5′ UTR length of zero (Figure 1E). Approximately 17% of the 5′ UTRs were found to be between 100 and 293 nts in length, suggesting that these sequences may harbor additional regulatory functions (Figure 1E). The 5′ UTR sequences greater than 10 nts in length, were also analyzed for a ribosome binding site motif using MEME as previously described (59). A conserved A/G rich, 11 nt sequence was found in 97.6% of the UTRs analyzed. Furthermore, this motif was primarily located near the ORF, with a median distance of 8 nts upstream of the annotated starts of translation (Figure 1F). Northern blot analyses validated the presence of the 195 nt 5′ UTR upstream the four-gene glp operon. Northern blot probes specific to the BB_0240 UTR (red probe) and coding (black probe) regions, both revealed a ∼4500 nt transcript (Figure 1G). These data demonstrated the presence of a large transcript that included the 195 nt 5′ UTR, consistent with the predicted combined size of the ORFs contained in glp operon (4300 nts) and several putative internal transcripts. The UTR probe also detected a small RNA transcript (∼195 nts), consistent with the size of the 5′ UTR.
Correlating TSSs with BbIVET putative promoters
The global identification of B. burgdorferi TSSs not only contributed understanding to the B. burgdorferi transcriptome but resulted in a 5′ end map of the annotated and un-annotated transcripts expressed by the spirochete during growth in culture, which could provide further insight into the sequences recovered from BbIVET. Of the 233 unique Bbive sequences, 119 were found to be associated with culture-expressed TSSs (Figure 2A and Supplementary Table S2), suggesting that these promoters are active both in culture and during infection. Bbive-TSSs were classified into the following categories: 39 primary, 37 secondary, 159 internal, 19 antisense and 2 orphan (Figure 2B). In some cases multiple TSSs were associated with the same Bbive and some TSSs were classified into multiple categories. Together these data validated the significance of the Bbive sequences located in previously un-annotated genomic positions. Moreover, the comparative analysis revealed that 114 Bbives were not associated with any detected TSS from the in culture derived 5′RNA-seq data set, suggesting that these sequences may be primarily active during murine infection.
Validation of BbIVET-associated transcripts
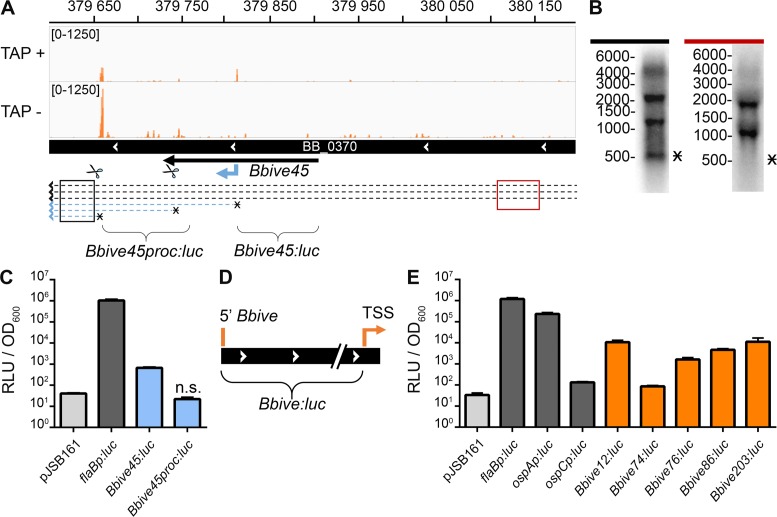
Fifty-one percent of the Bbives were associated to TSSs discovered by 5′RNA-seq, indicating that these transcripts are expressed both in culture and in the mammalian host. Although some of these sequences may provide housekeeping functions, the finding that a large percentage of the Bbives fall into this category does not exclude the possibility that these promoters, and the transcripts that they control, contribute to B. burgdorferi pathogenesis. Indeed 5′RNA-seq identified TSSs for a number of genes known to be critical for B. burgdorferi infectivity including ospC, bosR and vlsE (Supplementary Table S3) (5). Validation of a subset of Bbive-TSSs was carried out using Northern blot analyses and promoter fusion assays. Bbive45 is an intragenic sequence that maps within the chromosomal gene BB_0370 and is associated with an internal TSS and proximal to two processed 5′ ends (Figure 3A and Supplementary Table S2). Northern blot analysis using an oligonucleotide probe complementary to the sequence downstream of both the internal TSS and the processed 5′ ends revealed an ∼500 nt transcript, which could be attributed to one or more of these 5′ ends (Figure 3B, black probe). In contrast, this transcript was absent when a probe targeted against sequence upstream of the three 5′ ends was used (Figure 3B, red probe). Of note, bacterial transcriptomes are complex as a result of compact gene organization as well as overlapping and antisense orientated transcript expression (65). As expected due to this complexity, additional transcripts were also detected in these Northern blots, which may represent the ∼1.2 kb BB_0370 ORF itself and/or additional read-through transcripts initiated further upstream.
Figure 3.
Validation of BbIVET-associated RNA transcripts. (A) Deep-sequencing screen shot for a Bbive-internal TSS, displaying only the sequenced 5′ nucleotide, of overlaid biological replicates treated with (TAP+) and without (TAP−) tobacco acid pyrophosphatase. Read count ranges are shown in the upper left of each frame. The chromosome nucleotide coordinates, relative orientation of the BB_0370 ORF (wide black bar), Bbive45 sequence (thin black arrow), internal TSS (blue bent arrow), processed 5′ ends (scissors), putative transcripts (broken line arrows), Northern probe locations (black and red boxes) and luciferase fusion regions (brackets) are indicated. The predicted transcripts of interest are marked with an asterisk. (B) Northern blot analyses of the Bbive45 internal TSS, as described in the Figure 1 legend. Probes located upstream (red) and downstream (black) of the putative internal TSS, are indicated. Marker nucleotide sizes are indicated to the left of the blots. (C) Bbive45 promoter activity and specificity. B. burgdorferi clones harboring specific promoter fusions were grown to mid-log phase, and incubated with 750 μM D-luciferin. Relative luciferase units (RLU) were normalized to cell density by OD600. Data represent the mean ±SD from three biological replicates shown in log scale and were analyzed relative to pJSB161 with the two-tailed Student's t-test. Unless otherwise indicated all fusion constructs demonstrated significantly greater RLUs than the promoterless control, pJSB161 (P ≤ 0.01). n.s., not significantly greater RLU relative to pJSB161. (D) Schematic representation of Bbive luciferase fusions. The bracket designates the sequence selected for promoter fusion to luciferase in pJSB161. Relative orientation of the genome region (wide black bar with white arrows), 5′ boundary of Bbive sequence (orange line) and TSS (orange bent arrow) are indicated. (E) A variety of Bbives with associated TSSs have promoter activity in culture. Spirochetes containing control promoters (flaBp, ospCp and ospAp) and specific Bbives, fused to luciferase, were grown to mid-log phase, and analyzed as described above.
The DNA sequence located directly upstream of a TSS is expected to function as a promoter and initiate transcription, while the sequence directly upstream of a processed end should not. To validate the ability of 5′RNA-seq to distinguish TSSs and processed 5′ ends, as well as confirm the ability of Bbive45 to function as a promoter, the promoter activities of sequences upstream of the internal TSS or the processed 5′ ends in BB_0370 were measured using luciferase transcriptional fusions. Upstream regions of the internal TSS or the dominant processed 5′ end were cloned in front of the promoterless, B. burgdorferi codon optimized firefly luciferase gene, in the B. burgdorferi shuttle vector pJSB161 (53). This generated promoter fusions Bbive45:luc and Bbive45proc:luc, respectively (Figure 3A). The Bbive45:luc fusion, which carries the putative promoter for the internal TSS of interest, demonstrated a significant increase in luciferase activity relative to the promoterless reporter gene (Figure 3C). These data indicated that this sequence functions as a promoter albeit to a lesser extent than the B. burgdorferi promoter for the constitutive flagellar gene flaB. In contrast, the Bbive45proc:luc construct did not exhibit any detectable promoter activity (Figure 3C). Together these data support the ability of 5′RNA-seq to identify and distinguish TSSs and processed 5′ ends and confirm a BbIVET identified sequence functions as a promoter.
The luciferase transcriptional fusion validation approach was expanded to a larger subset of Bbive-TSSs. DNA including the Bbive sequence to the TSS (Figure 3D), for 5 Bbives were fused to luciferase in pJSB161. Bbive12, 74, 76, 86 and 203 were found to be associated with TSSs, which were classified as primary, internal or antisense (Table 2). The activities of the Bbive promoters were compared to that of the known constitutive flaB promoter and two environmentally regulated control promoters, ospAp and ospCp. The genes encoding for the critical virulence outer surface proteins, OspA and OspC, demonstrate inverse patterns of expression during the infectious cycle of B. burgdorferi (8–12). The ospA gene displays low expression in the mammalian host and high expression in culture and the unfed tick. In contrast, ospC exhibits high expression during mammalian infection and low expression in culture (29) and the unfed tick. All five Bbive-TSS promoter fusions resulted in significant levels of luciferase activity compared to the promoterless control (Figure 3E). Furthermore, the promoter activities of all of the sequences except for Bbive74 were found to be significantly higher than that of the mammalian induced ospCp. Although 5′RNA-seq measures steady state levels of RNA transcripts and promoter fusions quantify amounts of RNA polymerase activity, there was a correlative trend of promoter strength to 5′RNA-seq read counts. The greater the average TAP+ read count determined by 5′RNA-seq, the greater the average relative luciferase units measured by promoter fusion (Table 2). Overall these data provided experimental support for Bbive-TSS defined promoters across a diversity of annotated and un-annotated transcripts.
Table 2. BbIVET-associated TSS read counts and luciferase promoter fusion relative activity.
| Promotera | Replicon | TSS Positionb | Strand | Categoryc | Category Details (Annotation)d | Avg. TAP+ Read Counte | AVG RLUf |
|---|---|---|---|---|---|---|---|
| flaBp | chr | 148715 | - | primary | P:BB_0147 (flagellin) | 9.39E+05 | 1.19E+06 |
| ospAp | lp54 | 9421 | + | primary | P:BB_A15 (ospA, outer surface protein A) | 1.94E+04 | 2.31E+05 |
| ospCp | cp26 | 16883 | + | primary | P:BB_B19 (outer surface protein C) | 37.5 | 134.4 |
| Bbive12a | chr | 174219 | - | internal, primary | I:BB_0172 (von Willebrand factor type A domain-containing protein), P:BB_0171 (hypothetical protein) | 1.42E+04 | 1.07E+04 |
| Bbive45 | chr | 379813 | - | internal | I:BB_0370 (tyrS, tyrosine–tRNA ligase) | 243 | 663.1 |
| Bbive74 | chr | 647833 | + | antisense | A:BB_0620 (beta-glucosidase) | 194 | 85.9 |
| Bbive76 | chr | 659711 | + | antisense | A:BB_0629 (PTS system fructose-specific transporter subunit IIABC) | 362 | 1.62E+03 |
| Bbive86 | chr | 746049 | + | internal, primary | I:BB_0709 (hypothetical protein), P:BB_0710 (dnaG, pseudo) | 1.58E+03 | 4.66E+03 |
| Bbive203 | chr | 146579 | - | internal, primary | I:BB_0146 (glycine/betaine ABC transporter ATP-binding protein), P:BB_0145 (glycine/betaine ABC transporter permease) | 6.71E+03 | 1.12E+04 |
aControl promoters and Borrelia burgdorferi in vivo expressed sequence identification numbers.
bReplicon specific nucleotide position of 5′RNA-seq identified TSS.
cTSS category assignment.
dAnnotation of the TSS associated ORF (50,90), where single letter abbreviations indicate the annotation for the TSS category.
eAverage normalized read counts from 5′RNA-seq TAP+ libraries, for a given TSS.
fPromoter activity for each TSS, as determined by luciferase activity in RLU, for spirochetes grown to log phase in liquid culture.
Activity of novel B. burgdorferi promoters during murine infection
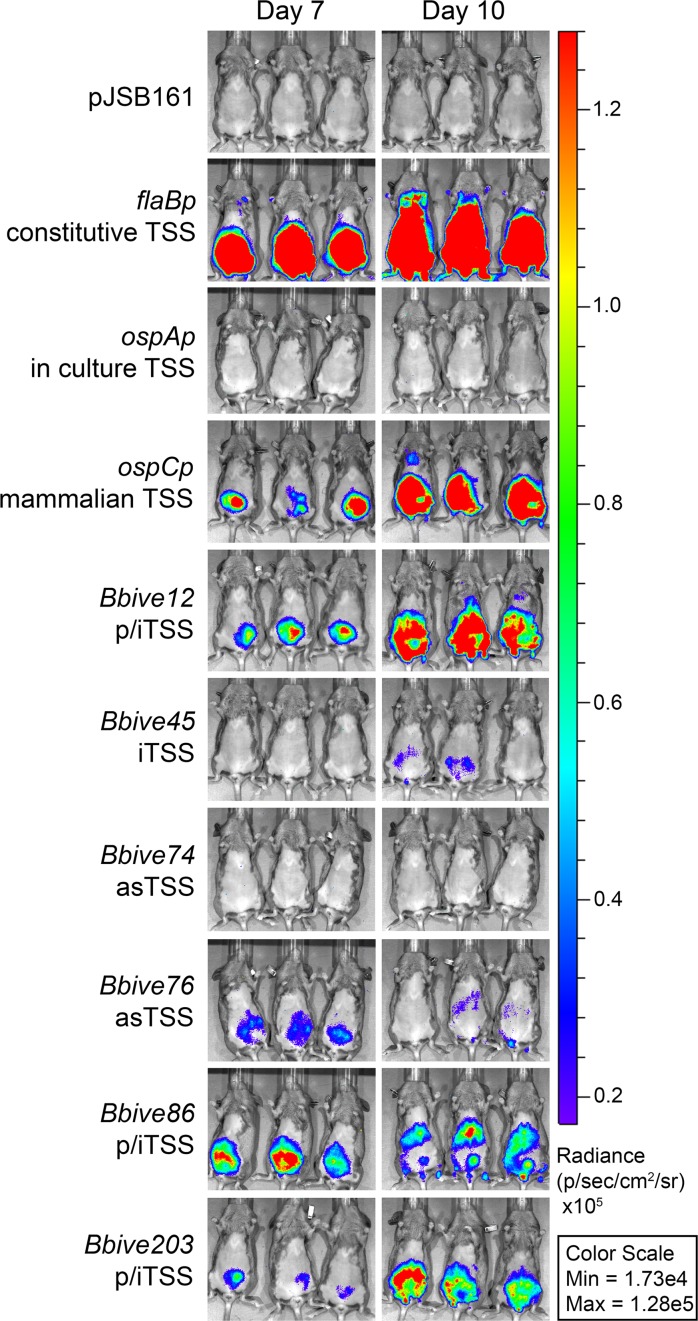
Luciferase-promoter fusion constructs can be used to assay promoter activity in real time during an active infection. Therefore, the activities of the six Bbive-TSS defined promoters were examined during murine infection using IVIS. Spirochetes carrying the individual Bbive:luc fusions (Table 2) or the promoterless, flaBp, ospAp or ospCp control constructs were inoculated intraperitoneally into groups of three mice at a dose of 1 × 105 bacteria. Bacterial bioluminescence was assessed in the live infected mice, following delivery of D-luciferin, by photon emission using IVIS over a 10 day time course (Figure 4). The 10-day time point was experimentally determined to have the highest sensitivity of detection using B. burgdorferi carrying the luciferase gene driven by the constitutive promoter, flaBp (Supplementary Figure S6). B. burgdorferi infection of all mice was confirmed by reisolation of spirochetes from tissues. Mice infected with spirochetes harboring the promoterless luciferase construct (pJSB161) served as the control for background levels of bioluminescence. The ospAp and ospCp luciferase fusion activities in the mouse were as expected based on their known patterns of expression, off and on, respectively (Figure 4) (8–12). All B. burgdorferi clones carrying the Bbive luciferase fusions, with the exception of Bbive74, demonstrated luciferase activity at day 7 and/or day 10 post-infection (Figure 4), but not at earlier time points (data not shown). Variations in the levels of bioluminescence detected for each B. burgdorferi clone at the two time points likely reflected sequence-and/or time-dependent regulation of the Bbive-TSS promoters during infection. The Bbive74:luc fusion demonstrated low but significant promoter activity in spirochetes grown in culture (Figure 3E) but no detectable bioluminescence in the mouse (Figure 4). Cumulatively, these data demonstrate culture and mouse infection activity of a panel of B. burgdorferi promoters defined by BbIVET-identified sequences and the associated TSSs for a diversity of transcript categories. Furthermore, these data show that global TSS identification by 5′RNA-seq provides significant insight into the B. burgdorferi transcriptome that validates un-annotated sequences discovered through BbIVET.
Figure 4.
Culture-expressed Bbive-TSSs demonstrate activity in the mammalian host. C3H/HeN mice infected intraperitoneally with 1 × 105B. burgdorferi containing promoterless pJSB161, control promoters (flaBp, ospCp or ospAp) or specific Bbive luciferase fusions as indicated (left). Mice were injected intraperitoneally with 150 mg/kg D-luciferin 7 and 10 days post infection and imaged with the IVISTM 50 Imaging System (IVIS) using a 5 min exposure. Images were normalized to the same p/s range of 1.73e4 to 1.128e5 and displayed on the same color spectrum scale (right). Data are representative of two independent biological replicates. asTSS, antisense TSS; p/iTSS, primary or internal TSS; iTSS, internal TSS.
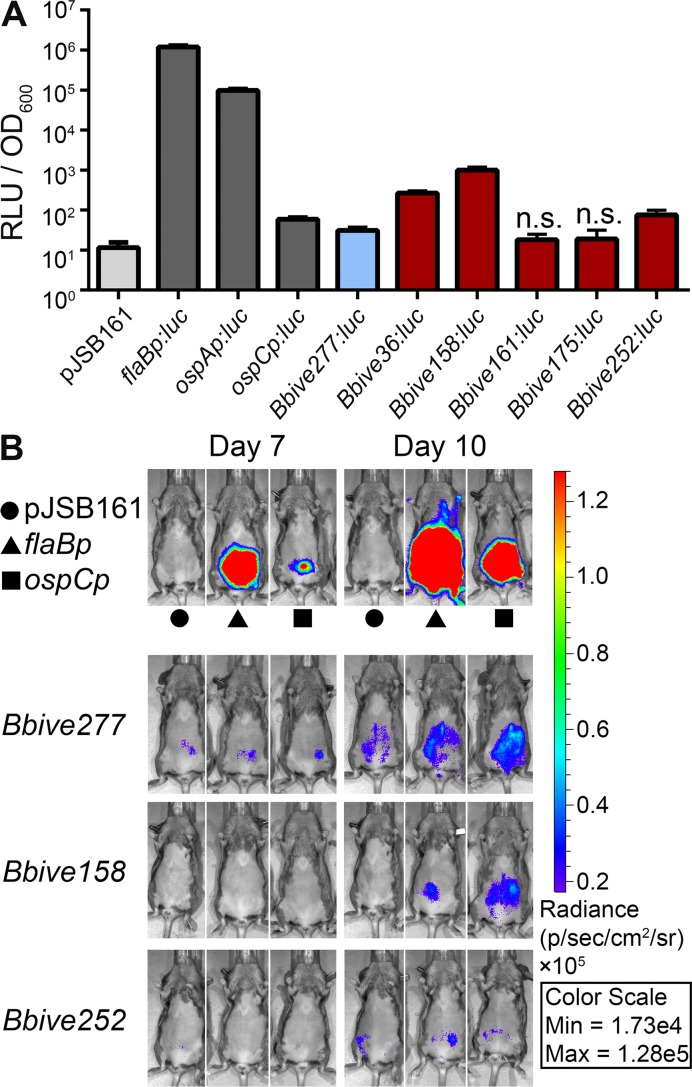
Unlike other pathogen transcriptome studies (66–68) that benefit from direct collection of bacterial cells from infected animals for RNA-seq, low spirochete numbers present during a B. burgdorferi infection requires innovative approaches to identify B. burgdorferi transcripts that are expressed in the mammalian environment. Comparative analysis of the BbIVET and 5′RNA-seq data sets revealed 114 Bbives with no associated culture-specific TSSs (Supplementary Table S2). Furthermore, considering the MQ0-analysis (Supplementary Table S5) only 5 of the 114 Bbives contain un-mappable TSSs. These sequences harbored sufficient promoter activity to be captured in the BbIVET screen, suggesting they may control transcripts that are specifically expressed during murine infection. Therefore, we generated luciferase promoter fusions of Bbive36, 158, 161, 175 and 252 that had no associated TSS by 5′RNA-seq. In addition, we tested Bbive277, which had a low-coverage TSS. For these experiments the entire Bbive sequence that was originally recovered from BbIVET (Supplementary Table S2) was fused to luciferase and evaluated for luminescence both in culture and during murine infection. Similar to the ospC promoter, Bbive277, 36, 158 and 252 exhibited promoter activity in culture above that of the promoterless control but significantly less than that of the flaB and ospA promoters (Figure 5A). These data demonstrated that these promoters display activity in culture despite the lack of associated TSSs. However, no significant promoter activity was detected for Bbive161 or Bbive175 during growth in culture (Figure 5A). To assess the promoter activities of these Bbive sequences in the mammalian host, groups of mice were infected with 1 × 105 spirochetes carrying each luciferase fusion construct and evaluated for luminescence using IVIS. Three of the six Bbive sequences, Bbive277, 158 and 252, displayed detectable luciferase activity at days 7 and 10 post infection (Figure 5B and data not shown), suggesting that these promoters are active during murine infection, albeit at a levels lower than that of the flaB and ospC promoters at these time points of infection. None of the tested Bbive sequences were found to be specifically active in the mammalian host, as Bbive 277, 158 and 252 demonstrated promoter activity in culture (Figure 5A). However, these Bbive sequences appear to represent promoters for novel un-annotated RNA transcripts. Bbive158 is located internal to the annotated pseudo gene BB_K02a on lp36. Bbive252 lies antisense to the hypothetical protein coding gene BB_F14 on lp28-1. Bbive277 maps internal to chromosomal gene BB_0319, which encodes a riboflavin binding protein that may be a component of a riboflavin ABC transporter (69). Future studies are focused on elucidation of the functions of these transcripts in B. burgdorferi biology. For the sequences that demonstrated no detectable promoter activity in this assay, further analysis is required to distinguish between the possibility that these Bbive sequences lack promoter activity completely or that the amount of transcription driven by these sequences falls below the threshold of sensitivity by IVIS. In sum, our data provide the first evidence of B. burgdorferi expression of antisense and intragenic transcripts in the mammalian host environment.
Figure 5.
Novel B. burgdorferi RNA transcripts expressed during murine infection. (A) A subset of Bbives that lack or have low read count associated TSSs demonstrate low promoter activity in culture. Bbive sequences were fused to luciferase in pJSB161 and measured for luciferase activity, as described in Figure 3 legend. Data represent the mean ± SD from three biological replicates shown in log scale and were analyzed relative to pJSB161 using the two-tailed Student's t-test. Unless otherwise indicated all fusion constructs demonstrated significantly greater RLUs than the promoterless control, pJSB161, (P ≤ 0.01). n.s., not significantly greater RLU relative to pJSB161. (B) Bioluminescence of Bbive sequences in the murine host. C3H/HeN mice infected intraperitoneally with 1 × 105B. burgdorferi containing controls and specific Bbive luciferase fusions, analyzed by IVIS as described in Figure 4 legend. Symbols indicate specific controls for each time point. Data are representative of two biological replicates. Note that the same color spectrum scale is used between figures.
DISCUSSION
A global understanding of the transcriptome of a pathogen in environments of interest can provide key insight into patterns of gene expression and possible modes of gene regulation, which are critical for elucidating molecular mechanisms of pathogenesis. B. burgdorferi requires coordinated mechanisms of gene regulation to survive throughout its enzootic life cycle (6); however, the molecular events that contribute to this process, particularly during mammalian infection have remained largely unknown. Clearly the mechanisms of B. burgdorferi adaptation for survival in the tick vector are critical for environmental persistence as well as pathogenesis of the spirochete. Nonetheless, the work presented herein focused on elucidation of B. burgdorferi transcripts expressed in the mammalian host using a mouse model of infection. We overcame the challenges of the paucity of spirochetes present in infected mouse tissues to direct transcriptome analysis through BbIVET (29). BbIVET identified 233 unique putative promoter sequences, a majority of which mapped to un-annotated antisense and intragenic positions in the genome. The BbIVET library likely did not provide complete coverage of the B. burgdorferi genome. However, there were several instances where we recovered the same Bbive from multiple mice and multiple, unique Bbives were recovered from the same mouse (29). Therefore, we are confident that BbIVET allowed discovery of unique and novel promoter regions across the chromosome and plasmid replicons, which could potentially contribute to Lyme disease pathogenesis. To gain further insight into the significance of the Bbive sequences we applied a genome-wide 5′ end RNA-seq approach using B. burgdorferi grown in culture. Transcriptome complexity and experimental technicalities pose additional challenges to the direct measurement of strand-specific RNA transcripts by conventional methods such as RT-qPCR. Therefore, we utilized luciferase transcriptional fusions, which provided a unique approach to evaluate promoter activity of novel B. burgdorferi transcripts during an active infection. Comparative analysis of BbIVET and 5′RNA-seq has validated 5′ ends, characterized a variety of categories of RNA transcripts and discovered novel transcripts that contribute to the B. burgdorferi transcriptome during mammalian infection.
5′ end RNA-seq has been shown to be a valuable transcript discovery tool for a variety of organisms, and the differential application of TAP, or similar enzymes, to globally define TSSs and processed 5′ ends has been comprehensively validated through many independent studies (35,37,42,57). Although 5′ ends may be inferred from total RNA-seq data (70), unlike 5′RNA-seq, these studies are unable to distinguish between TSSs and processed 5′ ends and therefore only provide estimations of transcript boundaries (35). Our application of 5′RNA-seq provides a snap shot of 6042 unique, potential TSSs across the B. burgdorferi genome during log phase growth in culture. Our 5′ end identification method was designed to exclude excessive sequencing background noise, but still report the 5′ ends for a variety of transcripts in 5′RNA-seq that have low expression in culture, such as the mammalian-induced ospC gene. We acknowledge that bias in the library preparation at either the 5′ linker ligation or the amplification steps, or unintentional 5′ phosphate loss during manipulation of the RNA prior to sequencing could potentially lead to inaccuracies in the classification of 5′ ends, particularly for sequences with TAP+:TAP− ratios close to the 2-fold enrichment cut off. Degradation of RNA during manipulations for 5′ end library preparations likely contributed, at least in some subset, to the high number of processed 5′ ends reported herein. It is also possible that in some instances a TSS was not captured in our libraries due to a high rate of mRNA turnover. Here, we provided several means of validation of 5′ end identification in B. burgdorferi. Initial comparison of the 5′RNA-seq data to published 5′ RACE- or primer extension-defined TSSs demonstrated high correlation in the data. This is consistent with the 87% correlation reported for a similar comparison performed in Helicobacter pylori (35). It is possible that highly structured regions of the transcriptome may pose technical challenges to 5′ RACE and primer extension as well as 5′RNA-seq, which may contribute to the differences between the published TSSs and the 5′RNA-seq data. Additionally, some 5′ RACE and primer extension approaches are unable to distinguish between TSSs and stable processed transcripts. Therefore, we provided bioinformatics evidence of conserved promoter elements upstream of the 5′RNA-seq identified TSSs as well as used promoter fusions and Northern blotting to further validate the 5′RNA-seq data.
As many 5′RNA-seq data sets have revealed in recent years, bacterial transcriptomes are rich in un-annotated sequences (65). 5′RNA-seq in B. burgdorferi led to the discovery of initiation sites for transcripts that span across, between, antisense and internal to annotated ORFs. Although TSS validation by Northern blot analysis may be obscured by transcriptional complexity, we have used multiple Northern blot probes targeting regions of interest to more confidently validate the presence of a transcript downstream of 5′ ends. Furthermore, our results indicate that transcriptional complexity may confound transcript boundary analysis techniques such as reverse transcriptase-PCR that is used to amplify across annotated gene gaps for evidence of operons. For example, the 5′RNA-seq data identified additional putative primary TSSs within the recently described operons, BB_0360-BB_0364 (71), BB_0215-BB_0218 (72) and BB_0404-BB_0406 (73). These findings do not rule out the possibility that these gene clusters are co-transcribed, but suggest that multiple transcripts initiate within these loci that may not be detected by traditional methods. Indeed, sorting out exact transcript maps, particularly in complex annotated regions, proves complicated. Even more challenging, is definitive determination of the origin of transcription for the identified processed 5′ ends. In these cases, we did not attempt to classify processed 5′ ends based on location and rather only reported specific nucleotide positions of all instances. Future targeted experimental approaches are required to determine the TSS for each processed 5′ end.
Principally 5′RNA-seq in B. burgdorferi was conducted to better understand and distinguish mammalian specific promoters discovered through BbIVET. However, in establishing this TSS map we discovered numerous novelties of B. burgdorferi transcription. Of most note, was the observation that adenine and cytosine demonstrated equivalent frequencies of occurrence as the initiating nucleotide for transcription at internal TSSs. It has been recognized that nucleoside triphosphate concentrations within the cell can dictate initiation of transcription based on the +1 nucleotide availability, as was first shown for ribosomal RNA (74). B. burgdorferi lacks the enzymes for de novo purine biosynthesis, and in turn must salvage these molecules from the host environment for nucleotide synthesis (75–77). Considering the concentrations of available nucleotides and nucleotide precursors likely differ depending on the spirochete's immediate environment (54,76,78), nucleotide availability may serve as one fundamental mechanism of transcriptional regulation. Indeed, the identity of the transcription start site nucleotide and changes in intracellular ATP and GTP levels have been shown to critically alter gene expression during starvation in Bacillus subtilis (79).
We have demonstrated strong conservation of the classical Pribnow box among the promoter regions of the primary, secondary and antisense TSSs. Collectively, the internal TSSs lacked a conserved promoter sequence, similar to what has been documented for Hfx. volcanii (80), but in contrast to findings in other bacterial species (35,42). However, the analysis of 7.5% of internal TSS promoter elements with TAP+ read counts of greater than 500, demonstrated 41.0% with a conserved Pribnow box. These data raise the possibility that expression of the majority of the B. burgdorferi intragenic transcripts may be controlled by mechanisms that differ from known means of sigma 70-dependent regulation. Alternatively, it is possible that in some instances the spirochete RNA polymerase initiates off-target, in what has been referred to as pervasive transcription. However, this does not diminish the biological relevance of these events, as evolutionary pressures could result in the functional roles of such transcripts (81).
We identified 85 leaderless mRNAs, having a UTR length of less than 10 nts. Typically leaderless transcripts in bacteria are thought to be translated at low efficiency due to the absence of a canonical ribosome binding site (82). However, in E. coli under certain stress conditions specialized ribosomes, lacking the anti-Shine–Dalgarno sequence, have been shown to proficiently translate leaderless mRNAs (83). An abundance of leaderless transcripts have recently been discovered in the genomes of Mycobacterium smegmatis and Mycobacterium tuberculosis (36,41), which have been found to be translated as efficiently as leadered transcripts (41). While the majority of B. burgdorferi leaderless transcripts are annotated as hypothetical proteins, it is feasible that these sequences may be involved in stress adaptation or that ribosome recognition of leaderless mRNA targets in B. burgdorferi may occur by a yet-to-be described mechanism, as proposed for mycobacterial species (41). It is also possible that mis-annotation of an ORF and/or the presence of an alternative start codon could have resulted in inaccurate identification of a transcript as leaderless.
Cis-regulatory RNA elements, such as riboswitches and attenuators present in 5′ UTRs, may control mRNA transcription and/or translation (84,85). To date, the only functionally characterized example of 5′ mRNA regulation in B. burgdorferi is the mechanism of post-transcriptional regulation of the alternative sigma factor rpoS by the temperature induced trans-acting sRNA dsrA (86). No riboswitch mediated gene regulation mechanisms have been discovered in B. burgdorferi, and among all spirochetes only one example of a riboswitch has been described, a thiamine sensor in Treponema denticola (87). Several studies have investigated the contribution of cyclic-di-GMP, the second messenger and common riboswitch ligand, in B. burgdorferi environmental sensing (88,89). Recent data demonstrate that the four-gene (BB_0240-BB_0243) glp operon, involved in glycerol transport and metabolism (89–91), is regulated by cyclic-di-GMP (c-di-GMP) (88,89,92). By Northern blotting we validate the presence of a long UTR in the glp operon. Additionally, the UTR probe alone revealed a ∼200 nt transcript. These data suggest the presence of a terminated transcript within the 5′ UTR of the glp operon, which may indicate a cis-regulatory RNA element (62) and provide support for the notion that long 5′ UTR sequences in B. burgdorferi may harbor additional regulatory information.
Of the 233 total unique Bbive sequences, 119 associated with 5′RNA-seq identified TSSs. This group is comprised of transcripts expressed both in culture and the mammalian host, some of which may be B. burgdorferi mammalian infection-induced and/or infection-essential RNA species. Among these transcripts, there are at least five genes that have been previously shown to be required for B. burgdorferi pathogenesis through a signature-tagged mutagenesis screen, including BB_0145 (27). Herein, we demonstrated that the Bbive203-TSS promoter associated with infection-relevant BB_0145 is active both in culture and during infection. These data highlight the importance of further analysis of these transcripts despite the finding that their expression is not limited to the mammalian host environment. Only one Bbive-TSS, Bbive74, failed to demonstration promoter activity during infection. This TSS was located only 29 nts downstream the 5′ end of the 125 bp Bbive74 sequence. Based on the cloning strategy of promoter fusions for Bbive-TSSs, 96 bp of Bbive74 were not tested for promoter activity and could conceivably contain an infection-specific promoter and TSS. Alternatively, the activity of this transcript in the mouse could fall below the level of detection by IVIS.
A subset of Bbives are associated with low-coverage TSSs, as determined by 5′RNA-seq. In fact, 53 Bbive-TSSs displayed an average TAP+ read count less than 50. Herein we demonstrate that Bbive277, which has an average TAP+ read count of 44.5 and just above the threshold cutoff of calling TSSs in 5′RNA-seq, is active during infection. Some portion of these in culture, lowly expressed Bbives may be induced in the mammalian host. For instance, the primary TSS for the mammalian infection specific ospC mRNA had an average TAP+ read count of 37.5 in culture, and demonstrated low promoter activity in culture, but strong promoter activity during murine infection. Notably, exact quantifications of these instances cannot be empirical determined by the in vivo imaging approaches reported herein.
Comparative analysis of the BbIVET and 5′RNA-seq data sets identified 114 Bbives that did not associate with culture-expressed TSSs. However, none of these putative mammalian-specific Bbives have activity specific to mammalian infection by luciferase transcriptional fusion. Bbive36, 158 and 252 showed low levels of luciferase activity in culture despite having no associated TSSs by 5′RNA-seq. This could reflect the possible low stability of the transcripts in culture leading to their absence from the 5′RNA-seq libraries and/or differences in sensitivity among the techniques. This does not, however, rule out the possibility that of the 114 Bbives in this category, some are primarily active during mammalian infection. Indeed Bbive161 and 175 had no detectable promoter activity in culture and no TSS detected by 5′RNA-seq. However, these sequences were sufficiently active during murine infection to drive expression of pncA through BbIVET. Likely, we did not observe luciferase activity for these sequences by IVIS due to the sensitivity of the currently available IVIS machine. Indeed, even with our strongest promoter (flaBp) we only detect robust luciferase activity at days 7 and 10 post infection by IVIS. Furthermore, unlike what has recently been shown for detection of ospC p activity by IVIS (93), using our experimental system we did not detect ospC p activity above background until 7 days post infection. Importantly, transcript levels do not necessarily correlate to biological relevance. Indeed, our group has recently demonstrated by quantitative RT-PCR analysis of B. burgdorferi infected mouse tissues at 10 days post infection that the transcript level of the BbIVET-identified and infection-essential gene bb0318 is ∼40-fold lower than that of the ospC transcript ((55) and unpublished data, P.P.A. and M.W.J.). Together, our findings indicate the sensitivity of BbIVET to identify low activity promoters and the potential for discovering B. burgdorferi infection-essential transcripts through BbIVET. These data demonstrate, for the first time, the expression of various types of previously un-annotated RNAs during B. burgdorferi infection of mice, including antisense and internal transcripts. Together our findings provide a framework for future studies focused on defining the contributions of these novel transcripts to the molecular mechanisms of B. burgdorferi pathogenesis.
CONCLUSIONS
The data sets described herein provide a significant resource for the spirochete community and others interested in bacterial transcriptomes. The 5′RNA-seq data have been uploaded to an interactive browser, available for viewing at www.ucf.edu/research/interactive-genomics/lyme-disease. This work provides the first examples of B. burgdorferi expression of non-annotated RNAs during mammalian infection and lays the foundation for mechanistic studies focused on elucidation of the contributions of RNA-based regulation to B. burgdorferi pathogenesis. With the continued advancement of high throughput sequencing methods and publication of ever increasing numbers of 5′ end deep sequencing data sets, the bottleneck in research innovation becomes validation and prioritization of novel transcripts of interest. This is particularly important for bacterial pathogens and understanding the biological significance of genetically intractable transcripts such as antisense and internal RNAs. We have used a unique synergistic approach of combining 5′RNA-seq and IVET, along with luciferase promoter fusions, to validate the expression of primary, antisense and internal transcripts during an active infection. This work encompasses the first application of 5′RNA-seq to validate a global promoter-activity based screen. The infection-model discovery and validation approach that we present is broadly applicable to any pathogen of interest providing (i) strand specific validation of novel transcripts expressed during infection, (ii) identification of transcripts of interest for mechanistic studies and (iii) transcriptional analysis in pathogens that do not achieve high loads during infection. In sum, this work contributes significant insight into the transcriptome of B. burgdorferi and provides an innovative approach for analysis of 5′ end RNA-seq data from medically relevant pathogens.
AVAILABILITY
The complete BbIVET sequences, including Bbive boundaries and those that are associated to TSSs are provided in Supplementary Table S2. The 5′RNA-seq data have been deposited to SRA at NCBI (Accesssion numbers: SRR4423918, SRR4423919, SRR4423920, SRR4423921). A complete list of 5′ ends identified in this study with genomic location and predicted annotation is provided in Supplementary Tables S3, S4 and S5. An interactive genome browser containing the 5′RNA-seq data described herein can be found at www.ucf.edu/research/interactive-genomics/lyme-disease.
Acknowledgments
The authors would like to Dr Jon Blevins and Dr Michael Norgard for generously providing the pJSB161 and pJSB175 plasmids, Dr Nikhat Praveen for technical suggestions related to IVIS and Dr Kyle Rohde for use of his plate reader. The authors would like to thank Dr Travis Jewett and members of the M. Jewett lab for insightful discussions and critical analysis. The authors would like to thank Philipp Rescheneder and Alex Lazin for assistance with bioinformatics, the UCF L.E.A.R.N. students, Carlos Tapia and Brianna King, for technical support. The authors would like to thank the UCF NAF animal care staff, Craig Anderson and the UCF COM HIT team. The authors wish to acknowledge the Vienna BioCenter Core Facilities (VBCF) for providing initial assistance in RNA-seq, and John Marchica and Subramaniam Shyamalagovindarajan at Sanford Burnham Prebys Medical Discovery Institute for performing the Illumina sequencing.
Author contributions: I.B., M.L. and R.S. developed the 5′ end RNA-seq method. P.P.A. generated the B. burgdorferi 5′RNA-seq libraries. P.P.A., M.L., R.S. and M.W.J. designed experiments. P.P.A. and C.F.A performed experiments. N.P. and P.P.A. performed bioinformatics analyses. P.P.A., C.F.A., N.P., M.L., I.B. and M.W.J. interpreted experimental results. P.P.A. and M.W.J. wrote the manuscript. All authors critiqued and edited the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Allergy and Infectious Diseases of the National Institutes of Health [R01AI099094 to M.W.J.]; National Research Fund for Tick-Borne Diseases [to M.W.J.]; University of Central Florida College of Medicine [to M.W.J.]; Austrian Science Fund [FWF F4301 and F4308 to R.S.]; University of Vienna [to R.S.]. Funding for open access charge: National Institute of Allergy and Infectious Diseases of the National Institutes of Health [R01AI099094 to M.W.J.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Adeolu M., Gupta R.S. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex) Antonie Van Leeuwenhoek. 2014;105:1049–1072. doi: 10.1007/s10482-014-0164-x. [DOI] [PubMed] [Google Scholar]
- 2.Schotthoefer A.M., Frost H.M. Ecology and Epidemiology of Lyme Borreliosis. Clin. Lab. Med. 2015;35:723–743. doi: 10.1016/j.cll.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Radolf J.D., Caimano M.J., Stevenson B., Hu L.T. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilly K., Rosa P.A., Stewart P.E. Biology of infection with Borrelia burgdorferi. Infect. Dis. Clin. North Am. 2008;22:217–234. doi: 10.1016/j.idc.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groshong A.M., Blevins J.S. Insights into the biology of Borrelia burgdorferi gained through the application of molecular genetics. Adv. Appl. Microbiol. 2014;86:41–143. doi: 10.1016/B978-0-12-800262-9.00002-0. [DOI] [PubMed] [Google Scholar]
- 6.Samuels D.S. Gene regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 7.Iyer R., Caimano M.J., Luthra A., Axline D., Jr, Corona A., Iacobas D.A., Radolf J.D., Schwartz I. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol. Microbiol. 2015;95:509–538. doi: 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwan T.G. Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi. Biochem. Soc. Trans. 2003;31:108–112. doi: 10.1042/bst0310108. [DOI] [PubMed] [Google Scholar]
- 9.Schwan T.G., Piesman J. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava S.Y., de Silva A.M. Reciprocal expression of ospA and ospC in single cells of Borrelia burgdorferi. J. Bacteriol. 2008;190:3429–3433. doi: 10.1128/JB.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery R.R., Malawista S.E., Feen K.J., Bockenstedt L.K. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwan T.G., Piesman J., Golde W.T., Dolan M.C., Rosa P.A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westermann A.J., Forstner K.U., Amman F., Barquist L., Chao Y., Schulte L.N., Muller L., Reinhardt R., Stadler P.F., Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 14.Fiedler T., Sugareva V., Patenge N., Kreikemeyer B. Insights into Streptococcus pyogenes pathogenesis from transcriptome studies. Future Microbiol. 2010;5:1675–1694. doi: 10.2217/fmb.10.128. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao A., Zhu J. Genetic tools to study gene expression during bacterial pathogen infection. Adv. Appl. Microbiol. 2009;67:297–314. doi: 10.1016/S0065-2164(08)01009-5. [DOI] [PubMed] [Google Scholar]
- 16.La M.V., Raoult D., Renesto P. Regulation of whole bacterial pathogen transcription within infected hosts. FEMS Microbiol. Rev. 2008;32:440–460. doi: 10.1111/j.1574-6976.2008.00103.x. [DOI] [PubMed] [Google Scholar]
- 17.Sturdevant D.E., Virtaneva K., Martens C., Bozinov D., Ogundare O., Castro N., Kanakabandi K., Beare P.A., Omsland A., Carlson J.H., et al. Host-microbe interaction systems biology: lifecycle transcriptomics and comparative genomics. Future Microbiol. 2010;5:205–219. doi: 10.2217/fmb.09.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang F.T., Nelson F.K., Fikrig E. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 2002;196:275–280. doi: 10.1084/jem.20020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojaimi C., Brooks C., Casjens S., Rosa P., Elias A., Barbour A., Jasinskas A., Benach J., Katona L., Radolf J., et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revel A.T., Talaat A.M., Norgard M.V. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livengood J.A., Schmit V.L., Gilmore R.D., Jr Global transcriptome analysis of Borrelia burgdorferi during association with human neuroglial cells. Infect. Immun. 2008;76:298–307. doi: 10.1128/IAI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokarz R., Anderton J.M., Katona L.I., Benach J.L. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks C.S., Hefty P.S., Jolliff S.E., Akins D.R. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caimano M.J., Iyer R., Eggers C.H., Gonzalez C., Morton E.A., Gilbert M.A., Schwartz I., Radolf J.D. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimhan S., Caimano M.J., Liang F.T., Santiago F., Laskowski M., Philipp M.T., Pachner A.R., Radolf J.D., Fikrig E. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15953–15958. doi: 10.1073/pnas.2432412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Coleman A.S., Anguita J., Pal U. A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. Plos Pathog. 2009;5:e1000326. doi: 10.1371/journal.ppat.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin T., Gao L., Zhang C., Odeh E., Jacobs M.B., Coutte L., Chaconas G., Philipp M.T., Norris S.J. Analysis of an Ordered, Comprehensive STM Mutant Library in Infectious Borrelia burgdorferi: Insights into the Genes Required for Mouse Infectivity. PloS One. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troy E.B., Lin T., Gao L., Lazinski D.W., Lundt M., Camilli A., Norris S.J., Hu L.T. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol. Microbiol. 2016;101:1003–1023. doi: 10.1111/mmi.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis T.C., Jain S., Linowski A.K., Rike K., Bestor A., Rosa P.A., Halpern M., Kurhanewicz S., Jewett M.W. In vivo expression technology identifies a novel virulence factor critical for Borrelia burgdorferi persistence in mice. Plos Pathog. 2014;10:e1004260. doi: 10.1371/journal.ppat.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanin A., Sava I., Bao Y., Huebner J., Hartke A., Auffray Y., Sauvageot N. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PloS One. 2010;5:e11879. doi: 10.1371/journal.pone.0011879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson R.W., Giddens S.R. Development and application of in vivo expression technology (IVET) for analysing microbial gene expression in complex environments. Infect. Disord. Drug Targets. 2006;6:207–240. doi: 10.2174/187152606778249944. [DOI] [PubMed] [Google Scholar]
- 32.Lee S.W., Cooksey D.A. Genes expressed in Pseudomonas putida during colonization of a plant-pathogenic fungus. Appl. Environ. Microbiol. 2000;66:2764–2772. doi: 10.1128/aem.66.7.2764-2772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendez J., Reimundo P., Perez-Pascual D., Navais R., Gomez E., Guijarro J.A. A novel cdsAB operon is involved in the uptake of L-cysteine and participates in the pathogenesis of Yersinia ruckeri. J. Bacteriol. 2011;193:944–951. doi: 10.1128/JB.01058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slauch J.M., Camilli A. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 2000;326:73–96. doi: 10.1016/s0076-6879(00)26047-3. [DOI] [PubMed] [Google Scholar]
- 35.Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermuller J., Reinhardt R., et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 36.Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuss A.M., Heroven A.K., Waldmann B., Reinkensmeier J., Jarek M., Beckstette M., Dersch P. Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet. 2015;11:e1005087. doi: 10.1371/journal.pgen.1005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosinski-Chupin I., Sauvage E., Sismeiro O., Villain A., Da Cunha V., Caliot M.E., Dillies M.A., Trieu-Cuot P., Bouloc P., Lartigue M.F., et al. Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics. 2015;16:419. doi: 10.1186/s12864-015-1583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papenfort K., Forstner K.U., Cong J.P., Sharma C.M., Bassler B.L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dugar G., Herbig A., Forstner K.U., Heidrich N., Reinhardt R., Nieselt K., Sharma C.M. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., Gawande R., Ahmad R., Sarracino D.A., Ioerger T.R., et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elias A.F., Stewart P.E., Grimm D., Caimano M.J., Eggers C.H., Tilly K., Bono J.L., Akins D.R., Radolf J.D., Schwan T.G., et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rego R.O., Bestor A., Rosa P.A. Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 2011;193:1161–1171. doi: 10.1128/JB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuels D.S. Methods in Molecular Biology. In: Nickoloff JA, editor. Electroporation Protocols for Microorgansisms. Vol. 47. Totowa, N.J.: Humana Press, Inc; 1995. pp. 253–259. [Google Scholar]
- 46.Barbour A.G. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 47.Rosa P.A., Hogan D. In: Proceeding of the First International Conference on Tick Borne Pathogens at the Host-Vector Interface. Munderloh UG, Kurtti TJ, editors. Paul, Minnesota: University of Minnesota, St; 1992. pp. 95–103. [Google Scholar]
- 48.Drecktrah D., Lybecker M., Popitsch N., Rescheneder P., Hall L.S., Samuels D.S. The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. Plos Pathog. 2015;11:e1005160. doi: 10.1371/journal.ppat.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sedlazeck F.J., Rescheneder P., von Haeseler A. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 2013;29:2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- 50.Casjens S., Palmer N., van Vugt R., Huang W.M., Stevenson B., Rosa P., Lathigra R., Sutton G., Peterson J., Dodson R.J., et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 51.Di L., Pagan P.E., Packer D., Martin C.L., Akther S., Ramrattan G., Mongodin E.F., Fraser C.M., Schutzer S.E., Luft B.J., et al. BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinformatics. 2014;15:233. doi: 10.1186/1471-2105-15-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Streit S., Michalski C.W., Erkan M., Kleeff J., Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- 53.Blevins J.S., Revel A.T., Smith A.H., Bachlani G.N., Norgard M.V. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 2007;73:1501–1513. doi: 10.1128/AEM.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jewett M.W., Lawrence K., Bestor A.C., Tilly K., Grimm D., Shaw P., VanRaden M., Gherardini F., Rosa P.A. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 2007;64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Showman A.C., Aranjuez G., Adams P.P., Jewett M.W. Gene bb0318 is critical for the oxidative stress response and infectivity of Borrelia burgdorferi. Infect. Immun. 2016;84:3141–3151. doi: 10.1128/IAI.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shepherd J., Ibba M. Bacterial transfer RNAs. FEMS Microbiol. Rev. 2015;39:280–300. doi: 10.1093/femsre/fuv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroger C., Dillon S.C., Cameron A.D., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hebrard M., Handler K., et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendoza-Vargas A., Olvera L., Olvera M., Grande R., Vega-Alvarado L., Taboada B., Jimenez-Jacinto V., Salgado H., Juarez K., Contreras-Moreira B., et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PloS One. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 60.Kortmann J., Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 61.Tucker B.J., Breaker R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 62.Dar D., Shamir M., Mellin J.R., Koutero M., Stern-Ginossar N., Cossart P., Sorek R. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016;352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Irnov I., Sharma C.M., Vogel J., Winkler W.C. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wurtzel O., Sesto N., Mellin J.R., Karunker I., Edelheit S., Becavin C., Archambaud C., Cossart P., Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stazic D., Voss B. The complexity of bacterial transcriptomes. J. Biotechnol. 2015;232:69–78. doi: 10.1016/j.jbiotec.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 66.Mandlik A., Livny J., Robins W.P., Ritchie J.M., Mekalanos J.J., Waldor M.K. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller C., Cacaci M., Sauvageot N., Sanguinetti M., Rattei T., Eder T., Giard J.C., Kalinowski J., Hain T., Hartke A. The intraperitoneal transcriptome of the opportunistic pathogen Enterococcus faecalis in mice. PloS One. 2015;10:e0126143. doi: 10.1371/journal.pone.0126143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan Y., Su S., Meng X., Ji X., Qu Y., Liu Z., Wang X., Cui Y., Deng Z., Zhou D., et al. Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PloS One. 2013;8:e74495. doi: 10.1371/journal.pone.0074495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deka R.K., Brautigam C.A., Biddy B.A., Liu W.Z., Norgard M.V. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio. 2013;4 doi: 10.1128/mBio.00615-12. doi:10.1128/mBio.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arnold W.K., Savage C.R., Brissette C.A., Seshu J., Livny J., Stevenson B. RNA-Seq of Borrelia burgdorferi in multiple phases of growth reveals insights into the dynamics of gene expression, transcriptome architecture and noncoding RNAs. PloS One. 2016;11:e0164165. doi: 10.1371/journal.pone.0164165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sultan S.Z., Pitzer J.E., Miller M.R., Motaleb M.A. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol. Microbiol. 2010;77:128–142. doi: 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brautigam C.A., Ouyang Z., Deka R.K., Norgard M.V. Sequence, biophysical, and structural analyses of the PstS lipoprotein (BB0215) from Borrelia burgdorferi reveal a likely binding component of an ABC-type phosphate transporter. Protein Sci. 2014;23:200–212. doi: 10.1002/pro.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kung F., Kaur S., Smith A.A., Yang X., Wilder C.N., Sharma K., Buyuktanir O., Pal U. A Borrelia burgdorferi surface-exposed transmembrane protein lacking detectable immune responses supports pathogen persistence and constitutes a vaccine target. J. Infect. Dis. 2016;213:1786–1795. doi: 10.1093/infdis/jiw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaal T., Bartlett M.S., Ross W., Turnbough C.L., Jr, Gourse R.L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 75.Jain S., Sutchu S., Rosa P.A., Byram R., Jewett M.W. Borrelia burgdorferi harbors a transport system essential for purine salvage and mammalian infection. Infect. Immun. 2012;80:3086–3093. doi: 10.1128/IAI.00514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jewett M.W., Lawrence K.A., Bestor A., Byram R., Gherardini F., Rosa P.A. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 2009;191:6231–6241. doi: 10.1128/JB.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence K.A., Jewett M.W., Rosa P.A., Gherardini F.C. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 2009;72:1517–1529. doi: 10.1111/j.1365-2958.2009.06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jain S., Showman A.C., Jewett M.W. Molecular dissection of a Borrelia burgdorferi in vivo essential purine transport system. Infect. Immun. 2015;83:2224–2233. doi: 10.1128/IAI.02859-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krasny L., Tiserova H., Jonak J., Rejman D., Sanderova H. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 2008;69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 80.Babski J., Haas K.A., Nather-Schindler D., Pfeiffer F., Forstner K.U., Hammelmann M., Hilker R., Becker A., Sharma C.M., Marchfelder A., et al. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq) BMC Genomics. 2016;17:629. doi: 10.1186/s12864-016-2920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lybecker M., Bilusic I., Raghavan R. Pervasive transcription: detecting functional RNAs in bacteria. Transcription. 2014;5:e944039. doi: 10.4161/21541272.2014.944039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moll I., Grill S., Gualerzi C.O., Blasi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 83.Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith A.M., Fuchs R.T., Grundy F.J., Henkin T.M. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biol. 2010;7:104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henkin T.M., Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 86.Lybecker M.C., Samuels D.S. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 87.Bian J., Shen H., Tu Y., Yu A., Li C. The riboswitch regulates a thiamine pyrophosphate ABC transporter of the oral spirochete Treponema denticola. J. Bacteriol. 2011;193:3912–3922. doi: 10.1128/JB.00386-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caimano M.J., Dunham-Ems S., Allard A.M., Cassera M.B., Kenedy M., Radolf J.D. Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect. Immun. 2015;83:3043–3060. doi: 10.1128/IAI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He M., Ouyang Z., Troxell B., Xu H., Moh A., Piesman J., Norgard M.V., Gomelsky M., Yang X.F. Cyclic di-GMP is essential for the survival of the lyme disease spirochete in ticks. Plos Pathog. 2011;7:e1002133. doi: 10.1371/journal.ppat.1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fraser C.M., Casjens S., Huang W.M., Sutton G.G., Clayton R., Lathigra R., White O., Ketchum K.A., Dodson R., Hickey E.K., et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 91.Pappas C.J., Iyer R., Petzke M.M., Caimano M.J., Radolf J.D., Schwartz I. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. Plos Pathog. 2011;7:e1002102. doi: 10.1371/journal.ppat.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sudarsan N., Lee E.R., Weinberg Z., Moy R.H., Kim J.N., Link K.H., Breaker R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skare J.T., Shaw D.K., Trzeciakowski J.P., Hyde J.A. In vivo imaging demonstrates that Borrelia burgdorferi ospC is uniquely expressed temporally and spatially throughout experimental infection. PloS one. 2016;11:e0162501. doi: 10.1371/journal.pone.0162501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jonsson M., Noppa L., Barbour A.G., Bergstrom S. Heterogeneity of outer membrane proteins in Borrelia burgdorferi: comparison of osp. operons of three isolates of different geographic origins. Infect. Immun. 1992;60:1845–1853. doi: 10.1128/iai.60.5.1845-1853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rhodes R.G., Coy W., Nelson D.R. Chitobiose utilization in Borrelia burgdorferi is dually regulated by RpoD and RpoS. BMC Microbiol. 2009;9:108. doi: 10.1186/1471-2180-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Lackum K., Ollison K.M., Bykowski T., Nowalk A.J., Hughes J.L., Carroll J.A., Zuckert W.R., Stevenson B. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's mammal-tick infectious cycle. Microbiology. 2007;153:1361–1371. doi: 10.1099/mic.0.2006/003350-0. [DOI] [PubMed] [Google Scholar]
- 97.Motaleb M.A., Sultan S.Z., Miller M.R., Li C., Charon N.W. CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. J. Bacteriol. 2011;193:3332–3341. doi: 10.1128/JB.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jutras B.L., Jones G.S., Verma A., Brown N.A., Antonicello A.D., Chenail A.M., Stevenson B. Posttranscriptional self-regulation by the Lyme disease bacterium's BpuR DNA/RNA-binding protein. J. Bacteriol. 2013;195:4915–4923. doi: 10.1128/JB.00819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H., Raji A., Theisen M., Hansen P.R., Marconi R.T. bdrF2 of Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J. Bacteriol. 2005;187:175–184. doi: 10.1128/JB.187.1.175-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marconi R.T., Samuels D.S., Garon C.F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Margolis N., Hogan D., Tilly K., Rosa P.A. Plasmid location of Borrelia purine biosynthesis gene homologs. J. Bacteriol. 1994;176:6427–6432. doi: 10.1128/jb.176.21.6427-6432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Padula S.J., Sampieri A., Dias F., Szczepanski A., Ryan R.W. Molecular characterization and expression of p23 (OspC) from a North American strain of Borrelia burgdorferi. Infect. Immun. 1993;61:5097–5105. doi: 10.1128/iai.61.12.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]