Abstract
Alternative splicing of terminal exons increases transcript and protein diversity. How physiological and pathological stimuli regulate the choice between alternative terminal exons is, however, largely unknown. Here, we show that Brahma (BRM), the ATPase subunit of the hSWI/SNF chromatin-remodeling complex interacts with BRCA1/BARD1, which ubiquitinates the 50 kDa subunit of the 3′ end processing factor CstF. This results in the inhibition of transcript cleavage at the proximal poly(A) site and a shift towards inclusion of the distal terminal exon. Upon oxidative stress, BRM is depleted, cleavage inhibition is released, and inclusion of the proximal last exon is favoored. Our findings elucidate a novel regulatory mechanism, distinct from the modulation of transcription elongation by BRM that controls alternative splicing of internal exons.
INTRODUCTION
Pre-mRNA splicing and 3′ end processing are essential steps for the expression of the vast majority of metazoan genes. Recently, RNAseq analysis showed that not only alternative splicing (AS) but also alternative polyadenylation (APA) is more frequent and complex than previously anticipated (1–4). The choice of alternative poly(A) sites generates different 3′ UTRs that can affect translation, stability and localization of the mRNA. Alternative pre-mRNA processing changes the length of the 3′ UTR during cell differentiation contributing to the regulation of gene expression (5,6). When coupled to the inclusion of an alternative last exon (ALE), alternative polyadenylation leads to the generation of mRNA variants that differ in their 3′ UTR and that may encode proteins with different C-terminal regions.
Whereas the molecular details of pre-mRNA 3′ end processing are rather well known, how the choice of APA sites is regulated is only partially understood. The mature 3′ ends of most eukaryotic mRNAs are generated by endonucleolytic cleavage of the primary transcript followed by the addition of a poly(A) tail to the upstream cleavage product (1,7). Maturation of the 3′ end is executed by a large multicomponent complex that is assembled in a cooperative manner on specific cis-acting sequence elements in the pre-mRNA (8,9). In mammalian cells, the cleavage and polyadenylation specificity factor (CPSF) recognizes the consensus hexanucleotide AAUAAA, whereas the cleavage stimulation factor (CstF), a hexameric complex of subunits of 77, 64 and 50 kDa (10,11), binds a more degenerate GU- or U-rich element downstream of the poly(A) site. Both CPSF and CstF interact with RNA polymerase II (RNAPII) at the promoter and appear to remain associated with it during elongation (12,13). CstF50 and CstF77 interact specifically with the carboxy-terminal domain (CTD) of RNAPII largest subunit. CstF50 was also shown to bind the BARD1/BRCA1 ubiquitin ligase after DNA damage, resulting in the inhibition of 3′ end processing (14). This observation and the fact that APA is modulated in development, differentiation and neuronal activation (reviewed in (9)) indicates that 3′ end processing can be regulated in response to physiological and pathological stimuli.
Oxidative stress is a widely occurring phenomenon in biological systems, and is due to an imbalance between the intracellular production or influx of reactive oxygen species (ROS) and the availability of antioxidant compounds, such as glutathione. Oxidative stress has been implicated in the etiology of many neurodegenerative disorders, including Alzheimer's and Parkinson's diseases (15). At the cell level, oxidative stress elicits a wide spectrum of responses ranging from proliferation to growth arrest, senescence, or cell death. The particular outcome reflects the balance between a variety of intracellular stress signalling pathways that are activated in response to the oxidative insult and that ultimately modulate gene expression.
We recently described that exposure of human neuroblastoma SH-SY5Y cells to different sources of ROS leads to genome-wide alternative splicing changes, modifying the relative proportion of alternatively spliced forms (16). Here, we show that oxidative stress specifically affects the choice of ALEs increasing the production of transcripts variants terminating at a more proximal poly(A). Oxidative stress induces the transcriptional downregulation of Brahma (BRM), one of the two alternative ATPase subunits of the SWI/SNF complex. We find that in normal condition BRM is enriched on the proximal ALE. In addition we observe the accumulation of BARD1, a protein that forms a functional heterodimer with BRCA1, which has E3 ubiquitin-ligase activity and interacts with the 50 kDa subunit of CstF inhibiting 3′ end processing (17). Consistent with these observations, we detect an ubiquitinated pool of CstF50 and show that ubiquitination is mediated by BARD1/BRCA1. Taken together, our results suggest that the presence of BRM on the proximal exon leads to the BARD1/BRCA1-mediated ubiquitination of CstF50 and the inhibition of 3′ end processing at the proximal poly(A). This in turn allows transcription to proceed to the distal terminal exon.
MATERIALS AND METHODS
Cell culture and transfections
Human neuroblastoma SH-SY5Y, SH-SY5Y/SOD1, SH-SY5Y/SOD1(G93A) and HEK293T cells were cultured in D-MEM High Glucose medium (Gibco, Invitrogen), 10% fetal bovine serum (FBS), 2.5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Euroclone) at 37°C with 5% CO2. Paraquat (Sigma-Aldrich) treatment of SH-SY5Y cells was carried out as described (16). Resveratrol (Sigma-Aldrich) was dissolved in DMSO, and cells were treated with 10 μM resveratrol for 24 h. Control cells were treated with vehicle. Adult dermal fibroblats (ATCC, PCS-201-012) were cultured D-MEM High Glucose medium (Gibco, Invitrogen), 10% fetal bovine serum (FBS), 5 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Euroclone) at 37°C with 5% CO2. Treatment with 2 mM paraquat was performed for 48 h, while treatment with 0.2 mM H2O2 (Sigma-Aldrich) was performed for 24 h.
Cells were transfected using polyethylenimine (PEI, Sigma-Aldrich) according to the manufacturer's instructions. Stably transfected cells were obtained by selection with 1 μg/ml puromycin (Sigma-Aldrich). For depletion of BRCA1, HEK293T cells were transfected twice (day 0 and day 3) in six-well plates with 75 pmol of the corresponding Silencer® select siRNAs and Negative control siRNA #1 (Ambion). Transfection was carried out using Lipofectamine® RNAiMax reagent (Invitrogen) following manufacturer's instructions. Cells were expanded and lysed at day 6.
For BRM-K755A overexpression, cells were electroporated with Amaxa® nucleofector® system (Lonza) using Amaxa® Cell Line Nucleofector® Kit V according to the manufacturer's instructions.
Intracellular ROS were measured with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA; Sigma-Aldrich, D6883). Cells were exposed to H2DCF-DA (20 μM) during the last 30 min of culture, then collected and washed with PBS. A blank sample (cells not exposed to H2DCF-DA) was also prepared. The H2DCF-DA fluorescence was measured by flow cytometry after addition of propidium iodide (PI) to the samples. Only the cellular population of PI impermeable cells was considered for measuring the fluorescence intensity of H2DCF-DA (18).
Plasmids and antibodies
The cDNAs encoding the N-terminal flag-tagged human BRM and N-terminal flag-tagged BRG1 proteins were a gift from B. Emerson. These cDNAs were subcloned into the SmaI–SalI (New England Biolabs) sites of the pAD5-CMV-Wpre-PGK-Puro expression vector (a kind gift from S. Philipsen). The mutation K755A (19) was introduced using a two-step mutagenic PCR procedure using Phusion polymerase (Finnzymes). The interfering shRNA oligonucleotides targeting human SMARCA2 and SMARCA4 transcripts were designed using the SiDesign Center (Dharmacon). The shRNA primers were cloned into pSUPuro vector. pSUPuro, and the pSUPuro ß2 T-Cell Receptor Beta used as an unrelated control shRNA were gifts from M.D. Ruepp. BARD1 and BRCA1 constructs were a gift of N. Chiba. The cDNA encoding CstF50 was subcloned from pcDNA3 HA-CstF50 (a gift from M.D. Ruepp) into a p3XFLAG-myc-CMV26 removing the myc tag. The histidine-tagged Ubiquitin was a gift of M.L. Guerrini. All constructs were verified by sequencing (BMR Genomics). All oligonucleotide sequences are listed in Supplementary Table S2. The commercial antibodies used are listed in the Supplementary Table S1. Non-immune rabbit IgGs (Millipore) were used as a control in the immunoprecipitation assays.
Luciferase assay
SH-SY5Y cells were transiently co-transfected with the indicated pGL2 luciferase reporter plasmids and with the Renilla-encoding pRL-TK plasmid (Promega Inc.). Twenty-four hours after transfection, cells were lysed and luciferase activity quantified using the Dual Luciferase Reporter kit (Promega Inc.) and a Berthold luminometer (Berthold Technologies). For the luciferase experiments, paraquat was added 3 h after transfection.
Bioinformatic analysis
Transcripts characterized by alternative splicing events at their 3′ end were detected by R scripting using Bioconductor 2.12 packages GenomicRanges, TxDb.Hsapiens.UCSC.hg19.knownGene and HuExExonProbesetLocation (www.bioconductor.org). Transcripts were extracted from TxDb.Hsapiens.UCSC.hg19.knownGene (80922 transcripts). After removal of transcripts lacking a link to Entrez Gene Identifier (20), 71 350 transcripts (26 5661 exons), associated to 22 932 EG, were left for further analysis. Subsequently, we selected all genes associated to the presence of alternative splicing even at the 3′ end (12 839 genes, 58 451 transcripts, 26 608 exons involved in ALE). Affymetrix Human Exon 1.0 ST Array (HuEx-1_0-st) exon-level probe sets chromosomal locations were extracted from HuExExonProbesetLocation (21). Only exon-level probesets associated to the Affymetrix core annotation were considered (284805 exon-level probesets). These exon-level probesets mapped on 12839 genes (59 986 UCSC transcripts, 230 112 exons). Out of the 230 112 exons 22 983 were associated to ALE. Alternatively spliced exon-level probesets were retrieved by Lenzken et al. (16). Four hyndred six exon-level probesets mapping on 418 exons were associated to 262 genes (1191 UCSC transcripts, 4844 exons). Within the 418 exons 89 exons (78 UCSC genes) were involved in ALE.
RNA analysis
RNA preparation and RT-PCR reactions were performed as described (16). The sequences of the exon-specific primers are listed in Supplementary Table S2. Assay conditions were optimized for each gene with respect to primer annealing temperatures, primer concentrations, and MgCl2 concentrations. Quantification was performed with Bioanalyzer 2100 (Agilent Technologies).
Chromatin immunoprecipitation and qPCR analysis
107 SH-SY5Y cells were cross-linked with 1% formaldehyde (Sigma-Aldrich), and quenched with the addition of 125 mM glycine (Sigma-Aldrich). Cell pellets were lysed in Lysis Buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitors (Roche) for 1 hour on ice, and then sonicated with a Branson 250 sonifier (Branson Inc.) to obtain chromatin fragments of ∼400 nt. Aliquots corresponding to 5 × 106 cells were flash-frozen in liquid nitrogen and then maintained at −80°C. For each immunoprecipitation, 1% of the total genomic DNA was saved as input DNA. The chromatin solution was precleared at 4°C for 1 h with sepharose beads. Immunoprecipitation was carried out using the Chromatin IP Assay Kit (Millipore), following manufacturer's instructions. Bound material was eluted with 500 μl of Elution Buffer (1% SDS, 100 mM NaHCO3) for 1 h at room temperature. Crosslinking was reversed at 65°C overnight, with the addition of 250 mM NaCl and 2 μg/ml Proteinase K (Sigma). DNA was purified by phenol:choloform:isoamyl alcohol (Sigma-Aldrich) extraction and ethanol precipitation. For ChIP-ReChIP experiments, eluates from the first ChIP assays were diluted in the ChIP Dilution buffer (Millipore) to reduce the SDS concentration to 0.1% (w/v). Then, the second ChIP was performed. Immunoprecipitated DNA was analyzed in triplicate by qPCR using the SYBR Green method with Mesa Green qPCR master mix (Biosense) in an ABI PRISM 7500 (Applied Biosystem). The primers used for the qPCRs are listed in the Supplemental Table S2. Data were normalized by the Fold Enrichment Method as follows: Relative Enrichment = 2 – (ΔCt antibody- ΔCt IgG).
Micrococcal nuclease assays
107 SH-SY5Y cells were synchronized in G0 by serum starvation as assessed by FACS analysis (FACSCalibur, BD Biosciences). Cells were cross-linked with 1% (v/v) formaldehyde and quenched with the addition of 125 mM glycine. Cell pellets were lysed in Lysis Buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitors (Roche). Nuclei were disrupted by five bursts of sonication. Aliquots of 5 × 106 cells were flash-frozen in liquid nitrogen and then maintained at −80°C. One undigested aliquot was used as control. Digestion was performed with 20 U of Micrococcal Nuclease (Fermentas) in 1 mM final CaCl2 and MgCl2 for 30 min at room temperature, then stopped with 20 mM EDTA. Crosslinking was reversed at 65°C overnight, with the addition of 250 mM NaCl and 2 μg/ml Proteinase K followed by phenol:choloform:isoamyl alcohol extraction and ethanol precipitation. qPCR analysis was performed on 4 ng. The primers used for the qPCRs are listed in Supplemental Table S2. Each sample was analyzed in triplicate, and the average Ct was calculated for each primer set. In order to determine the Relative Nucleosome Occupancy associated with each primer set, the following equation was used: Relative Nucleosome Occupancy = 10(Ct Untreated – Ct MNase).
Nuclear extracts preparation and co-immunoprecipitations
Nuclear protein extracts (NEs) were prepared using according to the Lamond protocol (www.Lamondlab.com). For co-immunoprecipitations, 300 μg of NEs were first precleared with protein A sepharose (GE) for 2 h at 4°C and then incubated with the indicated antibodies or with rabbit IgG as a control. The antibodies were coupled to protein A-Sepharose beads following manufacturer's instructions. Immunoprecipitations were carried out for 2 h at 4°C on a rotating wheel. Beads were then washed three times with a solution composed of 10 mM Tris–HCl, 100 mM NaCl, 0.01% Nonidet P-40, and 0.04% bovine serum albumin (BSA). The proteins bound to the beads were eluted by boiling the samples at 95°C for 10 min. Aliquots were analyzed by SDS-PAGE and immunoblotting.
For ubiquitin immunoprecipitation, cells were lysed in 1% SDS lysis buffer (50 mM Tris–HCl pH 7.4, 0.5 mM EDTA, 1% SDS) and further boiled for an additional 10 min. Lysates were clarified by centrifugation at 16 000 g for 10 min. Supernatant was diluted 10 times with a buffer composed of 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton and complete Protease Inhibitor Cocktail (Roche). Immunoprecipitation was performed with anti-Multi Ubiquitin mAb-Agarose (MBL) overnight at 4°C. The precipitates were washed three times and the samples were resolved on 6% SDS-PAGE.
For Flag-tagged CstF50 immunoprecipitation, cells were lysed as for ubiquitin immunoprecipitation. Immunoprecipitation was performed with ANTI-FLAG® M2 Affinity Gel (Sigma) overnight at 4°C. The precipitates were washed three times and eluted in a rotating wheel for 1 h at 4°C with a buffer containing 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton, complete Protease Inhibitor Cocktail (Roche) and 3× flag-peptide (5 μg/ml). The samples were resolved on 7% SDS-PAGE.
For myc-tagged BARD1 immunoprecipitation, cells were lysed in 50 mM Tris–HCl, pH 7.4, with 150 mM NaCl, 1 mM EDTA and 1% Triton X-100. Lysates were incubated overnight at 4°C with anti-myc antibody (71D10 Cell Signalling). The complexes were coupled to protein A-Sepharose beads following manufacturer's instructions. Immunoprecipitations were carried out for 2 h at 4°C on a rotating wheel. Beads were then washed three times with a solution containing 10 mM Tris–HCl, 100 mM NaCl, 0.05% Nonidet P-40, and 0.04% BSA. The proteins bound to the beads were eluted by boiling the samples at 95°C for 10 min. The sample were resolved on 7% SDS-PAGE.
Quantification of western blots was performed by using NIH ImageJ software the Image Studio software (Odissey FC-Licor) or Image Studio v1.1.
Statistical analysis
Statistical analysis of RT-PCR experiments was performed either using a one-way ANOVA test followed by a post-hoc Tukey–Kramer multiple comparison test, or a two-tailed, paired t-test. Analysis of FACS experiments was carried out using a one-way ANOVA test followed by a post-hoc Tukey–Kramer multiple comparison test. Analysis of western blot data was performed using a two-tailed, paired t-test.
RESULTS
BRM is down-regulated in response to oxidative stress
Oxidative stress has long been implicated in neuronal cell death that is associated with neurodegenerative disorders. For example, exposure of cells to paraquat (N,N’-dimethyl-4,4′-bipyridinium dichloride, PQ), a neurotoxic herbicide, increases the production of the superoxide anion (22), and has been linked to the incidence of Parkinson's disease. Moreover, the second most frequent cause of hereditary Amyotrophic Lateral Sclerosis (ALS) are mutations in the cytoplasmic superoxide dismutase 1 (SOD1) protein and have been shown to increases intracellular ROS levels (23).
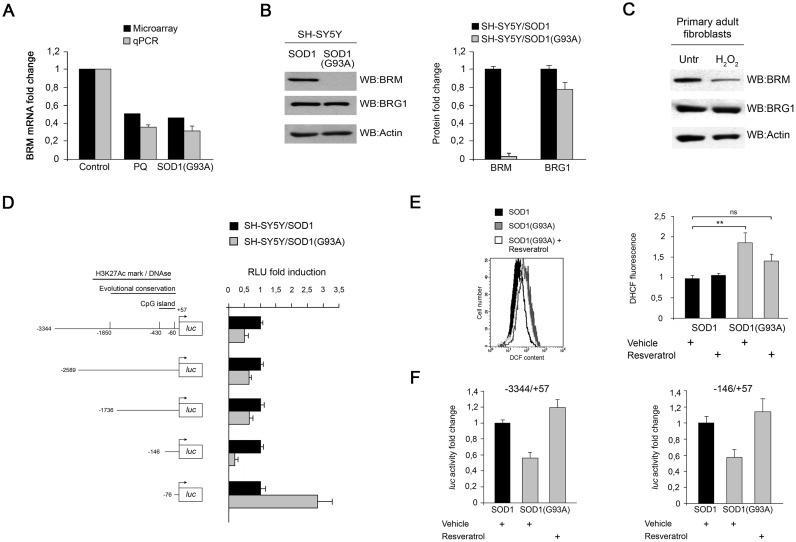
We recently showed that treatment with PQ or expression of the mutant SOD1(G93A) protein cause extensive changes in both mRNA expression and alternative splicing in human neuroblastoma SH-SY5Y cells (16). Among the most down-regulated genes was SMARCA2, the gene encoding BRM (Figure 1A and Supplementary Figure S1). SOD1(G93A) expression almost abrogated BRM expression at the protein level, while expression of the hSWI/SNF alternative ATPase subunit BRG1 was not significantly affected (Figure 1B). Similarly, treatment of SH-SY5Y cells with PQ strongly reduced BRM mRNA and protein levels without affecting BRG1 expression (Supplementary Figure S1). To test a possible causal link between oxidative stress and BRM expression, we exposed primary adult fibroblasts to different sources of ROS. Treatment with H2O2 (Figure 1C) or with PQ (Supplementary Figure S1) effectively reduced BRM expression.
Figure 1.
Oxidative stress impairs BRM expression. (A) Both PQ and mutant SOD1(G93A) expression reduce the level of the SMARCA2 transcript. qPCR validation of the gene-level microarray data of BRM expression in the indicated cell lines. Assays were performed in triplicate from three biological replicates. (B) Left panel: representative western blot analysis of BRM and BRG1 in SH-SY5Y cells stably expressing wild type SOD1 or the mutant SOD1(G93A) protein. Right panel: quantification of BRM and BRG1 protein levels from three independent biological replicates. Error bars represent standard deviation. (C) Oxidative stress inhibits BRM expression in human fibroblasts. Representative western blot analysis of BRM and BRG1 in primary fibroblasts treated with 0.2 mM H2O2 for 24 h. (D) Promoter activity of the 3.4 kb fragment containing the SMARCA2 regulatory region and its deletion constructs in the indicated cell lines. Cells were transiently co-transfected with the indicated luciferase reporter plasmids and with the Renilla luciferase-encoding pRL-TK plasmid. Renilla luciferase activity was used to normalize the transfection efficiency. Results are expressed as fold induction relative to controls. Error bars represent standard deviations calculated on three independent experiments. Left panel: schematic representation of the SMARCA2-promoter luciferase reporter plasmids used in transient transfection assays. (E) Expression of mutant SOD1(G93A) protein induces oxidative stress. Cells were incubated with 20 μM H2DCF-DA for 30 min at 37°C and assayed by FACS as described under ‘Materials and Methods.’ Resveratrol was added 18 h prior to H2DHCF-DA incubation. Left panel: FACS profiles. Black filled, SH-SY5Y/SOD1 cells; grey filled, SH-SY5Y/SOD1(G93A) cells; empty, resveratrol-treated SH-SY5Y/SOD1(G93A) cells. Right panel: quantification of H2DHCF-DA fluorescence. Statistical analysis was carried out using a one-way ANOVA test followed by a post-hoc Tukey–Kramer multiple comparison test. **P < 0.01. Error bars represent standard deviations calculated on three independent experiments. (F) Resveratrol treatment restores promoter activity of the −3344/+57 fragment and of the −146/+57 minimal promoter in SH-SY5Y(SOD1) cells. Results are expressed as fold induction relative to vehicle-treated cells. Error bars represent standard deviations calculated on three independent experiments.
We set out to identify regulatory elements in the SMARCA2 promoter that respond to oxidative stress. Inspection of the −3444/+57 region identified features associated to promoter sequences (Supplementary Figure S2): peaks of evolutionary conservation, a CpG island, a putative DNase hypersensitive region, and peaks of enrichment of acetylation of histone 3 lysine 27 (H3KAc27). A high GC-content and the absence of a detectable TATA-box suggested that the promoter belongs to the class of GC promoters. We then cloned a 3.4 kb genomic fragment (from position –3344 to +57) into a promoter-less luciferase reporter. This region conferred sensitivity to PQ (Supplementary Figure S2) and showed reduced luciferase expression when transfected in SH-SY5Y/SOD1(G93A) cells (Figure 1D). Further 5′ deletion analysis pointed to the −146/+57 fragment as critical for the oxidative stress response (Figure 1D). Addition of resveratrol, a natural antioxidant, to SH-SY5Y/SOD1(G93A) cells reduced ROS production (Figure 1E) and restored luciferase expression to both the long 3344/+57 and the minimal -146/+57 promoter constructs (Figure 1F).
Overall these results indicate that SMARCA2 transcription and hence BRM expression are reduced by oxidative stress.
Selection of the proximal alternative last exon is favored in BRM-depleted cell
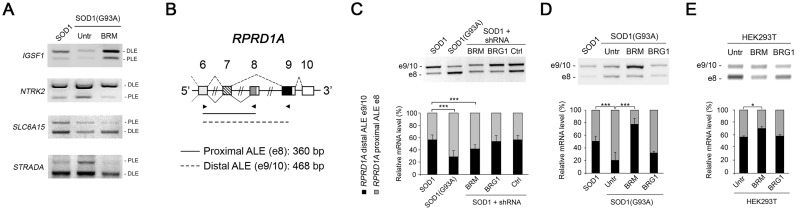
Using splicing-sensitive microarrays we had previously detected a large number of AS changes (262 genes, involving 418 exons) in SH-SY5Y/SOD1(G93A) cells (16). Within this dataset, 89 exons (in 78 genes) appeared as ALEs. We validated by PCR six of these genes: in five out of six genes, the distal ALE was favoured in the presence of BRM (i.e. in SH-SY5Y/SOD1 cells), whereas the proximal ALE was preferred when BRM was expressed at low level as in SH-SY5Y/SOD1(G93A) cells (Figure 2A, and Supplementary Figure S3).
Figure 2.
BRM inhibits inclusion of the proximal ALE. (A) Alternative splicing pattern of the ALEs of four genes affected by BRM expression. The splicing pattern in the indicated cell lines was analyzed by RT-PCR and gel electrophoresis. For quantifications see Supplementary Figure S3. PLE: proximal ALE, DLE: distal ALE. (B) Schematic representation of the relevant region of the RPRD1A gene indicating the positions of the PCR primers (arrowheads), the splicing pattern and the size of the expected PCR products. (C) Stable knock-down of BRM, but not of BRG1, favors e8 inclusion in SOD1 cells. (D) Stable overexpression of BRM, but not of BRG1, favors e10 inclusion in SOD1(G93A) cells. (E) Transient overexpression of BRM in HEK293T cells favors e10 inclusion. C-E. Upper panels: The splicing pattern in the indicated cell lines was analyzed by RT-PCR and gel electrophoresis. Lower panel: Quantification of proximal and distal ALE usage in the indicated cell lines. Data are representative of six independent experiments. Error bars represent standard deviation. *P < 0.05 and ***P < 0.001.
We chose the RPRD1A gene for mechanistic analysis because of the ease of PCR quantification of the ALE choice. As shown in Figure 2B, alternative splicing of the RPRD1A primary transcript generates two mRNA variants with different terminal exons. When exon 8 (e8) is chosen as proximal ALE, a shorter mRNA is produced. In contrast, a longer mRNA isoform is generated when e8 is skipped and cleavage and polyadenylation occur in the distal ALE exon 10 (e10). Exon e8 was preferentially skipped in SOD1 cells, where BRM is expressed, while it was preferentially included in SOD1(G93A) cells in which BRM is hardly detected (Figure 2C and D). An inverse correlation between BRM expression and e8 skipping was also found in SOD1 cells where BRM expression was silenced with a specific shRNA (Figure 2C, and Supplementary Figure S4 for expression levels). Moreover, when BRM was exogenously expressed in the SOD1(G93A) background, e8 was mostly skipped and the mRNA variant ending in e10 was favoured (Figure 2D, and Supplementary Figure S4 for expression levels). In contrast, neither silencing of BRG1 in SOD1 cells nor its overexpression in SOD1(G93A) cells affected the ratio of the two mRNA isoforms (Figure 2C and D). The transient transfection of expression constructs for BRM and BRG1 in HEK293T cells further confirmed the BRM-dependent inhibition of e8 selection (Figure 2E and Supplementary Figure S4 for expression levels). Similar results in the different cell lines were obtained for the SLC6A15 gene (Supplementary Figure S3).
Overall, these results indicate that a high expression level of BRM favours the skipping of the proximal ALE of RPRD1A.
BRM accumulates on the skipped proximal ALE
A well-known example of selection of alternative poly(A) sites located in different terminal exons is provided by the immunoglobulin mu (Igμ) gene. The regulation of this processing event was shown to depend on the different levels of the 3′ end processing factor CstF during the maturation from B cell to a plasma cell (24,25). We therefore tested the expression levels of the core cleavage and polyadenylation factors in SH-SY5Y/SOD1 and in SH-SY5Y/SOD1(G93A) cells, but did not detect significant differences in the levels of CPSF, CstF and CF Im subunits (data not shown).
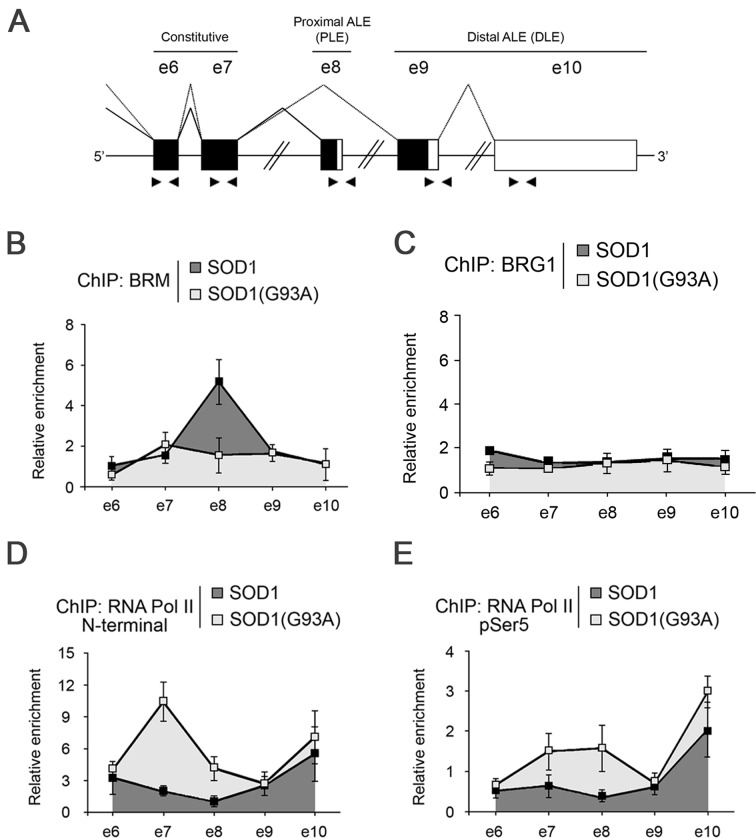
BRM was previously shown to accumulate on the variant exon v5 of the CD44 gene and to promote its inclusion by modulating the elongation rate of RNAPII, and by interacting with the splicing factor Sam68 (26). We thus explored the possibility that BRM could contribute to the choice of ALE by regulating the elongation rate of the polymerase. First, we characterized the distribution of BRM and BRG1 along the terminal region of the RPRD1A gene from e6 to e10 by chromatin immunoprecipitation (ChIP, Figure 3A). In SOD1(G93A) cells, where it was hardly expressed, BRM was distributed equally on all exons. In contrast, in SOD1 cells BRM accumulated on the proximal ALE (Figure 3B). BRG1, on the other hand, was distributed equally on all exons in both the cellular lines (Figure 3C). Next, we analysed the distribution of the elongating polymerase, using the N20 antibody that recognizes the N-terminal region of RNAPII. An increased density of RNAPII was detected on e7 and e8 in SOD1(G93A) cells and in both cell lines on e10 (Figure 3D). We also performed ChIP assays with the H14 antibody to RNAPII phosphorylated at Ser5 of the CTD, a modification associated with a paused polymerase (27,28) (Figure 3E). We detected a slight accumulation of p-Ser5 RNAPII on e8 in SOD1(G93A) cells. In addition, we observed a high density on e10 in both cell lines, where this exon is used as terminal exon, albeit with different efficiency. This result is reminiscent of the observed pausing of RNAPII on terminal exons in yeast (29).
Figure 3.
BRM localizes on the excluded proximal ALE of RPRD1A gene. (A) Schematic representation of the relevant region of the RPRD1A gene indicating the splicing pattern and the positions of the qPCR primers used for ChIP analysis (arrowheads). (B–E) BRM, BRG1, and RNAPII on the RPRD1A gene. ChIP assays were performed with the indicated antibodies on bulk chromatin from SH-SY5Y/SOD1 and SH-SY5Y/SOD1(G93A) cells. The graphs show the recruitment of the indicated proteins relative to the values obtained with the non-immune sera. Data are representative of three independent experiments. Error bars represent s.e.m.
Our findings indicate a modest inverse correlation between the accumulation of BRM and the accumulation of RNAPII on included exons. Thus, the contribution of RNAPII pausing to the inclusion of the proximal exon appears small, at variance with what previously observed for the inclusion of internal exons (26).
BRM does not require its ATPase activity to affect ALE choice
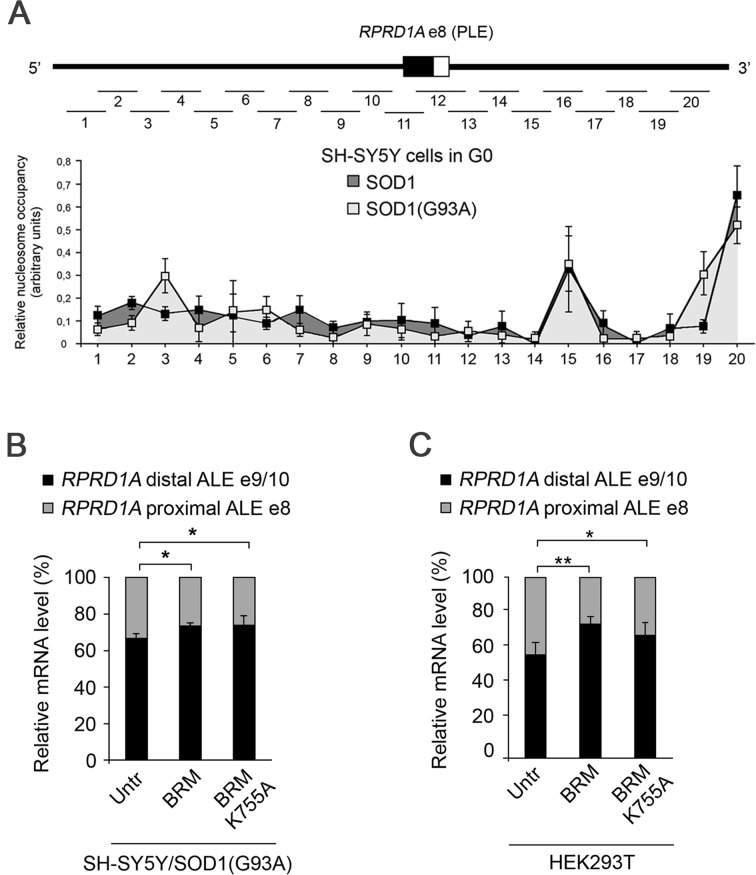
The SWI/SNF chromatin-remodeling complex destabilizes histone-DNA interactions and alters nucleosome positions. Recent studies indicate that nucleosome positioning may influence inclusion of internal exon sequences (30,31). We therefore wondered whether BRM might contribute to the choice of ALE by moving nucleosomes around the proximal last exon. We thus identified nucleosome positions and occupancy in SOD1 and SOD1(G93A) cells, using micrococcal nuclease (MNase) digestion in cells synchronized in G0. We used mononucleosomal DNA fragments (150 bp) generated by MNase as a template for qPCR with overlapping primer sets across a ∼1 kb region centered on e8 of the human RPRD1A genomic locus. As shown in Figure 4A, the MNase digestion pattern did not significantly differ between the two cell lines, indicating that the level of BRM protein had no influence on nucleosome distribution in the interrogated region of the RPRD1A gene.
Figure 4.
BRM ATPase activity is not required for ALE choice. (A) Nucleosome occupancy profile of the relevant region of the RPRD1A gene in G0-synchronized cells. Lines indicate the positions of the qPCR primers. Nucleosome positioning was analyzed with MNase digestion for the 33 605 100–33 606 100 region of human chromosome 18 containing the RPRD1A gene in SH-SY5Y/SOD1 and in SH-SY5Y/SOD1(G93A) cells. Data are representative of three independent biological replicates. Error bars represent s.e.m. (B and C) BRM ATPase activity is not required for exon 8 skipping. The splicing pattern was analyzed by RT-PCR and quantified as percentage of proximal and distal ALEs. Data are representative of six independent experiments. Error bars represent standard deviations. *P < 0.05 and **P < 0.01.
To further exclude that the chromatin remodeling activity of BRM-containing SWI/SNF complexes may contribute to e8 skipping in SOD1 cells, we analysed the alternative splicing pattern in SH-SY5Y cells stably expressing a BRM mutant carrying the K755A substitution in the ATPase domain that abrogates the chromatin remodeling activity (19) (Figure 4B, and Supplementary Figure S4 for expression levels) we also analysed HEK293T cells transiently transfected with the same construct (Figure 4C, and Supplementary Figure S4 for expression levels). Similarly to the wild type, expression of the mutant BRM stimulated exon 8 skipping and the selection of the distal ALE.
Taken together these data indicate that the chromatin remodeling function of BRM is not required for the inhibition of the inclusion of the proximal ALE in SOD1 cells. Therefore, we hypothesized that BRM might contribute to e8 skipping by negatively acting on pre-mRNA processing, possibly by recruiting an inhibitory complex.
BARD1 and CstF50 associate with BRM on the skipped proximal ALE
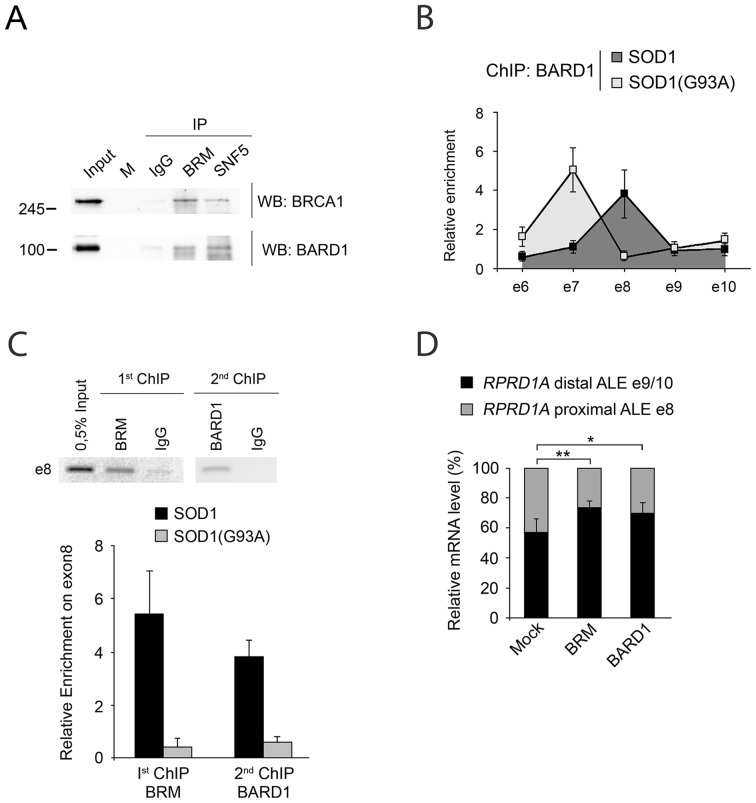
Previously, a BARD1-CstF complex was shown to inhibit 3′ processing of nascent transcripts deriving from premature transcription termination at sites of DNA damage (14,17,32). Therefore, we speculated that BARD1 (and its partner BRCA1) might also be involved in the inhibition of transcription termination at the proximal poly(A) site. Indeed, we found that endogenous BRCA1 and BARD1 in HEK293T cells co-immunoprecipitate with SNF5 and BRM (Figure 5A), indicating that both proteins can associate with the SWI/SNF complex.
Figure 5.
BRM and BARD1 accumulate on the excluded proximal ALE. (A) BARD1 and BRCA1 associate with the SWI/SNF complex. Immunoprecipitations were carried out with the indicated antibodies (IP) from HEK293T nuclear extracts. Filters were sequentially probed with the indicated antibodies (WB). (B) BARD1 is enriched on the RPRD1A gene. ChIP assays were performed with the indicated antibodies on bulk chromatin from SH-SY5Y/SOD1 and SH-SY5Y/SOD1(G93A) cells. The graphs show the recruitment of the indicated proteins relative to the values obtained with the non-immune Ig. Data are representative of three independent experiments. Variability is expressed as SEM. (C) ChIP-reChIP was performed for BRM and BARD, and analyzed by PCR (upper panel) and qPCR (lower panel). Data are representative of three independent experiments. Error bars represent standard deviations. (D) Transient overexpression of BRM, and BARD1 in HEK293T cells promotes skipping of the proximal ALE. Error bars represent standard deviations. *P < 0.05, **P < 0.01.
Next, we determined that in SOD1 cells, where BRM is present at higher level, BARD1 is recruited on the RPRD1A gene on e8 (Figure 5B). Conversely, in SOD1(G93A) cells BARD1 was enriched on e7. We then showed that BRM and BARD1 were present on the same DNA fragments by performing two successive ChIP assays (ChIP-reChIP) using anti-BRM followed by anti-BARD1 antibodies (Figure 5C). Finally, overexpression of BRM and BARD1 proteins (Supplementary Figure S4 for expression levels) favoured the inclusion of the distal ALE (Figure 5D).
Taken together, these data indicate that BARD1 and BRCA1 are integral to the process of ALE choice.
The BARD1/BRCA1 complex promotes the ubiquitination of CstF50
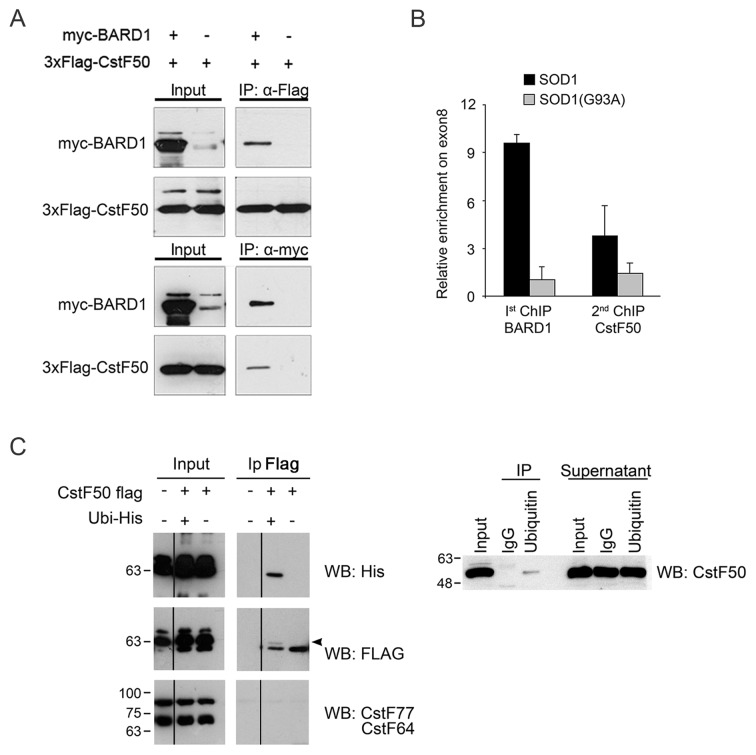
BARD1 and CstF50 were shown to interact both in vitro and in vivo (10,14,17). We confirmed this interaction by cotransfecting HEK293T cells with plasmids expressing myc-tagged BARD1 and Flag-tagged CstF50. Immunoprecipitation with either anti-myc or anti-Flag antibodies followed by western blotting demonstrated that BARD1 and CstF50 interact physically (Figure 6A). Moreover, ChIP-reChIP experiments using anti-BARD1 followed by CstF50 antibodies demonstrated that BARD1 and CstF50 co-assemble on e8 of RPRD1A in SOD1 cells, where the distal ALE is preferentially selected (Figure 6B).
Figure 6.
A fraction of CstF50 is ubiquitinated. (A) BARD1 and CstF50 physically interact. Cell lysates prepared from HEK293T cells and transfected with the indicated plasmids were immunoprecipitated with anti-FLAG (upper panel) or anti-Myc (lower panel) coniugated beads. Input (10% of the extract) and coimmunoprecipitated proteins were analyzed by western blots with the indicated antisera. (B) Both BARD1 and CstF50 are enriched on the RPRD1A gene. Quantitative PCR analysis of ChIP-reChIP assays for BARD1 and CstF50. Data are representative of three independent experiments. Error bars represent standard deviations. (C) CstF50 is ubiquitinated in HEK cells overexpressing his-tagged ubiquitin. HEK293T cells were cotransfected with plasmids expressing Flag-tagged CsF50 and His-ubiquitin. Total extracts were boiled in 1% of SDS lysis buffer, diluted tenfold, and immunoprecipitated with anti-FLAG coniugated beads (IP). Input (1% of the extract) and immunoprecipitated proteins were analyzed by western blotting with the indicated antisera. The arrowhead indicates the His-tagged CstF50 protein. The shift is consistent with a monoubiquitination. (The samples were run on the same gel but the lanes were rearranged for clarity). (D) A fraction of CstF50 is ubiquitinated in unperturbed HEK cells. Whole cell extracts were prepared from HEK293T cells and immunoprecipitated with an anti-ubiquitin conjugated resin. Input (1% of the extract) and coimmunoprecipitated proteins were analyzed by western blotting with an anti-CstF50 antiserum.
Since BRCA1 has E3-ubiquitin ligase activity (33), we wondered whether the interaction between BARD1 and CstF50 may lead to CstF50 ubiquitination. HEK293T cells were contransfected with plasmids encoding Flag-CstF50 and histidine-tagged ubiquitin. Indeed, CstF50 immunoprecipitated with an anti Flag-antibody and tested positive to the His-specific antibody (Figure 6C). We further verified if a fraction of CstF is always ubiquitinated in unperturbed cells. To this end, cells were lysed under denaturing conditions and the ubiquitinated protein fraction was immunoprecipitated with an anti-ubiquitin antibody. Western blot analysis with an anti-CstF50 antibody confirmed the ubiquitination of CstF50 (Figure 6D).
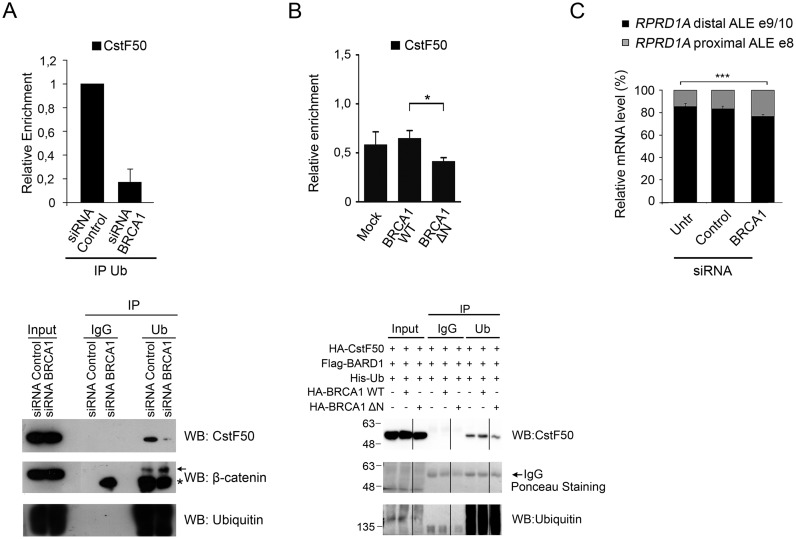
We next investigated whether the BRCA1 E3 ubiquitin ligase activity was responsible for the ubiquitination of CstF50. To this end, BRCA1 was transiently silenced with a specific siRNA. Three days after transfection the level of the BRCA1 protein was reduced to ∼25% (Supplementary Figure S4). Immunoprecipitation of the ubiquitinated protein fraction with an anti-ubiquitin antibody from BRCA1-silenced cell extracts showed a reduction in the amount of ubiquitinated CstF50 protein (Figure 7A). Moreover, expression of deltaN-BRCA1 protein, lacking the amino-terminal RING domain (34), reduced the amount of ubiquitinated CstF50 (Figure 7B). Together these results confirm the BRCA1/BARD1-mediated ubiquitination of CstF50.
Figure 7.
Ubiquitination of CstF50 is catalyzed by BRCA1. (A) BRCA1 knockdown impairs CstF50 ubiquitination. Whole cell extracts from HEK293T cells transfected with control or BRCA1 siRNAs were immunoprecipitated with a control or an anti-ubiquitin resin. Western blots were analysed with the indicated antibodies. β-catenin was used as control for BRCA1-independent ubiquitination and is highlighted by an arrow. The asterisk indicates non-specific bands. Upper panel: Quantification of CstF50 ubiquitination upon BRCA1 silencing. Quantification was performed on three independent immunoprecipitation experiments. Immunoprecipitated CstF50 protein was normalized on β-catenin. Error bars represent standard deviations. Lower panel: representative western blot. (B) Overexpression of a catalytically inactive mutant BRCA1 protein impairs CstF50 ubiquitination. HEK293T cells were cotransfected with plasmids expressing CstF50, BARD1 and either wild type BRCA1 or the BRCA1-ΔN mutant. Ubiquitinated proteins were immunoprecipitated with an anti-ubiquitin resin as described in ‘Materials and Methods’ and analysed by western blotting with the indicated antisera. Upper panel: Quantification was performed on three independent immunoprecipitation experiments. Immunoprecipitated CstF50 protein was normalized on IgG bands that are indicated in the Ponceau Staining. Error bars represent standard deviations. *P < 0.05. Lower panel: representative western blot. The arrow indicates the bands corresponding to the IgGs. (C) BRCA1 knockdown favors inclusion of the distal ALE. Quantification of the amount of proximal and distal ALE usage in the indicated cell lines. Data are representative of three independent experiments. Error bars represent standard deviation. ***P < 0.001.
Finally, we determined the effect BRCA1 silencing on pre-mRNA splicing. As shown in Figure 7C, siRNA-mediated knockdown of endogenous BRCA1 resulted in the preferred inclusion of the proximal ALE.
Overall these results suggest a model in which BRM positioned on the proximal ALE mediates the recruitment of the BARD1/BRCA1 complex, which in turn ubiquitinates CstF50 impairing 3′ end processing at the proximal poly(A) site. This allows transcription to proceed to the distal terminal exon.
DISCUSSION
Oxidative stress is a common biological event, and has been associated with cancer, neurodegenerative diseases and aging. We previously reported that oxidative stress affects splice site selection (16). Here, we show that it affects in particular the choice of ALEs, generally promoting the inclusion of the proximal ALE; it does so by inducing a severe decrease in the expression of the SWI/SNF subunit BRM. Our data also provide evidence for a regulatory mechanism that partially inhibits 3′ end processing at the proximal poly(A) site.
Recently, knock-down of Brm and of other SWI/SNF subunits was shown to affect splicing-associated polyadenylation in Drosophila S2 cells by an unknown mechanism (35). We suggest that the involvement of BRM-containing SWI/SNF complexes in the regulation of pre-mRNA processing is similar in Drosophila and mammalian cell, and thus is conserved in evolution, and possibly not restricted to the choice of ALE choice following oxidative stress.
We specifically analyse the ALE choice on RPRD1A gene, which generates two main mature transcripts: one including exons 1–7 followed by exon 8 (the proximal ALE) and the polyA, and one where exons 1–7 are followed by exons 9–10 and polyA. Both mature transcripts are present in cells, whether undergoing oxidative stress or not, but their relative abundance changes. We find that BRM is associated with RPRD1A e8 in normal conditions, whereas oxidative stress strongly reduces BRM expression and its association with e8. BRM and SNF5 (another core subunit of the SWI/SNF complex) co-immunoprecipitate the BRCA1/BARD1 E3 ubiquitin ligase. Indeed, our ChIP and ChIP-ReChIP experiments reveal that BRM, BARD1 and CstF co-accumulate on e8 in SOD1 cells, in which the choice of the distal ALE is promoted. Likewise, overexpression of BRM and BARD1 proteins promoted the choice of the distal ALE. Further, we show that BRCA1/BARD1 mediates the ubiquitination of CstF50. Mutation or silencing of BRCA1 reduces CstF50 ubiquitination, and promoted the choice of the proximal ALE. We thus propose that the presence of BRM on the proximal ALE promotes the recruitment of the BARD1/BRCA1 ubiquitin ligase, which by ubiquitinating CstF50 causes the inhibition of 3′ end processing at the proximal poly(A) site and allows the continuation of transcription through exons 9–10. A fraction of the pool of CstF50 is always monoubiquitinated, which is consistent with the fact that some transcripts incorporating the distal ALE are always present.
Our findings are consistent with previous knowledge. The coupling of transcription and 3′ end processing is well documented (reviewed in (1,9)). RNAPII and other transcription factors interact with components of the 3′-end-processing machinery (36–39)). Generally, these interactions appear to promote the use of the proximal poly(A) site, since this is transcribed first and is encountered first by the 3′-end-processing machinery. Cleavage and polyadenylation at the proximal poly(A) site is favoured by a reduction of the elongation rate of the transcribing polymerase (40). In this respect, it not surprising that the mechanism we describe restrains the cleavage of the nascent transcript.
In vivo BARD1 can interact with BRCA1 (41) and the BARD1/BRCA1 heterodimer has E3 ubiquitin ligase activity (33). Moreover, BRCA1 interacts with the SWI/SNF chromatin remodeling complex (42). Finally, the interaction between CstF50 and BARD1 in UV-treated cells was shown to inhibit erroneous 3′ processing of nascent, truncated transcripts deriving from premature transcription termination at sites of DNA damage (14,17,32).
BRM has already been implicated in alternative splicing: it was previously shown to promote inclusion of variant internal exons of the CD44 gene by inducing a transient pausing of the transcribing RNAPII (26). However, in Drosophila, Brm-containing SWI/SNF complexes, while inducing RNAPII pausing, can negatively regulate splicing (43). In this case, RNAPII stalling, and the consequent splicing reduction, were not induced by Brm but were rather dependent on SNF5. Instead, Brm remodeling activity was shown to be necessary for the subsequent chromatin remodeling following the release of the stalled polymerase (43).
In line with the report by Zraly et al. (43), we found that BRM negatively affects the inclusion of the proximal ALE. This effect does not require its ATPase activity. Moreover, we did not detect differences in the distribution of nucleosomes on the proximal ALE. These findings suggest that the choice of ALE is not correlated with the nucleosome remodeling activity of BRM. We also observed that RNAPII is not greatly enriched on the skipped proximal ALE. Instead, we observed an inverse correlation between the accumulation of BRM and RNAPII particularly on the distal ALE, where there was a distinct enrichment of RNAPII but BRM was virtually absent. Our data indicate that in the case of the choice of alternative terminal exons, the modulation of the elongation rate of RNAPII is not the main role of BRM, and rather implicate a different molecular mechanism.
The choice of alternative poly(A) sites located in different terminal exons occurs in conjunction with splicing, and splicing factors are known to influence 3′ processing (reviewed in (44)). Thus, the mechanism we describe is certainly not the unique determinant of the choice between proximal and distal terminal exons. Moreover, we cannot exclude that BRM and BRCA1/BARD1 may also affect binding of splicing factors to the terminal exon. Future work will determine how the interplay between chromatin remodeling complexes, splicing and polyadenylation factors eventually determines which terminal exon will be included in the mature transcript.
Acknowledgments
We gratefully acknowledge B.M. Emerson, I. Irminger-Finger, T. Banerjee, M.L. Guerrini, J.D. Parvin, M. Pilyugin, M.-D. Ruepp, O. Mühlemann, N. Chiba and W. Keller for reagents. We are grateful to S. Polo for help with ubiquitination analysis and to M. Lupi for technical assistance.
Footnotes
Present address: Gabriele A. Fontana, Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, 4058 Basel, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MIUR-PRIN [20083R593R_003]; Swiss National Science Foundation Sinergia [CRSII3 136222 to G.F.]. Funding for open access charge: University of Milan, Fondo ricerca di Ateneo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA (New York, N.Y.) 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danckwardt S., Hentze M.W., Kulozik A.E. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 10.Edwards R.A., Lee M.S., Tsutakawa S.E., Williams R.S., Nazeer I., Kleiman F.E., Tainer J.A., Glover J.N. The BARD1 C-terminal domain structure and interactions with polyadenylation factor CstF-50. Biochemistry. 2008;47:11446–11456. doi: 10.1021/bi801115g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai Y., Auperin T.C., Chou C.Y., Chang G.G., Manley J.L., Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 12.Dantonel J.C., Murthy K.G., Manley J.L., Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 13.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 14.Kleiman F.E., Manley J.L. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 15.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 16.Lenzken S.C., Romeo V., Zolezzi F., Cordero F., Lamorte G., Bonanno D., Biancolini D., Cozzolino M., Pesaresi M.G., Maracchioni A., et al. Mutant SOD1 and mitochondrial damage alter expression and splicing of genes controlling neuritogenesis in models of neurodegeneration. Hum. Mutat. 2011;32:168–182. doi: 10.1002/humu.21394. [DOI] [PubMed] [Google Scholar]
- 17.Kleiman F.E., Manley J.L. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 18.Rizzardini M., Mangolini A., Lupi M., Ubezio P., Bendotti C., Cantoni L. Low levels of ALS-linked Cu/Zn superoxide dismutase increase the production of reactive oxygen species and cause mitochondrial damage and death in motor neuron-like cells. J. Neurol. Sci. 2005;232:95–103. doi: 10.1016/j.jns.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Richmond E., Peterson C.L. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanges R., Cordero F., Calogero R.A. oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics (Oxford, England) 2007;23:3406–3408. doi: 10.1093/bioinformatics/btm469. [DOI] [PubMed] [Google Scholar]
- 22.Cocheme H.M., Murphy M.P. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 23.Ciriolo M.R., De Martino A., Lafavia E., Rossi L., Carri M.T., Rotilio G. Cu,Zn-superoxide dismutase-dependent apoptosis induced by nitric oxide in neuronal cells. J. Biol. Chem. 2000;275:5065–5072. doi: 10.1074/jbc.275.7.5065. [DOI] [PubMed] [Google Scholar]
- 24.Takagaki Y., Seipelt R.L., Peterson M.L., Manley J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 25.Takagaki Y., Manley J.L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 26.Batsche E., Yaniv M., Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 27.Morris D.P., Michelotti G.A., Schwinn D.A. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J. Biol. Chem. 2005;280:31368–31377. doi: 10.1074/jbc.M501546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehm A.K., Saunders A., Werner J., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrillo Oesterreich F., Preibisch S., Neugebauer K.M. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcarcel J., Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz S., Meshorer E., Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 32.Mirkin N., Fonseca D., Mohammed S., Cevher M.A., Manley J.L., Kleiman F.E. The 3′ processing factor CstF functions in the DNA repair response. Nucleic Acids Res. 2008;36:1792–1804. doi: 10.1093/nar/gkn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mallery D.L., Vandenberg C.J., Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiba N., Parvin J.D. The BRCA1 and BARD1 association with the RNA polymerase II holoenzyme. Cancer Res. 2002;62:4222–4228. [PubMed] [Google Scholar]
- 35.Waldholm J., Wang Z., Brodin D., Tyagi A., Yu S., Theopold U., Farrants A.K., Visa N. SWI/SNF regulates the alternative processing of a specific subset of pre-mRNAs in Drosophila melanogaster. BMC Mol. Biol. 2011;12:46. doi: 10.1186/1471-2199-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y., Di Giammartino D.C., Taylor D., Sarkeshik A., Rice W.J., Yates J.R., 3rd, Frank J., Manley J.L. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagaike T., Logan C., Hotta I., Rozenblatt-Rosen O., Meyerson M., Manley J.L. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozenblatt-Rosen O., Nagaike T., Francis J.M., Kaneko S., Glatt K.A., Hughes C.M., LaFramboise T., Manley J.L., Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. U.S.A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katahira J., Okuzaki D., Inoue H., Yoneda Y., Maehara K., Ohkawa Y. Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 2013;14:7060–7072. doi: 10.1093/nar/gkt414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto P.A., Henriques T., Freitas M.O., Martins T., Domingues R.G., Wyrzykowska P.S., Coelho P.A., Carmo A.M., Sunkel C.E., Proudfoot N.J., et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L.C., Wang Z.W., Tsan J.T., Spillman M.A., Phung A., Xu X.L., Yang M.C., Hwang L.Y., Bowcock A.M., Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 42.Bochar D.A., Wang L., Beniya H., Kinev A., Xue Y., Lane W.S., Wang W., Kashanchi F., Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 43.Zraly C.B., Dingwall A.K. The chromatin remodeling and mRNA splicing functions of the Brahma (SWI/SNF) complex are mediated by the SNR1/SNF5 regulatory subunit. Nucleic Acids Res. 2012;40:5975–5987. doi: 10.1093/nar/gks288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millevoi S., Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]