Abstract
Our current knowledge about the mechanisms of miRNA silencing is restricted to few lineages such as vertebrates, arthropods, nematodes and land plants. miRNA-mediated silencing in bilaterian animals is dependent on the proteins of the GW182 family. Here, we dissect the function of GW182 protein in the cnidarian Nematostella, separated by 600 million years from other Metazoa. Using cultured human cells, we show that Nematostella GW182 recruits the CCR4-NOT deadenylation complexes via its tryptophan-containing motifs, thereby inhibiting translation and promoting mRNA decay. Further, similarly to bilaterians, GW182 in Nematostella is recruited to the miRNA repression complex via interaction with Argonaute proteins, and functions downstream to repress mRNA. Thus, our work suggests that this mechanism of miRNA-mediated silencing was already active in the last common ancestor of Cnidaria and Bilateria.
INTRODUCTION
miRNAs are small, 21–23 nucleotide-long non-coding RNAs targeting more than half of all genes in mammals (1). Mutations disrupting production of miRNAs result in lethality or sterility, pointing to their integral role in gene expression (2). Because miRNA are involved in regulating numerous developmental and cellular processes in plants and animals, understanding the mechanisms and evolution of their function is important to fundamental science.
miRNAs function as guides, by pairing with complementary sites present in their target transcripts and recruiting a complex of proteins that ultimately down-regulates mRNA (3–5). In members of the Bilateria, a group that comprises the vast majority of existing animal species, this is achieved by both translational repression and RNA decay via deadenylation. In vertebrates, Drosophila and C. elegans, which represent evolutionary diverse groups of Bilateria, those activities are mediated by scaffold proteins of the metazoan-specific GW182/TNRC6 family (Figure 1A). A major role of those flexible proteins is to interact with: (i) the Argonaute proteins (AGOs), which carry the guide miRNAs, and thus mediate recruitment of the entire repression complex to the target mRNA, based on partial complementarity; and (ii) the CCR4-NOT deadenylation complex that enables mRNA deadenylation and translational repression. In contrast to bilaterians, in land plants, miRNAs bind to their targets via nearly-perfect matches and the Argonaute carrying the miRNA usually cleaves the target (6,7). In addition to cleavage, miRNA-mediated translational repression happens also in plants, but unlike in bilaterians it does not involve GW182 and still requires a nearly-perfect complementarity between the miRNA and its target (8,9). As the modes of miRNA action are so different between plants and bilaterian animals, their study in other clades is important for shedding light on the evolution of this mechanism. The starlet sea anemone Nematostella vectensis, a representative of Cnidaria (sea anemones, corals, jellyfish and hydrozoans), the sister phylum of Bilateria, was recently shown to frequently cleave its miRNA targets via nearly perfect matches (10). While this mechanism exhibits unexpected resemblance to the mode of action of plant miRNAs, Nematostella and other cnidarians also possess genes encoding for GW182 homologs (Figure 1A; (11)). The existence of GW182 proteins in Cnidaria raises the question whether they play a role in post-transcriptional regulation in this basally-branching animal phylum that separated from Bilateria ∼600 million years ago (MYA).
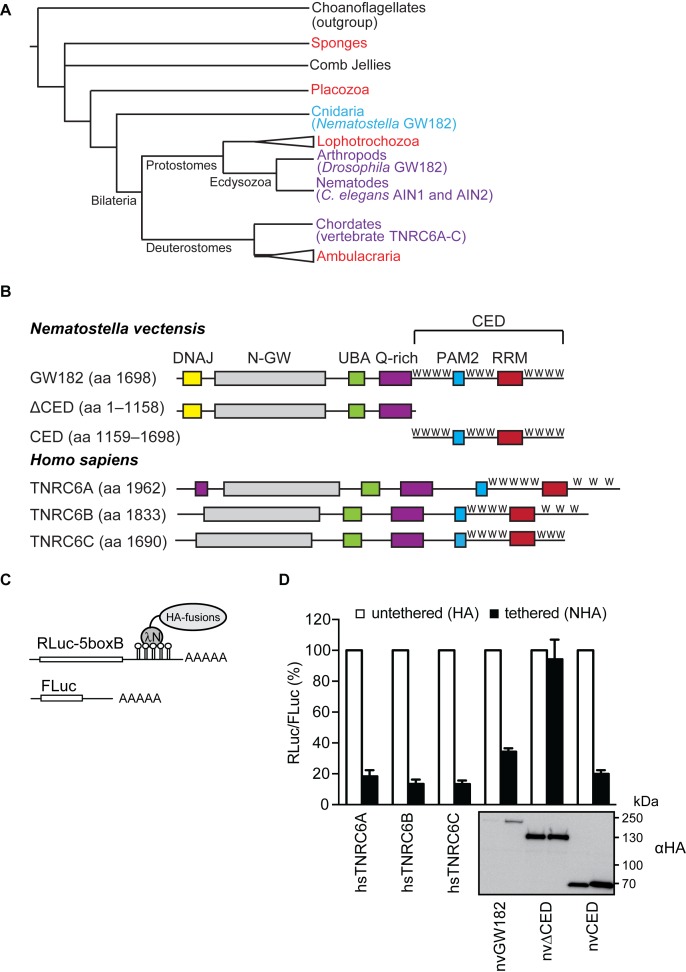
Figure 1.
nvGW182 represses tethered mRNA via its C-terminal effector domain (CED). (A) Schematic phylogenetic tree of Metazoa at the phylum level [based on (53,54)]. Phyla where GW182 proteins were functionally and biochemically studied appear in purple and the studied protein appear within brackets. Phyla where GW182 homologs are found in the genome but have not been functionally studied yet appear in red. Cnidaria appear in light blue. Phylogenetic groups of multiple phyla are indicated by a triangle. The illustrated polytomy of sponges and comb jellies is due to the current uncertainty regarding their relative phylogenetic positions. (B) Schematic representation of Nematostella vectensis GW182 (nvGW182), its human homologs TNRC6A, B and C, and their deletion mutants. DNAJ domain (yellow); N-GW: GW-repeat–rich region (grey); UBA: ubiquitin associated domain (green); PAM2: PABP associated motif 2 (blue); RRM: RNA-recognition motif (red). The C-terminal effector domain (CED) is formed RRM, PAM2 and the unstructured flanking regions with tryptophan-containing motifs, or W-motifs (W). The numbers correspond to the amino acid positions. (C) Schematic representation of reporter constructs used in tethering experiments: RLuc-boxB encodes for Renilla luciferase (RLuc) coding sequence fused to a 3′UTR with five boxB sites that specifically bind to λN peptide; FLuc encodes for Firefly luciferase (FLuc) coding sequence without boxB sites and serves as a control (55). (D) Repression of RLuc-boxB mRNA by NHA-nvGW182 and its deletion mutants. Human HEK293 cells were co-transfected with plasmids encoding RLuc-boxB, FLuc and full-length NHA-nvGW182 or indicated NHA-nvGW182 deletion mutants. As positive controls, plasmids encoding human NHA-TNRC6A, NHA-TNRC6B and NHA-TNRC6C were co-transfected. As negative controls, plasmids encoding untethered HA-fusions were used. RLuc activity was normalized to that of FLuc and presented as a percentage of RLuc produced in the presence of the corresponding untethered HA-fusions (open bars). Values represent means ±SD from 5 to 6 experiments. Expression levels of HA-fusion proteins were estimated by Western blotting with antibodies directed against HA-peptide.
Here, we show that the GW182 protein of Nematostella (nvGW182) interacts with AGO proteins and is able to repress mRNA. Its function in repression is mediated by tryptophan-containing motifs that recruit the CCR4-NOT deadenylation complex and lead to translational repression and mRNA decay. Our finding suggests that the mechanism of miRNA silencing via the CCR4-NOT complex is conserved for at least 600 million years.
MATERIALS AND METHODS
Cell culture, transfections and luciferase assay
Human HEK293 and HeLa cells were grown in Dulbecco's modified Eagle's medium with GlutaMAX™ supplement (DMEM+ Gluta MAX, GIBCO) with 10% FCS. Transfections were done in 24- and 96-well plates with polyethylenimine (PEI) using a 1:5 ratio of DNA:PEI. In tethering experiments, HEK293 cells were transfected with 2 ng RLuc-5BoxB and 30 ng FLuc (transfection control), and 10 ng HA- or NHA-fusion constructs per well of a 96-well plate. When indicated, increasing amounts of plasmids encoding CNOT6cat and CNOT7cat (50, 100 and 200 ng) were co-transfected. Transfections containing less than 200 ng of CNOTcat plasmids were topped up to 200 ng with LacZ-encoding plasmid. For other formats, the amounts of plasmids were adjusted proportionally. Cells were lysed 24 h post transfection. In rescue experiments, HeLa rtTA (control) and Hela rtTA- TNRC6-KD cells were transfected in a 96-well plate format.
For rescue experiments, HeLa line expressing TNRC6A&B-targeting shRNA under doxycycline-inducible promoter was generated using recombinase-mediated cassette exchange strategy. The shRNA sequences, corresponding to the previously used siRNAs (12) (5′-GCCUAAUCUCCGUGCUCAATT-3′, 5′-GGCCUUGUAUUGCCAGCAATT-3′) were cloned into pBIF3-miRNA-diGFP (13) and stably integrated between FRT sites of HeLa-11ht line (14), resulting in the HeLa-11ht-TNRC6A&B line. The parental HeLa-11ht line was used as ‘no knockdown’ control. TNRC6 knockdown was induced for 2 days with doxycyclin (1 μg/ml). On day 2, doxycyline was removed and cells were transfected with 0.5 ng FLuc/RLuc-hmga2-wt or FLuc/RLuc-hmga2-mut dox-inducible let7 reporters (12,15), and increasing amounts (25–200 ng/well) of the indicated rescue constructs. LacZ-encoding plasmid was used as a filler, to top up each transfection to the same the total amount of DNA. On day 3, expression of the reporters was induced with doxycyclin (1 μg/ml) for 4 h prior to cell lysis.
Luciferase activities were measured with a homemade luciferase reporter assay system. Forty five microliters of FLuc reagent (75 mM Hepes pH 8.0, 0.1 mM EDTA, 4 mM MgSO4, 530 μM ATP, 270 μM Coenzyme A, 470 μM DTT and 470 μM luciferin) and 45 μl of RLuc reagent (2.2 mM Na2EDTA, 220 mM K3PO4 pH 5.1, 0.44 mg/ml BSA, 1.1 M NaCl, 1.3 mM NaN3 and 0.6 μg/ml colenterazine) reagents per sample were used to measure luciferase activities.
DNA constructs
Tethering reporter plasmids RLuc-5BoxB and FLuc, NHA- and HA-AGO2 (16); let-7 reporter plasmids FLuc/RLuc-hmga2-wt and FLuc/RLuc-hmga2-mut (12,15); TNRC6 proteins tagged with HA, NHA (20) and GST (12) have been described previously. Plasmids encoding CNOT6cat and CNOT7cat were generated by re-cloning the corresponding ORFs from previously described plasmids (21) in pEBG-derived vector bearing a 3xHA-tag instead of GST (pEBH). pEBG (Addgene 22227) was a gift from David Baltimore (22). Coding sequences of nvAGO1 and nvAGO2 were PCR-amplified from Nematostella cDNA and cloned into pEBH vector (deposited to Addgene 82434, 82435) and pCiNeo vectors bearing an HA- or NHA-tag (16).
Plasmids encoding the HA-, NHA- and GST-tagged nvGW182 and its deletion mutants were PCR-amplified, using synthetic humanized nvGW182 coding sequence as a template, and cloned into pCiNeo vectors bearing an HA- or NHA-tag (16) and pEBG vectors (deposited to Addgene 82418–82426). pEBG constructs were used in GST pull-downs, and pCiNeo constructs in tethering and rescue experiments. The nvGW182 deletion mutants correspond to the following regions: nvΔDNAJ (aa 62–1698), nvΔCED (aa 1–1158), nvΔCED1 (aa 441–1158), nvΔCED2 (aa 497–1158), nvΔCED3 (aa 565–1158), nvΔCED4 (aa 643–1158), nvCED (aa 1159–1698), nvDNAJ (aa 1–61). Point mutations W→A in nvGW182 and nvCED W-motifs (deposited to Addgene 82427–82429) were introduced by site-directed mutagenesis (23). The positions of mutated W-motifs are the following: W1167A, W1183A, W1227A, W1309A, W1391A, W1453A, W1574A, W1589A, W1627A, W1638A, W1657A.
Human/Nematostella GW182 chimeric constructs were generated by PCR-based fusion of shRNA-resistant TNRC6A N-terminal region (aa 1–1169, Q8NDV7-2) with nvCED (aa 1159–1698), wt or 11W11A and cloning the resulting hybrid coding sequences into pCiNeo-NHA (deposited to Addgene 82430, 82431) and pEBG vector (deposited to Addgene 82432, 82433).
Pull-down assays, immunoprecipitation, Western blotting and mass spectrometry
For GST pull-down assays, HEK293 cells grown in 10 cm dishes were transfected with 0.2–5 μg plasmids expressing GST-fusions. Cells were lysed 24 h post-transfection and GST-fusions were pulled down as described (12). Liquid chromatography-tandem mass spectrometry analysis was performed with in-solution digested affinity purified protein samples on a Q Exactive mass spectrometer (Thermo Scientific) as previously described (24). Label-free quantitation (LFQ) was performed using MaxQuant Analysis Software (25,26), and statistic analysis using Perseus Software. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (27) partner repository with the data set identifier PXD004668.
For αAGO1 and αCNOT9 immunoprecipitation (IP) and Western blot in Nematostella, custom antibodies were raised against the synthetic peptide CMMDRDKEAGNDNSS derived from Nematostella AGO1 (nvAGO1) and a recombinant fragment spanning amino acid positions 1301–1698 of nvGW182 in rabbit and guinea pig, respectively (GenScript, USA). Ten micrograms of αnvAGO1 or αCNOT9 antibodies, coupled with protein A magnetic beads (Bio-Rad), and 500 μg of lysate from Nematostella embryos 48 h post-fertilization were used per each IP. Embryos were lysed in the lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 25 mM EDTA, 0.5% NP-40, 1 mM DTT, complete Ultra (Roche) and Set III (Merck) protease inhibitor cocktails), after IP the beads were washed with the wash buffer [50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 0.05% NP-40, complete Ultra (Roche) and Set III (Merck) protease inhibitor cocktails] and bound proteins were eluted with the SDS-PAGE sample buffer.
The following primary antibodies were used for Western blotting on human cells: αAGO2 (Ascenion GmbH 11A9) 1:4000; αCNOT7 (Abnova) 1:2000; αCNOT9 (Proteintech) 1:5000; αGST-HRP conjugated (Abcam) 1:5000; αalpha-TUBULIN (Sigma T5168) 1:4000; and αHA (Roche 3F10) 1:5000. For Nematostella samples the following antibodies were used in Western blotting and IP: αCNOT9 (OriGene) 1:1000; αnvGW182 (GenScript, described above) 1:1000; αnvAGO1 (GenScript, described above) 1:1000.
RNA extraction and qRT-PCR
For qRT-PCR analysis, cells were lysed with cytoplasmic RNA lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM KCl, 0.5% Triton X-100, 0.2 mM Pefabloc), clarified and split for luciferase and qRT-PCR assay. RNA was isolated with TriFast FL (Peqlab), treated with RQ1 DNase I, and reverse-transcribed using the Maxima first strand cDNA synthesis kit (Thermo Fisher). RLuc and FLuc were quantified using previously described primers (15) and sensiFAST SYBR No ROX qPCR kit (Bioline). Relative RLuc/FLuc expression levels were calculated using ΔΔCt method.
RESULTS
Nematostella GW182 protein represses mRNA via W-motifs in its C-terminal domain
Proteins of the GW182 family show relatively low global sequence conservation, but are characterized by a conserved domain organization that includes an N-terminal GW-rich region, an ubiquitin-associated (UBA) domain, a glutamine-rich (Q-rich) region, PABP-associated motif 2 (PAM2) and a C-terminal RNA recognition motif, flanked by tryptophan-motifs-containing regions [W; Figure 1B; (3)]. Nematostella GW182, in addition, contains DNAJ domain. The N-terminal GW repeats bind AGOs (19,28–31), and the C-terminal part of the protein (CED) recruits the CCR4-NOT and PAN2/PAN3 deadenylation complexes to repress mRNA (12,32,33). In our previous work, we identified tryptophan-containing WG/S/T and G/S/TW repeats (called W-motifs for their invariant residues) as the key silencing elements, both necessary and sufficient for recruitment of deadenylases and mRNA silencing (12). The structural aspects of this recruitment were dissected in further studies by Filipowicz, Conti, Weichenrieder and Izaurralde groups (34,35). Importantly, function of these motifs was shown to be conserved between human and Drosophila (12). As nvGW182 contains similar motifs in its C-terminal region, we reasoned that it may function via a similar mechanism.
To test if function of nvGW182 in mRNA repression is conserved, we analyzed its effect on mRNA in transfected HEK293 cells using an RNA–protein tethering assay. Tethering was achieved by co-expressing Renilla luciferase mRNA containing five boxB hairpins in its 3′-UTR (RLuc-5boxB; Figure 1C), and nvGW182 protein or its deletion mutants fused with HA-tag and lambda phage N peptide, recognizing the boxB sites (16,36). As untethered controls that were not expected to repress RLuc-5boxB, we used analogous HA-fusions lacking N peptide. As expected, human homologs TNRC6A, B and C, used as positive controls, led to efficient repression of tethered mRNA (Figure 1D; ∼10- to 5-fold). Importantly, we observed that tethering of nvGW182 leads to ∼3.5-fold repression. To identify the domains of nvGW182 that function in repression, we generated C- and N-terminal deletions (nvΔCED and nvCED, respectively) and tested their ability to repress tethered mRNA. Consistently with the prior mapping studies in human and Drosophila (17–20), we observed that nvCED effectively represses mRNA (∼5-fold), while nvΔCED has practically no effect (Figure 1D).
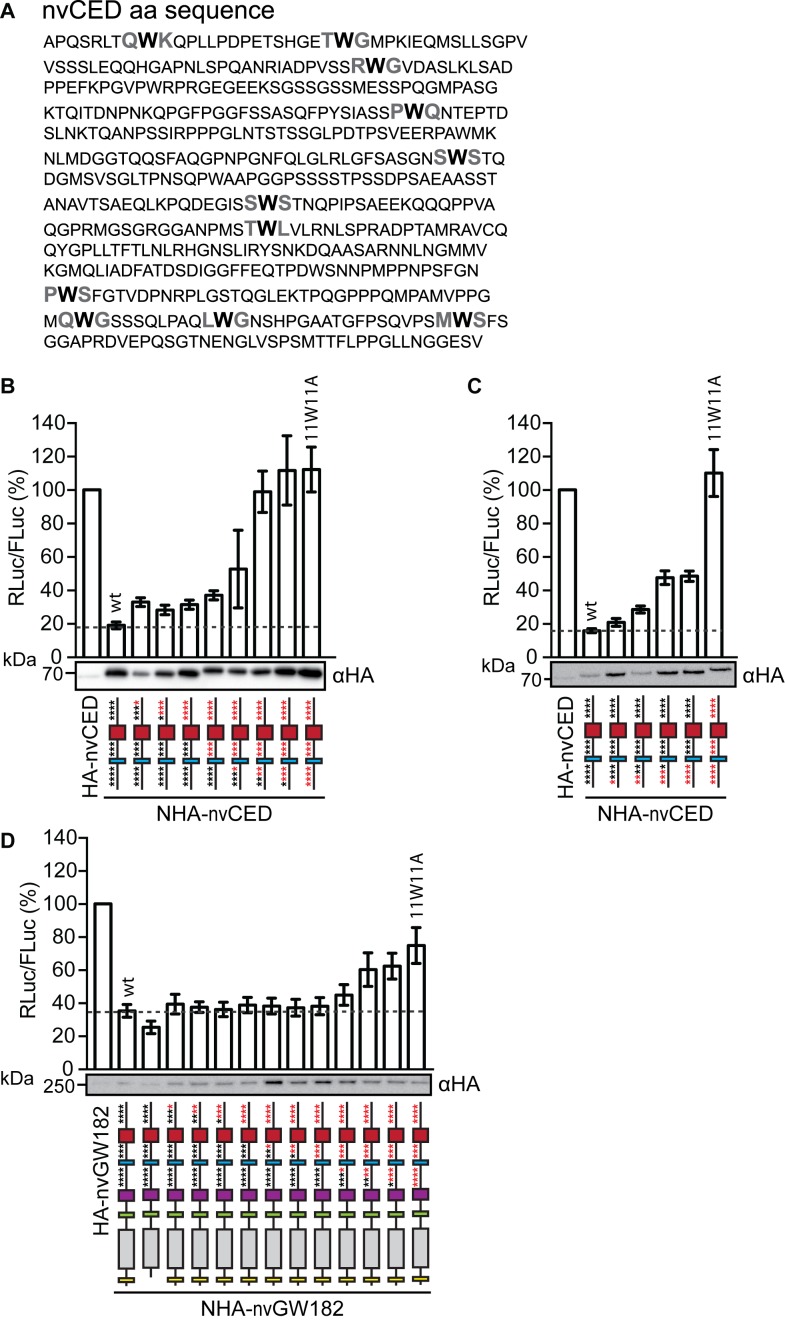
Next, we tested if nvCED mediates repression via presumptive W-motifs (Figure 2A; W-motifs are shown in bold). For that, we mutated W residues in W-motifs (W → A) starting from the C-terminus and analyzed the effect of these mutations on expression of the tethered mRNA (Figure 2B). We observed that mutations of W-motifs indeed led to alleviation of repression by nvCED. As most alleviation was achieved with mutating the four N-terminal W-motifs, we decided to test if these motifs are the most critical for repression. For that, we generated nvCED with sequential mutations of the four N-terminal W-motifs (Figure 2C). These mutations lead to partial alleviation of repression (∼2.5-fold), and repression was completely abrogated only when all eleven motifs were mutated (11W11A). These results suggest that W-motifs act in additive manner to mediate silencing. Importantly, W-motifs were also required for mRNA silencing in the context of the full-length nvGW182 (Figure 2D). Western blot analysis of the NHA-nvCED and NHA-nvGW182 confirmed that the differences in their repressive potential cannot be attributed to the expression levels (Figure 2B–D).
Figure 2.
nvGW182 represses tethered mRNA via W-motifs. (A) Sequence of the nvGW182 CED (nvCED) with tryptophans in presumptive W-motifs shown in bold. (B) Mutations of W residues (W→A) in W-motifs alleviate repression by the nvCED. Plasmids encoding either wt NHA-nvCED or its indicated mutants were co-transfected into HEK293 cells, together with RL-5BoxB and FL. As negative control, untethered HA-nvCED was used. Mutants 1W1A through 11W11A contain up to eleven mutated W residues (for details, see Materials and Methods). Schematic representations of nvCED constructs are drawn below the corresponding bars. Asterisks indicate presumptive W-motifs; W residues are shown in black and W→A mutations in red. Data were analyzed and presented as in Figure 1D (means ±SD, n = 4–10). Expression of NHA-fusion proteins was estimated by Western blotting. (C) W-motifs additively contribute to repression by tethered nvCED. Experiment was performed as in (B), except that nvCED with four N-terminal W-motifs mutated were used (n = 4). (D) W-motifs contribute to repression by the full-length nvGW182. Experiment was performed as in (B), except that full-length nvGW182 was used instead of nvCED domain (n = 4–12).
Nematostella GW182 recruits the CCR4-NOT deadenylation complex via its W-motifs to stimulate mRNA decay and repress translation
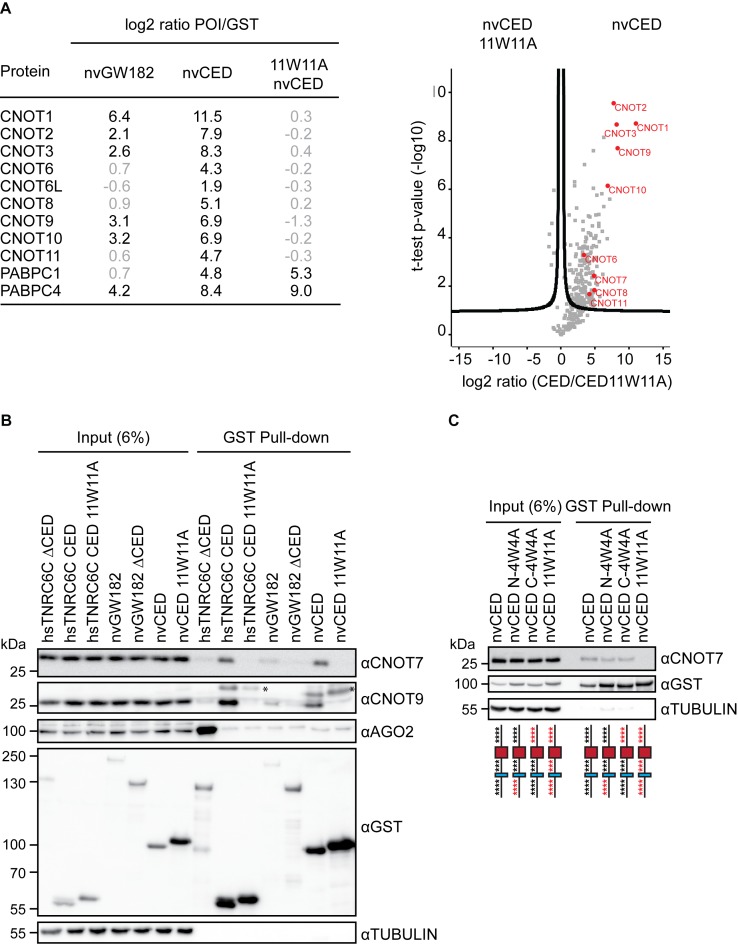
To understand how nvGW182 mediates repression, we decided to identify proteins that interact with nvGW182 and its repressive domain nvCED. We expressed either full-length GST-nvGW182 or its deletion mutants (GST-nvCED, GST-nvΔCED, GST-nvDNAJ) in HEK293 cells, pulled down with a GST tag and analyzed by mass spectrometry (Figure 3A; Supplementary Table S1). As silencing was mediated by W-motifs present in nvCED (Figure 2B and C), the 11W11A nvCED mutant was included as a negative control. Similarly to their mammalian counterparts (12,32,33,37–39), the full-length nvGW182 and wt CED associated with components of the human CCR4-NOT complex, CNOT1, CNOT2, CNOT3, CNOT9 and PABP proteins. Importantly, interactions with the CCR4-NOT subunits were lost in nvCED 11W11A mutant, demonstrating that nvGW182 recruits deadenylases via W-motifs. As both wt and 11W11A mutant nvCED proteins contain the PAM2, they both interacted with PABP. We have not detected PAN2/PAN3 deadenylation complex among interactors of nvGW182, although it was previously found to bind human and Drosophila GW182 (12,33). Thus, interaction with PAN2/PAN3 might not be conserved in Nematostella. Alternatively, it is possible that PAN2/PAN3 was not detected in the nvCED pull-downs because it is a weaker interactor than CCR4-NOT. To validate the results of the mass spectrometry, we analyzed the content of GST pull-downs by Western blotting. Indeed, both CNOT7/CAF1 and CNOT9 interacted with the wt nvCED, but not with the 11W11A mutant variant (Figure 3B). When only four N- or C-terminal W-motifs were mutated (N-4W4A and C-4W4A), nvCED retained most of its affinity to the CNOT7 (Figure 3C), suggesting an additive role of W-motifs in the CCR4-NOT recruitment.
Figure 3.
nvGW182 recruits subunits of the CCR4-NOT complex via its W-motifs. (A) Quantitative mass spectrometric analysis of proteins associated with nvGW182. The GST-nvGW182 full-length constructs, the nvCED domain only, as wild type and 11W11A mutant variant, were expressed in HEK293 cells and affinity purified using GST. GST alone was used as a negative control. On the right, relevant interactors for each variant are listed along with their corresponding enrichment factor (the log2 ratio of label-free quantification values (LFQ intensity) over corresponding LFQs detected in pull-down of GST tag alone). On the left, volcano plot showing proteins with differential binding to nvCED and nvCED11W11A variant. The logarithmic ratios of protein intensities were plotted against negative logarithmic P-values of a two samples t-test (two sided). A hyperbolic curve separates significantly enriched proteins from common binders. Selected interactors recruited via W-motifs of nvCED are labeled in red (FDR ≤ 0.05, n = 2). (B) Validation of CCR4-NOT-nvCED interactions identified in (A). GST-fusions of indicated proteins were expressed in HEK293 cells, and inputs (6%) and GST-pulldowns were analyzed by Western blotting using the indicated antibodies. Human TNRC6CΔCED was used as positive control for AGO2 binding. TNRC6C CED and TNRC6C 7W7A CED were used as positive and negative controls for CNOT7 and CNOT9 binding. Asterisks (*) indicate unspecific bands. (C) W-motifs function additively to recruit the CCR4-NOT complex. The experiment was performed as in (B), except that nvCED with four N-terminal (N-4W4A) or C-terminal (C-4W4A) W-motifs mutated were used for pull-downs. nvCED and nvCED 11W11A were used as positive and negative controls for CNOT7 binding, respectively. Schematic representations of nvCED domains are shown below the corresponding lane. Asterisks indicate W-motifs; W residues are shown in black and W→A mutations in red.
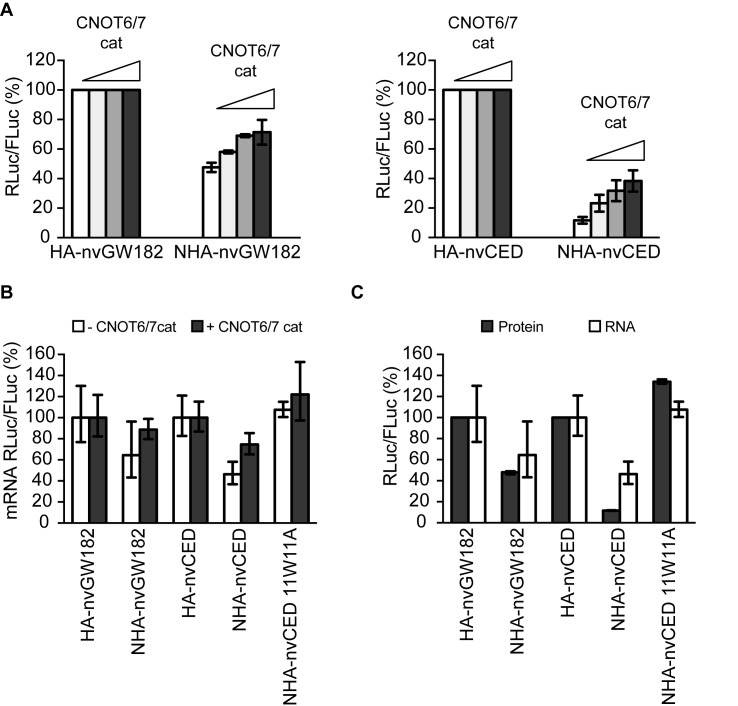
The CCR4-NOT complex includes two deadenylation enzymes, CNOT6/CNOT6L and CNOT7. To test if deadenylation activity of the recruited CCR4-NOT complex contributes to nvGW182-mediated repression, we made use of the catalytically inactive mutants CNOT6cat and CNOT7cat (40). These mutants function as dominant negative and their overexpression leads to mRNA stabilization because of decrease in deadenylation rates. Therefore, if function of nvGW182 involves CCR4-NOT-mediated deadenylation, we expect that interfering with CNOT6 and CNOT7 deadenylation activity would affect repression by nvGW182. Indeed, when co-expressing increasing amounts of CNOT6cat and CNOT7cat, we observed partial alleviation of repression by nvGW182 and nvCED (Figure 4A) accompanied by stabilization of tethered mRNA (Figure 4B). In bilaterians GW182 mediates not only mRNA deadenylation via CCR4-NOT, but also translational repression (12,34). To test if Nematostella protein functions similarly, we compared the effects of nvGW182 and nvCED on the levels of tethered RLuc-boxB reporter mRNA and its protein product, measured by qRT-PCR and luciferase assay (Figure 4C). This analysis confirmed that inhibition of protein production (filled bars) cannot be fully explained by changes in mRNA levels (open bars), suggesting that nvGW182 affects both mRNA stability and translation. Importantly, no silencing was observed with the 11W11A nvCED mutant, defective in CCR4-NOT recruitment, indicating the role for W-motifs in both mRNA decay and translational repression. Altogether, these results suggest that the mechanism by which GW182 protein represses mRNA may be conserved between Nematostella, flies and mammals.
Figure 4.
nvGW182 represses translation and mediates mRNA degradation via CNOT6/7 deadenylases. (A) The deadenylation catalytic activity of the CCR4-NOT complex contributes to nvGW182-mediated silencing. HEK293 cells were co-transfected with RLuc-boxB, FLuc, tethered NHA-nvGW182 or NHA-nvCED, and increasing amounts of CNOT6 and CNOT7 catalytic mutants (CNOTcat). Untethered HA-fusions were used as negative controls (means ±SD, n = 3-6). (B) Interfering with the CCR4-NOT deadenylation activity leads to stabilization of nvGW182-bound mRNAs. RLuc and FLuc mRNA from nvGW182-tethering experiments shown in (A) were estimated by qRT-PCR. Filled bars: +CNOTcat; open bars: -CNOTcat. RLuc values were normalized to FLuc mRNA. Untethered HA-fusions were used as negative controls (means ±SD, n = 2). (C) nvGW182 silences tethered mRNA via translational repression and stimulation of mRNA decay. RLuc and FLuc mRNA and protein levels from nvGW182-tethering experiments were estimated by qRT-PCR (RNA, open bars) and luciferase assays (protein, filled bars). Data were normalized and presented as in (A and B).
Nematostella GW182 interacts with Nematostella AGOs, but not human AGO2
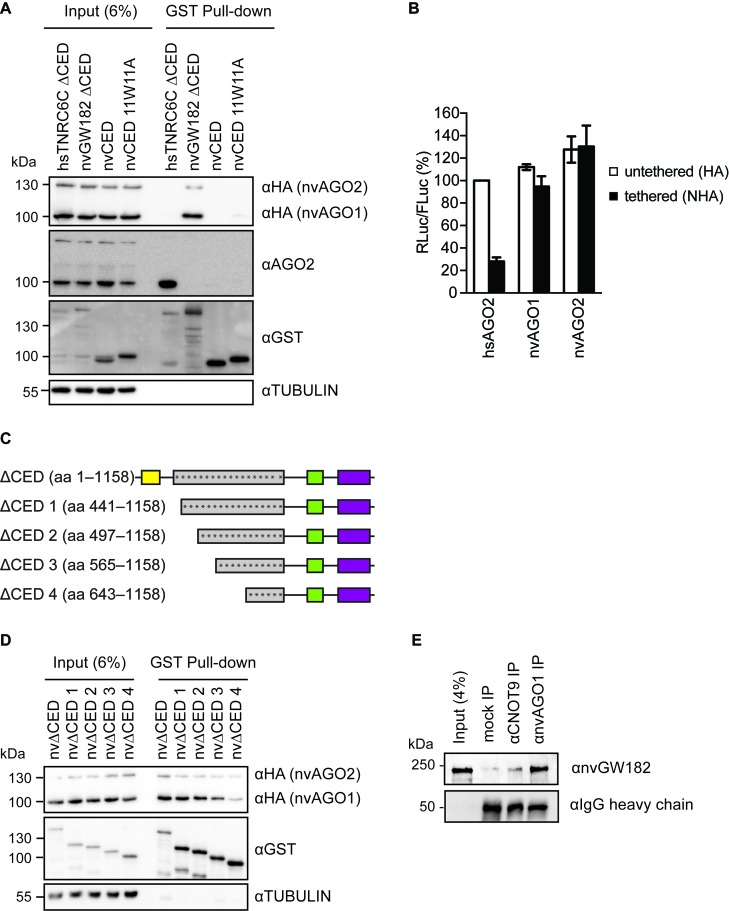
In order to function in the miRNA silencing pathway, GW182 proteins need to be recruited to an mRNA via direct interaction with AGO:miRNA complex, which is happening via the N-terminal GW repeats (19,28–31). Intriguingly, mass spectrometry performed on the GST pull-downs of nvGW182 or its N-terminal regions (nvΔCED, nvDNAJ) did not detect any significant interaction with human AGO proteins (Supplementary Table S1). To confirm this result, we performed Western blotting on the GST pull-downs with αAGO2 antibody, using N-terminal domain of the human TNRC6C (TNRC6C ΔCED) as a positive control (Figures 3B and 5A). Indeed, we detected strong interaction between AGO2 and human TNRC6C ΔCED, but not with nvGW182 domains overexpressed in HEK293 cells. The possible explanations of this result are that (i) nvGW182 binds only Nematostella, but not human AGOs, because of low conservation of binding sites or (ii) nvGW182 is recruited to mRNA via a different RNA-binding protein. To discriminate between these possibilities, we overexpressed HA-tagged Nematostella AGOs, nvAGO1 and nvAGO2, in human HEK293 cells, together with GST-nvGW182 domains. We then tested for the presence of nvAGO1 and nvAGO2 in the GST pull-down, by Western blotting with αHA antibody (Figure 5A). We found that nvAGOs interact with the N-terminal domain of nvGW182 protein (nvGW182 ΔCED), but not with its human homolog (TNRC6C ΔCED). These results suggest that GW182-AGO interaction regions are not conserved between human and Nematostella. Consistently with this result, Nematostella AGOs were not able to repress tethered mRNA, unlike human AGO2 (hsAGO2) used as a positive control (Figure 5B).
Figure 5.
GW182-AGO interaction regions are not conserved between human and Nematostella. (A) nvGW182 binds nvAGOs, but not human AGO2. GST-fusions of nvGW182 or its domains were co-expressed with HA-tagged nvAGO1 and nvAGO2 in HEK293 cells, pulled down with GST tag, and inputs (6%) and GST-pulldowns were analyzed by Western blotting using the indicated antibodies. Human TNRC6CΔCED was used as positive control for human AGO2 binding. (B) Nematostella AGOs are not able to repress tethered mRNA in human cells. Human HEK293 cells were co-transfected with RLuc-boxB, FLuc and NHA-nvAGO1 or NHA-nvAGO2. Human AGO2 (NHA-hsAGO2) was used as a positive control. As negative controls, plasmids encoding untethered HA-fusions were used (means ±SD, n = 3). (C) Schematic representation of nvΔCED deletion mutants used in (D). The numbers correspond to the amino acid positions. Asterisks represent putative AGO-binding motifs. (D) Multiple regions of nvΔCED contribute to nvAGOs binding. GST-fusions of nvΔCED deletion mutants (C) were co-expressed with HA-nvAGOs in HEK293 cells, and inputs (6%) and GST-pulldowns were analyzed by Western blotting using the indicated antibodies. (D) AGO, GW182 and the CCR4-NOT complex natively interact in Nematostella. Nematostella embryonic lysates were used in IP with αnvAGO1 and αCNOT9 antibody, and the co-immunoprecipitated proteins were analyzed by Western blotting with αnvGW182 antibody. Unspecific rabbit IgG was used as a negative control.
Previous works (30,41) have shown that human GW182 proteins have multiple regions capable of AGO binding. Interaction with AGO is mediated by tryptophan residues, but these AGO-binding motifs are functionally distinct from the CCR4-NOT-binding W-motifs (12,31). To test if nvGW182 also possesses multiple AGO-binding regions, we generated a set of N-terminal deletion mutants containing different amount of potential AGO-binding tryptophans (asterisks; Figure 5C). We used GST pull-down assay to assess the ability of the generated nvGW182 deletions to recruit nvAGOs (Figure 5D). Deletion of N-terminal portions lead to gradual reduction of nvAGOs signal, suggesting that nvGW182 recruits nvAGOs via multiple regions, similarly to human orthologs.
While our results provide strong evidence that Nematostella GW182 can interact with human CCR4-NOT and Nematostella AGOs, they were obtained under overexpression conditions and in human cell culture. To test whether those interactions also occur naturally in the sea anemone itself, we performed immunoprecipitation (IP) from Nematostella embryonic lysates, using a custom antibody raised against nvAGO1 and a commercially available antibody against a region of human CNOT9 that is 94% identical to its Nematostella homolog. Both IPs were then tested by Western blot, and showed the presence of nvGW182 in nvAGO1 and CNOT9 IP, but not in control mock IP performed with unspecific rabbit IgGs (Figure 5E). We concluded that AGO, GW182 and CNOT9 interact with one another in Nematostella like in mammals (12,32–35,37).
W-motifs of Nematostella GW182 contribute to bona fide miRNA-mediated silencing
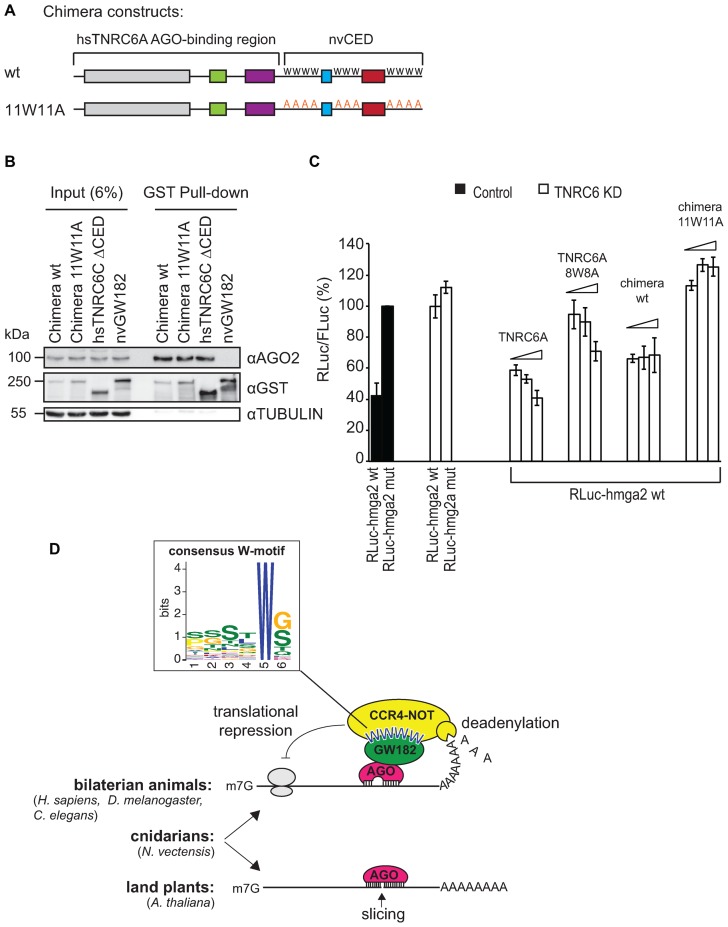
As nvGW182 does not interact with human AGOs, it cannot be used to rescue the depletion of TNRC6 proteins. To overcome this issue and show that nvGW182 can function in miRNA silencing, we generated a chimeric construct containing the N-terminal AGO-binding domain of human TNRC6A protein and C-terminal CCR4-NOT-binding domain of nvGW182 (Figure 6A). This chimeric construct was generated in two versions: wt and 11W11A mutant that does not interact with CCR4-NOT. We tested the constructs in GST pull-downs to make sure that both wt and 11W11A chimeras are able to bind human AGO2 with the same efficiency (Figure 6B).
Figure 6.
Human/Nematostella GW182 chimera partially rescues miRNA silencing in human HeLa cells. (A) Schematic representation of human/Nematostella GW182 protein chimeras (h/nvGW182) used in (B) and (C). AGO-binding N-terminal region of human TNRC6A was fused to nvCED, either wild type or 11W11A mutant. (B) h/nvGW182 chimera binds endogenous human AGO2. GST-fusions of the indicated proteins and their domains were expressed in HEK293 cells, and inputs (6%) and GST-pulldowns were analyzed by Western blotting using the indicated antibodies. (C) W-motifs of h/nvGW182 chimera are required to rescue depletion of endogenous TNRC6 proteins. HeLa cell line carrying stably integrated inducible shRNAs construct against endogenous TNRC6A and B was used for TNRC6 knockdown experiments (open bars); HeLa cells not expressing TNRC6-directed shRNAs was used as a control (filled bars). Expression of shRNAs was induced for 2 days prior to transfection of miRNA reporters and rescue constructs. Cells were transfected with RLuc-hmga2 reporter containing let-7 sites or its mutant version (RLuc-hmga2 mut), and increasing amounts of plasmids expressing h/nvGW182 chimera or its 11W11A mutant. TNRC6A was used as a positive control, TNRC6A 8W8A was used as a negative control. Values represent percentages of RLuc activity produced by hmga2-mut reporter without TNRC6 depletion (means ±SD, n = 3). (D) Speculative model illustrating the mechanisms of miRNA-mediated silencing in bilaterian animals (recruitment of the CCR4-NOT complex via W-motifs of GW182), land plants (slicing by AGO and translational repression via an unknown mechanism that depends on AGO), and cnidarians (both slicing and GW182-dependent mechanism). The close-up shows the graphic representation of the W-motif that recruits the deadenylation complexes and is conserved in human, Drosophila and Nematostella. The consensus was derived with the MEME suite (42), see also Supplementary Figure S1.
Next we analyzed if chimeric human/Nematostella GW182 can function in a bona fide miRNA repression assay. For that, we depleted HeLa cells of the endogenous TNRC6A and B using stably integrated inducible shRNA constructs, and tested chimeric GW182 proteins for their ability to rescue miRNA repression. To assess miRNA-mediated silencing, cells were co-transfected with the Fluc/RLuc-hmga2 reporter, targeted by let-7 endogenously expressed in HeLa cells (Figure 6C). In control cells let-7 efficiently repressed wild-type Rluc-hmga2 mRNA, when compared with the mutant reporter from which let-7-binding sites were deleted (compare Rluc-hmga2 wt with RLuc-hmga2 mut, filled bars). Upon depletion of endogenous TNRC6 proteins (open bars), repression of the hmga2 reporter was alleviated. As shown previously, transfection of a plasmid encoding wt TNRC6A resistant to shRNA rescued the repression, and its unfuctional mutant TNRC6A 8W8A could not rescue (12). Intriguingly, the chimeric protein carrying nvCED was also able to partially rescue miRNA-mediated silencing (wild-type chimera). Importantly, its function in miRNA silencing was dependent on the CCR4-NOT-recruiting W-motifs, as 11W11A mutant was not able to complement the knockdown of TNRC6 proteins. To summarize, these data suggest that W-motifs of nvGW182 mediate miRNA silencing in a genetic rescue experiment.
Thus, our experiments support the model where the mechanism of miRNA silencing via W-motifs is conserved between cnidarians and bilaterians (Figure 6D). To derive the consensus sequence of the CCR4-NOT-recruiting W-motif, shared between bilaterians and cnidarians, we applied the MEME motif-searching algorithm (42) on the experimentally validated W-motifs in human, Drosophila (12) and Nematostella GW182 proteins (Figure 2B, C and D). The resulting consensus sequence of W-motif (Figure 6D, Supplementary Figure S1) is rich in S, T and G residues, surrounding the invariant W.
DISCUSSION
Conservation of the mechanisms of miRNA silencing
In an earlier study, we have shown that Nematostella possesses the miRNA pathway (10). Unlike mammals and flies and similar to plants, Nematostella miRNAs tend to have high complementarity to their targets and mediate target slicing. This finding suggested that slicing might be the ancestral mode of action of miRNAs in animals. However, unlike plants, Nematostella possesses a GW182 homolog, a protein required for miRNA-mediated deadenylation and translational repression in Bilateria. Using human cultured cells and Nematostella embryonic lysates, we show that nvGW182 functions via a similar mechanism as its bilaterian homologs: it recruits the CCR4-NOT deadenylation complex via its W-motifs, to affect both mRNA stability and translation. Moreover, this conclusion is supported by the high conservation of the DDX6 helicase in Nematostella (72% identity and 85% similarity between human and Nematostella): this helicase was shown to contribute to miRNA-mediated translational inhibition in mammals (34,35,43).
The question remains, why Nematostella miRNAs employ two different mechanisms of action – AGO-mediated slicing and AGO/GW182/CCR4-NOT-mediated deadenylation and translational repression. It is unlikely that only one of two Nematostella AGOs possesses slicing activity, and the second relies on nvGW182-dependent mechanism, because both nvAGO1 and nvAGO2 possess the five residues previously found to be important for AGO slicing activity in bilaterians (11,44,45). The most likely possibility is that GW182-mediated mechanism is employed by a subset of Nematostella miRNAs, which are only partially complementary to their targets and therefore cannot mediate slicing. An alternative explanation is that the Nematostella miRNA-mediated silencing system employs each of the two modes of action under different conditions or for different targets. A possible way of switching the mode of action would be modification of AGO catalytic site, as such modification can interfere with the slicing activity. Of note, plant miRNAs are also able to mediate both slicing and translational repression, although it involves non-GW182 family SUO protein (46). These results suggest that less stringent and reversible mechanism of function—mRNA deadenylation and translational repression—might be advantageous and therefore preserved by evolution.
In previous work, we demonstrated that W-motifs are both necessary and sufficient to recruit the deadenylation complexes and repress tethered mRNA and therefore play a key role in miRNA silencing in human and Drosophila (12). Here, we show that the function of these potent elements is conserved in over 600 million years of evolution and derive the consensus sequence of the W-motif, shared between bilaterians and cnidarians (Figure 6D). Interestingly, we observed that these motifs are rich in serine and threonine residues, which makes them hotspots for phosphorylation (47,48). Such reversible phosphorylation of W-motifs in TNRC6/GW182 proteins could interfere with their ability to recruit the CCR4-NOT complex and, thus, represent an interesting mechanism for regulation of miRNA silencing in response to various stimuli.
Co-evolution of AGO and GW182 proteins
The fact that nvGW182 can interact with the human CCR4-NOT complex through its CED but fails to recognize human AGO2, points to a scenario where two domains of GW182 experienced very different evolutionary trajectories: the CED remained relatively conserved, as is indicated by the fact that it is the only part in nvGW182 that shows sequence homology to bilaterian GW182 proteins (11). The relative conservation of W-motifs in the CED might be the result of the very high conservation of the CCR4-NOT complex that is connected to numerous cellular pathways and hence has very low sequence flexibility (49). In striking contrast, the N-terminal part of the GW182 protein, which was shown to be crucial for AGO recognition [Figure 5A and D; (19,28–31)], evolved so rapidly that no sequence homology can be detected between this part of vertebrate TNRC6 and Nematostella GW182. The fast evolution of the GW182 N-terminal domain resulted in the inability of Nematostella AGOs to bind to human TNRC6 and vice versa. However, the ability of nvGW182 to bind nvAGOs and CNOT9 (as we also verified in Nematostella, Figure 5E), and the well-documented ability of bilaterian GW182/TNRC6 proteins to bind their respective AGOs and the CCR4-NOT complex (12,32–35,37,50,51), support a parsimonious view that the last common ancestor of Cnidaria and Bilateria carried at least one GW182 and one AGO and those were forming a miRNA-mediated silencing complex together with units of the CCR4-NOT complex. Further, it is likely that AGOs and the N-terminal domain of GW182 proteins have gone through separate co-evolutionary processes in the cnidarian and bilaterian lineages that are the reason for our observed lack of inter-lineage interaction. This escalating process might have started from a mildly disadvantageous mutation in one of the proteins that reduced the AGO-GW182 affinity and hence made a compensatory mutation in the partner protein advantageous (52).
In this work, we provide new insight into the evolution of the miRNA-mediated silencing mechanisms, using a cnidarian species, Nematostella vectensis, separated by more than half a billion years from the vast majority of Metazoa. We show that the core component of the miRNA pathway GW182 is conserved in cnidarians and is able to inhibit translation and stimulate mRNA decay by recruiting the CCR4-NOT deadenylation complex. An important task for the future is to understand how the two mechanisms of miRNA silencing—slicing and translational regulation/mRNA deadenylation—interact with each other.
Acknowledgments
The authors thank Dr Kai Schönig (Central Institute of Mental Health, Mannheim, Germany) for his kind gift of HeLa-11ht line (14) and pBIF3-miRNA-diGFP plasmid (13), Dr Ann-Bin Shyu (University of Texas Health Science Center at Houston, USA) for CNOT6- and CNOT7-encoding plasmids, Dr. Julien Bethune (Heidelberg University Biochemistry Center, Germany) for FLuc/RLuc-hmga2 miRNA reporters (15). This work was supported by the European Commission CIG grant (631482) to M.C. and a European Research Council starting grant (637456) to Y.M. M.M. is a recipient of the DAAD PhD fellowship. C.C.M. is supported by the Peter and Traudl Engelhorn PhD fellowship.
Authors contributions: M.M. performed the experiments presented in Figures 1–5D, 6; M.K. and M.S. contributed with the mass spectrometry analysis (Figure 3A and Supplementary Table S1); C.C.M. generated the HeLa-11ht-TNRC6A&B line; D.B contributed with cloning; R.A. performed the IP with Nematostella lysates shown in Figure 5E, N.G. amplified nvAGO coding sequences from cDNA and tested αnvAGO1 and αnvGW182 antibody; V.M. optimized Nematostella lysate IP experiments. M.M., Y.M. and M.C. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission CIG grant [631482 to M.C.]; European Research Council starting grant [637456 to Y.M.]; M.M. is a recipient of the DAAD PhD fellowship; Peter and Traudl Engelhorn PhD fellowship [to C.C.M.]. Funding for open access charge: CIG [631482 to M.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 4.Iwakawa H.O., Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 6.Baumberger N., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axtell M.J., Westholm J.O., Lai E.C. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakawa H.O., Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q., Wang F., Axtell M.J. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell. 2014;26:741–753. doi: 10.1105/tpc.113.120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran Y., Fredman D., Praher D., Li X.Z., Wee L.M., Rentzsch F., Zamore P.D., Technau U., Seitz H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014;24:651–663. doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran Y., Praher D., Fredman D., Technau U. The evolution of microRNA pathway protein components in Cnidaria. Mol. Biol. Evol. 2013;30:2541–2552. doi: 10.1093/molbev/mst159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger S.M., Pesold B., Reber S., Schonig K., Berger A.J., Weidenfeld I., Miao J., Berger M.R., Gruss O.J., Bartsch D. Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs. Nucleic Acids Res. 2010;38:e168. doi: 10.1093/nar/gkq616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidenfeld I., Gossen M., Low R., Kentner D., Berger S., Gorlich D., Bartsch D., Bujard H., Schonig K. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37:e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bethune J., Artus-Revel C.G., Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chekulaeva M., Filipowicz W., Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazzaretti D., Tournier I., Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zipprich J.T., Bhattacharyya S., Mathys H., Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang T.C., Yamashita A., Chen C.Y., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka M., Gupta R., Mayer B.J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol. Cell. Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanack C., Moroni M., Lima W.C., Wende H., Kirchner M., Adelfinger L., Schrenk-Siemens K., Tappe-Theodor A., Wetzel C., Kuich P.H., et al. GABA blocks pathological but not acute TRPV1 pain signals. Cell. 2015;160:759–770. doi: 10.1016/j.cell.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 26.Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 30.Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chekulaeva M., Parker R., Filipowicz W. The GW/WG repeats of Drosophila GW182 function as effector motifs for miRNA-mediated repression. Nucleic Acids Res. 2010;38:6673–6683. doi: 10.1093/nar/gkq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 33.Braun J.E., Huntzinger E., Fauser M., Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Mathys H., Basquin J., Ozgur S., Czarnocki-Cieciura M., Bonneau F., Aartse A., Dziembowski A., Nowotny M., Conti E., Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., Boland A., Kuzuoglu-Ozturk D., Bawankar P., Loh B., Chang C.T., Weichenrieder O., Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell. 2014;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 36.Gehring N.H., Neu-Yilik G., Schell T., Hentze M.W., Kulozik A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 37.Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zekri L., Huntzinger E., Heimstadt S., Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinek M., Fabian M.R., Coyle S.M., Sonenberg N., Doudna J.A. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C.Y., Zheng D., Xia Z., Shyu A.B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lian S.L., Li S., Abadal G.X., Pauley B.A., Fritzler M.J., Chan E.K. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 43.Rouya C., Siddiqui N., Morita M., Duchaine T.F., Fabian M.R., Sonenberg N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014;20:1398–1409. doi: 10.1261/rna.045302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 45.Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L. The making of a slicer: Activation of human Argonaute-1. Cell Rep. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L., Wu G., Poethig R.S. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:315–320. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang K.L., Chadee A.B., Chen C.Y., Zhang Y., Shyu A.B. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19:295–305. doi: 10.1261/rna.037317.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collart M.A., Panasenko O.O. The Ccr4–not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 50.Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 51.Kuzuoglu-Ozturk D., Huntzinger E., Schmidt S., Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 2012;40:5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pazos F., Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27:2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pisani D., Pett W., Dohrmann M., Feuda R., Rota-Stabelli O., Philippe H., Lartillot N., Worheide G. Genomic data do not support comb jellies as the sister group to all other animals. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15402–15407. doi: 10.1073/pnas.1518127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan J.F., Pang K., Schnitzler C.E., Nguyen A.D., Moreland R.T., Simmons D.K., Koch B.J., Francis W.R., Havlak P., Program N.C.S., et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]