Abstract
Human diseases are often diagnosed by determining levels of relevant enzymes and treated by enzyme inhibitors. We describe an assay suitable for both ultrasensitive enzyme quantification and quantitative inhibitor screening with unpurified enzymes. In the DNA-linked Inhibitor ANtibody Assay (DIANA), the target enzyme is captured by an immobilized antibody, probed with a small-molecule inhibitor attached to a reporter DNA and detected by quantitative PCR. We validate the approach using the putative cancer markers prostate-specific membrane antigen and carbonic anhydrase IX. We show that DIANA has a linear range of up to six logs and it selectively detects zeptomoles of targets in complex biological samples. DIANA's wide dynamic range permits determination of target enzyme inhibition constants using a single inhibitor concentration. DIANA also enables quantitative screening of small-molecule enzyme inhibitors using microliters of human blood serum containing picograms of target enzyme. DIANA's performance characteristics make it a superior tool for disease detection and drug discovery.
INTRODUCTION
Many human diseases are diagnosed and monitored based on selective protein quantification in biological samples, for which the gold standard is sandwich ELISA (1,2), in which an analyte is captured by an immobilized antibody, probed with a second enzyme-linked antibody and quantified via a reaction catalyzed by the linked enzyme. To increase sensitivity, sandwich immunoassays have been developed using DNA-linked antibodies allowing detection by quantitative polymerase chain reaction (qPCR) (3–6).
Many clinically relevant proteins are enzymes that are directly involved in disease pathogenesis and thus represent promising drug targets (7) and many currently marketed drugs are indeed small-molecule enzyme inhibitors. Identifying inhibitors of relevant enzymes typically involves screening small-molecule libraries (8) to find compounds capable of displacing an active site probe or directly influencing the enzyme reaction kinetics (9). A major drawback of currently used protocols is that they usually require purified enzymes which can be difficult and costly to prepare.
Here we describe a multiwell plate-based assay suitable for enzyme detection in complex biological matrices that offers significantly greater sensitivity than sandwich ELISA and that allows screening of small-molecule inhibitors of target enzymes without the need to purify the target. In our DNA-linked Inhibitor ANtibody Assay (DIANA), the enzyme is captured by an immobilized antibody, probed with a detection probe consisting of a DNA oligonucleotide covalently linked to a small molecule that binds to the active site of the target enzyme and is subsequently quantified by qPCR (Figure 1). Dual recognition of the target enzyme by antibody and detection probe provides selectivity, while qPCR provides sensitivity and broad linear range. Since the probe binds to the target enzyme's active site, DIANA selectively detects only the active form of the enzyme, which is likely to be the more clinically relevant form. This novel assay for enzyme detection can also be used to screen for small-molecule inhibitors of those enzymes by assessing the ability of potential inhibitors to compete with the probe for binding to the active site. The sensitivity and selectivity of DIANA means that picogram amounts of unpurified target enzyme can be used, while the broad linear range means that inhibition constants (Ki) can be determined from single-well measurements, without the need for serial dilutions of inhibitor.
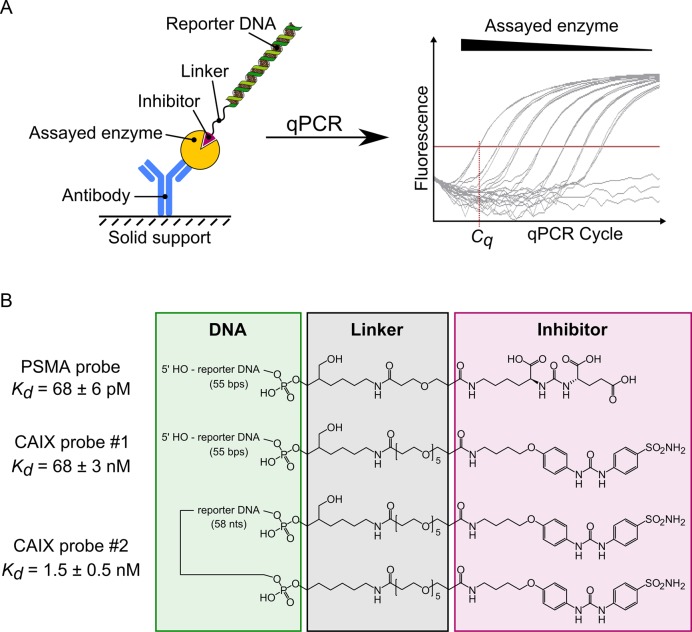
Figure 1.
Schematic representation of enzyme detection using DIANA. (A) A covalent conjugate comprising an oligonucleotide (reporter DNA) and small-molecule competitive inhibitor of the target enzyme is used as a detection probe. The probe binds to the active site of the enzyme, which has been captured on the solid support by binding to the immobilized antibody. The amount of detection probe bound to the target enzyme is measured by quantitative PCR in terms of the threshold cycle (Cq), which is indirectly proportional to the logarithm of probe concentration. (B) Structures of the detection probes used for quantification of PSMA and CAIX. Each probe consists of reporter DNA (green box) covalently attached via a linker region (black box) to a competitive inhibitor of PSMA or CAIX (magenta box). The PSMA probe and CAIX probe #1 contain one copy of inhibitor linked to the 3′ end of the double-stranded reporter DNA, whereas CAIX probe #2 contains two copies of the inhibitor linked to both ends of the single-stranded reporter DNA. Probe affinities (Kd values) were determined by titrating each probe into wells containing the corresponding captured enzyme.
We demonstrate the sensitivity and selectivity of DIANA by targeting two clinically relevant enzymes: prostate-specific membrane antigen (PSMA), also known as glutamate carboxypeptidase II (GCPII) and carbonic anhydrase IX (CAIX). Both enzymes are homodimeric integral membrane proteins (10,11) and tumor-associated antigens (12–14) suitable for use in targeted drug delivery (15,16) and diagnostics (17–19). Their direct involvement in pathogenesis of several diseases makes them attractive targets for drug discovery (20–25), and indeed high-affinity competitive inhibitors of both enzymes have been identified by rational design (24,26,27). However, PSMA inhibitors bear multiple negative charges and show poor bioavailability (28,29), while CAIX inhibitors bear a sulfonamide head and act non-selectively (24,30). Therefore, identifying new inhibitor scaffolds is essential for further development of drugs targeting diseases related to these enzymes. We provide evidence that DIANA may be an efficient system not only for identifying leads against these and other target enzymes, but also for selective and sensitive detection of these target enzymes for possible diagnostic purposes.
MATERIALS AND METHODS
Materials
Buffers
Tris buffered saline (TBS): 20 mM Tris (Promega; H5135), 150 mM NaCl (Penta; 16610–31000), pH = 7.4; TBST: TBS with 0.05% Tween 20 (Affymetrix; 20605); TBST’: TBS with 0.1% Tween 20; TBST'C: TBST’ with 500-fold diluted casein blocker (0.01% final, SDT; CBC1); TBST'CS: TBST’ with 500-fold diluted casein blocker (0.01% final) and 0.005% sodium dodecyl sulphate (SDS). HEPES: 100 mM HEPES (Sigma; H4034), 400 mM NaCl (Penta; 16610–31000), pH = 7.5; HEPESTC: HEPES with 0.01% Tween 20 and 500-fold diluted casein blocker (0.01% final); HEPEST'C: HEPES with 0.1% Tween 20 and 500-fold diluted casein blocker (0.01% final).
Detection probes
DNA oligonucleotide of the sequence CCT GCC AGT TGA GCA TTT TTA TCT GCC ACC TTC TCC ACC AGA CAA AAG CTG GAA A with 3′-terminally modified phosphate moiety by 6-amino-2-(hydroxymethyl)hexyl group (Generi-Biotech, OPC purification) was reacted with NHS-esters of the small-molecule ligands of PSMA and CAIX to prepare the monovalent detection probes. The resulting conjugates were then annealed with equimolar amount of DNA oligonucleotide of complementary sequence CCA GCT TTT GTC TGG TGG AGA AGG TGG CAG ATA AAA ATG CTC AAC TGG CAG G (Generi Biotech, standard purification) prior to use. DNA oligonucleotide of the sequence AAA CCT GCC AGT TGA GCA TTT TTA TCT GCC ACC TTC TCC ACC AGA CAA AAG CTG GAA A containing the 3′-terminal 6-amino-2-(hydroxymethyl) hexyl phosphate modification and the 5′-terminal 6-aminohexyl phosphate modification (custom synthesis Generi-Biotech, OPC purification) was reacted with NHS-esters of the small-molecule ligand of CAIX to prepare the bivalent CAIX detection probe. See Supplementary Data for more details.
Proteins
The recombinant extracellular portion of human PSMA (rhPSMA) was expressed and purified as previously described (31) (designated as rhGCPII) and its molar concentration was determined by amino acid analysis using a Biochrom30 device (Biochrom) according to the manufacturer's instructions. The mass concentration was derived from the molar concentration, whereby the molecular weight of 88.7 kDa for rhPSMA was determined by MALDI-TOF. For long-term storage, purified rhPSMA diluted in TBST’ was aliquoted, flash frozen in liquid nitrogen and kept at −80°C.
Human colon cancer cell line HT-29 endogenously expressing CAIX was used as a source of CAIX in DIANA experiments. Cells grown in Petri dishes at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Sigma; D5671) supplemented with 10% Fetal bovine serum (FBS, Sigma; F7524) and 4 mM glutamine to 100% confluency were harvested by resuspending in the medium, and then transferred to a microtube and pelleted at 300 g for 2 min and washed twice with TBS. Afterward, the supernatant was removed and cells were lyzed by resuspending in 50 mM Tris, 100 mM NaCl, pH 7.4, with 1% octaethylene glycol monododecyl ether (C12E8, Affymetrix; O330) and 2 h incubation on ice. The crude lysate was centrifuged at 600 g for 15 min at 4°C, and the supernatant was transferred to a new tube and centrifuged at 15 000 g for 15 min at 4°C. The resulting supernatant, hereafter referred to as the lysate, was transferred to a new tube. The total protein concentration in the lysate was determined using Bio-Rad Protein assay, and the amount of CAIX was determined using Quantikine ELISA for human CAIX (R&D Systems; DCA900) according to the manufacturer's instructions. The lysate was diluted in TBST’ and kept in aliquots at −80°C for long-term storage.
Capture antibodies
Mouse monoclonal antibody 2G7, which selectively binds human PSMA (Kd ∼2.9 nM (32)), was prepared and purified as previously described (33). Purified mouse monoclonal antibody M75 selectively recognizing the PG domain of CAIX (34) (Kd ∼1.5 nM (35)), was a kind gift from Pavlína Řezáčová. For long-term storage, the antibodies were aliquoted in TBS or PBS, flash frozen in liquid nitrogen and kept at −80°C.
Blood samples
Samples were drawn from healthy volunteers, patients with histologically proven prostate carcinoma (PCa) or clear cell renal carcinoma (ccRCC) at Thomayer Hospital in Prague, with the agreement of the local ethical commission and with informed consent obtained from all subjects. See Supplementary Table S5 for a more detailed description of the individuals, including gender, age and diagnosis. In case of patients with diagnosed PCa or ccRCC, the blood samples were drawn in the morning before surgical removal of the tumor, i.e. radical prostatectomy, radical nephrectomy or partial nephrectomy. Blood was withdrawn into Vacuette® tubes (Greiner Bio-One) containing clot activator (# 456071; serum) or sodium citrate (# 456323; citrate plasma) and placed in the refrigerator. Within 8 h, tubes were centrifuged at 2000 g for 10 min with minimal deceleration and blood serum was transferred into a microtube and stored at −80°C until analysis. At the time of analysis, the serum samples were thawed on ice, mixed thoroughly and centrifuged at 5000 g for 15 min at 4°C to remove precipitate if formed.
Methods
General assay protocol
DIANA experiments were done according to this assay protocol. Any experimental conditions not described in this protocol, such as used buffers or used probe concentrations, as well as any divergences from this protocol, such as different incubation times, are described separately in sections describing particular experiments. To emphasize the possibilities of optimization of the duration of the protocol, we report both the incubation times employed in reported experiments and ranges of incubation times that were tested and did not influence the assay performance.
Capture antibody recognizing the protein of interest was immobilized onto the surface of wells of a FrameStar 480/96 multiwell plate (4titude; 4ti-0951) by applying 10 μl of the antibody in TBS at a concentration of 10 ng.μl−1 to the bottom of the wells and incubating for 60 min (range 30 to 120 min) at room temperature (RT). To avoid evaporation of the liquid in the wells, plates were covered during all incubations with general adhesive plate seals (4titude; 4ti-0510). The liquid was then tapped out, and the wells were washed three times with 200 μl TBS. Then, for PSMA detection, 100 μl casein blocking agent diluted 5-fold in TBS (‘casein blocker biotin free 5.5% w/v’; SDT; CBC1) was applied to the bottom of the wells, while for CAIX detection 10 mg/ml BSA (Sigma; A7906) in TBS was applied. Plates were then incubated overnight (range 2 to 24 h) at RT. The liquid was tapped out again, and the wells were washed three times with 200 μl TBST. Thereafter, 10 μl of the sample containing analyte was applied to the bottom of the wells and incubated; during this step the target enzyme is being captured by the immobilized antibody. The samples were typically diluted in appropriate buffer containing 0.1% Tween 20, and the time of incubation was 2 h (range 1 to 21 h) at RT. The liquid was then tapped out, and the wells were washed three times with 200 μl TBST. Subsequently, 10 μl of detection probe diluted in appropriate buffer (typically containing 0.01 or 0.1% Tween 20 and 0.01% casein blocker to minimize non-selective binding of the probe) was added to the bottom of wells, and samples were incubated for 45 min (range 15 to 120 min) at RT. The liquid was then tapped out again, and the wells were washed at least six times with 200 μl TBST. Finally, 10 μl of a qPCR reaction mixture composed of LC 480 Probes Master (Roche, cat. no. 04707494001; diluted to the final concentration recommended by the manufacturer), forward and reverse primer (CCA GCT TTT GTC TGG TGG AG and CCT GCC AGT TGA GCA TTT TT; final concentration 1 μmol.l−1 each) and fluorescent hydrolysis probe #87 from the Roche Universal Probe Library (LNA octamer sequence CTG CCA CC, cat. no. 04689127001; final concentration 100 nmol.l−1) was added and the plate was sealed with adhesive optical film (Roche, cat. no. 4729692001). The amount of detection probe was then quantified by qPCR: 3 min at 95°C; then 45 cycles of 10 s at 95°C, 30 s at 66°C and 30 s at 72°C; and finally 2 min at 37°C in a LightCycler 480 II qPCR instrument (Roche) with the excitation and emission filters adjusted to 465 nm and 510 nm respectively. Threshold cycles (Cq) were obtained from the measured fluorescence curves using the method of maxima of the second derivative in the Light Cycler 480 II Software (Roche). Based on measurements with a dilution series of the PSMA detection probe in water as a template, the linear range of quantification of the probe extended from at least 6 copies to 6 × 108 copies of the probe per well with over 90% amplification efficiency.
Determination of the Kd values of the probes
K d values were determined according to the general assay protocol by titrating captured enzymes with the appropriate detection probes. For the PSMA detection probe, 1 pg of rhPSMA diluted in 10 μl TBST’ was captured onto immobilized antibody 2G7 in each well. Thereafter, different concentrations of the detection probe in TBST’ were incubated with the captured rhPSMA. Resulting amounts of bound probe were then fitted using the GraFit v.5.0.11 (Erithacus Software Ltd.) with the function [EP] = Etot × Ptot/(Kd + Ptot), where [EP] is the amount of bound probe, Ptot is the analytical concentration of the probe during the incubation and Etot is the amount of captured enzyme, while Etot and Kd were parameters of the fit to be solved. The same procedure was repeated with the detection probe diluted in TBST’ buffer with added casein blocker (TBST'C), or with added casein blocker and SDS (TBST'CS). Finally, the dissociation constant of the probe was determined in the same manner with PSMA captured from 1 μl serum diluted 10-fold in TBST’ prior to capture (determined Kd values are listed in Supplementary Table S1).
For the monovalent CAIX detection probe, 0.78 ng of CAIX in HT-29 cell lysate (10 μg total protein) diluted in TBST’ was captured onto immobilized M75 antibody in each well. Afterward, different concentrations of the probe in HEPEST'C were incubated with the captured CAIX and resulting amounts of bound probe were used to determine the Kd value in the same way as for PSMA. For the bivalent CAIX detection probe, only 0.078 ng of CAIX in HT-29 cell lysate (1 μg total protein) diluted in TBST’ was captured onto immobilized M75 antibody, and subsequently, different concentrations of the probe in HEPEST'C were incubated with captured CAIX. The Kd value of the probe was also determined on captured endogenous CAIX from 1 μl serum diluted 10-fold in TBST’ in the same manner (determined Kd values are listed in Supplementary Table S2).
Quantification of PSMA and CAIX in test matrices
Quantification followed the general assay protocol. In case of PSMA detection, serial dilutions of rhPSMA in TBST’ were captured via immobilized antibody 2G7 and subsequently detected by PSMA detection probe diluted in TBST'CS to 50 pM concentration. In case of CAIX, serial dilutions of HT-29 cell lysate in TBST’ were captured via immobilized antibody M75 and subsequently detected by bivalent CAIX detection probe diluted in HEPEST'C to 500 pM concentration.
Quantification of PSMA in serum samples
Quantification followed the general assay protocol with minor modifications. Known amounts of rhPSMA in TBST’ were used as standards. Prior to analysis of the serum samples, 10% stock solution of Tween 20 was added to the samples to a final concentration of 0.1%. The samples were then diluted 10-fold in TBST’ and added to the wells coated with the antibody 2G7. After overnight incubation (∼21 h) and washing, the wells with standards were incubated with the PSMA detection probe in TBST'CS at a concentration of 100 pM, whereas the wells with serum samples were incubated with the PSMA probe at a concentration of 500 pM (to compensate for the different affinities of the probe toward the recombinant and endogenous proteins). The incubation of serum samples was much longer than incubations used with purified recombinant standards described in general assay protocol in order to minimize matrix effects seen in some samples when analyzing PSMA (non-linear assay response upon serum dilution in both DIANA and ELISA quantifications; see Supplementary Data). However, experiments with limited number of samples indicated that shorter incubation times within the ranges described in general assay protocol were possible without compromising the assay performance even when assaying complex biological matrices, such as blood serum, cell lysate or urine (data not shown).
The amount of PSMA in serum samples was computed by comparing the measured Cq to the standard curve, which was constructed as a linear regression fit of the plot of Cq values versus the logarithm of rhPSMA concentrations in standards (the standards covered the five-log range from 1 fg to 100 pg PSMA per well). To validate the obtained results, replicate wells with serum samples were incubated with the detection probe in the presence of 10 μM compound 16 (Ki = 0.34 ± 0.06 nM as determined by enzyme kinetics), which was designed to completely disrupt the selective binding of the probe to PSMA but not the non-selective binding to the surface.
Quantification of CAIX in serum samples
Quantification followed the general assay protocol with minor modifications. Serial dilutions of HT-29 cell lysate in TBST’ of known CAIX concentration (determined by Quantikine ELISA for human CAIX supplied by RnD Systems) were used as a standard. Undiluted serum samples to which 10% stock solution of Tween 20 was added to a final concentration of 0.1% were loaded into the wells coated with the antibody M75. After overnight incubation (∼21 h) and washing, wells were incubated with the bivalent CAIX detection probe in HEPEST'C at a concentration of 200 pM.
The amount of CAIX in serum samples was computed by comparing the measured Cq to the standard curve, which was constructed as a linear regression fit of the plot of Cq values versus logarithms of the CAIX concentrations in standards (the standards covered a five-log range from 7.8 fg to 780 pg CAIX per well). To validate the results, replicate wells with serum samples were incubated with the detection probe in the presence of 10 μM acetazolamide (Ki ∼25 nM (36)), which was designed to completely disrupt the selective binding of the probe to CAIX but not the non-selective binding to the surface.
Evaluation of enzyme inhibitors
Evaluation of inhibitors followed the general assay protocol. For testing of PSMA inhibitors, 250 pg rhPSMA diluted in 10 μl TBST’ was first incubated with immobilized capture antibody 2G7. Thereafter, a few wells were incubated with the PSMA detection probe diluted to 125 pM in TBST'C and the remaining wells were incubated with 125 pM probe in TBST'C in the presence of various tested compounds (listed in Supplementary Table S12) at 100 μM concentration. After extensive washing, the amount of bound probe in each well was quantified by qPCR and the ΔCq values for each compound were computed as the difference between the Cq value in well(s) incubated with the particular compound and mean Cq value of wells incubated without any compound. The Ki values of the compounds were computed from their tested concentration and the corresponding ΔCq values according to the equation Ki = (2−ΔCq/(1 – 2−ΔCq)) × Itot/(1 + (Ptot/Kd)), where Itot is the total concentration of the tested compound (100 μM), Ptot is the total concentration of probe (125 pM) and Kd is the dissociation constant of the probe (∼125 pM). The Ki values of the same compounds toward PSMA naturally present in human serum were determined in the same manner, but PSMA from 1 μl of sera diluted 10-fold in TBST’ (containing ∼6 pg PSMA) was captured via immobilized antibody 2G7 in each well. Then, few wells were incubated with the PSMA detection probe diluted to 70 pM in TBST’ (Kd ∼200 pM) while the remaining wells were incubated with 70 pM probe in TBST’ in the presence of tested compounds at either 100 nM or 100 μM concentration.
For testing of CAIX inhibitors, CAIX from HT-29 cell lysate diluted in TBST’ (5 μg total protein containing 390 pg CAIX) was captured via immobilized antibody M75 in each well and a few wells were incubated with the bivalent detection probe diluted to 500 pM in HEPESTC with 10% DMSO, while the remaining wells were incubated with a mixture of 500 pM probe in the same buffer in the presence of various tested compounds (listed in Supplementary Table S17) at either 1 μM or 100 μM concentration. The Ki values of the compounds were computed from their tested concentrations and corresponding ΔCq values according to the equation: Ki = ((1 – 2 × Raff)/(1 – Raff – (Raff2 + (1 – 2 × Raff) × 2−ΔCq)0.5) – 1) × Itot/(1 + (Ptot/Kd2)), where Raff is the ratio of Kd2 of the double-binding probe to Kd1 of the single-binding probe (Raff = Kd2/Kd1; Kd1∼ 70 nM, Kd2 ∼ 1.5 nM; Supplementary Table S2). The Ki values of the same compounds toward CAIX naturally present in human serum were determined in the same manner, but CAIX from 10 μl of human serum (containing ∼6 pg CAIX) with Tween 20 added to a concentration of 0.1% was captured via immobilized antibody M75 in each well. Then, few wells were incubated with the bivalent detection probe diluted to 180 pM in HEPESTC with 10% DMSO, while the remaining wells were incubated with the 180 pM probe in the same buffer in the presence of tested compounds at either 1 or 100 μM concentration.
PSMA enzymatic assay
The rate of hydrolysis of pteroyl-di-L-glutamate (PteGlu2) in the presence of serial dilutions of various tested compounds was monitored by HPLC and was used to determine Ki values of the compounds toward PSMA. The procedure was carried out as previously described (37).
CAIX enzymatic assay
The rate of hydration of carbon dioxide in the presence of serial dilutions of compound 10 was monitored by detecting color changes of pH indicator and was used to determine the Ki value of the compound 10 toward CAIX. The procedure was carried out as previously described (38). The Ki values of carborane compounds were taken from ref. (38), and the Ki value of acetazolamide from ref. (36).
Quantification of PSMA by ELISA
ELISA was carried out as described previously (39) with some modifications. Briefly, 2G7 antibody was diluted to 5 ng/μl in 100 mM borate solution, pH 9.5 and 100 μl were loaded to each well of 96-well Maxisorp plate (Nunc, 437 111) and incubated for 1 h at RT. After washing with TBS (3 × 200 μl), the wells were blocked with 250 μl casein blocker (SDT; cat. no. CBC1) diluted 5-fold in TBS for 22 h at RT, followed by TBST wash (3 × 200 μl). Then, 100 μl of 10-fold diluted citrate plasma samples (n = 34) or serum samples when no citrate samples were available (n = 2) either in TBST’ or in TBST’ with 4.5 mM ethylenediaminetetraacetic acid (EDTA) were loaded to the wells, and the samples were incubated for 16 h at RT. As standard, rhPSMA (31) serially diluted in TBST’ to concentrations ranging from 1 pg to 1 ng per well was used. After subsequent TBST wash (3 × 200 μl), 100 μl biotinylated mouse monoclonal antibody J591 (40) recognizing GCPII diluted in TBST’ to 0.25 ng/μl was added and incubated for 1 h at RT. After washing out the unbound biotinylated J591 (3 × 200 μl TBST), 100 μl NeutrAvidin conjugated with horseradish peroxidase (Thermo Scientific) diluted in TBST’ to 0.5 ng/μl was added and incubated for 30 min at RT, followed by a TBST wash (5 × 200 μl). Finally, chemiluminescence in 170 μl of luminol substrate (prepared by mixing 0.1 ml 1 M Tris–HCl, pH 8.8; 10 μl 250 mM luminol in DMSO; 22.2 μl 90 mM 4-iodophenol in DMSO and filled to 1 ml with distilled water; 0.6 μl of 30% H2O2 was added shortly before use) was measured on a Tecan Infinite M1000 instrument.
To control for false-positive signal due to the presence of heterophilic interfering antibodies, the samples were diluted in replicate wells in TBST’ with 4.5 mM EDTA. The EDTA denatures PSMA, which leads to the loss of binding of the antibodies to PSMA. The remaining signal, if any, corresponds to the signal from interfering antibodies. The PSMA concentration was therefore determined as the difference between wells with and without EDTA.
Statistical analysis
Two-tailed Mann Whitney U-test was used to compare mean PSMA and CAIX serum levels between the groups of individuals. The calculations were run in the past software (41). Because the expression of genes (and consequently also the serum levels) are considered to be log-normally distributed (42), median values and 16th and 84th percentiles for serum levels determined in the sample set were indicated (rather than mean values and standard deviations) to describe the scatter of the values. The non-parametric rank statistical test was chosen because we do not expect the values to be normally distributed. While the amount of available serum samples was limited, the sample size of 12 individuals in each group was chosen to provide reasonable power of the statistical test.
RESULTS
Preparation and characterization of detection probes
To prepare detection probe that binds to the target enzyme active site and allows quantification by qPCR (Figure 1A), we derivatized known competitive inhibitors of PSMA (43) and CAIX (24) to carry a linker bearing a terminal NHS-ester. These derivatives were then reacted with the primary amine group of custom-synthesized DNA oligonucleotides (see Figure 1B for structures and Kd values). Interestingly, attaching the oligonucleotide to the inhibitors improved their binding affinity several-fold (see Supplementary Data for more details on probe synthesis and testing). Because the affinity of the CAIX probe was three orders of magnitude lower than that of the PSMA probe, we conjugated two CAIX inhibitors to a single oligonucleotide, increasing overall affinity nearly 50-fold. This substantial increase may be because each inhibitor of a single probe can bind to one active site in the CAIX homodimer, as described previously for a bivalent small-molecule CAIX inhibitor (16). We used only this bivalent probe #2 in all CAIX experiments.
Quantification of PSMA and CAIX in test matrices
Quantification of PSMA and CAIX consisted of several steps. First, the capture antibody was immobilized onto the bottom of wells in a 96-well polypropylene PCR plate, and wells were then blocked with appropriate reagent. Next, sample was added to the wells, and target enzyme from the sample was selectively bound by the capture antibody. Then the appropriate detection probe was added and allowed to bind to the active site of the target enzyme. Finally, after extensive washing, the amount of bound probe was quantified by qPCR.
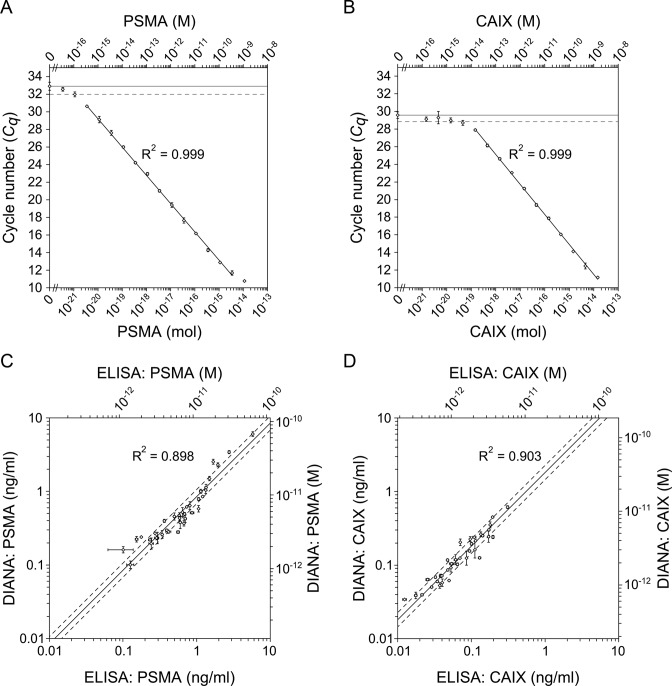
To assess the dynamic range and sensitivity of DIANA, we analyzed serial dilutions of enzyme standards diluted in buffered solution; the same standards were later used to quantify both targets in human sera. As a standard for PSMA quantification, we used purified recombinant human PSMA (rhPSMA). The linear range of detection extended over six orders of magnitude, from 3.6 zeptomoles to 3.6 femtomoles (320 ag to 320 pg) in a 10-μl sample. The lowest detected amount, defined as the amount of analyte that gave a signal higher than the blank signal plus two s.d., was 1.1 zeptomole (100 ag). The total range of detection thus extended over seven orders of magnitude from 1.1 zeptomole to 11 femtomoles (100 ag to 1 ng; Figure 2A and Supplementary Table S3). As a standard for CAIX quantification, we used lysate from cultured HT-29 cells; the CAIX concentration in the stock lysate was determined by Quantikine ELISA for human CAIX (RnD Systems). The linear range of detection extended over five orders of magnitude from 140 zeptomoles to 14 femtomoles (7.8 fg to 780 pg) in a 10-μl sample. The lowest detected amount was 46 zeptomoles (2.5 fg), corresponding to a total range of detection over five orders of magnitude from 46 zeptomoles to 14 femtomoles (2.5 fg to 780 pg; Figure 2B and Supplementary Table S4).
Figure 2.
Ultrasensitive detection of PSMA and CAIX by DIANA. (A and B) Plots of average Cq values versus amount of human recombinant purified PSMA standard diluted in buffered solution (A), or average Cq values versus amount of human CAIX present in HT-29 cell lysate diluted in buffered solution (B). Upper x-axes indicate molar concentration, while lower x-axes indicate the corresponding molar amount per well. Horizontal lines show the average background signal; dashed horizontal lines show the average background signal plus 2 s.d. Error bars show s.d. of quadruplicate measurements (A and B). (C and D) Plots of concentrations of PSMA (C) and CAIX (D) determined using DIANA in 36 human serum samples versus concentrations determined using ELISA in the same serum samples (CAIX) or in control citrate plasma samples (PSMA) drawn from the same individuals on the same occasion. Lines show the linear regression fit of log-transformed concentrations; dashed lines indicate values 1.25-fold higher or lower than the linear regression. Error bars show s.d. of triplicate (C) or duplicate (D) measurements.
Quantification of PSMA and CAIX in human serum
After establishing DIANA performance against target enzymes using purified enzyme or cell lysate, we examined its performance against clinical samples of human serum from 12 healthy men, 12 men with Prostate carcinoma (PCa) and 10 men and 2 women with clear cell renal cell carcinoma (ccRCC). Undiluted samples were assayed for CAIX, while samples were diluted 10-fold before assaying PSMA (for details on testing of the assay linearity and on the recovery of spiked protein standards see Supplementary Data). Thus, the serum volumes used per well were 1 μl for PSMA and 10 μl for CAIX.
Measured PSMA concentrations in serum ranged from 0.10 to 6.0 ng/ml, with a median value of 0.45 ng/ml (16th and 84th percentiles were 0.23 and 1.1 ng/ml, respectively; Supplementary Table S9). Mean concentration did not differ significantly among the three groups, suggesting that serum PSMA is not elevated in either ccRCC or PCa (Supplementary Figure S1a). This contrasts with a previous report of elevated PSMA protein levels in serum from PCa patients as determined by semiquantitative SELDI assay (17). Measured CAIX concentrations ranged from 0.034 to 0.61 ng/ml, with a median value of 0.12 ng/ml (16th and 84th percentiles were 0.059 and 0.25 ng/ml, respectively; Supplementary Table S10). Mean CAIX concentration was significantly lower in healthy individuals than in patients with ccRCC (two tailed Mann-Whitney test, P = 0.033) or PCa (P = 0.021; Supplementary Figure S1b). This is the first report showing the elevated level of CAIX in patients with prostate cancer. The elevated level of CAIX in serum of ccRCC patients is in line with previous reports (18,19).
To further validate the serum levels obtained by DIANA, we reanalyzed the samples by sandwich ELISA. We are not aware of any commercially available ELISA validated for quantification of PSMA in blood serum or plasma, so we used our previously reported ELISA protocol (39) with some modifications. We used this ELISA to determine PSMA concentrations in control citrate plasma samples drawn from the same individuals on the same occasion as samples used in DIANA determinations. PSMA concentrations determined by ELISA showed excellent agreement with values obtained by DIANA (R2 = 0.898; Figure 2C). Similarly, we validated CAIX concentrations by reanalyzing the serum samples with a commercially available Quantikine ELISA for human CAIX and CAIX concentrations determined by both methods showed excellent agreement (R2 = 0.903; Figure 2D). Reference ELISA thus confirmed the accuracy of serum levels of both enzymes determined by DIANA, and it corroborated our observations of PSMA and CAIX levels in clinical samples.
Next, we used the serum samples to examine the sensitivity and selectivity of DIANA by attempting to outcompete the binding of the detection probe using known competitive inhibitors of the target enzymes. We hypothesized that these inhibitors would quantitatively outcompete the selective binding of the probe to the target enzyme, without affecting possible non-selective binding of the probe to other proteins. Therefore, we reasoned that PSMA or CAIX concentrations determined in wells containing a suitable concentration of competitive inhibitor should provide an estimate of the limit of detection, reflecting non-selective binding of the probe. In this manner, we estimated the average limit of detection of PSMA in a 1-μl serum sample to be 0.8 pg/ml (range 0.4 to 2.4 pg/ml; Supplementary Table S9) and of CAIX in a 10-μl serum sample to be 1.1 pg/ml (range 0.7 to 2.8 pg/ml; Supplementary Table S10). These detection limits were much lower than the actual enzyme concentrations in the set of clinical samples analyzed; the limits for PSMA were on average 870-fold lower than the actual PSMA concentration (range 140- to 2800-fold), while the limits for CAIX were on average 210-fold lower than the actual CAIX concentrations (range 12- to 930-fold).
Using DIANA to evaluate inhibitors of target enzymes
Determination of inhibitor binding affinities (expressed as Ki values) followed the same general protocol as enzyme quantification, with slight modifications (Figure 3A). An identical amount of target enzyme was captured onto the immobilized antibody in each well; this amount was within the linear range of detection. Some wells were incubated with detection probe alone, and the rest were incubated with a mixture of the probe and a known concentration of putative inhibitor, present in molar excess with respect to the enzyme. Inhibitor affinity (Ki value) was calculated from the inhibitor concentration and from the difference in the number of PCR cycles needed to amplify the reporter DNA (ΔCq) in wells incubated in the presence or absence of inhibitor. Given the broad linear range of the assay, we were able to calculate the Ki value for each inhibitor from each well after assuming that the monovalent PSMA probe bound non-cooperatively to its target, while the bivalent CAIX probe bound cooperatively (see Supplementary Data for more details). These calculations are presented in detail in the next two sections.
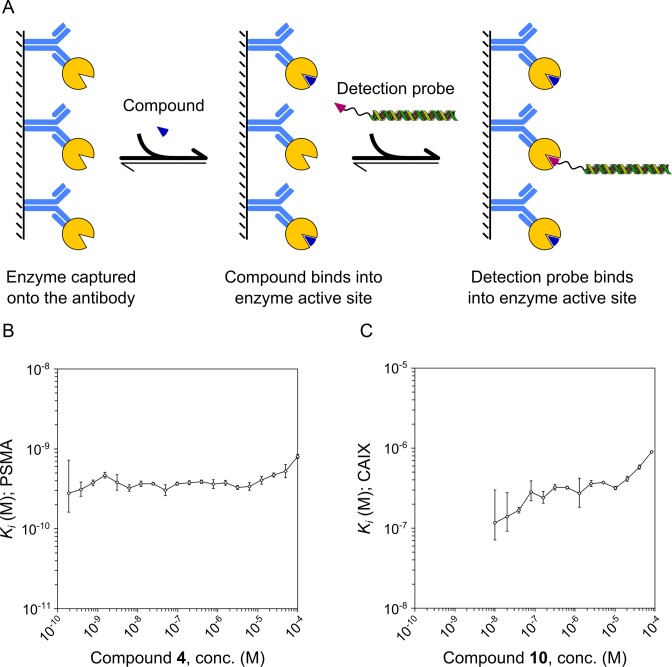
Figure 3.
Determination of inhibition constants from single inhibitor concentrations by DIANA and its validation by inhibitor titration. (A) Detection probe was incubated with target enzyme in the presence of putative inhibitor. The binding of the test compound resulted in lower amount of the bound probe. The amount of bound probe together with concentration of compound was then used to determine the inhibition constant at each dilution of the compound. (B) Purified PSMA standard was titrated with PSMA inhibitor (compound 4) and detected using the monovalent detection probe against PSMA. Measured inhibition constants were constant over more than five orders of magnitude of inhibitor concentration. (C) CAIX from cell lysate was titrated with CAIX inhibitor (compound 10) and detected using the bivalent detection probe against CAIX. Measured inhibition constants were constant over more than two orders of magnitude. The lowest inhibitor concentrations were lower than their respective Ki values, which leads to insufficient inhibition and therefore larger errors of determinations. Ki values determined at the highest inhibitor concentrations are higher than the average because of background signal. Error bars show s.d. of triplicate (B) or duplicate (C) measurements.
Evaluation of PSMA inhibitors
To examine the accuracy of our model to determine Ki values of PSMA inhibitors using single inhibitor concentrations (Supplementary Figure S2a), we incubated captured rhPSMA with serial dilutions of competitive inhibitor (compound 4; Ki = 1.1 ± 0.1 nM based on enzyme kinetics) ranging from 100 pM to 100 μM. Then we determined the Ki from each measured data point (Supplementary Table S13). Measured Ki values were constant over the entire dilution series except at the extremes (Figure 3B); mean Ki calculated over the range from 200 pM to 50 μM was 0.37 ± 0.06 nM. This indicates that DIANA can accurately determine Ki values from single concentrations of small-molecule inhibitors over the broad linear range of the assay, covering six orders of magnitude in the case of PSMA.
To further verify this, we determined Ki values of 41 known competitive inhibitors of PSMA (listed in Supplementary Table S12) using single-well measurements at a constant inhibitor concentration (100 μM). We then compared DIANA-determined values with those using an HPLC-based substrate cleavage assay (37) which requires testing serial dilutions of inhibitors (see Figure 4A). The two methods showed excellent agreement (R2 = 0.991) over the entire range of Ki determinations covering seven orders of magnitude from mid-picomolar to mid-micromolar values (Supplementary Table S15). This confirms that DIANA can accurately measure Ki values from a single inhibitor concentration.
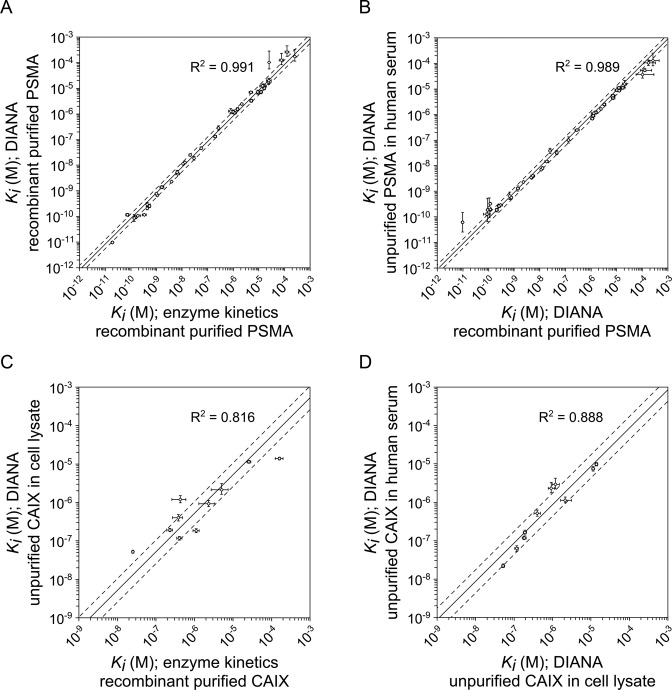
Figure 4.
Validation of single-well determination of inhibition constants of PSMA and CAIX inhibitors using DIANA. (A) Plot of Ki values of 41 PSMA inhibitors determined by titrating recombinant purified PSMA with inhibitor and measuring kinetics (x-axis) versus Ki values determined by DIANA from a single well containing 100 μM inhibitor and recombinant purified PSMA (y-axis). (B) Plot of Ki values of 41 PSMA inhibitors determined by DIANA from a single well containing 100 μM inhibitor with recombinant purified PSMA (x-axis) versus Ki values determined by DIANA from two measurements with unpurified endogenous PSMA in 1 μl of human serum in the presence of either 100 μM or 100 nM inhibitor (y-axis). (C) Plot of Ki values of 10 CAIX inhibitors determined by titrating recombinant purified CAIX with inhibitor and measuring kinetics (x-axis) versus Ki values determined by DIANA from two measurements with unpurified CAIX in cell lysate in the presence of either 100 μM or 1 μM inhibitor (y-axis). (D) Plot of Ki values of 10 CAIX inhibitors determined by DIANA from two measurements with unpurified CAIX in cell lysate in the presence of either 100 μM or 1 μM inhibitor (x-axis) and by DIANA from two measurements with unpurified endogenous CAIX in 10 μl of human serum in the presence of either 100 μM or 1 μM inhibitor (y-axis). (A–D) Lines show linear regression of log-transformed values; dashed lines indicate values that are 1.5-fold (PSMA) or 2-fold (CAIX) higher or lower than the linear fit. Error bars show s.d. of duplicate measurements in the case of DIANA, or s.e. of the titration in the case of enzyme kinetics.
The high selectivity and sensitivity of DIANA allowed us to determine Ki values for the 41 known PSMA inhibitors using only 1 μl of human serum (containing ∼6 pg of PSMA) rather than purified rhPSMA. Because of the low amount of PSMA present, we tested inhibitor concentrations of 100 μM and 100 nM in order to cover the range of Ki values. The values measured in this way agreed well with those determined using rhPSMA (Figure 4B), over the entire range of Ki values (R2 = 0.989; Supplementary Table S16).
To evaluate the robustness of DIANA, we examined the standard deviations of the assay and evaluated its response to different buffer compositions and solvents. Standard errors in the presence or absence of inhibitors were typically lower than 0.15, corresponding to a CV of 11%. Together with signal-to-background ratios of up to 22 qPCR cycles, these standard errors translated into a Z’ factor of over 0.9. Such Z’ values were obtained not only when assaying purified PSMA standard but also when assaying PSMA in human serum. The expected precision of the determined Ki values is shown in Supplementary Figure S2a. We found no significant variation in the signal-to-background ratio when we screened different concentrations of detergent, casein blocker or, most notably, organic solvents such as methanol, acetonitrile and DMSO at concentrations of up to 10%. These findings suggest that DIANA can serve as a highly versatile screening platform for testing small-molecule libraries dissolved in 100% DMSO (Supplementary Table S14).
To explore the possibility of omitting the capture antibody, which would expand the assay to include target enzymes for which specific antibodies are unavailable, we tested a panel of PSMA inhibitors against recombinant biotinylated PSMA captured on neutravidin. This led to nearly identical results as using untagged PSMA captured by an immobilized antibody (Supplementary Figure S3). Since the detection probe also binds the PSMA homolog GCPIII (44) (with Kd of ∼5 nM), we tested a subset of these PSMA inhibitors against recombinant biotinylated GCPIII captured on neutravidin and obtained Ki values corresponding to those obtained by conventional enzyme kinetics assays (Supplementary Figure S4; experimental details on assaying both proteins are described in Supplementary Data). The binding of the probe to both enzymes together with capturing of their tagged variants onto neutravidin thus enabled us to rapidly develop screening assay for both targets, but such promiscuity also may have compromised the selectivity of the PSMA detection in biological matrices. To examine the selectivity of the DIANA protocol which employs an immobilized anti-PSMA antibody 2G7 and which we used previously to quantify untagged PSMA, we assayed different amounts of GCPIII using this protocol. We found that the presence of GCPIII did not interfere with PSMA quantification even at very high concentrations (Supplementary Table S22). The selectivity ratio toward PSMA was more than six orders of magnitude, which suggests that using a highly selective capture antibody ensures detection of only the desired target, even when the probe can bind other proteins in the sample.
Evaluation of CAIX inhibitors
Determination of Ki values was more complex for CAIX inhibitors than for PSMA inhibitors because we used the bivalent probe against CAIX, which presumably binds cooperatively to the target. Therefore we altered the model for calculating Ki values (Supplementary Figure S2b). To test the accuracy of the model, we incubated CAIX captured from a lysate of HT-29 cells with serial dilutions of a competitive CAIX inhibitor (compound 10; Ki = 408 ± 64 nM based on enzyme kinetics) ranging from 0.6 nM to 80 μM, and we determined the Ki from each measured data point (Supplementary Table S18). We obtained a mean Ki of 323 ± 54 nM over a range of inhibitor concentrations spanning more than two orders of magnitude, from 80 nM to 20 μM. This validated our assumption that the detection probe binds CAIX cooperatively (Figure 3C).
This inhibitor concentration range over two orders of magnitude useful for determining Ki from single measurements is narrower than the concentration range giving a linear response for CAIX quantification. This result reflects (1) relatively low endogenous expression of CAIX by HT-29 cells, (2) higher non-selective binding of probe after CAIX capture from lysate versus capture from buffer or serum and (3) bivalence of the CAIX probe, which leads to a larger change in Cq with increasing inhibitor concentration than would be observed with a monovalent probe. These factors narrow the range of determined Ki values, but the last factor at the same increase precision and therefore sensitivity, which may make DIANA useful for identifying weak inhibitors (Supplementary Figure S2b).
Using our validated model for calculating Ki, we determined the Ki values of 10 CAIX inhibitors (listed in Supplementary Table S17) by incubating the inhibitors at 1 μM and 100 μM with CAIX captured from cell lysate in the presence of 10% DMSO, which was necessary because of the low solubility of some inhibitors. We compared these values with those determined from enzyme kinetics (38) (Supplementary Table S19) and found good agreement (R2 = 0.816, Figure 4C) over the entire range of Ki values covering three orders of magnitude from mid-nanomolar to mid-micromolar values. We assumed that the solvent did not influence our results, since we found that DMSO at final concentrations up to 10% did not affect selective or non-selective binding of the detection probe.
Finally, we determined Ki values using 10 μl human serum (containing approximately 6 pg of CAIX) and inhibitor concentrations of 1 μM and 100 μM in the presence of 10% DMSO. The resulting Ki values agreed well with those obtained previously using CAIX from cell lysate (R2 = 0.888, Figure 4D). Despite the very small amount of CAIX in cell lysate and human serum, Z’ values were reproducibly higher than 0.8. These results confirm the applicability of the assay to screen enzyme inhibitors with low-abundance enzymes in different biological matrices.
DISCUSSION
Here, we report the development of DIANA assay suitable for enzyme detection and screening of enzyme inhibitors. This assay is analogous to the immuno-PCR assay described in (3), in which the target protein is captured by an immobilized antibody and then detected by another DNA-linked antibody. However, in DIANA assay, the DNA-linked antibody has been replaced by detection probe consisting of a DNA oligonucleotide covalently linked to a small molecule that binds to the active site of a target enzyme. The use of conjugates of DNA and small molecules as affinity reagents has been proposed more than 20 years ago (45) and they have been since then employed in a number of contexts. Nevertheless, we show that DIANA represents a novel and powerful way of using such conjugates with some important implications which are discussed in following paragraphs.
We show that DIANA is a straightforward and extremely sensitive method for quantifying enzymes suitable for complex biological matrices. We estimated the limits of detection for two integral membrane proteins, PSMA and CAIX, in human sera to be, respectively, 10 × 10−21 mol and 200 × 10−21 mol. Such sensitivity is comparable to that of the most sensitive sandwich immunoassays available for complex biological matrices, including sandwich ELISA (10 × 10-18 mol; e.g. Abbott Architect total PSA kit, IVD Ref. 7K70), immuno-PCR (3) (100 × 10-21 mol), immuno-PCR on gold nanoparticles (Bio-Barcode assay (4); 300 × 10−21 mol), proximity ligation assay (5) (1 × 10−21 mol), and proximity extension assay (6) (100 × 10−21 mol). Additionally, DIANA is advantageous when assaying clinical samples: the synthetic nature of the detection probe means that the DIANA sandwich cannot be cross-linked by interfering antibodies present in human blood, which occasionally cause false-positive results in sandwich immunoassays (46,47).
The amount of target enzyme is quantified via quantitative PCR (qPCR), which has broad linear range and is extremely sensitive. This is achieved by geometrical amplification of the template DNA during PCR cycles. However, this is connected with slightly lower precision than by other linear readouts, such as colorimetric or chemiluminiscent detection. Depending on the amount of template, a typical qPCR machine has standard deviation of 0.10–0.25 cycle, which translates into coefficient of variation of 7–19% compared to few percent achieved in ELISA readout. Although such precision should be sufficient for most diagnostic tests, we suggest that in cases when small changes in enzyme levels need to be detected, either replicates could be run or other more precise DNA detection techniques could be employed, such as isothermal rolling circle amplification (48) or commercially available digital PCR.
The binding of the detection probe to the enzyme active site, the extremely high sensitivity and wide linear range of DIANA also make it well-suited for screening of competitive inhibitors of target enzymes. The sensitivity enables testing of inhibitors using tiny amounts of unpurified targets, and the wide range enables determination of Ki from a single well, minimizing the amount of reagents required. DIANA can identify both tight and weak inhibitors. The highest inhibitor affinity that can be accurately determined is limited by the assay sensitivity and therefore lies approximately in the Ki range of five to six orders of magnitude below the Kd of the probe. DIANA also correctly identifies inhibitors that bind even more tightly and ranks them as the most tightly binding in the screen, though it underestimates their affinities. The ability of DIANA to identify weak inhibitors is limited only by their solubility; thus, every inhibitor at a concentration at or above its Ki should be detectable in principle. Target binding by the detection probe does not compete with target binding by weak inhibitors, since the probe is used at concentrations below its Kd regardless of the magnitude of the Kd, which is made possible by ultrasensitive qPCR detection. Indeed, in the present study, we detected PSMA inhibitors with an affinity 106-fold lower than that of the detection probe, even when the inhibitor was present at a concentration similar to or lower than its Ki. The solid-phase assay format also allows testing of fluorescent or colored compounds, since they are washed out before analysis and therefore cannot interfere with the qPCR readout.
As with other ultrasensitive solid-phase assays, the sensitivity of DIANA is limited by non-selective adsorption of the detection probe, since every single probe molecule bound to the surface, including those that are non-selectively adsorbed, is detected by qPCR. In our hands, this non-selective background binding was low but detectable even at subnanomolar probe concentrations and in the presence of casein blocker and it increased with increasing probe concentration. This implies that a high-affinity ligand is essential for ultrasensitivity as lower probe affinity cannot be compensated for by increasing probe concentration.
We showed that a probe with subnanomolar affinity enabled detection of zeptomole amounts of PSMA. However, such potent ligands have been identified for a limited number of targets. As we showed here for CAIX, tight-binding probes can be prepared from weaker ligands by including several copies of the ligand moiety in order to induce multiple binding of the probe, though this strategy is likely to work only for multimeric enzymes and/or enzymes bearing at least two distinct active sites. Otherwise, the sensitivity will decrease in proportion to the decreasing affinity of the ligand, which may limit the use of weaker ligands for DIANA-based diagnostics. However, these ligands may still be useful for DIANA-based inhibitor screening, since in this case the assay's linear range is more important than its sensitivity. While loss of sensitivity also means loss of dynamic range, the range may be partially restored by capturing larger amounts of target enzyme. We therefore predict that even ligands that bind with micromolar affinity may be useful for DIANA-based inhibitor screening, offering linear response over several orders of magnitude.
The ability to use subnanomolar concentrations of probe without losing sensitivity or dynamic range, regardless of probe affinity, means that extremely small amounts of probe are needed. As nanomole amounts of probe are easily synthesized from milligram amounts of ligand, preparing sufficient probe for billions of measurements is straightforward. Since the probe is stable to multiple freeze-thaw cycles, we expect that a single probe synthesis will yield enough material for any size of project, facilitating replication studies, reagent sharing and long-term archiving. We obtained similar assay performance in evaluating enzyme inhibitors when we captured PSMA via an immobilized antibody as when we omitted the antibody and instead captured recombinant biotinylated PSMA onto neutravidin. This shows that DIANA can be adapted to screen inhibitors of target enzymes for which specific antibodies are unavailable; in these cases, immobilization can be achieved by derivatizing the surface to bind an affinity tag recombinantly fused to the target enzyme, i.e. biotin or his-tag. This eliminates the need for antibody while still allowing unpurified target to be used. This approach will enable rapid development of platforms for testing compounds against panels of enzymes and quantitative evaluation of their selectivity, which will be facilitated by the use of detection probes prepared from class-specific inhibitors, such as pepstatin (49), staurosporin (50) or acetazolamide (30), together with production and capture of tagged enzymes.
Detection probes prepared from promiscuous inhibitors are still useful for detection of enzymes in complex biological matrices, in such cases a selective antibody is used for capturing of the enzyme which complements the assay selectivity. By testing recombinant purified enzyme standards, we showed that in combination with suitable antibody we selectively detected PSMA over its closest homolog GCPIII even when using detection probe binding to both targets. Selectivity ratio of at least six orders of magnitude was achieved, even though the selectivity ratio of the probe itself was only about 60-fold. Moreover, despite both proteins share approximately 70% amino acid sequence identity without any dissimilar regions, there is a number of selective anti-PSMA antibodies available (32). This shows that it is possible to develop selective DIANA assay even for highly similar enzymes for which a selective inhibitor is not available. Similarly, we used probe prepared from non-selective inhibitor of several human carbonic anhydrases (24) for CAIX detection. There are 15 human carbonic anhydrases and 12 of them are enzymatically active and many potent yet non-selective inhibitors are known, while the development of selective inhibitors still remain challenge of current medicinal chemistry (30). Nevertheless, the low sequence homology between the carbonic anhydrases allowed us to use anti-CAIX antibody M75 which recognizes the proteoglycan-like domain of CAIX (34) absent from any other human carbonic anhydrase. The selectivity of resulting DIANA sandwich was validated by the excellent agreement of CAIX serum levels detected using DIANA and values determined using the commercial CAIX ELISA (Quantikine, R&D Systems), which has been tested by the vendor for selectivity against all other human carbonic anhydrases.
In principle, DIANA is not limited to enzymes and could be applied to any functional protein, including receptors or transporters, for which a sufficiently potent small-molecule ligand is available. Additionally, because DIANA does not require purified target, it may be especially advantageous for transmembrane proteins and other proteins that are difficult to prepare and purify. Moreover, the unique ability of DIANA to determine the compound's Ki from a single tested concentration might be especially useful for high-throughput inhibitor screening or for profiling inhibitor selectivity against a panel of enzymes. Finally, DIANA's simple and straightforward protocol, similar to that of sandwich ELISA or immuno-PCR, and its compatibility with multiwell plates, make it suitable for automation and high throughput.
Acknowledgments
We acknowledge Pavlína Řezáčová for generously providing the CAIX inhibitors, the HT-29 cell line and M75 antibody; Klára Pospíšilová for determining the reference Ki value of compound 10; Neil Bander for generously providing the J591 antibody; Josef Lazar and Barbora Vorlová for critical reading of the manuscript; and Radko Souček and Jana Starková for excellent technical support.
Author contributions. V.N., P.Š. and J.K. conceived the project; V.N., P.Š., J.S. and J.K. designed the experiments; V.N., J.S., J.T., T.K., P.M. and V.V. performed the experiments; V.N., P.Š. and J.K. analyzed the data; V.N. and J.K. wrote the manuscript; and all authors contributed to the editing of the manuscript.
Footnotes
Present addresses:
Jiří Schimer, School of Chemistry & Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332-0400, USA.
Jan Tykvart, The Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario M5S 3E1, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education of the Czech Republic [NPU I project LO 1302]. Funding for open access charge: Ministry of Education of the Czech Republic [NPU I project LO 1302].
Conflict of interest statement. V.N., J.S., P.M., J.K. and P.Š have filed an international patent application (PCT/CZ2015/000084), which is owned by the Institute of Organic Chemistry and Biochemistry of the Academy of Sciences of the Czech Republic.
REFERENCES
- 1.Engvall E., Perlmann P. Enzyme-linked immunosorbent assay (Elisa) quantitative assay of immunoglobulin-G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 2.Lequin R.M. Enzyme immunoassay (EIA)/Enzyme-linked immunosorbent assay (ELISA) Clin. Chem. 2005;51:2415–2418. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- 3.Hendrickson E.R., Truby T.M., Joerger R.D., Majarian W.R., Ebersole R.C. High sensitivity multianalyte immunoassay using covalent DNA-labeled antibodies and polymerase chain reaction. Nucleic Acids Res. 1995;23:522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaxton C.S., Elghanian R., Thomas A.D., Stoeva S.I., Lee J.S., Smith N.D., Schaeffer A.J., Klocker H., Horninger W., Bartsch G., et al. Nanoparticle-based bio-barcode assay redefines ‘undetectable' PSA and biochemical recurrence after radical prostatectomy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredriksson S., Dixon W., Ji H., Koong A.C., Mindrinos M., Davis R.W. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg M., Eriksson A., Tran B., Assarsson E., Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson J.G. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 8.Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Brit. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inglese J., Johnson R.L., Simeonov A., Xia M.H., Zheng W., Austin C.P., Auld D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 10.Mesters J.R., Barinka C., Li W.X., Tsukamoto T., Majer P., Slusher B.S., Konvalinka J., Hilgenfeld R. Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer. EMBO J. 2006;25:1375–1384. doi: 10.1038/sj.emboj.7600969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alterio V., Hilvo M., Di Fiore A., Supuran C.T., Pan P.W., Parkkila S., Scaloni A., Pastorek J., Pastorekova S., Pedone C., et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16233–16238. doi: 10.1073/pnas.0908301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinoshita Y., Kuratsukuri K., Landas S., Imaida K., Rovito P.M., Wang C.Y., Haas G.P. Expression of prostate-specific membrane antigen in normal and malignant human tissues. World J. Surg. 2006;30:628–636. doi: 10.1007/s00268-005-0544-5. [DOI] [PubMed] [Google Scholar]
- 13.Wykoff C.C., Beasley N.J.P., Watson P.H., Turner K.J., Pastorek J., Sibtain A., Wilson G.D., Turley H., Talks K.L., Maxwell P.H., et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- 14.Zhou G.X., Ireland J., Rayman P., Finke J., Zhou M. Quantification of carbonic anhydrase IX expression in serum and tissue of renal cell carcinoma patients using enzyme-linked immunosorbent assay: prognostic and diagnostic potentials. Urology. 2010;75:257–261. doi: 10.1016/j.urology.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 15.Heck M.M., Retz M., D'Alessandria C., Rauscher I., Scheidhauer K., Maurer T., Storz E., Janssen F., Schottelius M., Wester H.J., et al. Systemic Radioligand Therapy with (177)Lu Labeled Prostate Specific Membrane Antigen Ligand for Imaging and Therapy in Patients with Metastatic Castration Resistant Prostate Cancer. J. Urol. 2016;196:382–391. doi: 10.1016/j.juro.2016.02.2969. [DOI] [PubMed] [Google Scholar]
- 16.Krall N., Pretto F., Neri D. A bivalent small molecule-drug conjugate directed against carbonic anhydrase IX can elicit complete tumour regression in mice. Chem. Sci. 2014;5:3640–3644. [Google Scholar]
- 17.Xiao Z., Adam B.L., Cazares L.H., Clements M.A., Davis J.W., Schellhammer P.F., Dalmasso E.A., Wright G.L. Quantitation of serum prostate-specific membrane antigen by a novel protein biochip immunoassay discriminates benign from malignant prostate disease. Cancer Res. 2001;61:6029–6033. [PubMed] [Google Scholar]
- 18.Zavada J., Zavadova Z., Zat'ovicova M., Hyrsl L., Kawaciuk I. Soluble form of carbonic anhydrase IX (CA IX) in the serum and urine of renal carcinoma patients. Brit. J. Cancer. 2003;89:1067–1071. doi: 10.1038/sj.bjc.6601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim S.H., Messenger M.P., Gregory W.M., Wind T.C., Vasudev N.S., Cartledge J., Thompson D., Selby P.J., Banks R.E. Prognostic utility of pre-operative circulating osteopontin, carbonic anhydrase IX and CRP in renal cell carcinoma. Brit. J. Cancer. 2012;107:1131–1137. doi: 10.1038/bjc.2012.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slusher B.S., Vornov J.J., Thomas A.G., Hurn P.D., Harukuni I., Bhardwaj A., Traystman R.J., Robinson M.B., Britton P., Lu X.C.M., et al. Selective inhibition of NAALADase, which converts NAAG to glutamate, reduces ischemic brain injury. Nat. Med. 1999;5:1396–1402. doi: 10.1038/70971. [DOI] [PubMed] [Google Scholar]
- 21.Bacich D.J., Wozniak K.M., Lu X.C.M., O'Keefe D.S., Callizot N., Heston W.D.W., Slusher B.S. Mice lacking glutamate carboxypeptidase II are protected from peripheral neuropathy and ischemic brain injury. J. Neurochem. 2005;95:314–323. doi: 10.1111/j.1471-4159.2005.03361.x. [DOI] [PubMed] [Google Scholar]
- 22.Olszewski R.T., Bzdega T., Neale J.H. mGluR3 and not mGluR2 receptors mediate the efficacy of NAAG peptidase inhibitor in validated model of schizophrenia. Schizophr. Res. 2012;136:160–161. doi: 10.1016/j.schres.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Chiche J., Ilc K., Laferriere J., Trottier E., Dayan F., Mazure N.M., Brahimi-Horn M.C., Pouyssegur J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69:358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 24.Pacchiano F., Carta F., McDonald P.C., Lou Y.M., Vullo D., Scozzafava A., Dedhar S., Supuran C.T. Ureido-substituted benzenesulfonamides potently inhibit carbonic anhydrase IX and show antimetastatic activity in a model of breast cancer metastasis. J. Med. Chem. 2011;54:1896–1902. doi: 10.1021/jm101541x. [DOI] [PubMed] [Google Scholar]
- 25.Lou Y.M., McDonald P.C., Oloumi A., Chia S., Ostlund C., Ahmadi A., Kyle A., Keller U.A.D., Leung S., Huntsman D., et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 2011;71:3364–3376. doi: 10.1158/0008-5472.CAN-10-4261. [DOI] [PubMed] [Google Scholar]
- 26.Jackson P.F., Cole D.C., Slusher B.S., Stetz S.L., Ross L.E., Donzanti B.A., Trainor D.A. Design, synthesis, and biological activity of a potent inhibitor of the neuropeptidase N-acetylated alpha-linked acidic dipeptidase. J. Med. Chem. 1996;39:619–622. doi: 10.1021/jm950801q. [DOI] [PubMed] [Google Scholar]
- 27.Kozikowski A.P., Nan F., Conti P., Zhang J.H., Ramadan E., Bzdega T., Wroblewska B., Neale J.H., Pshenichkin S., Wroblewski J.T. Design of remarkably simple, yet potent urea-based inhibitors of glutamate carboxypeptidase II (NAALADase) J. Med. Chem. 2001;44:298–301. doi: 10.1021/jm000406m. [DOI] [PubMed] [Google Scholar]
- 28.van der Post J.P., de Visser S.J., de Kam M.L., Woelfler M., Hilt D.C., Vornov J., Burak E.S., Bortey E., Slusher B.S., Limsakun T., et al. The central nervous system effects, pharmacokinetics and safety of the NAALADase-inhibitor GPI 5693. Brit. J. Clin. Pharmacol. 2005;60:128–136. doi: 10.1111/j.1365-2125.2005.02396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rais R., Wozniak K., Wu Y., Niwa M., Stathis M., Alt J., Giroux M., Sawa A., Rojas C., Slusher B.S. Selective CNS uptake of the GCP-II inhibitor 2-PMPA following intranasal administration. PLoS One. 2015;10:e0131861. doi: 10.1371/journal.pone.0131861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supuran C.T. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 31.Barinka C., Rinnova M., Sacha P., Rojas C., Majer P., Slusher B.S., Konvalinka J. Substrate specificity, inhibition and enzymological analysis of recombinant human glutamate carboxypeptidase II. J. Neurochem. 2002;80:477–487. doi: 10.1046/j.0022-3042.2001.00715.x. [DOI] [PubMed] [Google Scholar]
- 32.Tykvart J., Navratil V., Sedlak F., Corey E., Colombatti M., Fracasso G., Koukolik F., Barinka C., Sacha P., Konvalinka J. Comparative analysis of monoclonal antibodies against prostate-specific membrane antigen (PSMA) Prostate. 2014;74:1674–1690. doi: 10.1002/pros.22887. [DOI] [PubMed] [Google Scholar]
- 33.Knedlik T., Navratil V., Vik V., Pacik D., Sacha P., Konvalinka J. Detection and quantitation of glutamate carboxypeptidase II in human blood. Prostate. 2014;74:768–780. doi: 10.1002/pros.22796. [DOI] [PubMed] [Google Scholar]
- 34.Zavada J., Zavadova Z., Pastorek J., Biesova Z., Jezek J., Velek J. Human tumour-associated cell adhesion protein MN/CA IX: identification of M75 epitope and of the region mediating cell adhesion. Brit. J. Cancer. 2000;82:1808–1813. doi: 10.1054/bjoc.2000.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chrastina A., Zavada J., Parkkila S., Kaluz T., Kaluzova M., Rajcani J., Pastorek J., Pastorekova S. Biodistribution and pharmacokinetics of I-125-labeled monoclonal antibody M75 specific for carbonic anhydrase IX, an intrinsic marker of hypoxia, in nude mice xenografted with human colorectal carcinoma. Int. J. Cancer. 2003;105:873–881. doi: 10.1002/ijc.11142. [DOI] [PubMed] [Google Scholar]
- 36.Vullo D., Scozzafava A., Pastorekova S., Pastorek J., Supuran C.T. Carbonic anhydrase inhibitors: inhibition of the tumor-associated isozyme IX with fluorine-containing sulfonamides. The first subnanomolar CA IX inhibitor discovered. Bioorg. Med. Chem. Lett. 2004;14:2351–2356. doi: 10.1016/j.bmcl.2004.01.095. [DOI] [PubMed] [Google Scholar]
- 37.Tykvart J., Schimer J., Barinkova J., Pachl P., Postova-Slavetinska L., Majer P., Konvalinka J., Sacha P. Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery. Bioorg. Med. Chem. 2014;22:4099–4108. doi: 10.1016/j.bmc.2014.05.061. [DOI] [PubMed] [Google Scholar]
- 38.Brynda J., Mader P., Sicha V., Fabry M., Poncova K., Bakardiev M., Gruner B., Cigler P., Rezacova P. Carborane-based carbonic anhydrase inhibitors. Angew. Chem. Int. Edit. 2013;52:13760–13763. doi: 10.1002/anie.201307583. [DOI] [PubMed] [Google Scholar]
- 39.Sacha P., Knedlik T., Schimer J., Tykvart J., Parolek J., Navratil V., Dvorakova P., Sedlak F., Ulbrich K., Strohalm J., et al. iBodies: modular synthetic antibody mimetics based on hydrophilic polymers decorated with functional moieties. Angew. Chem. Int. Ed. Engl. 2016;55:2356–2360. doi: 10.1002/anie.201508642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H., Moy P., Kim S., Xia Y., Rajasekaran A., Navarro V., Knudsen B., Bander N.H. Monoclonal antibodies to the extracellular domain of prostate-specific membrane antigen also react with tumor vascular endothelium. Cancer Res. 1997;57:3629–3634. [PubMed] [Google Scholar]
- 41.Hammer O., Harper D.A.T. Paleontological Data Analysis. Malden: Blackwell Publishing; 2006. [Google Scholar]
- 42.Bengtsson M., Stahlberg A., Rorsman P., Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maresca K.P., Hillier S.M., Femia F.J., Keith D., Barone C., Joyal J.L., Zimmerman C.N., Kozikowski A.P., Barrett J.A., Eckelman W.C., et al. A series of halogenated heterodimeric inhibitors of prostate specific membrane antigen (PSMA) as radiolabeled probes for targeting prostate cancer. J. Med. Chem. 2009;52:347–357. doi: 10.1021/jm800994j. [DOI] [PubMed] [Google Scholar]
- 44.Hlouchova K., Barinka C., Konvalinka J., Lubkowski J. Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III. FEBS J. 2009;276:4448–4462. doi: 10.1111/j.1742-4658.2009.07152.x. [DOI] [PubMed] [Google Scholar]
- 45.Brenner S., Lerner R.A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry N., Sebe P., Cussenot O. Inappropriate treatment of prostate cancer caused by heterophilic antibody interference. Nat. Clin. Pract. Urol. 2009;6:164–167. doi: 10.1038/ncpuro1317. [DOI] [PubMed] [Google Scholar]
- 47.Poyet C., Hof D., Sulser T., Muntener M. Artificial prostate-specific antigen persistence after radical prostatectomy. J. Clin. Oncol. 2012;30:E62–E63. doi: 10.1200/JCO.2011.38.2788. [DOI] [PubMed] [Google Scholar]
- 48.Ali M.M., Li F., Zhang Z.Q., Zhang K.X., Kang D.K., Ankrum J.A., Le X.C., Zhao W.A. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 49.Rich D.H., Bernatowicz M.S., Agarwal N.S., Kawai M., Salituro F.G., Schmidt P.G. Inhibition of aspartic proteases by pepstatin and 3-Methylstatine derivatives of pepstatin - evidence for collected-substrate enzyme-inhibition. Biochemistry. 1985;24:3165–3173. doi: 10.1021/bi00334a014. [DOI] [PubMed] [Google Scholar]
- 50.Karaman M.W., Herrgard S., Treiber D.K., Gallant P., Atteridge C.E., Campbell B.T., Chan K.W., Ciceri P., Davis M.I., Edeen P.T., et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]