Abstract
MicroRNAs originate from primary transcripts containing hairpin structures. The levels of mature miR156 influence the leaf number prior to flowering in the life cycle of plants. To understand the molecular mechanism of biogenesis of primary miR156a (pri-miR156a) to mature miR156, a base-pair opening dynamics study was performed using model RNAs mimicking the cleavage site of wild type and B5 bulge-stabilizing mutant pri-miR156a constructs. We also determined the mature miR156 levels and measured leaf numbers at flowering of plants overexpressing the wild type and mutant constructs. Our results suggest that the stabilities and/or opening dynamics of the C15·G98 and U16·A97 base-pairs at the cleavage site are essential for formation of the active conformation and for efficient processing of pri-miR156a, and that mutations of the B5 bulge can modulate mature miR156 levels as well as miR156-driven leaf number phenotypes via changes in the base-pair stability of the cleavage site.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs that negatively regulate expression of their target genes via either sequence-specific mRNA degradation or translational repression (1). MiRNAs originate from primary transcripts (pri-miRNAs), which have hairpin structures composed of an upper stem with a terminal loop, an miRNA/miRNA* duplex, and a lower stem (see Figure 1A for miR156a). In plants, DICER-LIKE1 (DCL1), which forms a complex with HYPONASTIC LEAVES1 (HYL1) and SERRATE (SE), generates the mature miRNA from pri-miRNA (2). The miRNA/miRNA* duplex is methylated by HUA ENHANCER1 (HEN1) (3,4) and then exported to the cytoplasm to load into the ARGONAUTE complex (5). The miRNA then guides the complex to its target mRNA sequences via partial complementarity.
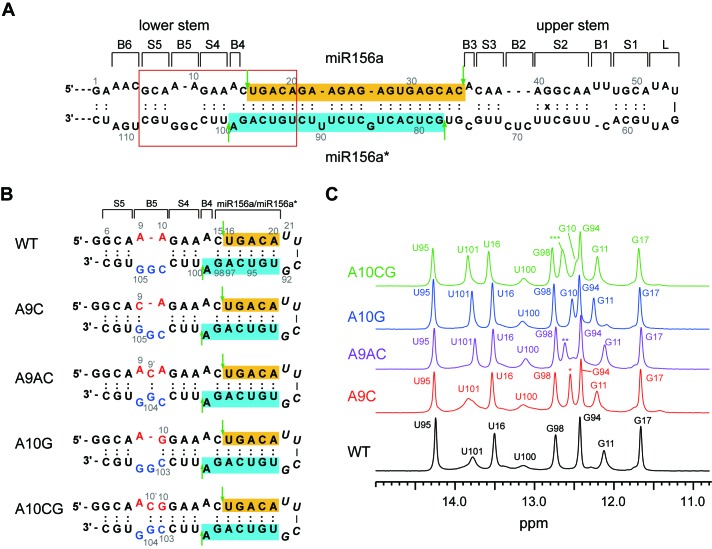
Figure 1.
Secondary structures of wild type and B5 stabilizing mutated pri-miR156a. (A) Secondary structures of primary miRNA156a and (B) model RNAs mimicking WT and mutant pri-miRNA156a constructs used in this study. Mature miR156 and miR156a* sequences are highlighted in gold and cyan, respectively. Green vertical arrows indicate the cleavage sites of pri-miR156a by DCL1. (C) 1D imino proton spectra of WT (black), A9C (red), A9AC (purple), A10G (blue) and A10CG (green) pri-miRNA156a at 23°C. The asterisks indicate the imino proton resonances which could not be exactly assigned. *: G105 or G107; **: G104 or G107; ***: G104 or G107.
Sequential cleavage of pri-miRNAs by DCL1 releases mature miRNAs. The secondary structure of pri-miRNA has important consequences for miRNA processing. For example, a ∼15-nucleotide segment in the lower stem is essential for processing of pri-miR172a (6,7), whereas its upper stem has only a weak effect. In pri-miR171a, closing a bulge adjacent to a cleavage site in the lower stem decreased the levels of mature miR171 (8). The 4–6 nucleotide below the miR390a/miR390a* duplex contribute to the efficiency and accuracy of miR390a processing (9). In contrast, the conserved upper stem of pri-miR319, including a terminal loop, plays a crucial role in miRNA processing (10). Thus, it is thought that the structural determinants important for miRNA processing are encoded in the miRNA itself.
The proper transition to the flowering phase is pivotal for ensuring reproductive success in plants. Ambient temperature-responsive miRNAs can affect the floral transition (11). Among them, miR156 and miR172 play important roles in regulating ambient temperature-responsive phenotypes before flowering (12,13). Plants overexpressing miR156 produced more leaves than wild-type (WT) plants before flowering via regulation of the expression of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family members (14,15). In contrast, plants overexpressing miR172 produced fewer leaves than WT plants before flowering by targeting genes encoding APETALA2-like transcription factors (16). Although the structural determinants for miR172 processing have been revealed (6,7), the molecular basis of miR156 processing and its effects on the leaf number phenotype of plants remain unknown.
RNAs generally undergo conformational transitions that exhibit distinct structural and dynamic features required for proper function. NMR hydrogen exchange studies provide information on the dynamic motions of the base-pairs. Here, to understand the molecular mechanism of biogenesis of pri-miR156a to mature miR156, the base-pair opening dynamics were studied using model RNAs mimicking the cleavage site in the lower stem of WT pri-miR156a and four pri-miR156a mutants, in which the B5 bulge was stabilized by additional base-pairing (Figure 1B). We also determined the mature miR156 levels and the leaf numbers at flowering of plants overexpressing the WT and B5-stabilizing mutant miR156a constructs, to investigate the in vivo effects of the mutations. This study reveals the unique dynamic features of the cleavage site that play an important role in the processing of pri-miR156a.
MATERIALS AND METHODS
Sample preparation for NMR
The RNA oligomers were purchased from M-biotech Inc. (Korean branch of IDT Inc., USA). The RNA oligomers were purified by reverse-phase HPLC and desalted using a Sephadex G-25 gel filtration column. The RNA samples were dissolved in 90% H2O/10% D2O aqueous solution containing 10 mM Tris-d11-HCl (pH 8.00 at 24.2°C) and 50 mM NaCl. The Tris-HCl concentration was increased from 10 mM to 200 mM by successive additions of a 500 mM Tris-d11-HCl stock solution. The pH of the sample dissolved in Tris-HCl buffer was calculated using the equation ΔpKa = −0.031 × ΔT (17).
NMR experiments
NMR experiments were performed on an Agilent DD2 700 MHz spectrometer (GNU, Jinju, Korea) using a cryogenic triple-resonance probe. One-dimensional (1D) NMR data were processed with either VNMR J (Agilent, CA, USA) or FELIX2004 (FELIXNMR, CA, USA), whereas 2D data were processed with NMRPIPE (18) and analyzed with Sparky (19). The imino protons in the WT and mutant miR156a constructs were assigned using watergate-NOESY spectra (mixing times of 120 and 250 ms). The apparent longitudinal relaxation rate constants of the imino protons (R1a = 1/T1a) were determined by semi-selective inversion recovery 1D NMR, where a semi-selective 180° inversion pulse was applied to the imino proton region (9–15.5 ppm) before the jump-return-echo water suppression pulse. The apparent relaxation rate constant of water (R1w) was determined by a selective inversion recovery experiment using a DANTE sequence for selective water inversion (20). The hydrogen exchange rate constants (kex) of the imino protons were measured by a water magnetization transfer, where a selective 180° pulse for water was applied, followed by a variable delay and then a 3-9-19 acquisition pulse was used to suppress the water signal (20,21). The intensities of each imino proton were measured with 20 different delay times ranging from 5 to 100 ms. The kex for the imino protons was determined by fitting the data to Equation (1):
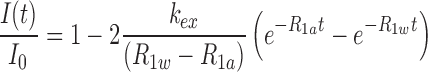 |
(1) |
where I0 and I(t) are the peak intensities of the imino proton in the water magnetization transfer experiments at times zero and t, respectively.
Hydrogen exchange theory
The formalism of base-catalyzed proton exchange has been extensively described (20,22–25). It assumes that the imino proton exchange from a base-pair consists of a two-step process requiring base-pair opening followed by proton transfer to a base catalyst. The rate constant for imino proton exchange (kex) is given by Equation (2):
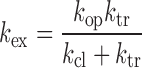 |
(2) |
where ktr is the imino proton transfer rate constant from the mononucleotide, and kop and kcl are the rate constants for base-pair opening and closing, respectively. In the base-pair, the exchange is catalyzed not only by the added base catalyst but also by the nitrogen of the complementary base, which acts as an intrinsic catalyst (24,25). The ktr value is calculated as:
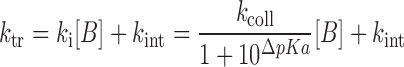 |
(3) |
where kB is the rate constant for imino proton transfer by a base catalyst, kint is the exchange rate constant catalyzed by an intrinsic base, kcoll is the collision rate constant, [B] is the base catalyst concentration and ΔpKa is the pKa difference between the imino proton and the base. Thus, the kex for the base-paired imino proton is represented by Equation (4):
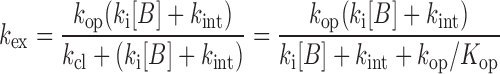 |
(4) |
where Kop ( = kop/kcl) is the equilibrium constant for base-pair opening. The apparent relaxation rate constant (R1a) for an imino proton is the sum of the R1 relaxation rate constant and the kex values contributed by intrinsic and external base catalysts, given by Equation (5):
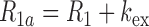 |
(5) |
The exchange rate constants for the imino protons studied here were measured using the water magnetization transfer method [using Equation (1)] or an indirect method [using Equation (5)] at various total concentrations of Tris-HCl buffer (10–200 mM). Re-organization of Equation (4) yields the following equation:
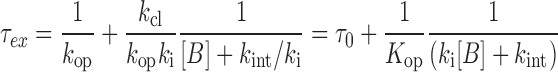 |
(6) |
where τex is the exchange time ( = 1/kex) and τ0 is the base-pair lifetime ( = 1/kop). Curve fitting the τex values of the imino protons as a function of the Tris (base form) concentration with Equation (6) gives the Kop, τ0 and kint values. The lifetime for base-pair opening (τopen) is calculated using the relation τopen = 1/kcl = Kopτ0. Under certain conditions where ki[B] is much larger than kint, Equation (6) simplifies to:
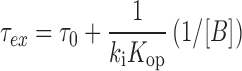 |
(7) |
The Gibbs free energy difference (ΔGobp) between the closed and open states is calculated from the equilibrium constant for base-pair opening using Equation (8):
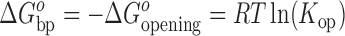 |
(8) |
where ΔGoopening is the Gibbs free energy change in the opening process, T is the absolute temperature and R is the universal gas constant. The activation energies for base-pair opening (ΔG‡op) and closing (ΔG‡cl) are related to the kop and kcl values, respectively, by the Arrhenius equation. The differences in activation energies for base-pair opening (ΔΔG‡op) and closing (ΔΔG‡cl) in WT and mutant RNAs are calculated using Equations (9) and (10), respectively:
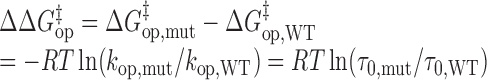 |
(9) |
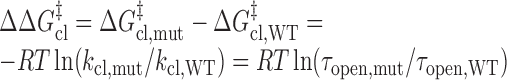 |
(10) |
where the subscripts, WT and mut, indicate the thermodynamic parameters of the WT and mutant RNAs, respectively.
Plant materials and measurement of leaf numbers
The Arabidopsis thaliana Columbia-0 (Col-0) was used for transformation. The plants were grown in soil at 23°C under long-day conditions (16 h light, 8 h dark) with a light intensity of 120 μmol/m2s. The number of primary rosette and cauline leaves of homozygous plants at flowering was measured. We first scored the leaf numbers of primary transformants in the T1 generation and selected a few representative lines that showed leaf numbers close to the median value of leaf numbers seen in the T1 population at flowering to isolate homozygous lines. At least 25 homozygous plants from each transgenic line were used to score the number of leaves at flowering.
Generation of constructs carrying structural variants
The constructs of wild-type and B5-stabilizing mutated pri-miR156a were synthesized (Bioneer, Daejeon, Korea). After sequence confirmation of individual constructs, these constructs were cloned into the pCHF3 vector (26), which contains the 35S promoter. The resulting plasmids were introduced into wild-type Arabidopsis plants by a modified floral dip method (27). Primary transformants were selected on Murashige Skoog (MS) media supplemented with kanamycin and transferred to soil on day 7 at 23°C.
MiRNA Northern blot analysis
We used an enhanced miRNA detection method by chemical cross-linking with N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC) (Sigma) (28). Total RNA was extracted from 8-day-old plants using Plant RNA Purification Reagent (Invitrogen). Total RNA (10 μg) was loaded onto 17% denaturing polyacrylamide gels containing 7 M urea and electrophoresed. The separated RNA was transferred to a Hybond-NX neutral nylon membrane (GE Healthcare), which was cross-linked with EDC. The membrane was hybridized with probes labeled at the 3′ end with [γ-32P] ATP using OptiKinase (USB Corp., USA). The hybridized membranes were exposed and analyzed using Fuji BAS FLA-7000 (FUJI, Japan). U6 was used as an internal control.
RESULTS
Hydrogen exchange of imino protons of WT pri-miR156a
Figure 1C shows the 1D imino proton spectra of the WT and mutant pri-miR156a constructs in NMR buffer containing 10 mM Tris-d11-HCl (pH 8.03) and 50 mM NaCl at 23°C. The imino proton resonance assignments of each cobstruct were made by the analysis of the NOESY spectra (Figure 2). We observed no chemical shift changes in the G98, U16, G17, U95 and G94 imino resonances, suggesting that these B5-stabilizing mutants did not affect the 3D structure of the miR156a/miR156a* duplex region (Figure 1C).
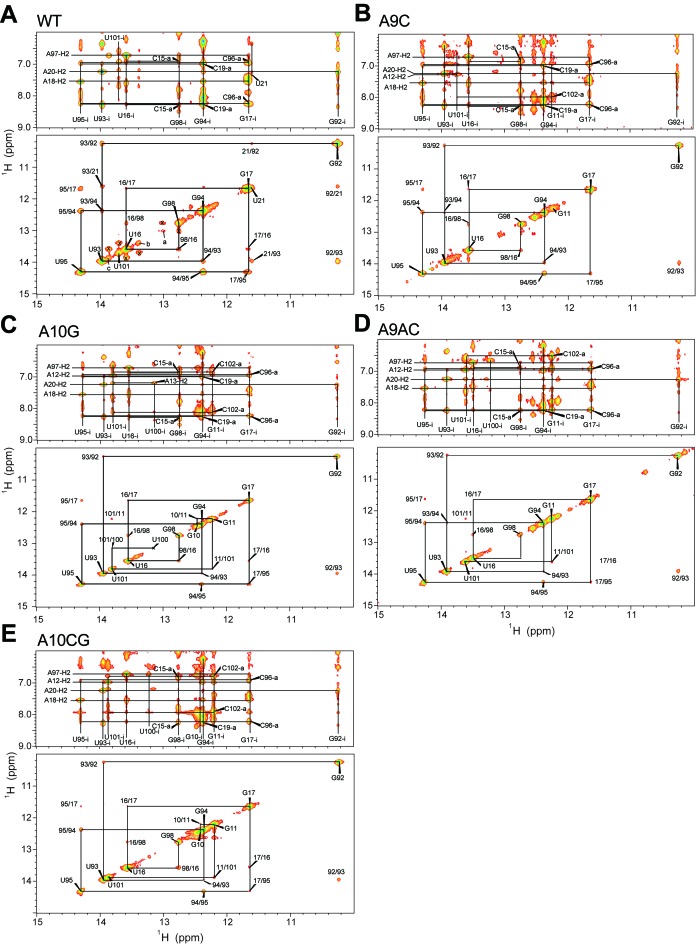
Figure 2.
Imino proton resonance assignments of WT and mutant pri-miR156a. Watergate NOESY spectra of (A) the WT, (B) A9C, (C) A10G, (D) A9AC and (E) A10CG pri-miR156a in 90% H2O/10% D2O containing 10 mM sodium phosphate (pH 8.0) and 50 mM NaCl. Solid lines indicate (upper) NOE cross-peaks between imino protons and their own and neighboring H2 or amino protons and (lower) imino-imino NOE connectivities. The letters ‘i’ and ‘a’ indicate the imino and amino proton resonances, respectively. The x symbols indicate the exchange NOE cross-peaks of the G98, U101 and U16 imino protons with the imino resonances a, b and c, respectively.
Hydrogen exchange rate constants (kex) were determined from water magnetization transfer experiments on the imino protons for the WT and mutant pri-miR156a (Table 1) (20,21). In the WT pri-miR156a, the G98 imino proton next to the B4 bulge has a kex of 5.0 s−1 at 16°C, which is significantly larger than those of the G17 and G94 imino protons in the miR156a/miR156a* duplex (Table 1). Similarly, the U16 imino proton has a 2-fold larger kex value than the U95 imino proton (Table 1). These results indicate that the B4 bulge significantly destabilized the neighboring C15·G98 and U16·A97 base-pairs. The B5 bulge also destabilized the neighboring base-pairs. Thus, the S4 stem between the B4 and B5 bulges is much more unstable than the miR156a/miR156a* duplex. For example, U100, which contains the other neighboring imino proton of the B4 bulge, shows a severely line-broadened resonance (Figure 1C) and thus its kex value could not be determined at 16°C. In addition, the G11 and U101 imino protons in the S4 stem have kex values of 13.2 and 89.1 s−1 at 16°C, respectively, as compared with the kex values (G98: 5.0 s−1 and U16: 7.5 s−1) of the miR156a/miR156a* duplex.
Table 1. Hydrogen exchange rate constants (kex, s−1) of the imino protons of the WT and mutant pri-miR156a constructs in 90% H2O/ 10% D2O buffer containing 10 mM TRIS-d11, 50 mM NaCl at 16°C (pH = 8.25) and 23°C (pH = 8.03).
| Temp | Imino | WT | A9C | A9AC | A10G | A10CG |
|---|---|---|---|---|---|---|
| 16°C | G94 | 0.7 ± 0.4 | 0.7 ± 0.4 | 0.8 ± 0.4 | 0.9 ± 0.5 | 0.8 ± 0.4 |
| U95 | 3.8 ± 0.3 | 3.3 ± 0.2 | 3.4 ± 0.2 | 4.3 ± 0.3 | 3.4 ± 0.4 | |
| G17 | 0.3 ± 0.2 | 0.4 ± 0.2 | 0.4 ± 0.2 | 0.3 ± 0.2 | 0.4 ± 0.3 | |
| U16 | 7.5 ± 0.4 | 5.1 ± 0.4 | 5.9 ± 0.2 | 8.5 ± 0.4 | 4.2 ± 0.5 | |
| G98 | 5.0 ± 0.6 | 3.8 ± 0.4 | 4.0 ± 0.3 | 4.4 ± 0.3 | 3.7 ± 0.5 | |
| U100 | n.d.a | n.d. | 198 ± 8 | 249 ± 64 | 194 ± 38 | |
| U101 | 89.1 ± 4.9 | 78.5 ± 5.3 | 13.4 ± 0.4 | 17.3 ± 0.7 | 13.0 ± 0.8 | |
| G11 | 13.2 ± 0.8 | 8.6 ± 0.7 | 4.9 ± 0.5 | 0.6 ± 0.4 | 0.9 ± 0.5 | |
| G10 | −b | − | − | 5.2 ± 0.7 | 1.0 ± 0.5 | |
| G104 | − | − | 26.6 ± 0.9 | − | 12.9 ± 1.4 | |
| G105 | − | 6.8 ± 0.6 | − | − | − | |
| 23°C | G94 | 1.0 ± 0.2 | 1.0 ± 0.4 | 1.4 ± 0.4 | 1.5 ± 1.0 | 0.9 ± 0.4d |
| U95 | 4.8 ± 0.3 | 4.5 ± 0.6 | 7.2 ± 0.5 | 8.9 ± 0.9 | 5.8 ± 0.7 | |
| G17 | 0.4 ± 0.2 | 0.5 ± 0.2 | 0.4 ± 0.3 | 0.5 ± 0.4 | 0.5 ± 0.2 | |
| U16 | 11.1 ± 0.4 | 7.8 ± 0.7 | 13.5 ± 0.6 | 19.0 ± 0.9 | 6.6 ± 0.9 | |
| G98 | 7.2 ± 0.5 | 5.4 ± 0.8 | 8.7 ± 0.5 | 10.0 ± 0.9 | 6.7 ± 0.9 | |
| U100 | n.a.c | n.a. | n.d. | n.d. | n.d. | |
| U101 | n.d. | n.d. | 39.8 ± 1.7 | 43.5 ± 1.2 | 26.7 ± 1.2 | |
| G11 | 32.5 ± 3.4 | 17.8 ± 2.8 | 11.1 ± 1.2 | 1.2 ± 0.7 | 1.0 ± 0.6 | |
| G10 | − | − | − | 11.2 ± 0.8 | 0.9 ± 0.4d | |
| G104 | − | − | 86.8 ± 12.4 | − | 16.1 ± 1.6 | |
| G105 | − | 15.0 ± 1.4 | − | − | − |
aNot determined because of severe line-broadening.
bNo imino proton resonance.
cNot available because the imino proton resonance disappeared.
dThe G10 and G94 resonances overlapped with each other.
Hydrogen exchange of imino protons of mutant pri-miR156a constructs
The A9C pairing mutation in the B5 bulge produced slightly smaller kex values for the G11 and U101 imino protons in the S4 stem compared to WT pri-miR156a at 16°C (Figure 3A). In the case of the A9AC, A10G and A10CG mutants, the G11 and U101 imino protons have significantly smaller kex values compared to WT pri-miR156a (Figure 3A). These results indicate that the pairing mutations in the B5 bulge stabilize the base-pairs in the neighboring S4 stem.
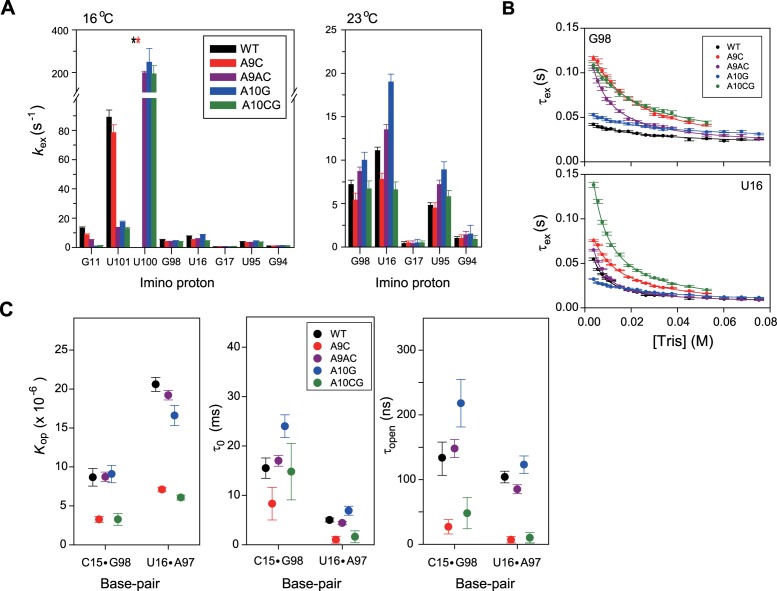
Figure 3.
Base-pair opening dynamics of WT and mutant pri-miR156a. (A) Hydrogen exchange rate constants (kex, s–1) of the imino protons of WT (black), A9C (red), A9AC (purple), A10G (blue) and A10CG (green) pri-miRNA156a at 16°C (left) and 23°C (right). Asterisks indicate that the imino proton resonances exhibited severe line-broadening. (B) Hydrogen exchange times (τex = 1/kex) for the G98 (upper) and U16 (lower) imino protons as a function of the Tris concentrations at 23°C. The solid lines are the best fits to Equation (6). (C) Equilibrium constants for base-pair opening (Kop), base-pair lifetimes (τ0) and lifetimes for base-pair opening (τopen) of WT (black), A9C (red), A9AC (purple), A10G (blue) and A10CG (green) pri-miRNA156a determined by the Tris-catalyzed NMR exchange experiments at 23°C.
These mutations also slightly affect the kex values of the miR156a/miR156a* duplex (Figure 3A). For a clearer comparison, the kex values were determined at 23°C. Interestingly, these B5-stabilizing mutations displayed distinct effects on the kex of the miR156a/miR156a* duplex. In the A9C mutant, the G98 and U16 imino protons have significantly smaller kex compared to WT pri-miR156a (Figure 3A). Similar results were also observed in the A10CG pri-miR156a (Figure 3A). Surprisingly, in the A9AC and A10G mutants, which stabilized the S4 stem, the kex of the G98 and U16 are slightly larger than those of WT pri-miR156a (Figure 3A). These results suggest that the stability of the S4 stem induced by the B5-stabilizing mutations showed no correlation with the kex of the the miR156a/miR156a* duplex.
Base-pair opening dynamics of WT and mutant pri-miR156a constructs
The equilibrium constants for base-pair opening (Kop) in the WT and mutant pri-miR156a constructs were determined from Tris-catalyzed imino proton exchange measurements at 23°C. The effects of [Tris] on τex ( = 1/kex) of the imino protons of the WT and mutant pri-miR156a were measured by inversion recovery experiments, and results for the G98 and U16 imino protons are shown in Figure 3B. From these data, the Kop (C15·G98: 8.5 × 10–6; U16·A97: 20.6 × 10–6) and base-pair lifetimes (τ0 = 1/kop) (C15·G98: 16 ms; U16·A97: 5 ms) in WT pri-miR156a were determined by curve fitting using Equation (6) (Table 2). These data can be used to calculate a lifetime for base-pair opening (τopen = 1/kcl) (C15·G98: 131 ns; U16·A97: 104 ns) using the relation, τopen = τ0Kop (Table 2).
Table 2. Base-pair dissociation constants (Kop), base-pair lifetimes (τ0 = 1/kop) and lifetimes for base-pair opening (τ open = 1/kcl) of the WT, A9C, A9AC, A10G and A10CG pri-miR156a constructs determined by the Tris-catalyzed NMR exchange experiments at 23°Ca.
| Base pair | Imino | WT | A9C | A9AC | A10G | A10CG | |
|---|---|---|---|---|---|---|---|
| C15·G98 | G98 | Kop (×10–6) | 8.45 ± 1.12 | 3.27 ± 0.37 | 8.72 ± 0.61 | 9.09 ± 1.38 | 3.27 ± 0.77 |
| τ0 (ms) | 15.5 ± 2.4 | 8.3 ± 3.3 | 17.0 ± 1.1 | 24.0 ± 2.3 | 14.8 ± 6.4 | ||
| τopen (ns) | 131 ± 27 | 27 ± 11 | 148 ± 14 | 218 ± 64 | 48 ± 24 | ||
| kint (×106) | 4.06 ± 1.05 | 2.22 ± 0.26 | 0.76 ± 0.10 | 3.36 ± 0.86 | 2.73 ± 0.60 | ||
| U16·A97 | U16 | Kop (×10–6) | 20.6 ± 0.9 | 7.10 ± 0.30 | 19.2 ± 0.6 | 16.4 ± 1.7 | 6.07 ± 0.31 |
| τ0 (ms) | 5.0 ± 0.4 | 1.0 ± 0.7 | 4.4 ± 0.3 | 7.6 ± 0.6 | 1.6 ± 1.2 | ||
| τopen (ns) | 104 ± 9 | 7 ± 5 | 85 ± 7 | 124 ± 16 | 10 ± 8 | ||
| kint (×106) | 0.37 ± 0.05 | 1.24 ± 0.07 | 0.26 ± 0.03 | 1.95 ± 0.23 | 0.60 ± 0.06 | ||
| G17·C96 | G17 | Kop (×10–6) | 0.42 ± 0.18 | 0.28 ± 0.09 | 0.46 ± 0.10 | 0.31 ± 0.15 | 0.33 ± 0.13 |
| τ0 (ms) | 145 ± 115 | 287 ± 79 | 132 ± 42 | 340 ± 143 | 333 ± 139 | ||
| τopen (ns) | 61 ± 48 | 80 ± 33 | 61 ± 23 | 107 ± 72 | 110 ± 64 | ||
| kint (×106) | 0.41 ± 0.39 | 0.61 ± 0.49 | 0.57 ± 0.41 | 0.97 ± 0.69 | 0.97 ± 0.69 | ||
| A18·U95 | U95 | Kop (×10–6) | 4.68 ± 0.84 | 2.34 ± 0.26 | 5.00 ± 0.54 | 1.91 ± 0.95 | 2.69 ± 0.34 |
| τ0 (ms) | 18 ± 4 | 15 ± 4 | 21 ± 2 | 14 ± 6 | 21 ± 4 | ||
| τopen (ns) | 84 ± 22 | 36 ± 9 | 102 ± 14 | 26 ± 14 | 57 ± 14 | ||
| kint (×106) | 1.30 ± 0.39 | 2.73 ± 0.34 | 1.05 ± 0.24 | 6.06 ± 2.27 | 1.89 ± 0.32 | ||
| C19·G94 | G94 | Kop (×10–6) | 1.93 ± 0.29 | 0.89 ± 0.17 | 1.41 ± 0.26 | 1.01 ± 0.43 | 1.45 ± 0.29 |
| τ0 (ms) | 194 ± 11 | 220 ± 31 | 160 ± 14 | 180 ± 49 | 143 ± 17 | ||
| τopen (ns) | 375 ± 61 | 197 ± 48 | 225 ± 45 | 182 ± 92 | 207 ± 48 | ||
| kint (×106) | 0.52 ± 0.20 | 0.22 ± 0.17 | 0.80 ± 0.32 | 1.03 ± 0.43 | 0.97 ± 0.33 |
aParameters used in the calculation: kcoll = 1.5 × 109 s–1, pKa(G-NH1) = 9.24, pKa (U-NH3) = 9.20, pKa (Tris, 23°C) = 8.47; Sample condition: 100 mM NaCl. [Tris]total = 10–300 mM, 23°C. The pH of all samples and buffers was adjusted to 8.00 at 24.1°C and then calibrated to 8.03 at 23°C using the following equation: pH(23°C) = pH(24.1°C)−0.031 × (23–24.1). The errors for these values were determined from the curve fitting using Equation (6).
The A9C pri-miR156a has 2.5- and 3-fold smaller Kop for the C15·G98 and U16·A97 base-pairs, respectively, compared to WT pri-miR156a (Figure 3C), although both RNAs have similar stabilities of the S4 stem (Figure 3A). In addition, these base-pairs have significantly shorter τ0 and τopen values in the A9C than WT pri-miR156a (Figure 3C). Similar results were observed for the A10CG pri-miR156a (Figure 3C). In contrast, in the A9AC and A10G pri-miR156a constructs, there were no significant effects on the base-pair opening dynamics of the the C15·G98 and U16·A97 base-pairs (Figure 3C). These results indicate that the A9AC and A10G mutations on the B5 bulge had little effect on the base-pair stabilities of C15·G98 and U16·A97, even though they led to greater stabilization of the S4 stem.
Base-pair opening thermodynamics of WT and mutant pri-miR156a constructs
The difference in Gibbs free energy between the closed and completely open states (ΔGobp) of the C15·G98 (–6.87 ± 0.14 kcal/mol) and U16·A97 (–6.35 ± 0.05 kcal/mol) base-pairs can be calculated from Kop using Equation (8). This approach revealed that the differences between the ΔGobp values of the C15·G98 and U16·A97 base-pairs in the A10G and WT pri-miR156a (ΔΔGobp) were only 0.04 and –0.13 kcal/mol, respectively (Table 3). Similar results were observed for the A9AC pri-miR156a (Table 3). However, in the A9C and A10CG pri-miR156a, the ΔΔGobp values of the C15·G98 and U16·A97 base-pairs were –0.7 to –0.5 kcal/mol (Table 3). These results indicate that the A9C and A10CG mutations on the B5 bulge led to greater stabilization of the C15·G98 and U16·A97 base-pairs, while the A9AC and A10G mutations had little effect on these base-pair stabilities.
Table 3. Gibbs free energy (kcal/mol) for base-pair stability of the WT and mutant pri-miR156a constructs at 23°C.
| Base pair | WT | A9C | A9AC | A10G | A10CG | |
|---|---|---|---|---|---|---|
| C15·G98 | ΔG°bp | –6.87 ± 0.14 | –7.44 ± 0.11 | –6.86 ± 0.07 | –6.83 ± 0.12 | –7.43 ± 0.24 |
| ΔΔG°bp | – | –0.57 | 0.01 | 0.04 | –0.56 | |
| ΔΔG‡op | – | –0.37 | 0.05 | 0.26 | –0.04 | |
| ΔΔG‡cl | – | –0.94 | 0.06 | 0.30 | –0.60 | |
| U16·A97 | ΔG°bp | –6.35 ± 0.05 | –6.98 ± 0.04 | –6.39 ± 0.03 | –6.48 ± 0.08 | –7.07 ± 0.05 |
| ΔΔG°bp | – | –0.63 | –0.04 | –0.13 | –0.72 | |
| ΔΔG‡op | – | –0.96 | –0.08 | –0.07 | –0.66 | |
| ΔΔG‡cl | – | –1.59 | –0.12 | –0.20 | –1.38 | |
| U16·A97 + C15·G98 | ΔG°bp | –13.22 | –14.42 | –13.25 | –13.31 | –14.50 |
| ΔΔG°bp | – | –1.20 | –0.03 | –0.09 | –1.28 | |
| ΔΔG‡op | – | –1.33 | –0.03 | 0.19 | –0.70 | |
| ΔΔG‡cl | – | –2.53 | –0.06 | 0.10 | –1.98 | |
| G17·C96 | ΔG°bp | –8.44 ± 0.43 | –8.67 ± 0.32 | –8.38 ± 0.22 | –8.61 ± 0.48 | –8.57 ± 0.39 |
| ΔΔG°bp | – | –0.23 | 0.06 | –0.17 | –0.13 | |
| ΔΔG‡op | – | 0.39 | –0.06 | 0.49 | 0.47 | |
| ΔΔG‡cl | – | 0.16 | 0.00 | 0.32 | 0.34 | |
| A18·U95 | ΔG°bp | –7.05 ± 0.18 | –7.45 ± 0.11 | –7.01 ± 0.07 | –7.57 ± 0.50 | –7.37 ± 0.13 |
| ΔΔG°bp | – | –0.40 | 0.04 | –0.52 | –0.32 | |
| ΔΔG‡op | – | –0.10 | 0.08 | –0.15 | 0.09 | |
| ΔΔG‡cl | – | –0.50 | 0.12 | –0.67 | –0.23 | |
| C19·G94 | ΔG°bp | –7.56 ± 0.15 | –8.00 ± 0.19 | –7.74 ± 0.18 | –7.94 ± 0.43 | –7.73 ± 0.20 |
| ΔΔG°bp | – | –0.44 | –0.18 | –0.38 | –0.17 | |
| ΔΔG‡op | – | 0.07 | –0.11 | –0.04 | –0.17 | |
| ΔΔG‡cl | – | –0.37 | –0.29 | –0.42 | –0.34 |
aΔG°bp = –ΔG°opening = RTln(Kop), T = 296.15 K.
bΔΔG°bp = ΔG°bp,mut – ΔG°bp,WT.
cΔΔG‡op = ΔG‡op,mut – ΔG‡op,WT = –RTln(kop,mut/kop,WT) = RTln(τ0,mut/τ0,WT), T = 296.15 K.
dΔΔG‡cl = ΔG‡cl,mut – ΔG‡cl,WT = –RTln(kcl,mut/kcl,WT) = RTln(τopen,mut/τopen,WT), T = 296.15 K.
The ΔG‡op and ΔG‡cl values represent energy differences between the closed and transition states, and the open and transition states, respectively. In the A9C and A10CG pri-miR156a, the C15·G98 and U16·A97 base-pairs had significantly lower activation energies for base-pair opening compared to WT, with ΔΔG‡cl of –0.6 to –1.4 kcal/mol (Table 3). However, the A9AC and A10G mutations had little effect on the activation energies for base-pair opening (Table 3).
Mature miR156 levels of plants overexpressing WT and mutant miR156a
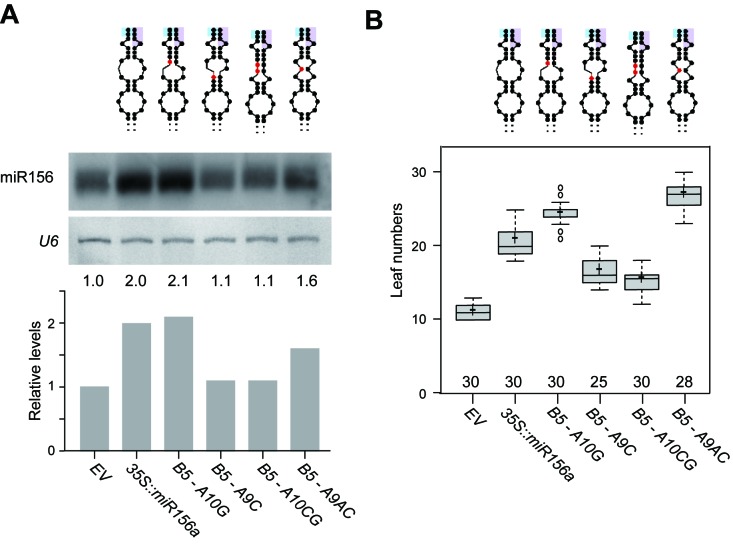
Next, to analyze the in vivo effects of the four B5-stabilizing mutations on pri-miR156a processing, we determined the levels of mature miR156 in plants overexpressing the WT pri-miR156a construct (35S::miR156a) or B5-stabilizing mutant constructs (B5-A9C, B5-A9AC, B5-A10G and B5-A10CG) and compared them with the empty vector control (EV) plants (Figure 4A). Small RNA gel blot analyses showed that the 35S::miR156a plants accumulated 2-fold larger amounts of mature miR156 than EV plants (Figure 4A). Interestingly, significantly lower miR156 levels were seen in B5-A9C and B5-A10CG plants, whereas B5-A10G and B5-A9AC plants had similar or higher levels of miR156, compared to 35S::miR156a plants, suggesting that the mature miR156 levels in B5-stabilizing mutated plants could be categorized into two groups. These results showed that the B5-stabilizing mutations in the lower stem of pri-miR156a had some correlation with miR156 processing.
Figure 4.
Mature miR156 levels and leaf numbers of plants overexpressing WT and mutant pri-miR156a. (A) Mature miR156 level in plants overexpressing WT or B5-stabilizing mutant miR156a grown under long-day conditions. U6 was used as a loading control. Introduced mutations are indicated in red in the secondary structure of pri-miR156a above the gel image. Partial sequences of miR156a and miR156a* are shaded in cyan and purple, respectively. (B) Leaf numbers of homozygous plants carrying WT or B5-stabilizing mutant miR156a. Leaf numbers are presented as a box-and-whiskers plot. Center lines show the medians, plus symbols indicate mean values and box limits indicate the 25th and 75th percentiles as determined by R software. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, and outliers are represented by dots. Numbers above the X-axis indicate the number of plants scored.
Leaf numbers at flowering of plants overexpressing WT and mutant miR156a
The leaf number before flowering is thought to have an impact on the flowering time of plants. Thus we analyzed the changes in leaf number of plants carrying WT or B5-stabilizing mutant miR156a at 23°C (Supplementary Figure S1). B5-A9C (16.5 leaves) and B5-A10CG plants (15.3 leaves) produced fewer leaves than 35S::miR156a plants (20.8 leaves) (Figure 4B), consistent with the decreased miR156 levels (Figure 4A). In contrast, B5-A10G (24.3 leaves) and B5-A9AC plants (26.9 leaves) produced more leaves than 35S::miR156a plants (Figure 4B). A significant correlation between leaf numbers and mature miR156 levels in plants overexpressing WT or B5-stabilizing mutant miR156a was observed with Pearson correlation values of 0.67 (P = 0.017) (data not shown).
DISCUSSION
NMR hydrogen exchange studies have been performed to probe the thermodynamics and kinetics of base-pair opening in a variety of nucleic acid systems. Our study shows that the stable G17·C96 base-pair has Kop values of 0.3–0.5 × 10–6 in all pri-miR156a constructs (Table 2). These values have orders of magnitude similar to the previous Kop values of the stable G·C base-pairs in the P1 duplex of Tetrahymena group I ribozyme (20), sarcin–ricin domain RNA (29), and various RNA duplexes (30). The Kop values of the A18·U95 base-pair also show a similar order of magnitude to those of stable A·U base-pairs previously reported (20,29,30). In the WT pri-miR156a, the C15·G98 base-pair next to the B4 bulge has a Kop of 8.5 × 10–6 at 23°C, which is 20-fold larger than that of the stable G17·C96 base-pair (Table 2). Similarly, the U16·A97 base-pair has a 4-fold larger Kop value than the A18·U95 base-pair (Table 2). It was reported that the G·U wobble pairing in the RNA duplex significantly affects the stabilities of neighboring base-pairs compared to the Watson–Crick base-pairing (20,31). These results indicate that the A14/A99 mismatch at the B4 bulge significantly destabilized the neighboring C15·G98 and U16·A97 base-pairs, like the previously reported G·U wobble pair.
In this study, we analyzed base-pair opening dynamics of the pri-miR156a lower stem and found that the stability of its secondary structure is important for miR156a processing. Our results are consistent with a previous finding that the change in base-pairing stability of a specific site on a pri-miRNA foldback affected miRNA processing (1). For instance, the substitution of a base at 5 nucleotides below the miR390a/miR390a* duplex, which showed lower calculated entropy, resulted in significantly reduced levels of miR390 (9). By contrast, the change of a base showing higher calculated entropy at the site did not induce any change in the levels of miR390 (9).
We also provide direct NMR evidence that the stability of the secondary structure of pri-miRNA is related to miRNA processing. The B5-stabilizing mutations affected the base-pair stabilities of the neighboring S4 stem. In A9AC, A10G and A10CG mutants, the G11 and U101 imino protons had much smaller kex values compared to WT pri-miR156a (Figure 3A). The A10CG mutation significantly decreased miR156 levels, whereas the A9AC and A10G mutations did not affect miR156 levels (Figure 4A). Interestingly, the A9C mutation reduced the efficiency of pri-miR156a processing (Figure 4A), even though it caused no significant effect on the base-pair stabilities of the S4 stem (Figure 3A), indicating that the base-pair stability of the S4 stem is not important for the cleavage of pri-miR156a.
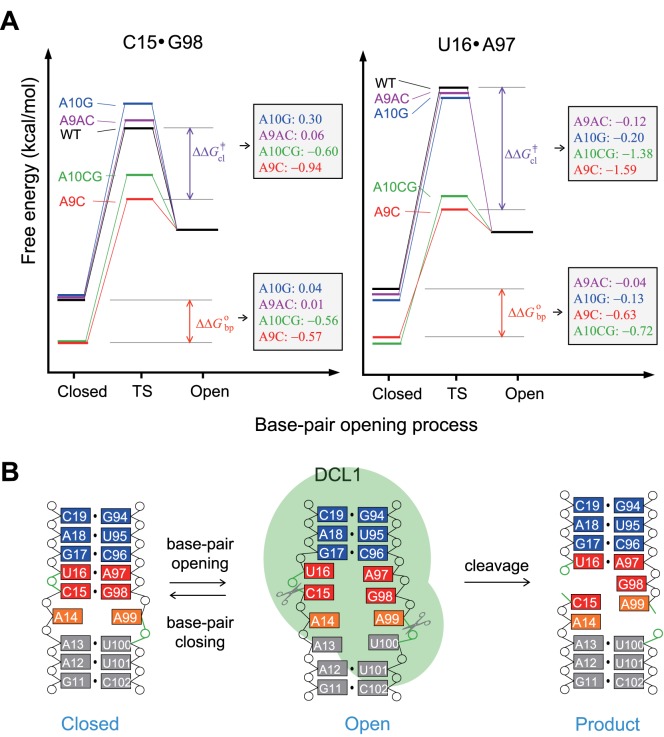
Instead, the base-pair stability at the cleavage site of pri-miR156a exhibited a strong correlation with mature miR156 levels and leaf number data. The A9C mutation induced greater stabilization of the C15·G98 and U16·A97 base-pairs with ΔΔGobp values of –0.57 and –0.63 kcal/mol, respectively (Figure 5A). In addition, the C15·G98 and U16·A97 base-pairs had the stable transition state for base-pair opening, with ΔΔG‡cl of –0.94 and –1.59 kcal/mol, respectively (Figure 5A). Similar results were observed for the A10CG mutants (Figure 5A). Interestingly, the B5-A9C and B5-A10CG plants showed reduced mature miR156 levels, comparing to the plants carrying the WT miR156a construct (Figure 4A), and flowered with fewer leaves than the 35S::miR156a plants (Figure 4B).
Figure 5.
Proposed mechanism for the cleavage reaction of pri-miR156a. (A) Schematic representations of the Gibbs free energy diagram of the base pair opening and closing of the C15·G98 (left) and U16·A97 (right) base-pairs in the WT (black), A9C (red), A9AC (purple), A10G (blue) and A10CG (green) pri-miR156a at 23°C. Red and purple arrows indicate differences in ΔG°bp and ΔG‡cl between the WT and A9C pri-miR156a, respectively. The ΔΔG°bp and ΔΔG‡cl values of A9C, A9AC, A10G and A10CG mutants are shown in the right box. (B) Suggested mechanism for the cleavage reaction of pri-miR156a during the biogenesis of miR156.
In contrast, the A9AC and A10G mutations did not significantly affect the stabilities and opening dynamics of the C15·G98 and U16·A97 base-pairs. Also, the B5-A9AC and B5-A10G plants showed similar or higher mature miR156 levels (Figure 4A) and flowered with more leaves than the 35S::miR156a plants (Figure 4B). Thus, based on the altered processing observed in the transgenic plants used in this study, we suggest that the stabilities and/or opening dynamics of the C15·G98 and U16·A97 base-pairs at the cleavage site are essential for efficient processing of pri-miR156a, to produce mature miR156.
In animals, pri-miRNAs are first processed by ribonuclease III (RNase III), the Drosha-DGCR8 complex, and then further cleaved by another RNase III, Dicer (32,33). In the case of prokaryotes, X-ray crystal structural studies of double-stranded RNA (dsRNA) complexes with RNase III revealed that some base-pairs near the cleavage sites are partially open to form an active RNA–protein–metal conformation for efficient phosphodiester hydrolysis (34). For example, in the dsRNA complexed with Aquifex aeolicus RNase III, the G·C base-pair at the R+1 position of the cleavage site is partially open, as evidenced by the observation that the heavy atom distances of G-N1↔C-N3 (3.5 Å) and G-O6↔C-N4 (3.6 Å) are longer than those of a Watson–Crick base-pair (2.7–2.9 Å) (34). In plants, the RNase III-like enzyme, DCL1 and two cofactors, HYL1 and SE, orchestrate this process (35). It seems that the information embedded in the secondary structures is important for miRNA generation via the recognition and cleavage by DCL1-HYL1-SE complex (1–4). Indeed, inefficient and inaccurate processing of pri-miR390a occurred in structural mutants of pri-miR390a, possibly due to a loss of interaction with HYL1 (1). Furthermore, an in vitro processing experiment using pri-miR171a containing an artificially created long unstructured segment below the miR171/miR171* duplex generated an aberrant miRNA (4). In our studies, the partially or fully open states of the C15·G98 and U16·A97 base-pairs adjacent to the cleavage site are thought to be essential for formation of the active DCL1-HYL1-SE conformation for pri-miR156a processing (Figure 5B). Using free energy differences calculated from imino proton exchange experiments (Figure 5A), we estimated the difference in energy that the C15·G98 and U16·A97 base-pairs in the WT and mutant pri-miR156a constructs require to reach a partially or fully open state from a closed state. These energy differences in the A9C and A10CG pri-miR156a constructs, which can be extrapolated from the ΔΔGobp values, are much larger than those of the WT, A10G and A9AC pri-miR156a constructs (Figure 5A). Thus, the pri-miR156a substrate containing more stable nucleotide combinations of these two base-pairs requires more energy to form the active conformation and be processed by the DCL1 complex. Based on this hypothesis, in the A9C and A10CG mutants, these two base-pairs are more stable than those of WT pri-miR156a, which explains why these mutants need more energy to form open base-pairs (Figure 5A). Conversely, the A9AC and A10G mutations caused little effect on the miR156 production because the active conformation could easily form, just like WT pri-miR156a (Figure 5A).
An important question is which protein recognizes the altered secondary structures of pri-miR156a at different temperatures. We propose that HYL1 is one of the promising candidates, as dimerized HYL1 scans through pri-miRNA to recognize the miRNA/miRNA* duplex. The dsRNA binding domain1 (dsRBD1) of HYL1 recognizes the miRNA/miRNA* duplex and binds to the duplex in a structure-dependent manner, not in a sequence-dependent manner (3). It is thus possible that A9C and A10CG mutations, which cause more stable C15·G98 and U16·A97 base-pairs adjacent to the cleavage site of pri-miR156a, affect the recognition of miR156a/miR156a* duplex by the dsRBD1 of HYL1, and ultimately lead to visible changes in mature miR156 production. Further study will be necessary to determine the molecular mechanism of the recognition of the temperature-dependent changes in the secondary structures of pri-miRNA during miRNA processing.
Based on our results, we conclude that precisely tuned base-pair stability at the cleavage site is essential for efficient processing of pri-miR156a. Mutations adjacent to the cleavage site can modulate mature miR156 levels as well as leaf number phenotypes via changes in the base-pair stability of the cleavage site (Figure 5B).
Acknowledgments
The authors thank the GNU Central Instrument Facility for the NMR experiments, Young Ja Kim for her technical assistance and Melissa Stauffer, of Scientific Editing Solutions, for editing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [2013R1A2A2A05003837 and 2012R1A4A1027750 (BRL) to J.-H.L.], which is funded by the Korean Government (MSIP); Creative Research Initiative Program [2008-0061988 to J.H.A]; Next-Generation BioGreen21 Program [SSAC, no. PJ01117701 to J.-H.L. (Rural Development Administration, Korea)]. Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 2.Kurihara Y., Takashi Y., Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., Ebright Y.W., Yu B., Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai H.L., Li Y.H., Hsieh W.P., Lin M.C., Ahn J.H., Wu S.H. HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell. 2014;26:2858–2872. doi: 10.1105/tpc.114.126722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulsen C., Vaucheret H., Brodersen P. Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell. 2013;25:22–37. doi: 10.1105/tpc.112.105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis miR172a precursor. Curr. Biol. 2010;20:49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 7.Werner S., Wollmann H., Schneeberger K., Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 8.Song L., Axtell M.J., Fedoroff N.V. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 9.Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:466–471. doi: 10.1073/pnas.0913203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bologna N.G., Schapire A.L., Zhai J., Chorostecki U., Boisbouvier J., Meyers B.C., Palatnik J.F. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013;23:1675–1689. doi: 10.1101/gr.153387.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H., Yoo S.J., Lee J.H., Kim W., Yoo S.K., Fitzgerald H., Carrington J.C., Ahn J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010;38:3081–3093. doi: 10.1093/nar/gkp1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C.M., Wang J.W. Regulation of flowering time by microRNAs. J. Genet. Genomics. 2013;40:211–215. doi: 10.1016/j.jgg.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Kim W., Ahn J.H. MicroRNA-target interactions: important signaling modules regulating flowering time in diverse plant species. Crit. Rev. Plant Sci. 2014;33:470–485. [Google Scholar]
- 14.Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G., Poethig R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aukerman M.J., Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan C. Buffers: A guide for the preparation and use of buffers in biological systems. Darmstadt: CalBiochem; 2003. [Google Scholar]
- 18.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Goddard T., Kneller D. SPARKY 3. San Francisco: University of California; 2003. [Google Scholar]
- 20.Lee J.-H., Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J.-H., Jucker F., Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS Lett. 2008;582:1835–1839. doi: 10.1016/j.febslet.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang J., Kang Y.-M., Park C.-J., Lee J.-H., Choi B.-S. Thermodynamics and kinetics for base pair opening in the DNA decamer duplexes containing cyclobutane-pyrimidine dimer. FEBS Lett. 2009;583:2037–2041. doi: 10.1016/j.febslet.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Bang J., Bae S.-H., Park C.-J., Lee J.-H., Choi B.-S. Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites. J. Am. Chem. Soc. 2008;130:17688–17696. doi: 10.1021/ja8038272. [DOI] [PubMed] [Google Scholar]
- 24.Leroy J.L., Bolo N., Figueroa N., Plateau P., Gueron M. Internal motions of transfer RNA: a study of exchanging protons by magnetic resonance. J. Biomol. Struct. Dyn. 1985;2:915–939. doi: 10.1080/07391102.1985.10507609. [DOI] [PubMed] [Google Scholar]
- 25.Gueron M., Leroy J.L. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- 27.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 28.Pall G.S., Hamilton A. J. Improved northern blot method for enhanced detection of small RNA. Nat. Protocol. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Jiang L., Michalczyk R., Russu I.M. Structural energetics and base-pair opening dynamics in sarcin-ricin domain RNA. Biochemistry. 2006;45:13606–13613. doi: 10.1021/bi060908n. [DOI] [PubMed] [Google Scholar]
- 30.Snoussi K., Leroy J.-L. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- 31.Varnai P., Canalia M., Leroy J.-L. Opening mechanism of G·T/U pairs in DNA and RNA duplexes: A combind study of imino proton exchange and molecular dynamics simulation. J. Am. Chem. Soc. 2004;126:14659–14667. doi: 10.1021/ja0470721. [DOI] [PubMed] [Google Scholar]
- 32.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 34.Gan J., Shaw G., Tropea J.E., Waugh D.S., Court D.L., Ji X. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol. Microbiol. 2008;67:143–164. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 35.Dong Z., Han M.H., Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]