Abstract
The development of Ribosome Profiling (RiboSeq) has revolutionized functional genomics. RiboSeq is based on capturing and sequencing of the mRNA fragments enclosed within the translating ribosome and it thereby provides a ‘snapshot’ of ribosome positions at the transcriptome wide level. Although the method is predominantly used for analysis of differential gene expression and discovery of novel translated ORFs, the RiboSeq data can also be a rich source of information about molecular mechanisms of polypeptide synthesis and translational control. This review will focus on how recent findings made with RiboSeq have revealed important details of the molecular mechanisms of translation in eukaryotes. These include mRNA translation sensitivity to drugs affecting translation initiation and elongation, the roles of upstream ORFs in response to stress, the dynamics of elongation and termination as well as details of intrinsic ribosome behavior on the mRNA after translation termination. As the RiboSeq method is still at a relatively early stage we will also discuss the implications of RiboSeq artifacts on data interpretation.
INTRODUCTION
It has been known for decades that the ribosome protects the portion of mRNA that is enclosed within its subunits from ribonuclease digestion (1–5). The advent of massively parallel sequencing enabled the development of the RiboSeq technique which is based on the sequencing of these protected mRNA fragments on a large scale. Initially it was demonstrated for yeast cells (6) and later was applied to other species. Since its inception, ribosome profiling has been used to examine many aspects of translation in humans (7) and mice (8), fishes (9), insects (10), nematodes (11), as well as plants (12), protozoa (13), bacteria (14) and virus-infected cells (15).
The fragments that the elongating ribosome protects are normally 18–34 nucleotides long depending on the organism, the ribosome conformation state and the experimental protocol applied (14,16–19). Usually, the treatment of cytoplasmic extracts with ribonucleases is followed by the isolation of 80S particles which contain Ribosome Protected Fragments (RPFs) or ribosome ‘footprints’. The positions of the mapped reads on the mRNA sequence display a 3 nucleotide periodicity reflecting the stepwise movement of the elongating ribosome over codons. This enables for the detection of translated ORFs and their reading-frame transitions (20). In the course of RPF preparation, the ribosomes need to be blocked at mRNA. This is frequently done with antibiotics that arrest elongation (cycloheximide, emetine, chloramphenicol, anisomycin) or flash freezing in liquid nitrogen. The isolated RPFs are used to generate cDNAs that are subsequently sequenced using massively parallel sequencing platforms. An important control is to measure in parallel the level of mRNAs present in the tested cells (RNAseq). To this end, the mRNA is isolated from a proportion of the cytoplasmic extract, prior to nuclease digestion, and then randomly cleaved by alkaline hydrolysis to yield mRNA fragments (typically around the same length as ribosome footprints). The ratio of RPFs to RNAseq fragments mapped to each transcript is often used as an estimate of its relative translation efficiency (TE). The ribosome profiling protocol is described in dedicated methodology publications (21,22) and commercial kits are currently available. Numerous ad hoc modifications of the protocols have been made.
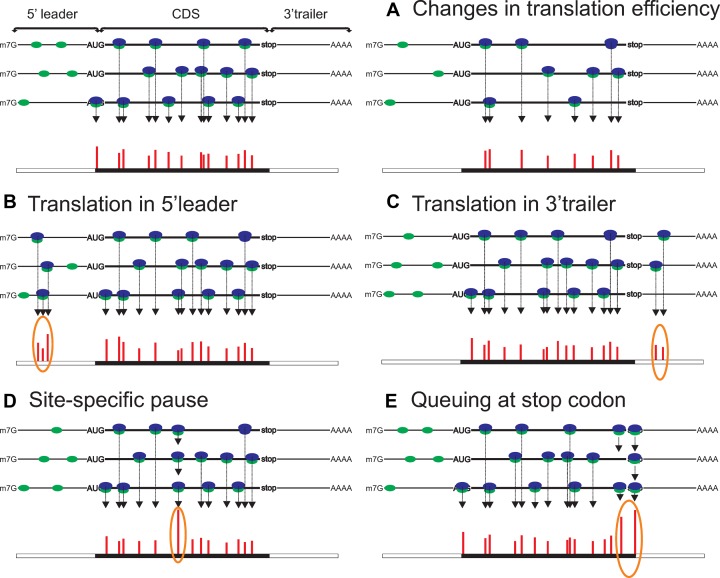
RiboSeq is not the first genome wide approach to study translation. Polysome profiling quantifies mRNAs that are bound by multiple ribosomes (polysomes), which are isolated by sucrose gradient centrifugation. Here, the RNA quantitation was initially achieved using microarrays (23–25), and later with massively parallel sequencing. It is assumed that the presence of mRNAs in polysomes reflects translation of the annotated coding ORFs (acORF), while in reality, ribosomes may translate other ORFs. RiboSeq provides a more direct estimate of protein synthesis as it generates position specific information. This allows a relative measure of the number of ribosomes occupying either a specific mRNA region, such as the acORF (Figure 1A), the 5′ leader (also known as 5′UTR) (Figure 1B), the 3′ trailer (also known as 3′UTR) (Figure 1C), or specific elongation pause sites (Figure 1D) or start and stop codons (Figure 1E).
Figure 1.
Types of alterations in ribosome density observed with ribosome profiling. (A) Changes in translation efficiency (TE), represented here by decrease of TE (compare with left part); Left panel represents control conditions and the right panel corresponds to changed conditions. (B) Translation in 5′ leader. (C) Presence of ribosomes in 3′ trailer. (D) Site specific pause originating from ribosomes stalled within acORF at a specific location. (E) Ribosomes paused at the stop codon and queued upstream ribosomes; green and blue shapes represent 40S complexes and 60S subunits respectively, only 80S ribosomes (40S+60S complexes) produce footprints using the conventional RiboSeq protocol. Red bars show the number of RPFs aligned to a particular location, due to biases of library preparation and sequencing the bar heights do not fully correspond to actual ribosome occupancies. Orange ovals highlight specific locations of ribosome footprints.
Novel applications of RiboSeq continue to emerge, recently RiboSeq was used for the analysis of translation in subcellular compartments (26,27), in certain cell types in the context of a whole organism (28,29) and for the detection of translation quantitative trait loci (30,31). Ribosome Profiling is now further empowered by a wide spectrum of computational resources. At RiboSeq.Org, GWIPS-viz (32,33) provides visualizations of genomic alignments for publicly available RiboSeq data aligned to the genomes of a dozen of organisms and RiboGalaxy (34) allows researchers to analyse their data on a cloud server via an internet browser using a variety of tools from such packages as RUST (35), riboSeqR (36) and RiboTools (37). Publicly available RiboSeq data also can be accessed at RPFdb (38). Furthermore, sORFs.org (39) collects ORFs whose translation is supported with RiboSeq data, and TISdb (40) is a collection of translation initiation sites predicted from RiboSeq data. Several stand alone tools were developed for differential gene expression analysis (anota (41), Babel (42), RiboDiff (43), Xtail (44)), and for the prediction of translated ORFs (RiboTaper (45), ORF-Rator (46) and riboHMM (47)). Ribomap (48) estimates footprint origins in the presence of multiple splice isoforms and repetitive regions and Rfoot (49) uses RiboSeq data to predict non-ribosome protected RNA sites. Additional RiboSeq specific pipelines include PROTEOFORMER (50) and RiboProfiling (51).
While the use of ribosome profiling has been reviewed extensively, see (52–59), this review focuses on how RiboSeq has been used to characterize the mechanisms of initiation, elongation, termination and ribosome recycling specifically in eukaryotic cells.
INITIATION
mRNA recruitment to the 43S preinitiation complex
The 43S pre-initiation complex (PIC) consists of a 40S ribosomal subunit loaded with Met-tRNA*eIF2*GTP (ternary complex, TC) and initiation factors eIF3, eIF1, eIF1A and eIF5. Its recruitment to mRNA is preceded by recognition of the mRNA m7G cap by initiation factor eIF4E. eIF4E operates as a part of the trimeric complex eIF4F (eIF4E, eIF4G and eIF4A) where eIF4G has a role as a scaffold protein and eIF4A is a helicase that unwinds base paired nucleotides within 5′ leaders of mRNA during 43S PIC scanning (for recent reviews see (60,61)).
The recognition of m7G cap by eIF4F is under stringent control. Some proteins can sequester eIF4E from eIF4G, and thus act as translational repressors. The most well studied translational repressors are mammalian eIF4E binding proteins (4EBPs). Hyper-phosphorylated 4EBPs do not bind to eIF4E, thereby allowing eIF4F engage in translation. The phosphorylation of 4EBP in mammalian cells is mostly carried out by the mTORC1 complex. mTORC1 includes mTOR kinase which plays a central role in proliferation and cell growth. Once mTORC1 is inactivated, rapid dephosphorylation of 4EBPs occurs; 4EBPs then sequester eIF4E from eIF4F thereby inhibiting cap-dependent translation (62–65).
The interactions of eIF4E with the mRNA cap and eIF4G were considered to be of paramount importance for the translation of almost all cellular mRNAs. Disruption of either interaction is believed to produce a global suppression effect on bulk protein synthesis. However, recent RiboSeq data suggests that disruption of the eIF4E–eIF4G interaction by 4EBP leads to a highly differential effect on translation and that global translation is less inhibited than may have been expected (66).
Two independent RiboSeq studies (67,68) examined the translational response to mTOR inactivation. Mammalian cells were briefly treated with inhibitors PP242, INK128 and Torin-1, which target the catalytic subunit of mTOR and thus activate 4EBPs more efficiently than the pioneer drug rapamycin, that acts allosterically (69). The global level of translation was significantly reduced, ∼40% relative to control conditions (68). Around 150 mRNAs were inhibited >4–8 folds relative to the global translation. The majority of these mRNAs (70) bear a 5′ Terminal Oligopyrimidine Tract (5′ TOP) motif. In addition to 5′TOP, other motifs termed Pyrimidine Tract Responsive Elements (PTREs) were proposed (67).This element, unlike 5′TOP motifs, possesses an invariant uridine flanked by pyrimidines and, importantly, does not reside at the +1 position of the 5′ leader. It was proposed that 5′TOP and PTRE motifs (and perhaps others) may act synergistically. Many ‘mTOR sensitive’ mRNAs encode components of the protein synthesis apparatus which are in particular demand during active growth and proliferation. This is consistent with observations that mTOR also regulates both rRNA and tRNA transcription (71–73). Two other classes of non-TOP ‘mTOR-sensitive’ mRNAs were identified recently (74). The first includes certain mRNAs with long 5′ leaders, which mostly encode survival- and proliferation promoting proteins (e.g. CCND3, ODC1, MCL1, BIRC5, MYC). The second class is a subset of mRNAs with very short 5′ leaders, which mostly encode proteins with mitochondrial functions. Interestingly, these mRNAs are enriched with TISU (Translation Initiator of Short 5′ UTR) initiation contexts, which provide unusual requirements for translation initiation components (75,76). These TISU elements are not the cause of the mTOR sensitivity, however (74). The fact that these ‘mTOR-sensitive’ mRNAs were not identified in prior RiboSeq studies was claimed to be due to technical reasons, including insufficient sequencing depth (74). Continuous translation of mRNAs under conditions of mTOR inhibition can be explained by residual levels of active eIF4F, but reliance on other ribosome recruitment factors is an intriguing possibility.
Although the role of eIF4E has been actively addressed with RiboSeq, many aspects of 43S PIC recruitment have yet to be elucidated. For instance, we do not know the role of the different initiation factor paralogs on the regulation of translation. In humans, there are two different eIF4F scaffolds, eIF4G1 and eIF4G3 (a third isoform, eIF4G2 a.k.a. Dap5, lacks an eIF4E binding site), three eIF4Es, and also three 4EBPs which may have different preferences to each other and to certain mRNAs. We also still have not delineated the importance of other executors of 4EBP phosphorylation, for example, cyclin-dependent kinase 1/cyclin B1 was reported to do so during mitosis (77). Finally, apart from ‘canonical’ 4EBPs there are a number of alternative factors which may regulate m7G cap recognition by eIF4E (reviewed in (78)). RiboSeq is ideal for exploring these understudied aspects of 43S PIC recruitment.
Old and new players that participate in scanning 5′ leaders
The 43S PIC scanning along 5′ leaders bearing secondary structures requires helicase activity (for review, see (79)) and eIF4A is believed to be a key component in mRNA leader structure unwinding.
A few studies addressed the effect of eIF4A inhibition on genome wide protein synthesis in mammalian cells. In two studies the inhibition was achieved by silvestrol, which belongs to the flavagline family of plant natural products (80,81). This drug increases binding of eIF4A to RNA (64) and thus impairs the activity of eIF4F. Genome wide analysis of ‘silvestrol-sensitive’ translationally downregulated mRNAs revealed that their 5′ leaders are usually long and structured. In both studies, the number of these mRNAs was relatively low (284 in case of (80)). Specifically, the 5′ leaders of ‘silvestrol-sensitive’ mRNAs were shown to be enriched in G-quadruplexes. These structures are considered to be efficient inhibitors of translation, and their unwinding upon scanning requires higher helicase activity. Interestingly, many ‘silvestrol-sensitive’ mRNAs encode oncogenes and cell-cycle regulators, which aligns well with the anti-proliferative properties of this drug. A more recent RiboSeq study with another flavagline closely related to silvestrol, rocaglamide A, led to a different conclusion. It was found that this drug specifically clamps eIF4A onto polypurine sequences, and secondary structures in 5′ leaders are only a minor determinant for rocaglamide A selectivity (82). Whether silvestrol and rocaglamide A possess different selectivity is not clear.
Recently, a study carried out RiboSeq after treating the cells with Pateamine A, which also targets eIF4A (83). The authors report that this treatment potently blocks translation and resulted in a widespread reduction in RPFs on coding sequences with a median inhibition of 22-fold. This effect was independent of the length of the 5′ leader. Pateamine A has a different mode of action than flavaglines in that it is responsible for inhibition of the interaction between eIF4A and eIF4G (as opposed to reducing its helicase activity (84)). It is of interest whether physiological modulation of mammalian eIF4A availability by means of PDCD4, which competes with eIF4G for eIF4A binding (85,86), would produce a pattern of regulation similar to these pharmacological treatments.
Whether other helicases and accessory proteins can complement or even substitute for eIF4A is unknown. RiboSeq was applied to yeast strains with mutations in DED1 (DDX3 is the mammalian orthologue) or TIF1 (EIF4A1 is the mammalian orthologue) (87). Both mutations resulted in a significant impairment on general translation as evidenced by a reduction of bulk polysomes. Footprint densities in DED1 mutants were significantly changed (relative to the average) for 814 transcripts (∼17% of expressed genes). The majority of these genes displayed a reduction in TE, indicating that >10% of all mRNAs revealed a dependence on Ded1 for efficient translation. The 5′ leaders of Ded1-dependent mRNAs are typically long and predicted to have a greater than average propensity to form secondary structures. In contrast, despite a similarly strong reduction in general translation observed for an eIF4A mutant, translation of only 36 mRNAs was hyperdependent on an eIF4A orthologue, implying that translation of most mRNAs was reduced comparably by the inactivation of eIF4A. Further analysis performed with reporter constructs suggests that the eIF4A orthologue, assisted by Ded1, is required for binding of the 43S PIC at the 5′ ends of mRNAs harboring cap-proximal RNA secondary structure.
Later this research was substantially developed by the same group to investigate the role of eIF4B, a cofactor of eIF4A which stimulates its helicase activity. To this end, the effect of TIF3 (eIF4B is the mammalian orthologue) mutation in yeast was investigated (88). Similar to Ded1 and TIF1, TIF3 depletion resulted in a strong decrease in bulk translation, with 111 hyperdependent and 48 hypodependent mRNAs identified. The change in gene expression was greater (and considerably different) for the TIF3 mutant than the TIF1 mutant which was unexpected given that TIF3 is a cofactor of TIF1 and that TIF3 is not an essential gene in yeast. In addition the subsets of eIF4B-hyperdependent (111 mRNA) and eIF4A-hyperdependent mRNAs (36 mRNAs) overlap by only three mRNAs. This implies that eIF4B may have an alternative function or acts independently of eIF4A, as many eIF4B-hyperdependent mRNAs have ‘standard’ requirements for eIF4A.
Translation initiation site (TIS) selection
Once the 43S PIC is successfully loaded onto mRNA and is in the scanning mode, it searches for an appropriate initiation codon. One of the general conclusions from the very first RiboSeq data was widespread alternative initiation both at AUGs and near-cognate start codons. This was initially observed by RPFs derived from the mRNA sequences which were annotated as 5′ leaders. Later, more specialized approaches were used to precisely map translation initiation sites genome wide. These approaches are based on the properties of particular antibiotics such as harringtonine (8), and later, lactimidomycin (89). Both of these drugs block elongation of de novo assembled 80S initiation complexes, whereas those 80S already engaged in elongation are believed to be refractory to these treatments. In another approach treatment with puromycin followed by cycloheximide was applied in order to release peptide-elongating ribosomes from mRNAs and then to block ribosomes during the first elongation step (90). These studies found that widespread non-AUG initiation occurs in eukaryotes. The occurrence of such initiation was not surprising as the ability of the translation initiation apparatus to recognize non-AUG codons as initiation sites is known from pioneering work by Kozak (91). However, its apparent frequency was unexpected as analysis using classic molecular biology techniques suggested that such initiation is generally inefficient.
Recent computational estimation of TIS strength from several published RiboSeq datasets provides potential explanations for this discrepancy (92). Using quantitative estimations of initiation efficiencies, AUG codons were confirmed to be much better initiators than near-cognate codons. The discrepancy between widespread occurrence of non-AUG initiation and its low efficiency is easily resolved once their locations are examined. In the vast majority of cases non-AUG initiation is observed upstream of AUG codons consistent with ‘leakiness’ of the former and ‘tightness’ of the latter.
The influence of nucleotide context surrounding a TIS on the efficiency of initiation is well documented (93,94), especially at positions −3 and +4 with respect to the first (+1) nucleotide of the start codon. However, the correlation of the context strength and TIS efficiency in RiboSeq studies was weak (92). This could be due to technical artifacts (see below), and/or it is also possible that the effect of the nucleotide context is confounded by the other mRNA elements that influence the efficiency of translation initiation. These may include specific secondary structures and RNA binding proteins interfering with the progression of scanning complexes, obscuring some potentially strong initiation codons and enhancing the weak ones. To conclude, even with the findings of the recent RiboSeq studies, at present we cannot reliably predict the efficiency or even the precise location of translation initiation in particular mRNAs from its sequence alone.
Regulation of translation by upstream open reading frames
Almost all RiboSeq studies reported higher footprint densities for 5′ leaders than for 3′ trailers indicating translation upstream of acORFs, although some can be attributed to the synthesis of N-terminally extended proteoforms (95). Upon various stress conditions, the percentage of RPFs at 5′ leaders, typically <1% (of all footprints aligned to the transcriptome) in mammalian cells, increases relative to acORFs. Translation of uORFs may generally be expected to be repressive. However, sometimes reciprocal changes in the translation of uORFs and acORFs can be observed at the level of individual genes, although this is not a general phenomenon. Alterations in ribosome density at uORFs also occur during conditions unrelated to stress e.g. during meiosis in yeast (96), cell cycle (97) and circadian oscillations in gene expression (28).
The events that trigger alterations in uORF translation are not fully understood. One possibility is that the activity or availability of those translation initiation factors responsible for initiation codon selection changes upon stress. In fact, upon oxygen and glucose deprivation (OGD), extensive translation regulation was observed for mRNAs encoding initiation factors eIF1, eIF5, eIF1A (and its paralogs), all of which are known as modulators of initiation codon selection. This may reflect an ongoing reprogramming of the translation initiation apparatus (98). It is not well known whether or how the activities of these initiation factors are regulated, except for the fact that both eIF1 and eIF5 are subject to post-translational modifications which may affect initiation codon selection (99–101). Intriguingly, both factors regulate their own translation (102,103).
Much more is known about regulation of eIF2, another principal player in TIS selection, and about the consequences of its regulation on translation. eIF2 forms the ternary complex (TC) with GTP and Met-tRNAi and is loaded onto the 40S ribosome to enable it to recognize a start codon, after which eIF2*GDP is released. GDP is then recycled to GTP by guanine exchange factor (GEF), eIF2B, to enable another round of initiation. Various stress conditions trigger the integrated stress response (ISR). Upon ISR, any of four kinases, HRI, PKR, PERK or GCN2, phosphorylate the alpha subunit of eIF2 at Ser51 (104). eIF2B forms a stable complex with phosphorylated eIF2 and is unable to rapidly recycle GTP. Phosphorylation of a modest number of eIF2 molecules rapidly reduces the limited pool of active eIF2B resulting in the general inhibition of protein synthesis. Some mRNAs are known to evade such translation repression, and the translation of their uORFs plays a critical role in this control.
To explore this regulation genome wide, a few research groups applied RiboSeq under conditions of ISR. One study found that upon robust phosphorylation of eIF2, triggered by treatment with arsenite, translation was greatly suppressed except for only a dozen mRNAs, all of which contained translated uORFs (105). Another study found that a very similar set of mRNAs were resistant to the unfolded protein response (UPR) induced with tunicamycin, a small molecule that blocks N-glycosylation and causes endoplasmic reticulum (ER) stress (106), which also results in eIF2 phosphorylation. Strikingly, differential translation upon UPR can be reversed upon treatment with a small molecule ISRIB, which potentiates the activity of eIF2B irrespective of eIF2's phosphorylation status (106–108).
In these studies, the number of preferentially translated mRNAs was low in comparison to all mRNAs with translated uORFs. The common characteristic of the resistant mRNAs is poor translation of acORFs and the presence of at least one efficiently translated uORF under normal conditions (105). Under stress conditions uORF translation is decreased whereas translation of the acORF is modestly enhanced or at least is not changed. Interestingly, in several cases (e.g. for DDIT3, IFRD1 and PPP1R15B mRNAs) a single uORF appeared to be sufficient for eIF2 mediated translation control (105,109). It is not clear why particular uORFs involved in the translation control are mediating regulation by eIF2 inactivation. It can be speculated that in addition to uORFs other properties of leaders are responsible for translation control upon eIF2 phosphorylation (105). It appears that TC deficiency does not influence TIS recognition directly, as no alterations in AUG versus non-AUG codon initiation were observed in general.
A recent computational study of available ribosome profiling data (110) that examined the relationship between the features of the 5′ leader and acORF TE concluded that while uORFs are generally repressive they are only minor determinants of global acORF TE. Thus, it is possible that only a minority of translated uORFs are strong repressors of acORF translation, while the majority are not repressive due to leaky scanning, 43S sliding and/or reinitiation (60,111,112). It is also likely that individual uORFs may work as translation regulators sensing specific conditions as exemplified by polyamine level sensing by uORFs in AMD1 (113) and AZIN1 (114) 5′ leaders, and by an uORF-based Mg2+ concentration sensor in a leader of TRPM7 mRNA, encoding a magnesium channel (115). It can be expected that there may be different modes of regulation, through TC levels, and/or through changes in start codon selection stringency (eIF1 and/or eIF5 levels). Moreover, any alterations in translation elongation, termination and recycling, discussed below, could also affect initiation at mRNAs containing translated uORFs. Given that about half of mammalian leaders possess uAUGs (116), and non-AUG initiation also occurs, a sophisticated interplay between all stages of translation may occur for uORF containing mRNAs (see (61) for a recent review).
TCP-Seq: RiboSeq of scanning and initiating ribosomes
An important limitation of conventional RiboSeq is the absence of information regarding the intrinsic behavior of scanning ribosomes. To overcome this, Archer et al. developed TCP-Seq (117), a variation of RiboSeq to detect the footprints protected by 40S ribosome complexes in addition to 80S ribosomes. It is based on formaldehyde crosslinking followed by RNase treatment and isolation of 40S/48S and 80S crosslinked intermediates. TCP-Seq was applied to yeast and its results supported a model where multiple ribosomes can scan the 5′ leader simultaneously. It was shown that scanning ribosomes pause within 5′ leaders without the apparent involvement of secondary structures, and that initiating ribosomes have at least three conformations (protecting fragments of 19, 29 and 37 nt). These footprints share the same 5′ end but differ at their 3′ ends, reflecting changes at the entry channel following start codon recognition. Furthermore, footprints from lingering 40S subunits at termination codons were also identified which may prove useful for identifying translation of overlapping ORFs. The development of TCP-Seq for mammalian cells could bring important insights into mammalian translation control, which is especially interesting as mammalian 5′ leaders are in general much longer than in yeast.
ELONGATION
Once initiation of translation is accomplished, the ribosome starts to synthesize protein. The rate of protein elongation in living cells has been estimated from experiments with reporter constructs, but until recently it was not possible to investigate it at a global level.
An elegant approach using RiboSeq was applied to calculate the average elongation rate in mammalian cells, where cells were pulse-treated with a drug to selectively stop new initiation followed by RiboSeq at several brief time intervals. This delay creates an area of low ribosome density immediately downstream of start codons whose length is proportional to the interval between the treatments. In this experiment, the mean elongation rate for mammalian cells was estimated to be 5.6 codons per second (8). This average speed was described to have little variance among all mRNAs irrespective of the length of their coding sequences and TE. Dana and Tuller, however, argued that the rate of elongation is slower at the beginning of coding regions, owing to differences in codon adaption to the tRNA pool (118,119).
Metagene profiles (an average profile from many transcripts) of many ribosome profiling studies show increased ribosome density at TIS and termination codons, and a consistent elevation of ribosome density in the first few hundred codons after which the density of ribosomes along the coding sequence is often uniform until the end of the acORF. However, under certain conditions this distribution can be altered. Indeed, the accumulation of ribosomes at around codon 65 on most mRNAs during severe heat stress was observed in both mouse and human cells (120). This was shown to be linked to chaperone (HSP70) function. Thus, the interaction of the chaperone machinery with the nascent peptide is an important factor that modulates the velocity of the ribosome along the mRNA. Similar findings were made for the translation response to proteotoxic stress induced by the amino acid analog l-azetidine-2-carboxylic acid (AZC) which competes with proline during amino acid incorporation (121). A case of global deviation from uniform ribosome distribution toward the end of the acORFs was observed in PC12 cells (interestingly—under normal conditions) (98). A metagene profile demonstrated a pattern of RPF close to the stop codon that most likely represents queuing of ribosomes. This queuing disappears upon OGD, which could occur if either the elongation rate reduces, or the rate of termination/recycling increases, or both (98).
The interaction of the signal recognition particle (SRP) with the nascent polypeptide as it emerges from the 60S ribosome subunit tunnel was speculated to cause pausing of translation. Pechmann et al. found that in vivo the co-translational recognition of the signal amino acid sequence (SS) or transmembrane domain (TMD) by the SRP is enhanced when the mRNA encoding the secretory polypeptide contains a cluster of non-optimal codons (∼35–40 codons) downstream of the SRP-binding site, i.e. at a distance sufficient to span the ribosomal exit tunnel (122). This led the authors to speculate that the moment of enhanced SRP interaction is controlled by a genetically programmed slowdown in elongation when the SS emerges from the ribosome and this slow-down is determined by a cluster of non-optimal or ‘slow’ codons. In contrast, a later RiboSeq study by the same group (123) suggests that in general elongation arrest is not associated with SRP binding. Instead, perhaps in order to avoid such arrest, SRPs are directed to ribosomes translating the mRNA even before translation of the SS/TMD encoding region in a large subset of the mRNAs recognized by the SRP. For some of the mRNAs this recognition was driven through signals contained in their mRNA 3′ trailers (123). The pausing of ribosomes however was still observed for mRNAs encoding non secretory proteins that are targeted by the SRP.
It is believed that for most mRNAs initiation is the rate limiting step of translation. While the advantage for codon bias is not obvious, it has been suggested, for example, that codon usage optimizes global, rather than local translation (124). Yet another possibility was recently provided by Presnyak et al. who showed that in yeast, the codon optimality index is a very strong predictor of mRNA stability, and that optimal codons in coding sequences are linked to faster decoding rates and vice versa (125). In the follow-up study, Radhakrishnan et al. provided a mechanistic insight into how elongation rates affect mRNA stability in yeast (126). This was linked to DEAD-box protein Dhh1p, which targets slowly translated mRNAs for decay. Strikingly, the effect of non-optimal codons on mRNA stability was shown to depend on their locations within acORF (the ‘polarity’ effect). In the context of reporter mRNAs, non-optimal codons show greater effects when placed closer to the acORF 5′ end. This was explained by the fact that more ribosomes accumulate upstream and thus recruit more Dhh1p, which triggers more efficient degradation. Therefore, prediction of mRNA half-life may be further improved by codon position-specific information.
Whatever the effect of local decoding rates on global translation might be, their alteration can influence cell fitness and phenotype. It was found that the loss of the anticodon wobble uridine (U34) modification (mcm5s2U) in a subset of tRNAs leads to ribosome pausing at cognate codons in Saccharomyces cerevisiae and Caenorhabditis elegans (127). Interestingly, upon loss of the U34 modification, decoding of only CAA and AAA triplets is affected (becomes slower), whereas other codons that are decoded by U34 bearing tRNAs are not affected. This anomaly elicits a widespread protein aggregation in vivo.
Availability of certain amino acids (and hence aminoacylated tRNAs) for protein synthesis may be limiting for certain tumors. Using Ribosome Profiling, Loayza-Punch et al. (128) demonstrated that proline levels are restrictive for certain kidney cancers because of insufficient levels of corresponding aminoacylated tRNAs. This is compensated with high levels of PYCR1, the key enzyme of the proline biosynthesis pathway, in these tumors. In support of this observation, knocking out PYCR1 impedes tumorigenic growth.
TERMINATION AND RECYCLING
Recent studies of translation termination and recycling in eukaryotes revealed that, as well as canonical release factors (eRFs), additional players are critically involved (111,129,130). Guydosh and Green (131) used RiboSeq to uncover the function of Dom34 in yeast. Dom34 (PELO and pelota are the human and fruit fly orthologues, respectively), is a distant homolog of release factor eRF1. Dom34 was shown to dissociate stalled ribosomes in vitro with participation of Hbs1 and Rli1 (132). However, its function in vivo was not clear. Comparison of RPFs from wild type cells with those from cells with Dom34 knocked out (dom34Δ) allowed the authors to discover increased ribosome occupancy at one particular position within the HAC1 mRNA (yeast orthologue of mammalian XBP1). HAC1/XBP1 mRNA is known to be subject to an unusual event - splicing in the cytoplasm that leads to the synthesis of a protein product encoded in an alternative frame relative to the unspliced variant (133,134). The observation of ribosomes stalled at the very 3′ end of the splicing intermediate in Dom34 knockout cells allowed the authors to conclude that Dom34 normally rescues such stalled ribosomes. Dom34 was found to be unable to rescue ribosomes if they pause within the coding sequence enriched in proline residues or a stretch of histidines when cells are deprived of this amino acid. Another intriguing observation was the presence of ribosomes within the 3′ trailers of some mRNAs in the knockout strain. They accumulated mostly at the border of the 3′ trailers and the polyA tail and did not reveal any triplet periodicity. Strikingly, only a small percentage (11%) of mRNA species revealed such 3′ trailers bound ribosomes, but specific features common to these mRNAs were not found. Thus, it seems likely that Dom34 also participates in termination/recycling on certain intact mRNAs.
These studies were substantially developed in research devoted to Rli1 (ABCE1 in mammals) (135). Rli1 is presumed to be the principal recycling factor in yeast cells. RiboSeq showed that in Rli1-depleted cells, 80S ribosomes accumulate at stop codons and within the 3′ trailers of all yeast genes and to a much greater extent than in dom34Δ cells. These ribosomes are very likely engaged in translation, as dozens of endogenous peptides potentially encoded within the 3′ trailers were detected. The authors presented compelling evidence that the ribosome density in 3′ trailers originates from reinitiation events rather than from readthrough of the acORF stop codons where RPFs would not be expected 3′ of the next in-frame stop codon.
The phenomenon of ribosome travelling along 3′ trailers remains mysterious. Miettinen and Björklund (136) showed that nucleases selected for RiboSeq affect the quantitative analysis of translatability of mRNAs. RPFs at the beginning of acORF were more abundant in RNAse I digests, than in micrococcal nuclease (MN) samples. Also curiously, digestion with MN yielded high amounts of RPFs localizing in 3′ trailers. RPFs derived from 3′ trailers have a different mean length in comparison to ‘normal’ RPFs suggesting that if these footprints derive from ribosomes, they may traverse 3′ trailers in a different conformation.
A well-known phenomenon that could explain the presence of some of these ribosome footprints in 3′ trailers is stop codon readthrough, where tRNAs are incorporated at stop codons. C-terminal extensions of proteins produced by readthrough may alter protein function (137) and localization (138). Instances of this rare recoding phenomenon have been observed in many organisms (see (139) for a recent review on recoding) including humans (140). However, this phenomenon was shown to be more frequent in Drosophila (10) and Anopheles mosquitoes (141). Extensive readthrough was revealed which was consistent with earlier predictions based on phylogenetic analysis of fruit fly genomes (142). RiboSeq, applied to Drosophila embryos and a macrophage-like lineage S2 cell line, revealed significant variations in readthrough efficiencies for the same mRNAs, which suggests that readthrough may be controlled in a cell type and development stage specific manner. The cause for the cell type-specific and mRNA specific readthrough is not known.
Stress dependent alterations in readthrough efficiencies were observed for mammalian cells upon OGD (98). For 19 mRNAs, relatively high RPF densities were observed downstream of acORF stop codons under normal conditions, and the triplet periodicity of the signal strongly suggests that ribosomes do translate these regions in the same reading frame. In all of these cases, the readthrough stop codon is UGA, known as the most prone to readthrough (138,140,143). Upon OGD, the efficiency of readthrough significantly decreased as early as 20 min. Although the cause of readthough is not clear, it may be speculated that it may relate to hydroxylation of either termination factors or the ribosomal proteins (144,145) which should be reduced in the absence of oxygen.
In some yeast strains, termination factor eRF3 is known to form prion aggregates and this phenotype is described as [PSI+]. Baudin-Baillieu et al. took advantage of this and carried out comparative ribosome profiling of [PSI+] and [PSI−] strains to explore the role of eRF3 (146). Readthrough in about 100 genes was identified in [PSI+] cells. Curiously, [PSI+] was also found to affect the accuracy of triplet decoding. Thus, the presence of this conformational form of termination factor eRF3 may have a broader impact on protein synthesis than just an effect on translation termination, however, this also could be due to secondary effects of deregulated protein synthesis.
Variations in genetic codes often arises as a result of stop-codon reassignment (139). Two independent studies (147,148) uncovered strikingly unusual decoding in the ciliate Condylostoma magnum, where all stop-codons are decoded as amino acids, except when they are in close proximity to polyA tails. Swart et al. (148) carried out ribosome profiling in C. magnum and experimentally confirmed that 3′ end dependent termination is indeed the case. Lobanov et al. used ribosome profiling to explore global translation in the ciliate Euplotes where numerous instances of ribosomal frameshifting have been reported (149,150). It appears that, like C. magnum, termination of translation takes place only on stop codons in close proximity to polyA tails, while internal stop codons trigger either +1 or +2 ribosomal frameshifting depending on the preceding codon (151). Most probably, the proximity of the polyA tail is a prerequisite for termination in these instances. It was speculated that the polyA binding protein (PABP) bound at the polyA tail is needed for the termination and when the stop codons are distant from polyA tails they function as sense codons or frameshifting triggers.
At least some ribosomes that successfully accomplish polypeptide synthesis are likely to be engaged in subsequent initiation on the same mRNA. Indeed, growing evidence supports the mRNA closed-loop model which was suggested a long time ago (152,153). It is now known that the interaction between eIF4G, bound to m7G cap via eIF4E, and PABP, bound to the polyA tail of mRNA, is responsible for the convergence of both mRNA termini. Intuitively, one feature which may account for the efficiency of closed-loop formation is the length of a particular mRNA. A recent RiboSeq study by Thompson et al. (154) provides evidence that the eukaryote-specific 40S ribosomal protein Asc1/RACK1 is required for efficient translation of mRNAs with short ORFs in S. cerevisiae. The effect of Asc1/RACK on length-dependent translation was reproduced with reporter constructs. This specificity to short mRNAs may be explained by the hypothesis that efficiency of short mRNA translation is more dependent on closed-loop complex formation. Accordingly, depletion of eIF4G mimics the translational defects of ASC1 mutants - where a similar dependence on mRNA length was revealed. How Asc1/RACK1 modulates translation mechanistically remains unclear.
RIBOSEQ ARTIFACTS
Notwithstanding its notable successes, it has become increasingly clear that RiboSeq data does not provide an unbiased representation of translation. Perhaps the most well-known reason for this is the sequence bias introduced during ribosome footprint library generation and its conversion to cDNA and subsequent sequencing. These procedures involve a number of reactions using enzymes with nucleotide sequence specificity. A recent survey of publicly available RiboSeq datasets revealed that in some of the datasets this sequencing bias has a greater influence on the distribution of reads across mRNAs than the identity of codons in the decoding center of the ribosome (35). Certain improvements have been made recently to reduce these biases with the optimization of protocol steps and use of enzymes with reduced sequence specificity.
A requirement of ribosome footprint generation is the use of RNases to digest the unprotected mRNA, which may degrade rRNA to different extents and display sequence specificity. A systematic study of digestion with various nucleases (I, A, S7 and T1) across different species found that T1 best preserved ribosome integrity while thoroughly digesting polysomes to monosomes (155). The weak pairwise correlations of the ribosome density profiles at individual transcripts obtained using four different RNases suggests that this could be a major distorting factor in RiboSeq data.
The pretreatment of cells with antibiotics,which is used to stall the ribosomes prior to cell lysis, generates another important artifact. Notably, due to the differences in experimental protocols, drug pretreatment times in different studies range from1 min to as long as 30 min. Gerashchenko and Gladlyshev reported that cycloheximide pretreatment was a major contributing factor to the observed increased read density at the start of coding sequences (156).This is likely an artifact due to cycloheximide inhibiting elongating ribosomes, but not initiating ones. An accumulation of ribosomes shortly downstream of initiation sites due to this phenomenon is therefore expected (Figure 2A). However, the increased density is not wholly caused by this artifact as it has also been observed in datasets produced without cycloheximide pretreatment (157). This increase of footprint density was earlier ascribed as evidence of slower elongation rates at these regions where codon bias was found to be different from other parts of coding regions (118).
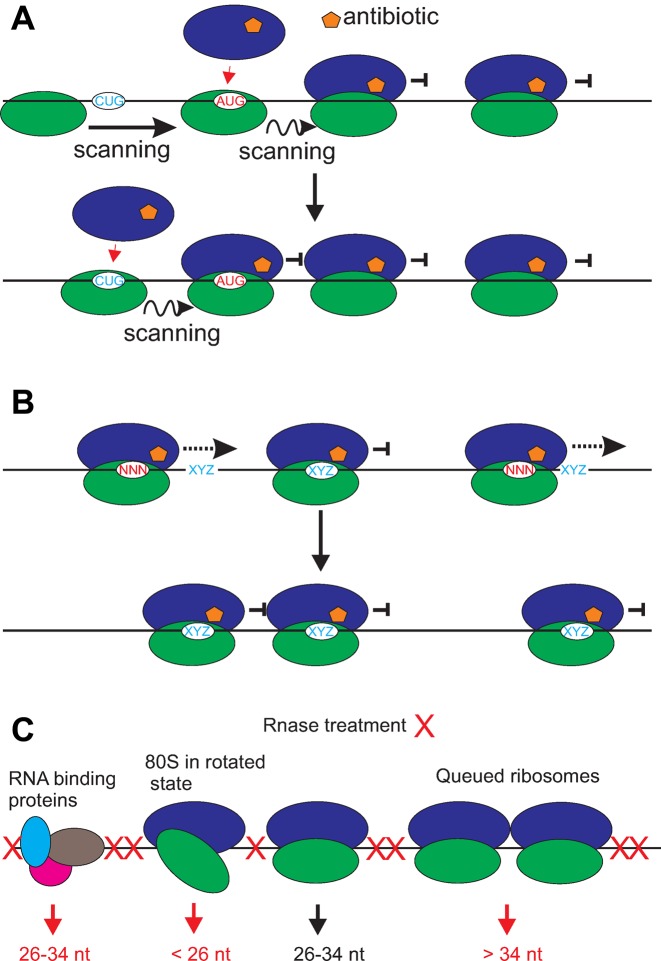
Figure 2.
Examples of some known and potential artifacts associated with ribosome profiling. (A) Antibiotic pretreatments may cause increased initiation on upstream translation initiation sites (TISs) because arrested elongating ribosomes prevent scanning. (B) Antibiotics may not block elongation completely (on NNN codon in the A-site) but allow slow elongation (until preferred XYZ codon is in the A-site), which affects the estimation of local decoding rates. (C) Ribosome protected fragments shorter than 26 and longer than 34 nt may be excluded upon size selection. In contrast, mRNA protected by RNA binding proteins may be selected as RPFs.
Pretreatments with cycloheximide, as well as with elongation inhibitors that preferentially block ribosomes at initiation codons, i.e. lactimidomycin and harringtonine, may have even more serious consequences on the rate of initiation itself and even on identity of codons identified as translation initiators. The successful initiation at a particular codon requires eIF1 dissociation from PIC and the initiation rate at a potential start codon increases if the PIC is slowed down at this codon (60). In this case, the arrest of a translating ribosome shortly downstream of a potential start codon would result in pausing of the next scanning PIC and artificially increase the initiation rate at codons which are not normally recognized as starts. At present it is difficult to estimate the significance of this potential artifact, therefore it is advisable to be cautious when interpreting the initiation rate estimates and the initiation sites obtained from ribosome profiling data. This potential artifact may artificially overstate the amount of TISs and translated uORFs (Figure 2A).
Furthermore, different antibiotics have a preference for blocking ribosomes at specific codons (16,158). Hussmann et al. have shown that after treatment with cycloheximide ribosomes may continue elongation in a sequence dependent manner which they demonstrated is dependent on cycloheximide concentration (159). Consequently, the read density at codons of the decoding center may not be representative of codon decoding rates in cycloheximide treated samples (Figure 2B). Importantly, a high codon specificity is also observed for the ‘initiation inhibitor’ harringtonine—it appears that this antibiotic preferentially arrests ribosomes at Lys, Arg or Tyr codons (160). This may seriously impact its implication for mapping TISs.
Although such artifacts are undesirable they are generally considered to be relatively unimportant for differential expression analysis as they are thought to occur in both conditions. However, for some conditions this may not be the case. The local footprint density is a product of the local codon decoding time and initiation rate. In the steady state the latter is equal to the rate of protein synthesis and if local decoding rates do not change, changes in footprint density for the entire mRNA equates to changes of TE. However, this may not always be the case—consider an mRNA that is occupied with stalled ribosomes, while the footprint density at such an mRNA would be high, the rate of protein synthesis would be close to zero. This limitation of footprint density as a proxy for the rate of protein synthesis can be observed in real data. In the ciliates Euplotes, sites of ribosomal frameshifting are more frequent in lowly expressed genes. Frameshifting is slow and leads to ribosome pauses at the frameshifting sites. Consequently, TE calculated as the number of footprints divided by the number RNA-seq reads is higher for lowly expressed genes (151). Thus, it is important to note again that RiboSeq only provides a measurement of the number of elongation ribosomes on an mRNA but does not say anything about the elongation rate of ribosomes. Because of this using the TE metric as a measure of protein production per mRNA should be used with some caution.
The current RiboSeq workflow usually consists of the exclusive selection of reads of an expected length. Although this has significant advantages in avoiding additional rRNA contamination, such a restricted selection is likely to conceal part of a translation response (16) (Figure 2C). For example, it is known that a pair of stalled ribosomes protect longer mRNA fragments from nuclease digestion (5).Thus, it is possible that selection of only ∼30 nt fragments deplete the data from some footprints left by queued stalled ribosomes. The claim that Shine-Dalgarno sequences is a major determinant of ribosome speed in bacteria (161) was reported to be an artifact of biased selection for longer footprints (162) since Shine-Dalgarno interactions with the ribosome lead to longer footprints (17). Although no such variability has been found in eukaryotes yet, this case signals for caution. In a similar vein, the enrichment of mRNA-seq transcripts by the selection of polyA tails may conceal part of the transcriptional data. mRNAs with no or short polyA tails may be underrepresented as well as those undergoing degradation. This may have significant implications, for example Presnyak et al. calculated the half-life of ADH1 to be 4.2 min and 31.7 min for polyA selected and rRNA depleted RNA, respectively (125). Thus, rRNA depletion as in (157) may be advantageous over polyA mRNA selection as it may provide a more accurate TE measure.
In a few studies, RNase treatment was applied to polysomal, rather than total ribosomal fractions (89,121,163).This protocol modification is presumably based on the widely-held belief that 80S ribosomes found in monosome fractions are not engaged in translation. However, a recent report from Heyer and Moore (164) found that in S. cerevisiae some 80S ribosomes in monosome fractions are engaged in translation. They were found mostly at uORFs and short acORFs. Importantly, mRNAs encoding low abundance regulatory proteins tend to be enriched in the monosome fraction. Therefore, the exclusion of the monosome fraction may deplete the data of footprints derived from those mRNAs being translated by single 80S.
Finally, not all RNA fragments identified during ribosome profiling may arise due to genuine translation. They may possibly occur because of other heavy RNPs, in which the RNA is protected from ribonucleases (Figure 2C). One signature of translation that can be used for discriminating genuine translation from such artifacts is triplet periodicity, however, triplet periodicity might be distorted when overlapping ORFs are translated simultaneously. Ingolia et al. (165) suggested a different approach—analysis of read length distribution. The small number of mRNAs with abnormal read length distribution and the large fraction of reads aligning to acORF suggests the extent of this artifact appears to be relatively low but it may be a significant cause of the reads aligning to RNAs annotated as non-coding (165).
PERSPECTIVES
Ribosome profiling has opened up a novel way for exploring global protein synthesis in vivo at great detail. Despite experimental difficulties, technical artifacts and discrepancies between the studies, the technique has demonstrated its utility and is certain to stay in the arsenal of omics tools. RiboSeq has undergone substantial improvements since its development but it is clearly not yet mature (35). Further optimization to minimize artifacts and starting material (ideally—to single cell level) is readily achievable. RiboSeq has been instrumental at addressing the mechanistic aspects of mRNA translation and it will continue to be used for this purpose. For example, we expect that ribosome profiling will be fruitful in combination with recently developed genome editing techniques that enables modification of known and putative translation factors to explore their roles in translation. RiboSeq is a powerful approach for studying genome wide effects of drug treatments. We expect therefore that it will be routinely used for exploring drug effects on gene expression, especially those that target translation. Finally, RiboSeq may be significantly enhanced by a combination with other omics approaches such as RNA-protein crosslinking which allows mapping of mRNA binding proteins to the transcriptome (166). The combination of the two may enable delineation of RNA binding proteins in translational regulation.
FUNDING
Russian Science Foundation 14-14-00127 to D.E.A., RBFR 14-04-00412 to I.N.S.; Science Foundation Ireland [12/IA/1335 to P.V.B.]. Funding for open access charge: Russian Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Castles J.J., Singer M.F. Degradation of polyuridylic acid by ribonuclease II: protection by ribosomes. J. Mol. Biol. 1969;40:1–17. doi: 10.1016/0022-2836(69)90292-7. [DOI] [PubMed] [Google Scholar]
- 2.Lazarowitz S.G., Robertson H.D. Initiator regions from the small size class of reovirus messenger RNA protected by rabbit reticulocyte ribosomes. J Biol Chem. 1977;252:7842–7849. [PubMed] [Google Scholar]
- 3.Kozak M. Nucleotide sequences of 5′-terminal ribosome-protected initiation regions from two reovirus messages. Nature. 1977;269:391–394. doi: 10.1038/269390a0. [DOI] [PubMed] [Google Scholar]
- 4.Steitz J.A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969;224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- 5.Wolin S.L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini A.A., Lee M.T., Giraldez A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadler M., Fire A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011;17:2063–2073. doi: 10.1261/rna.02890211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M.J., Wu S.H., Wu J.F., Lin W.D., Wu Y.C., Tsai T.Y., Tsai H.L., Wu S.H. Translational landscape of photomorphogenic Arabidopsis. Plant cell. 2013;25:3699–3710. doi: 10.1105/tpc.113.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caro F., Ahyong V., Betegon M., DeRisi J.L. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife. 2014;3 doi: 10.7554/eLife.04106. doi:10.7554/eLife.04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G., et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern-Ginossar N., Weisburd B., Michalski A., Le V.T., Hein M.Y., Huang S.X., Ma M., Shen B., Qian S.B., Hengel H., et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lareau L.F., Hite D.H., Hogan G.J., Brown P.O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor P.B., Li G.W., Weissman J.S., Atkins J.F., Baranov P.V. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013;29:1488–1491. doi: 10.1093/bioinformatics/btt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooijers K., Loayza-Puch F., Nijtmans L.G., Agami R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013;4:2886. doi: 10.1038/ncomms3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia N.T. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel A.M., Choudhury K.R., Firth A.E., Ingolia N.T., Atkins J.F., Baranov P.V. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid D.W., Shenolikar S., Nicchitta C.V. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods. 2015;91:69–74. doi: 10.1016/j.ymeth.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannes G., Carter M.S., Eisen M.B., Brown P.O., Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zong Q., Schummer M., Hood L., Morris D.R. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikulits W., Pradet-Balade B., Habermann B., Beug H., Garcia-Sanz J.A., Mullner E.W. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- 26.Jan C.H., Williams C.C., Weissman J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janich P., Arpat A.B., Castelo-Szekely V., Lopes M., Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25:1848–1859. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez C., Sims J.S., Hornstein N., Mela A., Garcia F., Lei L., Gass D.A., Amendolara B., Bruce J.N., Canoll P., et al. Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J. Neurosci. 2014;34:10924–10936. doi: 10.1523/JNEUROSCI.0084-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cenik C., Cenik E.S., Byeon G.W., Grubert F., Candille S.I., Spacek D., Alsallakh B., Tilgner H., Araya C.L., Tang H., et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015;25:1610–1621. doi: 10.1101/gr.193342.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel A.M., Ahern A.M., Donohue C.A., Baranov P.V. GWIPS-viz as a tool for exploring ribosome profiling evidence supporting the synthesis of alternative proteoforms. Proteomics. 2015;15:2410–2416. doi: 10.1002/pmic.201400603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel A.M., Fox G., A M.K., De Bo C., O'Connor P.B., Heaphy S.M., Mullan J.P., Donohue C.A., Higgins D.G., Baranov P.V. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014;42:D859–D864. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel A.M., Mullan J.P., Velayudhan V., O'Connor P.B., Donohue C.A., Baranov P.V. RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 2016;13:316–319. doi: 10.1080/15476286.2016.1141862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor P.B., Andreev D.E., Baranov P.V. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat. Commun. 2016;7:12915. doi: 10.1038/ncomms12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung B.Y., Hardcastle T.J., Jones J.D., Irigoyen N., Firth A.E., Baulcombe D.C., Brierley I. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA. 2015;21:1731–1745. doi: 10.1261/rna.052548.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legendre R., Baudin-Baillieu A., Hatin I., Namy O. RiboTools: a Galaxy toolbox for qualitative ribosome profiling analysis. Bioinformatics. 2015;31:2586–2588. doi: 10.1093/bioinformatics/btv174. [DOI] [PubMed] [Google Scholar]
- 38.Xie S.Q., Nie P., Wang Y., Wang H., Li H., Yang Z., Liu Y., Ren J., Xie Z. RPFdb: a database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res. 2016;44:D254–D258. doi: 10.1093/nar/gkv972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olexiouk V., Crappe J., Verbruggen S., Verhegen K., Martens L., Menschaert G. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016;44:D324–D329. doi: 10.1093/nar/gkv1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan J., Qian S.B. TISdb: a database for alternative translation initiation in mammalian cells. Nucleic Acids Res. 2014;42:D845–D850. doi: 10.1093/nar/gkt1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson O., Sonenberg N., Nadon R. anota: analysis of differential translation in genome-wide studies. Bioinformatics. 2011;27:1440–1441. doi: 10.1093/bioinformatics/btr146. [DOI] [PubMed] [Google Scholar]
- 42.Olshen A.B., Hsieh A.C., Stumpf C.R., Olshen R.A., Ruggero D., Taylor B.S. Assessing gene-level translational control from ribosome profiling. Bioinformatics. 2013;29:2995–3002. doi: 10.1093/bioinformatics/btt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Y., Karaletsos T., Drewe P., Sreedharan V., Kuo D., Singh K., Wendel H.G., Ratsch G. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics. 2016:btw585. doi: 10.1093/bioinformatics/btw585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Z., Zou Q., Liu Y., Yang X. Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 2016;7:11194. doi: 10.1038/ncomms11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 46.Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T., et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015;60:816–827. doi: 10.1016/j.molcel.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raj A., Wang S.H., Shim H., Harpak A., Li Y.I., Engelmann B., Stephens M., Gilad Y., Pritchard J.K. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife. 2016;5:e13328. doi: 10.7554/eLife.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H., McManus J., Kingsford C. Isoform-level ribosome occupancy estimation guided by transcript abundance with Ribomap. Bioinformatics. 2016;32:1880–1882. doi: 10.1093/bioinformatics/btw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji Z., Song R., Huang H., Regev A., Struhl K. Transcriptome-scale RNase-footprinting of RNA-protein complexes. Nat. Biotechnol. 2016;34:410–413. doi: 10.1038/nbt.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crappe J., Ndah E., Koch A., Steyaert S., Gawron D., De Keulenaer S., De Meester E., De Meyer T., Van Criekinge W., Van Damme P., et al. PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 2015;43:e29. doi: 10.1093/nar/gku1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popa A., Lebrigand K., Paquet A., Nottet N., Robbe-Sermesant K., Waldmann R., Barbry P. RiboProfiling: a bioconductor package for standard Ribo-seq pipeline processing. F1000Research. 2016;5:1309. doi: 10.12688/f1000research.8964.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mumtaz M.A., Couso J.P. Ribosomal profiling adds new coding sequences to the proteome. Biochem. Soc. Trans. 2015;43:1271–1276. doi: 10.1042/BST20150170. [DOI] [PubMed] [Google Scholar]
- 53.Bartholomaus A., Del Campo C., Ignatova Z. Mapping the non-standardized biases of ribosome profiling. Biol. Chem. 2016;397:23–35. doi: 10.1515/hsz-2015-0197. [DOI] [PubMed] [Google Scholar]
- 54.Brar G.A., Weissman J.S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015;16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern-Ginossar N. Decoding viral infection by ribosome profiling. J. Virol. 2015;89:6164–6166. doi: 10.1128/JVI.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 57.Kuersten S., Radek A., Vogel C., Penalva L.O. Translation regulation gets its ‘omics’ moment. Wiley interdiscipl. Rev. RNA. 2013;4:617–630. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michel A.M., Baranov P.V. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley interdiscipl. Rev. RNA. 2013;4:473–490. doi: 10.1002/wrna.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson R., Standart N. The awesome power of ribosome profiling. RNA. 2015;21:652–654. doi: 10.1261/rna.049908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 61.Hinnebusch A.G., Ivanov I.P., Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fonseca B.D., Smith E.M., Yelle N., Alain T., Bushell M., Pause A. The ever-evolving role of mTOR in translation. Semin. Cell Dev. Biol. 2014;36:102–112. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 63.Thoreen C.C. Many roads from mTOR to eIF4F. Biochem. Soc. Trans. 2013;41:913–916. doi: 10.1042/BST20130082. [DOI] [PubMed] [Google Scholar]
- 64.Cencic R., Carrier M., Galicia-Vazquez G., Bordeleau M.E., Sukarieh R., Bourdeau A., Brem B., Teodoro J.G., Greger H., Tremblay M.L., et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Proud C.G. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 66.Shatsky I.N., Dmitriev S.E., Andreev D.E., Terenin I.M. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit. Rev. Biochem. Mol. Biol. 2014;49:164–177. doi: 10.3109/10409238.2014.887051. [DOI] [PubMed] [Google Scholar]
- 67.Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J., et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi J., Chen J., Schreiber S.L., Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 70.Perry R.P., Meyuhas O. Translational control of ribosomal protein production in mammalian cells. Enzyme. 1990;44:83–92. doi: 10.1159/000468749. [DOI] [PubMed] [Google Scholar]
- 71.Mayer C., Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 72.Kantidakis T., Ramsbottom B.A., Birch J.L., Dowding S.N., White R.J. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsang C.K., Liu H., Zheng X.F. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gandin V., Masvidal L., Hulea L., Gravel S.P., Cargnello M., McLaughlan S., Cai Y., Balanathan P., Morita M., Rajakumar A., et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26:636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elfakess R., Sinvani H., Haimov O., Svitkin Y., Sonenberg N., Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39:7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sinvani H., Haimov O., Svitkin Y., Sonenberg N., Tamarkin-Ben-Harush A., Viollet B., Dikstein R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015;21:479–492. doi: 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 77.Shuda M., Velasquez C., Cheng E., Cordek D.G., Kwun H.J., Chang Y., Moore P.S. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5875–5882. doi: 10.1073/pnas.1505787112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Topisirovic I., Borden K.L. Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol. Histopathol. 2005;20:1275–1284. doi: 10.14670/HH-20.1275. [DOI] [PubMed] [Google Scholar]
- 79.Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W.C., Sonenberg N. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 80.Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L., Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J., et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iwasaki S., Floor S.N., Ingolia N.T. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Popa A., Lebrigand K., Barbry P., Waldmann R. Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics. 2016;17:52. doi: 10.1186/s12864-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 85.Goke A., Goke R., Knolle A., Trusheim H., Schmidt H., Wilmen A., Carmody R., Goke B., Chen Y.H. DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem. Biophys. Res. Commun. 2002;297:78–82. doi: 10.1016/s0006-291x(02)02129-0. [DOI] [PubMed] [Google Scholar]
- 86.Yang H.S., Jansen A.P., Komar A.A., Zheng X., Merrick W.C., Costes S., Lockett S.J., Sonenberg N., Colburn N.H. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sen N.D., Zhou F., Ingolia N.T., Hinnebusch A.G. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015;25:1196–1205. doi: 10.1101/gr.191601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sen N.D., Zhou F., Harris M.S., Ingolia N.T., Hinnebusch A.G. eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc. Natl. Acad. Sci. U.S.A. 2016;113:10464–10472. doi: 10.1073/pnas.1612398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee S., Liu B., Lee S., Huang S.X., Shen B., Qian S.B. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J., et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Michel A.M., Andreev D.E., Baranov P.V. Computational approach for calculating the probability of eukaryotic translation initiation from ribo-seq data that takes into account leaky scanning. BMC Bioinformatics. 2014;15:380. doi: 10.1186/s12859-014-0380-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014;10:748. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 95.Ivanov I.P., Firth A.E., Michel A.M., Atkins J.F., Baranov P.V. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stumpf C.R., Moreno M.V., Olshen A.B., Taylor B.S., Ruggero D. The translational landscape of the mammalian cell cycle. Mol. Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andreev D.E., O'Connor P.B., Zhdanov A.V., Dmitriev R.I., Shatsky I.N., Papkovsky D.B., Baranov P.V. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 2015;16:90. doi: 10.1186/s13059-015-0651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Homma M.K., Wada I., Suzuki T., Yamaki J., Krebs E.G., Homma Y. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15688–15693. doi: 10.1073/pnas.0506791102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zach L., Braunstein I., Stanhill A. Stress-induced start codon fidelity regulates arsenite-inducible regulatory particle-associated protein (AIRAP) translation. J. Biol. Chem. 2014;289:20706–20716. doi: 10.1074/jbc.M114.547828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Majumdar R., Bandyopadhyay A., Deng H., Maitra U. Phosphorylation of mammalian translation initiation factor 5 (eIF5) in vitro and in vivo. Nucleic Acids Res. 2002;30:1154–1162. doi: 10.1093/nar/30.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc. Natl. Acad. Sci. U.S.A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012;40:2898–2906. doi: 10.1093/nar/gkr1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baird T.D., Wek R.C. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andreev D.E., O'Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sidrauski C., McGeachy A.M., Ingolia N.T., Walter P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife. 2015;4 doi: 10.7554/eLife.05033. doi:10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B.R., Li H., Gamache K., Gallagher C.M., Ang K.K., Wilson C., et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sekine Y., Zyryanova A., Crespillo-Casado A., Fischer P.M., Harding H.P., Ron D. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science. 2015;348:1027–1030. doi: 10.1126/science.aaa6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palam L.R., Baird T.D., Wek R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chew G.L., Pauli A., Schier A.F. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 2016;7:11663. doi: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jackson R.J., Hellen C.U., Pestova T.V. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 112.Terenin I.M., Akulich K.A., Andreev D.E., Polyanskaya S.A., Shatsky I.N., Dmitriev S.E. Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis. Nucleic Acids Res. 2016;44:1882–1893. doi: 10.1093/nar/gkv1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ruan H., Shantz L.M., Pegg A.E., Morris D.R. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J. Biol. Chem. 1996;271:29576–29582. doi: 10.1074/jbc.271.47.29576. [DOI] [PubMed] [Google Scholar]
- 114.Ivanov I.P., Loughran G., Atkins J.F. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nikonorova I.A., Kornakov N.V., Dmitriev S.E., Vassilenko K.S., Ryazanov A.G. Identification of a Mg2+-sensitive ORF in the 5′-leader of TRPM7 magnesium channel mRNA. Nucleic Acids Res. 2014;42:12779–12788. doi: 10.1093/nar/gku951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kochetov A.V., Sarai A., Rogozin I.B., Shumny V.K., Kolchanov N.A. The role of alternative translation start sites in the generation of human protein diversity. Mol. Genet. Genomics. 2005;273:491–496. doi: 10.1007/s00438-005-1152-7. [DOI] [PubMed] [Google Scholar]
- 117.Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- 118.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 119.Dana A., Tuller T. Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput. Biol. 2012;8:e1002755. doi: 10.1371/journal.pcbi.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu B., Han Y., Qian S.B. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pechmann S., Chartron J.W., Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014;21:1100–1105. doi: 10.1038/nsmb.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chartron J.W., Hunt K.C., Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Radhakrishnan A., Chen Y.H., Martin S., Alhusaini N., Green R., Coller J. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016;167:122–132. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Loayza-Puch F., Rooijers K., Buil L.C., Zijlstra J., Oude Vrielink J.F., Lopes R., Ugalde A.P., van Breugel P., Hofland I., Wesseling J., et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- 129.Pisarev A.V., Hellen C.U., Pestova T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nurenberg E., Tampe R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem. Sci. 2013;38:64–74. doi: 10.1016/j.tibs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 131.Guydosh N.R., Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pisareva V.P., Skabkin M.A., Hellen C.U., Pestova T.V., Pisarev A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 134.Sidrauski C., Cox J.S., Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 135.Young D.J., Guydosh N.R., Zhang F., Hinnebusch A.G., Green R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miettinen T.P., Bjorklund M. Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions. Nucleic Acids Res. 2015;43:1019–1034. doi: 10.1093/nar/gku1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eswarappa S.M., Potdar A.A., Koch W.J., Fan Y., Vasu K., Lindner D., Willard B., Graham L.M., DiCorleto P.E., Fox P.L. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014;157:1605–1618. doi: 10.1016/j.cell.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stiebler A.C., Freitag J., Schink K.O., Stehlik T., Tillmann B.A., Ast J., Bolker M. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLoS Genet. 2014;10:e1004685. doi: 10.1371/journal.pgen.1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Baranov P.V., Atkins J.F., Yordanova M.M. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet. 2015;16:517–529. doi: 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- 140.Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014;42:8928–8938. doi: 10.1093/nar/gku608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jungreis I., Chan C.S., Waterhouse R.M., Fields G., Lin M.F., Kellis M. Evolutionary dynamics of abundant stop codon readthrough. Mol. Biol. Evol. 2016;33:3108–3132. doi: 10.1093/molbev/msw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jungreis I., Lin M.F., Spokony R., Chan C.S., Negre N., Victorsen A., White K.P., Kellis M. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gartner J., Thoms S. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014;3:e03640. doi: 10.7554/eLife.03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Feng T., Yamamoto A., Wilkins S.E., Sokolova E., Yates L.A., Munzel M., Singh P., Hopkinson R.J., Fischer R., Cockman M.E., et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell. 2014;53:645–654. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Loenarz C., Sekirnik R., Thalhammer A., Ge W., Spivakovsky E., Mackeen M.M., McDonough M.A., Cockman M.E., Kessler B.M., Ratcliffe P.J., et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baudin-Baillieu A., Legendre R., Kuchly C., Hatin I., Demais S., Mestdagh C., Gautheret D., Namy O. Genome-wide translational changes induced by the prion [PSI+] Cell Rep. 2014;8:439–448. doi: 10.1016/j.celrep.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 147.Heaphy S.M., Mariotti M., Gladyshev V.N., Atkins J.F., Baranov P.V. Novel Ciliate Genetic Code Variants Including the Reassignment of All Three Stop Codons to Sense Codons in Condylostoma magnum. Mol. Biol. Evol. 2016;33:2885–2889. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Swart E.C., Serra V., Petroni G., Nowacki M. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang R., Xiong J., Wang W., Miao W., Liang A. High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Scientific Rep. 2016;6:21139. doi: 10.1038/srep21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang R., Zhang Z., Du J., Fu Y., Liang A. Large-scale mass spectrometry-based analysis of Euplotes octocarinatus supports the high frequency of +1 programmed ribosomal frameshift. Scientific Rep. 2016;6:33020. doi: 10.1038/srep33020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lobanov A.V., Heaphy S.M., Turanov A.A., Gerashchenko M.V., Pucciarelli S., Devaraj R.R., Xie F., Petyuk V.A., Smith R.D., Klobutcher L.A., et al. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat. Struct. Mol. Biol. 2016 doi: 10.1038/nsmb.3330. doi:10.1038/nsmb.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 153.Tarun S.Z., Jr, Sachs A.B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 154.Thompson M.K., Rojas-Duran M.F., Gangaramani P., Gilbert W.V. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife. 2016;5:e11154. doi: 10.7554/eLife.11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gerashchenko M.V., Gladyshev V.N. Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw822. doi:10.1093/nar/gkw822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gerashchenko M.V., Gladyshev V.N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016;14:1787–1799. doi: 10.1016/j.celrep.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Requiao R.D., de Souza H.J., Rossetto S., Domitrovic T., Palhano F.L. Increased ribosome density associated to positively charged residues is evident in ribosome profiling experiments performed in the absence of translation inhibitors. RNA Biol. 2016;13:561–568. doi: 10.1080/15476286.2016.1172755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hussmann J.A., Patchett S., Johnson A., Sawyer S., Press W.H. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dmitriev S., Akulich K., Andreev D., Terenin I., Shatsky I. The peculiar mode of translation elongation inhibition by antitumor drug harringtonin. FEBS J. 2013;280:51–51. [Google Scholar]
- 161.Li G.W., Oh E., Weissman J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mohammad F., Woolstenhulme C.J., Green R., Buskirk A.R. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016;14:686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Heyer E.E., Moore M.J. Redefining the translational status of 80S monosomes. Cell. 2016;164:757–769. doi: 10.1016/j.cell.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 165.Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sauliere J., Le Hir H. CLIP-seq to discover transcriptome-wide imprinting of RNA binding proteins in living cells. Methods Mol. Biol. 2015;1296:151–160. doi: 10.1007/978-1-4939-2547-6_14. [DOI] [PubMed] [Google Scholar]