Abstract
Canonical growth factors act indirectly via receptor-mediated signal transduction pathways. Here, we report on an autonomous pathway in which a growth factor is internalized, has its localization regulated by phosphorylation, and ultimately uses intrinsic catalytic activity to effect epigenetic change. Angiogenin (ANG), a secreted vertebrate ribonuclease, is known to promote cell proliferation, leading to neovascularization as well as neuroprotection in mammals. Upon entering cells, ANG encounters the cytosolic ribonuclease inhibitor protein, which binds with femtomolar affinity. We find that protein kinase C and cyclin-dependent kinase phosphorylate ANG, enabling ANG to evade its inhibitor and enter the nucleus. After migrating to the nucleolus, ANG cleaves promoter-associated RNA, which prevents the recruitment of the nucleolar remodeling complex to the ribosomal DNA promoter. The ensuing derepression of rDNA transcription promotes cell proliferation. The biochemical basis for this unprecedented mechanism of signal transduction suggests new modalities for the treatment of cancers and neurological disorders.
INTRODUCTION
Angiogenesis, the process of establishing new blood vessels from pre-existing vasculatures, is essential for the growth and development of mammals. Over forty years ago, Folkman postulated that tumors orchestrate the composition of new blood vessels to nourish their growth (1,2). A few years later, Vallee and coworkers isolated a small protein from the conditioned medium of human adenocarcinoma cells and found that this protein, which he named ‘angiogenin’ (ANG), promotes neovascularization (3). This discovery was lauded widely as the first of a ‘substance that initiates the growth of any human organ’ (4). Subsequently, ANG levels in the serum of cancer patients were found to correlate with the progression of their tumors (5,6).
In addition to promoting neovascularization, ANG is neuroprotective (7,8). Moreover, ANG mutations are common in amyotrophic lateral sclerosis (ALS) patients (9,10). In accord, administration of ANG to an ALS mouse model improves motor functions and extends lifespan (11).
Despite its physiological and historical significance, the cellular mechanism by which ANG promotes cell proliferation is unclear. Remarkably, ANG belongs to the pancreatic-type ribonuclease (RNase) superfamily, exemplified by RNase A (12). These secretory proteins catalyze the cleavage of a phosphodiester bond on the 3′ side of cytidine or uridine residues in single-stranded RNA. ANG is the only RNase with angiogenic activity, and the only angiogenic factor with ribonucleolytic activity (13). Even though ANG shares 33% sequence identity with RNase A, including a conserved active-site triad (His14, Lys40, and His114), its ribonucleolytic activity toward di- and tetraribonucleotide substrates is 106-fold less than that of RNase A (14,15). Still, this low ribonucleolytic activity is essential for the promotion of neovascularization (16). The crystal structure of ANG revealed that Gln117 blocks the pyrimidine-binding pocket in the active site (17). A Q117G substitution increases the catalytic activity of ANG toward conventional substrates by 30-fold (18). Still, the Q117G substitution is not sufficient to endow ANG with catalytic activity comparable to that of RNase A, suggesting that ANG might have evolved to cleave a particular cellular RNA. The identity of that RNA substrate is unknown.
Secreted ANG enters cells via receptor-mediated endocytosis and ultimately accumulates in the nucleolus. There, ANG elicits angiogenesis from endothelial cells and exerts mitogenic effects on a variety cell types (5,19,20). To date, two cell-surface receptors have been identified for ANG: a putative 170-kDa protein and syndecan-4 (8,21). The 170-kDa protein was isolated with ANG-affinity chromatography, but its identity remains unclear. ANG is, however, known to interact with heparan sulfate moieties on syndecan-4, and treating cells with heparinase or adding exogenous heparin deters entry by ANG (22).
In the cytosol, ANG encounters an endogenous inhibitor protein. This protein, which is known as ‘ribonuclease inhibitor (RI)’, binds to ANG to form a complex with a Kd value in the low femtomolar range (23). Upregulating RI suppresses tumor growth and tumor microvessel density through suppression of ANG function (24). Conversely, ANG variants endowed with the ability to evade RI display enhanced angiogenic activity (25). Inside of cells, ANG exists in two pools: one is bound to RI in the cytosol and the other is unbound in the nucleus (26), where ANG manifests its angiogenic activity (19,27). How ANG, which is a secretory protein, evades cytosolic RI on its way to the nucleus is unknown.
ANG has a nuclear localization sequence (NLS), which equips the protein for nuclear import (27). After entering the nucleus, ANG accumulates in the nucleolus, which is the site of ribosome biogenesis. Within the nucleolus, ANG stimulates ribosomal DNA (rDNA) transcription (20,28). ANG-mediated rDNA transcription is essential for the angiogenesis induced by other proteins, including vascular endothelial growth factor and acidic/basic fibroblast growth factor (19). In cancer cells, ANG promotes the surplus production of rRNA, contributing to the uncontrolled growth and proliferation of cancer cells (20,29). How ANG manifests its ribonucleolytic activity to activate rDNA transcription is unclear.
Here, we reveal the biochemical basis for the biological activities of ANG. Specifically, we show that ANG catalyzes the cleavage of a particular phosphodiester bond in a nucleolar substrate, promoter-associated RNA (pRNA). That cleavage prevents the silencing of rDNA transcription by the nucleolar remodeling complex (NoRC). We also show that phosphorylation of key serine residues enables ANG to evade cytosolic RI and translocate to the nucleus. Thus, unlike canonical growth factors that deliver epigenetic information to DNA indirectly via receptor-mediated signal-transduction pathways, ANG is unique in conveying its proliferative signal from outside of the cell directly to a nucleic acid.
MATERIALS AND METHODS
General procedures
Production and purification of ANG and its variants (15), and of His6–TIP5510-611 (TAM) (30), immunofluorescence of TIP5 (human antibody from Abcam) (31), chromatin immunoprecipitation of the rDNA promoter (32) and RI-binding assays with variants of Q19C ANG (33,34) were performed as described previously.
Run-off transcription
A DNA template encoding the 97 bases of pRNA downstream from a T7 RNA polymerase promoter was obtained from Integrated DNA Technology (IDT). pRNA biosynthesis was accomplished with the AmpliScribe™ T7-Flash™ Transcription kit (Epicentre) in the presence of 3 μl of [α-32P]UTP. Transcribed pRNA was purified by excision from a urea polyacrylamide (8% w/v) gel. Modified pRNAs were made in a similar manner.
Gel-based assay of ribonucleolytic activity
In each reaction mixture, pRNA containing [α-32P]UTP (∼1000 cpm) was subjected at 37°C to either wild-type ANG (50 nM) or buffer alone (which was 50 mM Tris–HCl buffer, pH 7.5, containing 50 mM NaCl). At known times, a 10-μl aliquot was removed and quenched with 2 μl of 6× urea gel-loading dye. Samples were heated to 95°C and resolved by electrophoresis through a urea polyacrylamide (8% w/v) gel. The gel was dried and exposed to a phosphor screen for 24 h. Radiography of the screen was performed with a Typhoon LFA9000 phosphorimager (GE Healthcare Life Sciences).
Sequencing of pRNA fragments
Nonradioactive pRNA was made as described above. In a 40-μl reaction mixture, pRNA (1 μg) was treated with wild-type ANG (∼1 μM) for 10 min at 37°C. The reaction was quenched by the addition of proteinase K (Qiagen). Degraded pRNA fragments were resolved in a urea polyacrylamide (8% w/v) gel. The major pRNA fragment was purified as described above and then treated with alkaline phosphatase (New England Biolabs) to generate a 3′-hydroxyl terminus. A poly(A) tail was added to the 3′ end of the fragment by using poly(A) polymerase (New England Biolabs). A reverse transcription (RT) reaction was conducted using the RT primer listed in Supplementary Table S1. The ensuing cDNAs were amplified further by PCR and subjected to TOPO-cloning for Sanger sequencing with an M13 forward primer.
Cell culture
HeLa cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) containing fetal bovine serum (10% v/v) and penicillin/streptomycin (1% v/v) (Invitrogen) at 37°C under 5% (v/v) CO2(g). Human umbilical vein endothelial cells (HUVEC) were grown in EGM™-2 at 37°C under 5% (v/v) CO2(g). Cells (5 × 105) were plated in complete medium in 10-cm dishes. After 24 h, cells were washed with DMEM or endothelial basal medium-2 (EBM-2) and then incubated with wild-type ANG or a variant (to 1 μg/ml) for another 24 h (20). For kinase-inhibitor treatment, cells were pre-incubated with either bisindolylmaleimide I (4 μM) or roscovitine (14 μM) for 30 min prior to treatment with ANG. Cells were then harvested for the isolation of RNA or protein.
LNA-Antisense knockdown
HeLa cells were plated at 150 000 cells per well in a six-well plate. After 24 h, cells were transfected with 50 nM LNA–DNA gapmers (Sigma Chemical) in an antisense orientation (5′-CGTGtcgcgcgtcgcCTGG-3′, where locked nucleic acid (LNA) bases are in upper case and DNA bases are in lower case) using Lipofectamine® RNAiMAX (Thermo Scientific). After 48 h, the knockdown of pRNA was assessed by quantitative RT-PCR (qRT-PCR) as described below.
Quantification of cellular RNAs by qRT-PCR
Total cellular RNA was isolated by extraction with Trizol (Invitrogen). RNA was purified by phenol–chloroform extraction, followed by ethanol precipitation. RNA samples were treated with DNase I (Invitrogen) for 15 min at 37°C, and the enzymatic reaction was quenched by the addition of DNase-inactivation resin. RNA concentrations and purities were assessed with a NanoVue instrument (GE Healthcare Life Sciences). Purified cellular RNA (∼1 μg) was used in the reverse transcription reaction along with random hexamers from the SuperScript III Reverse Transcriptase kit (Invitrogen). A 1-μl solution of the ensuing cDNAs was used in qPCR reactions performed with the PerfeCTa SYBR Green FastMix cocktail (Quantabio). Amplified cDNAs were evaluated with an ABI Prism 7200 sequence detector (Perkin Elmer) using the primers listed in Supplementary Table S1. Data were analyzed by the comparative CT method (35) using the actin gene as an endogenous standard.
RNA immunoprecipitation
Cells were treated with FLAG–H114R ANG, and then washed with cold PBS. Cellular contents were then crosslinked by using 254-nm light at 1500 J/cm2. Cells were lysed in IP buffer (20 mM HEPES–KOH buffer, pH 7.5, containing 250 mM NaCl, 1 mM EDTA, 1% v/v NP-40, 10% v/v glycerol, and a protease-inhibitor cocktail) for 20 min at 4°C, and the cell lysate was subjected to centrifugation. The clarified lysate was treated with 500 pM RNase A and 2 μl of 2 units/μl DNase I (Invitrogen) for 10 min at 37°C, and then incubated with α-FLAG magnetic beads (Sigma Chemical). The beads were washed and then eluted three times with 150 ng/μl 3× FLAG peptide (APExBIO). The combined eluates were split into two portions: one for an immunoblot and the other for RT-PCR to identify co-precipitated RNAs. To determine its sequence, the PCR product was incorporated into a TOPO®TA vector (Invitrogen), which enables use of the M13 forward primer.
Immunoblots
Cells grown in a 10-cm dish were lysed with 1 ml of M-PER mammalian protein extraction reagent (Pierce) containing a protease-inhibitor cocktail. Protein (∼30 μg) was separated by SDS–PAGE (Bio-Rad), and the resulting gel was subjected to immunoblotting. Densitometry data were analyzed with ImageQuant TL software (GE Healthcare Life Sciences). For immunoprecipitation experiments, cells were lysed with IP buffer containing protease- and phosphatase-inhibitor cocktails. Clarified lysates were incubated with α-FLAG magnetic beads (Sigma Chemical), and the beads were washed three times. Samples were eluted with 30 μl of SDS gel-loading dye and processed further for immunoblotting.
Immunofluorescence
Prior to immunofluorescence experiments, 1 × 105 HeLa or HUVEC cells were plated in 0.2 ml of medium in an eight-well μ-chamber (Ibidi). After 24 h, cells were washed with serum-free medium (3 × 0.2 ml). FLAG–WT ANG and its variants (1 μg/ml) were added in serum-free medium, and the cells were incubated for 3 h. All subsequent steps were performed at room temperature and with PBS washes between steps. Cells were fixed with 4% (v/v) paraformaldehyde (Thermo Scientific) in PBS for 10 min and permeabilized using freshly prepared 0.1% (v/v) Triton X-100 (Sigma Chemical) in PBS for 10 min. Next, cells were incubated with blocking solution (Thermo Scientific) for 1 h, and then with mouse anti-FLAG monoclonal antibody (Sigma Chemical) for 1 h. After extensive washes, cells were incubated with goat anti-mouse Alexa Fluor 488 for 1 h, counterstaining with the nuclear probe Hoechst 33342 (Invitrogen) at 37°C during the final 5 min. Cells were imaged with an Eclipse TE2000-U laser scanning confocal microscope (Nikon) equipped with an AxioCam digital camera (Carl Zeiss).
Cell-proliferation assay
HUVEC cells in EGM™-2 were plated at 5000 cells per well in a 96-well microplate. After 24 h, cells were switched to EBM-2 containing ANG or a variant (1 μg/ml). At known times, growth medium was removed, and the cells were incubated with fluorescent dye CyQUANT® NF binding solution (Invitrogen). Fluorescence intensity was recorded on an M1000 fluorimeter (Tecan) with excitation at 485 nm and emission detection at 530 nm. Data were analyzed with Prism 5.0 software (GraphPad).
Tube-formation assay
HUVEC cells in EGM™-2 were plated at 125 000 cells per well in a six-well plate. After 24 h, cells were incubated with EBM-2 containing ANG or a variant (1 μg/ml) for another 24 h. On the same day, Matrigel was coated on a μ-Slide Angiogenesis (Ibidi) at 37°C for 1 h under 5% (v/v) CO2(g). Treated cells (1 × 104) were plated on the Matrigel, and the resulting slide was incubated at 37°C under 5% v/v CO2(g). After 2 h, phase-contrast images were acquired with an N-STORM Eclipse Ti-E inverted microscope (Nikon) using 10× magnification. For a z-stack series, images were constructed using NIS-Elements software. The junctions, tubules, and tubule lengths were quantified with the Angiogenesis Analyzer plugin for ImageJ software (NIH).
Gel-shift assay for protein·nucleic acid complexation
In each reaction mixture, pRNA (0.2 nM) labeled with [γ-32P]ATP was pre-incubated with His6–TIP5510-611 (5 μM) for 15 min before adding wild-type ANG or a variant (to 5 μM). At known times, a 7-μl aliquot was removed and quenched on ice by the addition of RI (to 5 μM). The samples were resolved by electrophoresis through a TBE polyacrylamide (4% w/v) gel. Radioactivity was detected with a phosphorimager as described above. Bound and unbound pRNA was quantified by densitometry with ImageQuant TL software, and values of Kd were determined with Prism 5.0 software.
In vitro assay of kinase activity
In each reaction mixture, wild-type ANG or a variant (20 μM) was incubated with a HeLa cell lysate (1 μg of total protein) containing protease- and phosphatase-inhibitor cocktails and [γ-32P]ATP. Reactions were allowed to proceed for 2 min at 30°C, and then quenched with SDS gel-loading dye. Samples were subjected to SDS–PAGE, and radioactivity was detected with a phosphorimager as described above.
RESULTS
ANG cleaves pRNA in vitro in a specific manner
Cell proliferation requires the continuous synthesis of ribosomes to support ongoing translation. The rate of ribosome biogenesis is limited by the transcription of DNA encoding rRNA (36). ANG is known to stimulate the transcription of rDNA in a manner that leads to endothelial cell proliferation and the induction of neovascularization (19,27,37).
The transcription of rDNA is regulated tightly (38,39). Importantly, rDNA transcription is silenced by NoRC, in which transcription termination factor I-interacting protein 5 (TIP5) is a large subunit and non-fermenting protein 2 homologue (SNF2h) is a small subunit (30,40). Upon the binding of NoRC to the rDNA promoter, the complex recruits histone deacetylase and DNA methyltransferase activity to remodel or maintain a repressive conformation of heterochromatin (32,41,42). The binding of NoRC to the rDNA promoter requires pRNA, which is a product of a long, processed non-coding RNA transcribed by RNA polymerase I from the intergenic space of rDNA. pRNA (∼200 nt) has a conserved stem–loop structure (97 nt) and a sequence that forms a triple helix with the rDNA promoter (32,43). TIP5 binds to pRNA via its TAM domain (44). Removing that domain abolishes the repression of rDNA transcription by NoRC, as does knocking-down or altering pRNA (31,43). Because pRNA-associated NoRC constrains rDNA transcription, we hypothesized that pRNA could be the key cellular target for the ribonucleolytic activity of ANG.
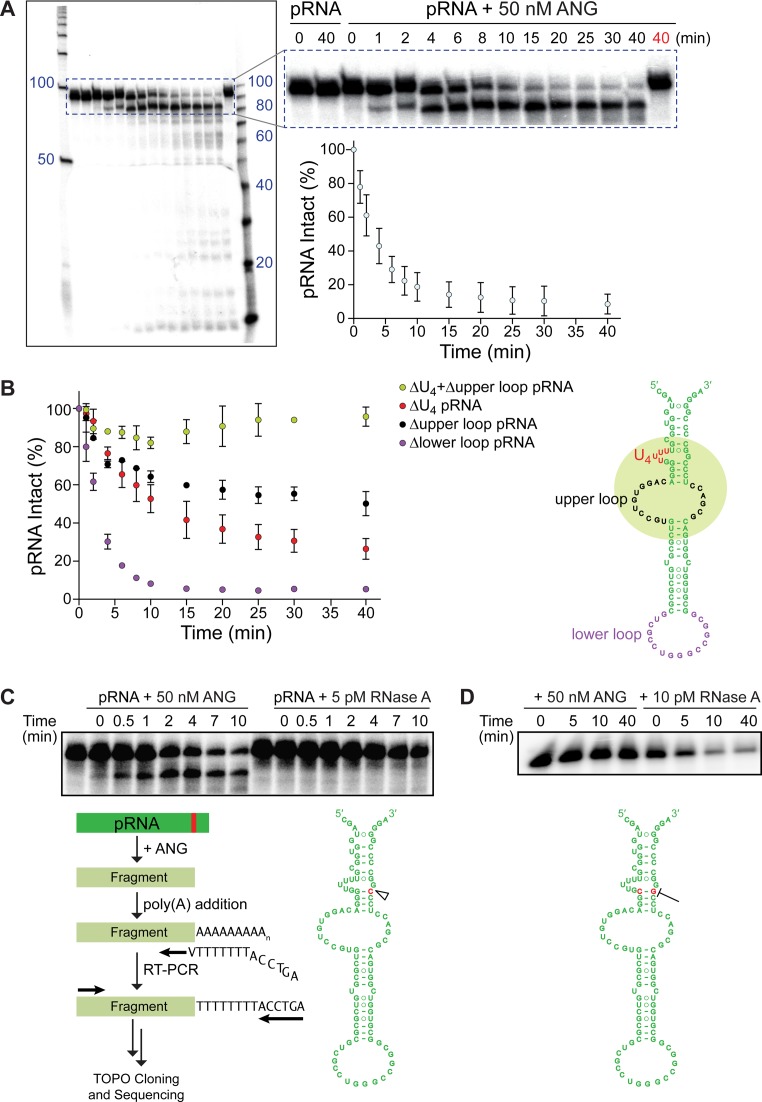
We examined pRNA as a substrate of ANG in vitro. To do so, we first incubated an internal 32P-labeled pRNA containing only the stem–loop structure (97 nt) with recombinant ANG and monitored its cleavage over time. Approximately 80% of the pRNA was cleaved after 10 min (Figure 1A). Next, we determined if any of the single-stranded RNA loops of pRNA are important for cleavage. Excision of the lower loop led to no change in cleavage (Figure 1B). In contrast, deleting a series of 4 uridine residues in pRNA decreased the ability of pRNA to serve as a substrate for ANG. This decrease was even greater upon removal of the upper loop. By examining different loops of pRNA, we determined that ANG relies primarily on the U4 region and the upper loop for the manifestation of its catalytic activity. Indeed, removing the putative binding region on pRNA obliterates catalysis by ANG (Figure 1B).
Figure 1.
ANG cleaves pRNA in vitro in a specific manner. (A) Autoradiogram of a urea–polyacrylamide gel and graph of ensuing densitometry data demonstrating that ANG cleaves 32P-labeled pRNA specifically and completely to generate a product of ∼85 nucleotides and, subsequently, smaller fragments. Heat-inactivated ANG was used at 40 min (red). (B) Graph showing that ANG cleavage of pRNA is affected by deletion of the U4 loop (red), the upper loop (black), and these two loops combined (green), but not the lower loop (purple). The green circle highlights the putative ANG-binding region on pRNA. Values represent the mean ± SD (n = 3, technical replicates). (C) Autoradiogram demonstrating that RNase A, unlike ANG, cleaves 32P-labeled pRNA non-specifically. Sequencing reveals that ANG cleaves pRNA specifically after residue C86 (red). (D) Autoradiogram showing that replacement of G18–C86 in 32P-labeled pRNA with C18–G86 eliminates cleavage by ANG but not RNase A.
We found that ANG cleaved pRNA at a specific site to give a product of ∼85 bases and, subsequently, smaller fragments. This specificity was distinct, as RNase A cleaved RNA without accumulating a single product (Figure 1C, Supplementary Figure S1). Importantly, the specificity of ANG was resistant to the excision of single-stranded RNA loops on pRNA (Supplementary Figure S1). To determine the cleavage site on pRNA by ANG, we isolated the ∼85-base product and determined its sequence. We found that a particular C–G phosphodiester bond near the 3′ end of pRNA is cleaved by ANG (Figure 1C). This finding is consistent with the known preference of ANG to cleave RNA between a pyrimidine and purine residue (15,45). Moreover, the ANG-cleavage site in pRNA could be conserved by evolution (Supplementary Figure S2).
We interrogated the putative C–G cleavage site in pRNA by mutagenesis. Based on the predicted secondary structure of pRNA, the cytidine residue participates in Watson–Crick base-pairs with a guanidine residue from the 5′ end. When we replaced the three G·C base pairs in pRNA with C·G, ANG no longer cleaved the substrate (Supplementary Figure S3). Further, a single C·G→G·C substitution at the putative cleavage site impaired ANG from degrading the substrate (Figure 1D). In contrast, RNase A degraded pRNA efficiently regardless of these modifications.
ANG degrades pRNA in cellulo
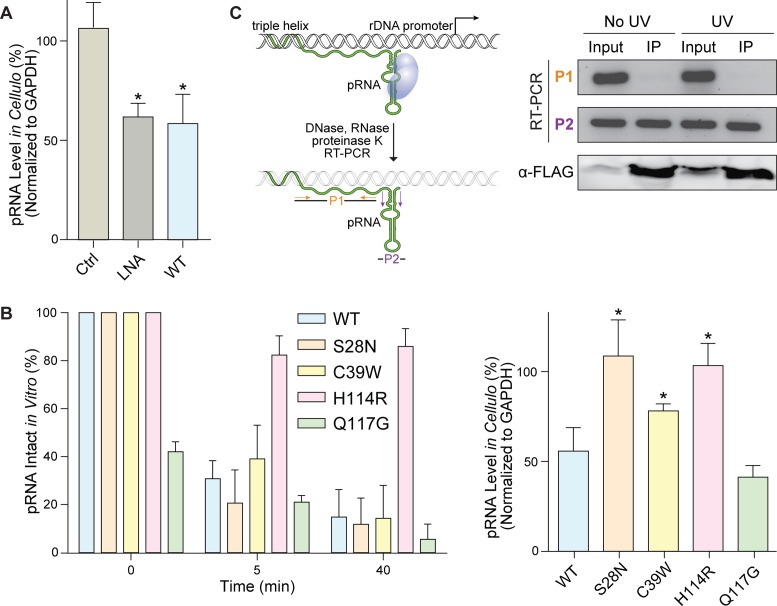
Having shown that pRNA is a substrate for ANG in vitro, we asked if ANG can access and degrade nucleolar pRNA in HeLa cells. Using qPCR to quantify the level of pRNA, we found that treatment with ANG reduced pRNA levels by 50%. This lower level is the same as that achieved with LNA-mediated knockdown of pRNA (Figure 2A). We used variants of ANG, including those found in ALS patients (S28N, C39W, and H114R), to probe enzymatic activity in cellulo (9,10,46,47). We found that substitutions that affect the nuclear localization (S28N) or thermostability (C39W) of ANG had no effect on its catalytic activity in vitro but diminished its catalytic activity in cellulo (Figure 2B). Replacing a critical active-site residue (H114R) led to no degradation of pRNA either in vitro or in cellulo. In contrast, a hyperactive variant (Q117G) degraded pRNA robustly.
Figure 2.
ANG degrades pRNA in cellulo. (A) Graph of qRT-PCR data indicating that ANG (1 μg/ml) reduces the level of pRNA in HeLa cells by 50%, which is the same level achieved with LNA-antisense knockdown done as described previously (31). Values represent the mean ± SD (n = 3, biological replicates). (B) Graphs showing that ANG variants have differential effects on pRNA levels in vitro (left) and in cellulo (right). S28N ANG, which is an active enzyme in vitro but defective in nuclear localization (46), leads to no change in pRNA level in cellulo. C39W ANG, which is unstable, reduces pRNA level by only 25% in cellulo. H114R ANG, which has a deleterious active-site substitution, leads to no change in pRNA levels in vitro or in cellulo. Q117G ANG, which has an advantageous active-site substitution, leads to highly reduced pRNA levels in vitro and in cellulo. Values represent the mean ± SD (n = 3, in vitro: technical replicates, in cellulo: biological replicates). Paired Student's t-test: differences were considered significant at *P < 0.05. (C) An RNA co-immunoprecipitation with FLAG–H114R ANG (1 μg/ml) demonstrates a direct interaction between ANG and pRNA in cellulo. EtBr-stained agarose gel of PCR products that were amplified from the pRNA region corresponding to primer 1 (P1) and primer 2 (P2). Only the P2-derived pRNA region was detected in the IP samples, indicating that this region was protected by FLAG–H114R ANG. The P1-derived pRNA region as well as other cellular RNAs were vulnerable to degradation by RNase A. The PCR product of P2 was sequenced and its identity was confirmed as the conserved stem-loop structure of pRNA.
We next sought evidence for a direct interaction between ANG and pRNA in cellulo. We installed a FLAG tag on ANG and used RNA immunoprecipitation to probe for the existence of an ANG·pRNA complex. Because wild-type ANG degraded pRNA (Figure 1), we used the inactive H114R variant. This variant was unable to cut pRNA (Figure 2B), but did bind to pRNA with nanomolar affinity (Supplementary Figure S4). Following treatment with the FLAG–H114R ANG variant, cellular RNAs were treated with RNase A to cleave unprotected RNAs. With or even without UV-mediated crosslinking, we were able to detect a fragment of pRNA that was protected by ANG, and confirmed its identity as the conserved stem–loop structure of pRNA by sequence analysis (Figure 2C).
ANG promotes dissociation of a TIP5·pRNA complex by pRNA cleavage
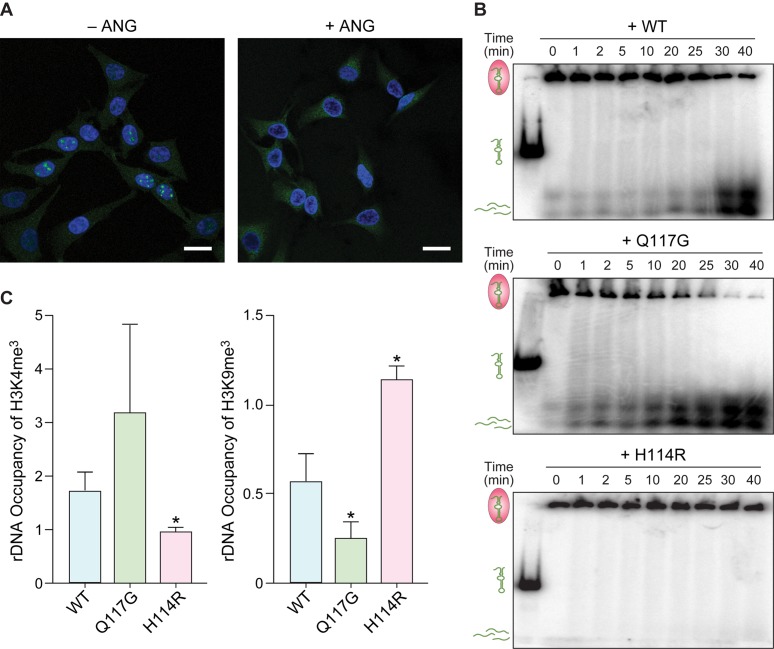
As described above, the silencing of rDNA transcription by NoRC is mediated by the binding of pRNA to TIP5 (31). The depletion of pRNA leads to TIP5 dissociation from the rDNA promoter and thereby activates rDNA transcription via chromatin remodeling. To demonstrate further the involvement of ANG in suppressing NoRC function, we used immunofluorescence microscopy to probe for the accumulation of TIP5 in nucleoli as an indicator of rDNA silencing in the presence and absence of ANG. Punctate staining of TIP5 in the nucleolus was observed in untreated cells but not in cells treated with ANG (Figure 3A). The absence of TIP5 in nucleoli is consistent with its being unable to bind to the rDNA promoter because pRNA had been cleaved by ANG and no longer remained at the rDNA promoter region.
Figure 3.
Cleavage of pRNA by ANG Promotes Dissociation of TIP5 in Cellulo. (A) Immunofluorescence images of nucleolar TIP5 (green) in HeLa cells indicating that ANG (1 μg/ml) limits the accumulation of TIP5 in the nucleolus. Blue: Hoechst 33342. Scale bar: 20 μm. (B) Autoradiograms of gel-shift assays indicating that ANG and the hyperactive Q117G variant degrade the pRNA within a TAM·pRNA complex in vitro; the inactive H114R variant does not. (C) Graphs of chromatin immunoprecipitation at the rDNA promoter revealing that treatment with ANG and the Q117G variant enrich the occupancy of H3K4me3 but decrease the occupancy of H3K9me3, which is a marker of repressive transcription. The H114R variant does not change H3K4me3 or H3K9me3 levels. Values represent the mean ± SD (n = 3, biological replicates). Paired Student's t-test: differences were considered significant at *P < 0.05.
pRNA could exist in a complex with TIP5 prior to interacting with ANG. Thus, angiogenic activity might require ANG to cleave pRNA within a TIP5·pRNA complex. To test for this ability, we reconstituted an in vitro model wherein an excess of the TAM domain of TIP5, which is necessary and sufficient to interact with pRNA (31,44), was incubated with the RNA substrate and then exposed to wild-type ANG and its Q117G and H114R variants (Figure 3B). Over the course of 40 min, a decrease in the level of the ensuing pRNA·TAM complex along with the accumulation of degraded pRNA was observed with wild-type ANG but not the H114R variant. Even more degradation was observed with the Q117G variant. Notably, the rate of pRNA cleavage by ANG was slower with pRNA in the bound state than in the unbound state (Figure 1A). Still, the data indicate that ANG can access its cleavage site on pRNA within a pRNA·TIP5 complex and that pRNA cleavage leads to dissociation of that complex. We note that, in cellulo, ANG could target nascent pRNA to prevent NoRC from anchoring to the rDNA promoter.
The assembly of NoRC facilitates heterochromatic histone modification, resulting in a reduction in the methylation of H3K4, which is a euchromatic histone mark. NoRC also promotes H3K9 methylation, which marks repressive transcription (49). Accordingly, we probed H3K4 and H3K9 methylation upon treating cells with ANG variants of varying enzymatic activity. We found that enrichment of histone-methylation occupancy at the rDNA promoter correlated with enzymatic activity (Figure 3C).
Chromatin immunoprecipitation (ChIP) of H3K4me3 at the rDNA promoter revealed a 2- and 3-fold increase in methylation upon treatment with ANG and the Q117G variant, respectively, but no change with the H114R variant relative to untreated cells (Figure 3C). Conversely, ChIP to probe methylation of H3K9, which marks repressive transcription, revealed that that the level of H3K9me3 at the rDNA promoter decreased upon treatment with either ANG or the Q117G variant, but not the H114R variant.
ANG is phosphorylated by PKC and CDK
The activation of rDNA transcription via the degradation of pRNA and consequent suppression of NoRC function can occur only if ANG gains access to the nucleus. A femtomolar inhibitor of ANG, RI lurks in the cytosol at low micromolar concentrations (50). We hypothesized that ANG undergoes a post-translational modification to evade RI.
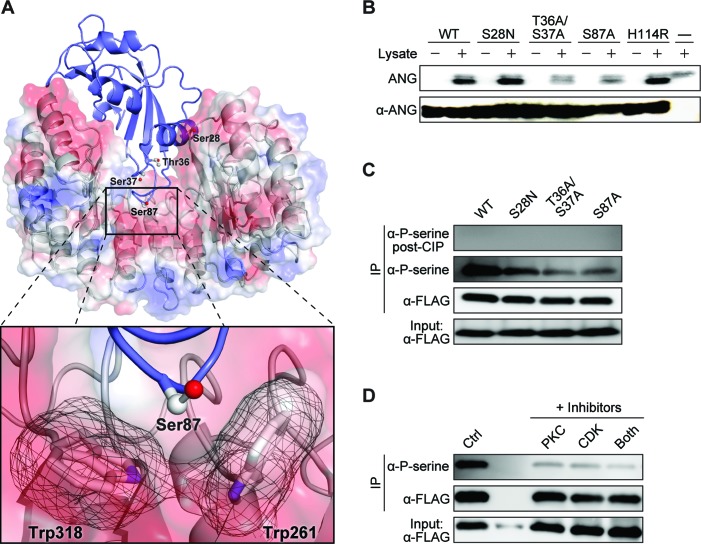
The extraordinarily tight binding between RI and ANG is due largely to favorable Coulombic interactions, as RI is highly anionic and ANG is highly cationic (51). Appending phosphoryl groups on ANG residues would generate Coulombic repulsion and diminish affinity for RI. Among the sixteen serine and threonine residues of ANG, four were of special interest to us: Ser87, which is at the molecular interface of the RI·ANG complex (52) and is in a known ‘hotspot’ for amino-acid substitutions that engender evasion of RI (25,53), and a cluster of three residues near the NLS: Ser28, Thr36 and Ser37 (Figure 4A).
Figure 4.
ANG is phosphorylated by PKC and CDK. (A) Structure of the human RI·ANG complex (PDB:1A4Y) (52). Putative phosphorylation sites in ANG (blue ribbon) are labeled and depicted in ball-and-stick. The Coulombic surface of RI (grey ribbon) is depicted with red = negative and blue = positive. Inset: Close contact between Ser87 of ANG and two tryptophan residues of RI. (B) Autoradiogram of a polyacrylamide gel demonstrating that ANG is phosphorylated upon incubation with a HeLa cell lysate and [γ-32P]ATP. Replacing Thr36/Ser37 or Ser87 with an alanine residue decreases phosphorylation. An immunoblot of the same gel shows consistent loading of ANG and its variants. (C) Immunoblots showing that FLAG–ANG (1 μg/ml) taken up by HeLa cells and isolated by immunoprecipitation (IP) with an anti-FLAG antibody (α-FLAG) is recognized by an anti-phosphoserine antibody (α-P-serine). Recognition is eliminated upon treatment with lambda protein phosphatase (LPP). IP of the S28N variant was reduced, and that of the T36A/S37A and S87A variants was reduced even more. (D) Immunoblots showing that small-molecule inhibitors of CDK or PKC reduce the phosphorylation of FLAG–ANG by HeLa cells. For kinase-inhibitor treatment, cells were pre-incubated with either bisindolylmaleimide (4 μM) or roscovitine (14 μM) for 30 min prior to the treatment with ANG. Representative immunoblots are shown in panels C and D, and independent biological replicates are shown in Supplementary Figure S5.
We discovered that ANG is phosphorylated by intracellular kinases. Incubation of wild-type ANG with a whole cell lysate and [γ-32P]ATP led to 32P-labeled ANG (Figure 4B). We used variants of ANG to determine the specificity of the phosphorylation reaction. None of the four residues is essential for the ribonucleolytic activity or conformational stability of ANG (54). We found that S28N ANG (which is linked to ALS) was phosphorylated in vitro approximately as extensively as was the wild-type enzyme (Figure 4B). In contrast, the T36A/S37A and S87A variants had a severe reduction in their phosphorylation. Then, we examined phosphorylation in cellulo. Wild-type ANG and variants with a FLAG tag were isolated by immunoprecipitation from HeLa cells after incubation for 4 h. The uniform intensity of the input samples indicated that neither the amino-acid substitutions nor the FLAG tag affected cellular uptake (Figure 4C, Supplementary Figures S5A and S5B), which occurred via the syndecan-4 receptor (Supplementary Figure S6). Likewise, the substitutions had inconsequential effects on thermostability (Supplementary Table S2). To detect phosphorylated species, we used antibodies to phosphoserine or phosphothreonine. No detection was observed with the phosphothreonine antibodies (data not shown). In contrast, immunoblots with phosphoserine antibodies displayed a strong band for wild-type ANG, indicating the existence of phosphorylated ANG in cellulo (Figure 4C, Supplementary Figures S5A and S5B). A weaker band was observed with the S28N variant, consistent with the phosphorylation of Ser28 by cellular kinases. A very weak band was detected for both the T36A/S37A and the S87A variants. These data indict Ser28, Ser37, and Ser87 as sites of phosphorylation in cellulo.
We identified kinases that phosphorylate ANG. For guidance, we analyzed the amino-acid sequence of ANG with the program NetPhos 2.0 (55). The computational results indicated that protein kinase C (PKC) and cyclin-dependent kinase (CDK) were likely kinases. We then treated cells with small-molecule inhibitors of PKC and CDK, namely, bisindolylmaleimide I and roscovitine (56,57). The inhibitors did not influence the cellular uptake of ANG, but did reduce its phosphorylation (Figure 4D, Supplementary Figures S5C and S5D). We conclude that ANG is a substrate for PKC and CDK.
Phosphorylation of ANG is essential for its nuclear translocation
We probed the effect of phosphorylating residues 28, 36/37 and 87 on the affinity of ANG for RI. To do so, we generated phosphomimetic variants and determined the value of Kd for the ensuing ANG·RI complexes. The femtomolar affinity of RI for wild-type ANG required month-long assays to detect complex dissociation (23). In comparison, the dissociation of the ANG variants was rapid, as monitored by using an assay in which the fluorescence of a fluorescein-labeled ANG variant decreases upon binding to RI (33,34). Both the T36D/S37D and the S87D variants had 107-fold lower affinity for RI than did wild-type ANG (Table 1). Moreover, the S28D/T36D/S37D/S87D variant exhibited a >109-fold reduction in affinity. These data are consistent with a mechanism in which phosphorylation of interface residues 37 and 87 enables ANG to evade RI.
Table 1. Values of Kd (± SD) of the complexes of RI with wild-type ANG and its phosphorylation-mimetics.
| ANG | Kd (nM)a | Fold change |
|---|---|---|
| Wild-type | 0.7 × 10–6b | 1 |
| T36D/S37D | 4.2 ± 0.5 | 6 × 106 |
| S87D | 8.7 ± 0.8 | 1.2 × 107 |
| T36D/S37D/S87D | 90 ± 5 | 1.3 × 109 |
| S28D/T36D/S37D/S87D | 150 ± 10 | 2.1 × 109 |
aValues represent the mean ± SD (n = 3, technical replicates).
b(23).
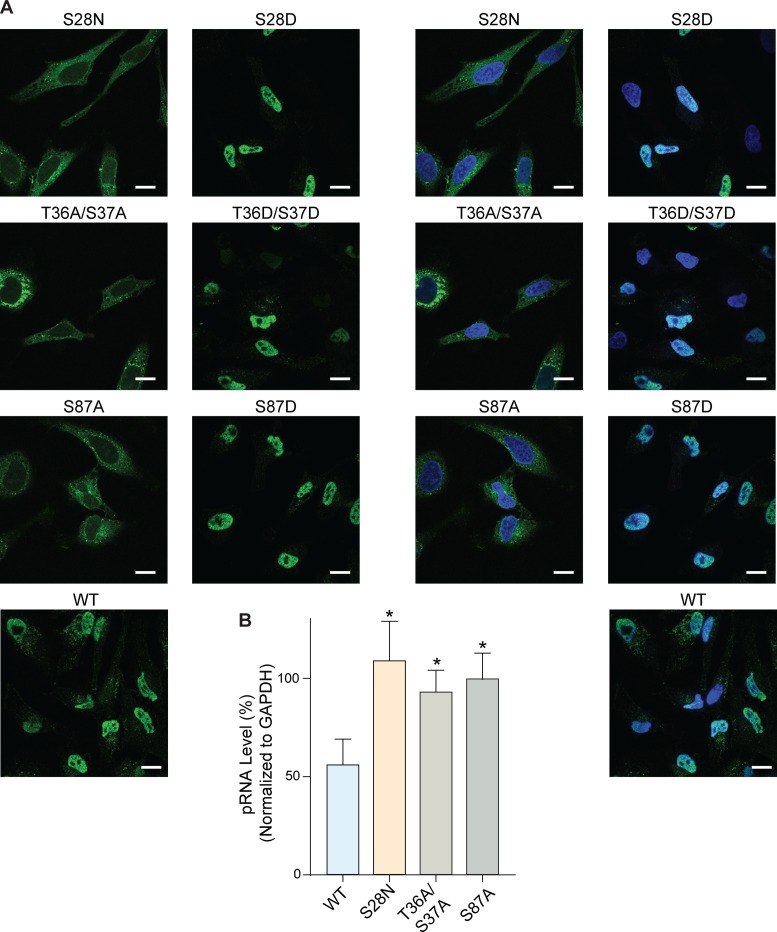
Finally, we used immunofluorescence microscopy to probe the effect of phosphorylating residues 28, 36/37 and 87 of FLAG-tagged ANG on its nuclear translocation. Immunofluorescence microscopy can be a reliable method for revealing the distribution of a protein within a cell (48). Nevertheless, this method necessitates the fixation and permeabilization of cells to enable immunoreagents to access all cellular compartments. Cell fixation, even under mild conditions, can promote the artifactual translocation of cationic peptides and proteins across cellular membranes (58–60). (Pendant fluorophores can elicit similar artifacts, even without cell fixation (61,62).) To maximize the likelihood of proper data interpretation, we used immunofluorescence microscopy only to compare the subcellular localization of ANG variants that contain one or two conservative substitutions.
With immunofluorescence microscopy, we found that alanine substitution at residues 28, 36/37 or 87 (which creates a phosphorylation defect) leads to the retention of ANG in the cytosol (Figure 5A). Conversely, aspartate substitution leads to its accumulation in the nucleus. Using pRNA degradation as a marker for the entry of ANG into the nucleus, we found that pRNA remained intact in cellulo upon treatment with S28N, T36A/S37A or S87A ANG (Figure 5B), even though these variants cleaved pRNA in vitro (data not shown). These data indicate that the phosphorylation of residues Ser28, Ser37 and Ser87 enables internalized ANG to enter the nucleus and degrade pRNA.
Figure 5.
Phosphorylation of ANG is essential for its nuclear translocation. (A) Immunofluorescence images of FLAG–ANG (1 μg/ml; green) in HeLa cells showing its nuclear localization. Blue: Hoechst 33342. Scale bar: 20 μm. FLAG–ANG variants with a defective phosphorylation site are cytosolic; FLAG–ANG variants with a phosphomimetic substitution are nuclear. Left: green channel only. Right: overlay. (B) Graph of qRT-PCR data indicating that ablating nuclear localization prevents ANG from cleaving pRNA in HeLa cells. Data for wild-type ANG and its S28N variant are from Figure 2A. Values represent the mean ± SD (n = 3, biological replicates). Paired Student's t-test: differences were considered significant at *P < 0.05.
Phosphorylation and pRNA cleavage are essential for ANG-mediated cell proliferation
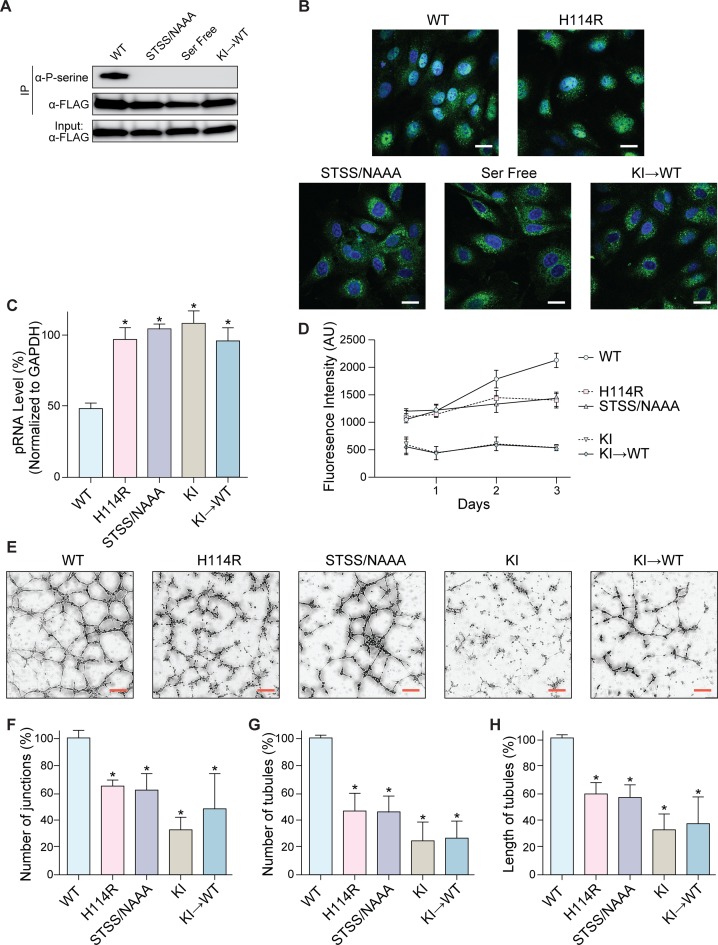
We found that ANG is phosphorylated at the same sites by PKCs and CDKs in endothelial cells as in HeLa cells (Figure 6A). Moreover, the extent of phosphorylation of the S28N/T36A/S37A/S87A variant (STSS/NAAA ANG) was indistinguishable from that of a variant in which all nine serine residues were replaced with alanine (Ser-Free ANG), indicating that no other serine residues are susceptible to phosphorylation. With immunofluorescence microscopy, we found that phosphorylation governed ANG entry into the nucleus of endothelial cells (Figure 6B). Once in the nucleus, ANG reached the nucleolus and cleaved pRNA (Figure 6C). Most importantly, these features—ANG phosphorylation and cleavage of pRNA—are essential for the biological activity of ANG. HUVEC cell-proliferation in basal medium containing ANG or a variant was stimulated by the wild-type enzyme, but not the inactive H114R or STSS/NAAA variant (Figure 6D). Moreover, cells pre-exposed to small-molecule inhibitors of CDKs and PKCs were quiescent, even after treatment with ANG. Using a tube-formation assay (63), we found that pre-incubating endothelial cells with ANG led to the assembly of capillary-like networks (Figure 6E). In contrast, cells pre-incubated with the H114R or STSS/NAAA variant constructed fewer junctions and tubules, and the tubules were of shorter length (Figure 6F–H). Likewise, ANG treatment did not revive network formation in cells pre-treated with kinase inhibitors.
Figure 6.
Phosphorylation and pRNA cleavage are essential for ANG-mediated cell proliferation. (A) Immunoblots showing that FLAG–ANG taken up and phosphorylated by HUVEC cells. Deletion of phosphorylation sites in variants (STSS/NAAA and Ser Free) or treatment of the wild-type enzyme with kinase inhibitors (KI→WT) decreases ANG phosphorylation. (B) Immunofluorescence images of FLAG–ANG (green) in HUVEC cells showing its nuclear localization. Deficient phosphorylation (STSS/NAAA, Ser Free, and KI→WT) restricts ANG to the cytosol. Blue: Hoechst 33342. Scale bar: 20 μm. (C) Graph of qRT-PCR data showing that wild-type ANG reduces the level of pRNA in HUVEC cells. pRNA level is not altered by the inactive H114R variant or upon deficient phosphorylation (STSS/NAAA, Ser Free, and KI→WT). Values represent the mean ± SD (n = 3, biological replicates). Paired Student's t-test: differences were considered significant at *P < 0.05. (D) Graph showing that ANG promotes the growth of HUVEC cells. Growth is not affected by the inactive H114R variant or upon deficient phosphorylation (STSS/NAAA). Treating HUVEC cells with kinase inhibitors arrests cell division and could not be rescued by treatment with wild-type ANG. Values represent the mean ± SD (n = 3, technical replicates). (E) Images of capillary-like tubules showing that wild-type ANG stimulates angiogenesis in cellulo. Potency is less from the inactive H114R variant or upon deficient phosphorylation (STSS/NAAA, KI, and KI→WT). Scale bar: 100 μm. (F–H) Graphs of key parameters extracted from tubule images like those in panel E. Values represent the mean ± SD (n = 3, biological replicates). Paired Student's t-test: differences were considered significant at *P < 0.05.
DISCUSSION
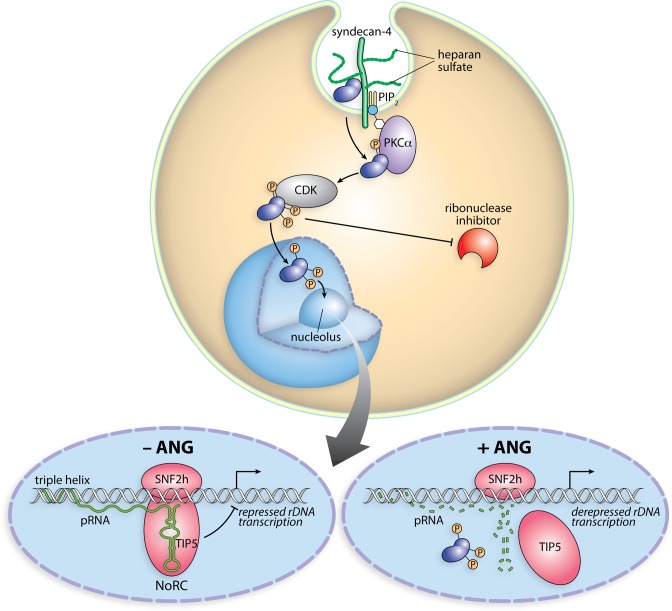
Members of the pancreatic-type RNase superfamily have evolved to be efficient non-specific catalysts of RNA degradation (64). Unlike its homologs, ANG has nearly unmeasurable ribonucleolytic activity towards model substrates (15,45). Moreover, whereas other pancreatic-type RNases function in the extracellular space or cytosol, ANG acts in the nucleus (19,27). Here we have revealed the cellular mechanism used by ANG to effect cell proliferation by virtue of specific ribonucleolytic activity. That mechanism is depicted in Figure 7.
Figure 7.
Scheme of the cellular action of ANG. Angiogenin binds to syndecan-4 on the cell surface and is internalized by endocytosis. A fraction translocates to the cytosol, where ANG is phosphorylated by PKC and CDK. Phosphorylation endows ANG with the ability to evade the ribonuclease inhibitor protein. Phosphorylated ANG translocates into the nucleus and accumulates in the nucleolus. There, ANG digests pRNA, leading to the dissociation of TIP5 from the rDNA promoter. The ensuing rDNA transcription fuels the proliferation of endothelial cells (neovascularization) and tumor cells (cancer progression).
Intracellular kinases enable ANG to manifest its ribonucleolytic activity. Like its homologs, ANG is a highly cationic protein that enters mammalian cells via endocytosis. A fraction of the ANG in endosomes escapes into the cytosol, where it encounters a potent inhibitor, RI. The RI·ANG complex is stabilized largely by favorable Coulombic interactions, as RI is highly anionic. The introduction of anionic phosphoryl groups into ANG generates Coulombic (as well as steric) repulsion with RI, resulting in weaker binding. We note that site-directed mutagenesis has been used to endow other pancreatic-type RNases with the ability to evade RI, and such variants are cytotoxic at the nanomolar level (53). In contrast, the ribonucleolytic activity of ANG is so low, that its evasion of RI does not lead to any apparent cytotoxicity.
ANG binds to the same region of pRNA as does TIP5. The reported Kd value of the TIP5·pRNA complex is 0.3 nM (31), whereas the Kd value of the H114R ANG·pRNA complex is 192 nM (Supplementary Figure S4). These disparate values indicate that TIP5 competes favorably with ANG for binding to pRNA. We note, moreover, that ANG is an enzyme and that its cleavage of a phosphodiester bond in pRNA is irreversible. Accordingly, only a transient interaction between ANG and pRNA is necessary for ANG to assert its biological activity. Thus, the cleavage of pRNA by ANG is apparent even in the presence of TIP5 (Figure 3B).
An angiogenin-binding element (ABE) near the rDNA promoter has been proposed to be responsible for driving the expression of genes encoding rRNA upon stimulus with ANG (65). We find that the affinity of ANG for pRNA is 3-fold higher than that for ABE (Supplementary Figure S4). Moreover, as noted above, the binding of ANG to pRNA leads to an irreversible event—phosphodiester bond cleavage. As ribonucleolytic activity is essential for ANG action (16), the cleavage of pRNA is likely to play a larger role in the action of ANG than does its binding to the ABE.
pRNA is not the only cellular substrate of ANG under all conditions. We have demonstrated that ANG can localize in the nucleolus and cleave a particular phosphodiester bond in pRNA, thereby inducing rDNA gene expression. This upregulation satisfies the rRNA demands of ongoing translation and promotes angiogenesis. In contrast, in cells that have suffered oxidative damage, ANG localizes in stress granules (Supplementary Figure S7) and cleaves a subset of tRNAs, thereby generating tRNA-derived, stress-induced small RNAs that stall translation and drive the production of anti-apoptotic proteins that inhibit cell death (66–68). The molecular basis for the localization of ANG in stress granules as a consequence of oxidative stress is unclear.
The mechanism of action of ANG is autonomous and unique. Typical growth factors bind to the extracellular face of membrane-bound receptors and rely upon changes in receptor conformation or valency to transduce a signal to the cytosol, often leading to kinase or phosphatase activity (69,70). The growth factors themselves do not enter cells. This action-at-a-distance model contrasts markedly with the mechanism of ANG action (Figure 7). Instead of conveying signals via other proteins, ANG has evolved to deliver its proliferative signal directly. We speculate that this mechanism of signal transduction is conserved in mammals (Supplementary Figure S2).
The route taken by ANG has other implications. In astrocytes, ANG is internalized after binding to syndecan-4, a transmembrane heparan sulfate proteoglycan (8,71). This receptor is also displayed by endothelial cells and involved in inflammatory reactions, wound healing, and angiogenesis (72). We found that this receptor also mediates the uptake of ANG into HeLa cells (Supplementary Figure S6). Because syndecan-4 is involved in the activation of PKCα (73), ANG could be phosphorylated immediately by this PKC isoform upon cytosolic entry.
Finally, we note a dichotomy: ANG is upregulated in transformed cells but is necessary to prevent neurodegeneration of nontransformed cells. Hence, drugs that target this pro-tumorigenic protein could also promote neurological damage, as loss-of-function mutations of ANG genes are associated with ALS (9,10). The understanding of ANG action provided in this work suggests new chemotherapeutic strategies. For example, Ser37 and Ser87 of ANG, which are phosphorylated in cellulo, make intimate contacts with Ile459 and Trp261/Trp318 of RI, respectively, in the RI·ANG complex (Figure 4A) (52). Replacing those large RI residues with smaller ones in endothelial cells could effect a ‘bump–hole’ strategy that leads to an RI variant capable of apprehending ANG even after its phosphorylation. Alternatively, expression of an uncleavable pRNA variant in endothelial cells could elicit a dominant negative phenotype that sequesters ANG and thereby decreases its activity. We believe that modulating kinase activity holds special promise. Antagonizing the activity of PKC or CDK in endothelial cells, but not astrocytes, could benefit ALS patients treated with exogenous ANG. Indeed, known drugs that inhibit these kinases in cancer patients could be acting, in part, by an unappreciated mechanism—decreasing the activity of endogenous ANG (Figure 7). For example, flavopiridol and enzastaurin are inhibitors of CDK and PCK, respectively, and both antagonize angiogenesis (74,75). Our data provide a plausible link for these two clinical paradigms.
Acknowledgments
We thank Dr E. Lund, Dr D.A. Wassarman, Dr K.M. Shokat, Dr B.E. Turk, R.J. Presler, C.H. Eller and K.A. Andersen for contributive discussions. We are grateful to Dr T.F.J. Martin for the use of his confocal microscope.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Advanced Opportunity/Graduate Research Scholar Fellowship and by Molecular Biosciences Training Grant [T32 GM007215 to T.T.H.] from the National Institutes of Health (NIH); NIH [R01 CA073808]. Funding for open access charge: NIH [R01 CA073808].
Conflict of interest statement. None declared.
REFERENCES
- 1.Folkman J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Cooke R. Dr. Folkman's War: Angiogenesis and the Struggle to Defeat Cancer. NY: Random House; 2001. [Google Scholar]
- 3.Fett J.W., Strydom D.J., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F., Vallee B.L. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 4.Schmeck H.M., Jr. The New York Times. NY: 1985. A1, B11. [Google Scholar]
- 5.Yoshioka N., Wang L., Kishimoto K., Tsuji T., Hu G.-f. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pina F., Botelho F., Lopes T., Lopes I., Figueiredo G., Portugal R., Ferro A., Cruz F., Barros H., Lunet N. Can serum angiogenin be used to improve the diagnostic performance in prostate cancer screening. Eur. J. Cancer Prev. 2014;23:166–172. doi: 10.1097/CEJ.0b013e3283647453. [DOI] [PubMed] [Google Scholar]
- 7.Folkman J., Klagsbrun M. Angiogenic factors. Science. 1987;235:442–447. doi: 10.1126/science.2432664. [DOI] [PubMed] [Google Scholar]
- 8.Skorupa A., King M.A., Aparicio I.M., Dussmann H., Coughlan K., Breen B., Kieran D., Concannon C.G., Marin P., Prehn J.H. Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J. Neurosci. 2012;32:5024–5038. doi: 10.1523/JNEUROSCI.6366-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenway M.J., Andersen P.M., Russ C., Ennis S., Cashman S., Donaghy C., Patterson V., Swingler R., Kieran D., Prehn J., et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 10.Padhi A.K., Jayaram B., Gomes J. Prediction of functional loss of human angiogenin mutants associated with ALS by molecular dynamics simulations. Sci. Rep. 2013;3:1225. doi: 10.1038/srep01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieran D., Sebastia J., Greenway M.J., King M.A., Connaughton D., Concannon C.G., Fenner B., Hardiman O., Prehn J.H. Control of motoneuron survival by angiogenin. J. Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strydom D.J., Fett J.W., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F., Vallee B.L. Amino acid sequence of human tumor derived angiogenin. Biochemistry. 1985;24:5486–5494. doi: 10.1021/bi00341a031. [DOI] [PubMed] [Google Scholar]
- 13.Riordan J.F. Angiogenin. Methods Enzymol. 2001;341:263–273. doi: 10.1016/s0076-6879(01)41157-8. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro R., Fett J.W., Strydom D.J., Vallee B.L. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 15.Leland P.A., Staniszewski K.E., Park C., Kelemen B.R., Raines R.T. The ribonucleolytic activity of angiogenin. Biochemistry. 2002;41:1343–1350. doi: 10.1021/bi0117899. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro R., Vallee B.L. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogen-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 17.Acharya K.R., Shapiro R., Allen S.C., Riordan J.F., Vallee B.L. Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2915–2919. doi: 10.1073/pnas.91.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo N., Shapiro R., Acharya K.R., Riordan J.F., Vallee B.L. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2920–2924. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishimoto K., Liu S., Tsuji T., Olson K.A., Hu G.-f. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- 20.Tsuji T., Sun Y., Kishimoto K., Olson K.A., Liu S., Hirukawa S., Hu G.-f. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- 21.Hu G.F., Riordan J.F., Vallee B.L. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo K.J., Hwang E., Min K.M., Jee J.G., Lee C.K., Hwang K.Y., Jeon Y.H., Chang S.I., Cheong H.K. The dual binding site of angiogenin and its inhibition mechanism: the crystal structure of the rat angiogenin-heparin complex. Chem. Commun. (Cambridge, England) 2014;50:12966–12969. doi: 10.1039/c4cc05175k. [DOI] [PubMed] [Google Scholar]
- 23.Lee F.S., Shapiro R., Vallee B.L. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Pan X.-Y., Shu J., Jiang R., Zhou Y.-J., Chen J.-X. Ribonuclease inhibitor up-regulation inhibits the growth and induces apoptosis in murine melanoma cells through repression of angiogenin and ILK/PI3K/AKT signaling pathway. Biochimie. 2014;103:89–100. doi: 10.1016/j.biochi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Dickson K.A., Kang D.K., Kwon Y.S., Kim J.C., Leland P.A., Kim B.M., Chang S.I., Raines R.T. Ribonuclease inhibitor regulates neovascularization by human angiogenin. Biochemistry. 2009;48:3804–3806. doi: 10.1021/bi9005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizzo E., Sarcinelli C., Sheng J., Fusco S., Formiggini F., Netti P., Yu W., D'Alessio G., Hu G.-f. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J. Cell Sci. 2013;126:4308–4319. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroianu J., Riordan J.F. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z.P., Tsuji T., Riordan J.F., Hu G.F. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem. Biophys. Res. Commun. 2002;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 29.Hu G., Xu C., Riordan J.F. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J. Cell. Biochem. 2000;76:452–462. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 30.Strohner R., Nemeth A., Jansa P., Hofmann-Rohrer U., Santoro R., Langst G., Grummt I. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz K.-M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavis L.D., Rutkoski T.J., Raines R.T. Tuning the pKa of fluorescein to optimize binding assays. Anal. Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomax J.E., Eller C.H., Raines R.T. Rational design and evaluation of mammalian ribonuclease cytotoxins. Methods Enzymol. 2012;502:273–290. doi: 10.1016/B978-0-12-416039-2.00014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 36.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer. Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 37.Sheng J., Yu W., Gao X., Xu Z., Hu G.-f. Angiogenin stimulates ribosomal RNA transcription by epigenetic activation of the ribosomal DNA promoter. J. Cell Physiol. 2014;229:521–529. doi: 10.1002/jcp.24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drygin D., Rice W.G., Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 39.Grummt I., Längst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim. Biophys. Acta. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Santoro R., Li J., Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 41.Guetg C., Scheifele F., Rosenthal F., Hottiger M.O., Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 42.Zillner K., Filarsky M., Rachow K., Weinberger M., Langst G., Nemeth A. Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res. 2013;41:5251–5262. doi: 10.1093/nar/gkt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer C., Neubert M., Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anosova I., Melnik S., Tripsianes K., Kateb F., Grummt I., Sattler M. A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes. Nucleic Acids Res. 2015;43:5208–5220. doi: 10.1093/nar/gkv365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curran T.P., Shapiro R., Riordan J.F. Alteration of the enzymatic specificity of human angiogenin by site-directed mutagenesis. Biochemistry. 1993;32:2307–2313. doi: 10.1021/bi00060a023. [DOI] [PubMed] [Google Scholar]
- 46.Wu D., Yu W., Kishikawa H., Folkerth R.D., Iafrate A.J., Shen Y., Xin W., Sims K., Hu G.-f. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann. Neurol. 2007;62:609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crabtree B., Thiyagarajan N., Prior S.H., Wilson P., Iyer S., Ferns T., Shapiro R., Brew K., Subramanian V., Acharya K.R. Characterization of human angiogenin variants implicated in amyotrophic lateral sclerosis. Biochemistry. 2007;46:11810–11818. doi: 10.1021/bi701333h. [DOI] [PubMed] [Google Scholar]
- 48.Stadler C., Rexhepaj E., Singan V.R., Murphy R.F., Pepperkok R., Uhlén M., Simpson J.C., Lundberg E. mmunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods. 2013;10:315–323. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]
- 49.Cong R., Das S., Ugrinova I., Kumar S., Mongelard F., Wong J., Bouvet P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012;40:9441–9454. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haigis M.C., Kurten E.L., Raines R.T. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickson K.A., Haigis M.C., Raines R.T. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papageorgiou A., Shapiro R., Acharya K. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 Å resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rutkoski T.J., Raines R.T. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr. Pharm. Biotechnol. 2008;9:185–189. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith B.D., Raines R.T. Genetic selection for critical residues in ribonucleases. J. Mol. Biol. 2006;362:459–478. doi: 10.1016/j.jmb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Blom N., Gammeltoft S., Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 56.Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F., et al. The bisindoylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 57.De Azevedo W.F., Leclerc S., Miejer L., Havlicek L., Strnad M., Kim S.-H. Inhibition of cyclin-dependent kinases by purine analogues. Crystal structure of human cdk2 complexed with roscovitine. Eur. J. Biochem. 1997;2443:518–526. doi: 10.1111/j.1432-1033.1997.0518a.x. [DOI] [PubMed] [Google Scholar]
- 58.Lundberg M., Johansson M. Positively charged DNA-binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 2002;291:367–371. doi: 10.1006/bbrc.2002.6450. [DOI] [PubMed] [Google Scholar]
- 59.Richard J.P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M.J., Chernomordik L.V., Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanisms of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 60.Boisguérin P., Deshayes S., Gait M.J., O'Donovan L., Godfrey C., Betts C.A., Wood M.J.A., Lebleu B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv. Drug Deliv. Rev. 2015;87:52–67. doi: 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szeto H.H., Schiller P.W., Zhao K., Luo G. Fluorescent dyes alter intracellular targeting and function of cell-penetrating tetrapeptides. FASEB J. 2006;19:118–120. doi: 10.1096/fj.04-1982fje. [DOI] [PubMed] [Google Scholar]
- 62.Puckett C.A., Barton J.K. Fluorescein redirects a ruthenium–octaarginine conjugate to the nucleus. J. Am. Chem. Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnaoutova I., Kleinman H.K. In vitro angiogenesis: Endothelial cell tube formation on gelled basement membrane extract. Nat. Protoc. 2010;5:628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 64.Raines R.T. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 65.Xu Z.P., Tsuji T., Riordan J.F., Hu G.F. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 66.Yamasaki S., Ivanov P., Hu G.-f, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ivanov P., O'Day E., Emara M.M., Wagner G., Lieberman J., Anderson P. G-Quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cantley L.C., Hunter T., Sever R., Thorner J. Signal Transduction: Principles, Pathways, and Processes. NY: Cold Spring Harbor Laboratory Press; 2014. [Google Scholar]
- 70.Lim W., Mayer B., Pawson T. Garland Science. NY: 2014. Cell Signaling. [Google Scholar]
- 71.Aparicio-Erriu I.M., Prehn J. Molecular mechanisms in amyotrophic lateral sclerosis: The role of angiogenin, a secreted RNase. Front. Neurosci. 2012;6:167. doi: 10.3389/fnins.2012.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Echtermeyer F., Streit M., Wilcox-Adelman S., Saoncella S., Denhez F., Detmar M., Goetinck P. Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J. Clin. Invest. 2001;107:R9–R14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Couchman J.R. Syndecans: Proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 2003;4:926–937. doi: 10.1038/nrm1257. [DOI] [PubMed] [Google Scholar]
- 74.Newcomb E.W. Flavopiridol: Pleiotropic biological effects enhance its anti-cancer activity. Anticancer Drugs. 2004;15:411–419. doi: 10.1097/01.cad.0000127332.06439.47. [DOI] [PubMed] [Google Scholar]
- 75.Chen Y.B., LaCasce A.S. Enzastaurin. Expert Opin. Investig. Drugs. 2008;17:939–944. doi: 10.1517/13543784.17.6.939. [DOI] [PubMed] [Google Scholar]