Abstract
Transcription termination events in bacteria often require the RNA helicase Rho. Typically, Rho promotes termination at the end of coding sequences, but it can also terminate transcription within leader regions to implement regulatory decisions. Rho-dependent termination requires initial recognition of a Rho utilization (rut) site on a nascent RNA by Rho's primary binding surface. However, it is presently unclear what factors determine the location of transcription termination, how RNA secondary structures influence this process and whether mechanistic differences distinguish constitutive from regulated Rho-dependent terminators. We previously demonstrated that the 5′ leader mRNA of the Salmonella corA gene can adopt two mutually exclusive conformations that dictate accessibility of a rut site to Rho. We now report that the corA leader also controls two subsequent steps of Rho-dependent termination. First, the RNA conformation that presents an accessible rut site promotes pausing of RNA polymerase (RNAP) at a single Rho-dependent termination site over 100 nt downstream. Second, an additional RNA stem-loop promotes Rho activity and controls the location at which Rho-dependent termination occurs, despite having no effect on initial Rho binding to the corA leader. Thus, the multi-step nature of Rho-dependent termination may facilitate regulation of a given coding region by multiple cytoplasmic signals.
INTRODUCTION
The hexameric RNA helicase and ATPase Rho triggers 20–30% of transcription termination events in bacteria (1,2). Most studied Rho-dependent terminators perform housekeeping roles that necessitate constitutive function, whereas others serve as platforms for gene regulation and thus function at varying efficiencies in response to intracellular signals. It remains to be determined whether there are mechanistic differences between constitutive and regulated Rho-dependent terminators. We previously identified a Rho-dependent terminator in the 5′ leader of the corA gene in Salmonella enterica serovar Typhimurium (3). Here, we report that, in addition to regulating accessibility of a Rho loading site, RNA secondary structures within the corA leader modulate two subsequent steps of Rho-dependent termination. Our findings suggest that the complex series of interactions between Rho, RNA and RNA polymerase (RNAP) required for transcription termination allows multiple regulatory signals to influence the fate of an elongating RNAP.
Initial recognition of an RNA substrate is achieved by a primary binding site (PBS) on an exposed surface of the Rho hexamer that is formed by the N-terminal domains of each Rho monomer (4–6). The PBS contains six (one per monomer) binding clefts that can each accommodate one YC dinucleotide (7), with adjacent YC dinucleotides separated by a spacer of at least 7 nt (8). As a result, regions of nascent RNA recognized by the PBS, designated Rho utilization (rut) sites, tend to be C-rich, unstructured and 60–90 nt long (9). Binding of the Rho PBS to a rut site directs the RNA into the ring-shaped protein's central channel, where it contacts Rho's secondary binding site (SBS) (7,10). This secondary interaction, which is transient and strongly dependent on the presence of adenosine triphosphate (ATP) (8,10), activates Rho's ATPase and helicase activities to allow translocation along the substrate RNA in a 5′ to 3′ direction (7,11). The PBS appears to remain bound to the rut site throughout this process, resulting in a ‘tethered tracking’ mechanism of translocation (8,12). A comprehensive model of how Rho navigates structured regions of RNA has yet to emerge (13–15).
Once a translocating Rho molecule catches up to a paused RNAP, it induces dissociation of the elongation complex (6,16,17). The location and efficiency of Rho-dependent transcription termination are thus dictated by the location and efficiency, respectively, of RNAP pausing. However, Rho can only access RNAP complexes paused within a particular ‘termination zone’ downstream of the rut site and upstream of regions in which translating ribosomes would impede Rho access to the RNA (14,18). In addition, pausing defines the kinetic window in which Rho can operate; in order to terminate transcription at a particular site, Rho must successfully recognize, load and translocate before a paused RNAP moves on. This timeframe is further constrained if rut site accessibility is modulated by factors such as RNA secondary structure. RNAP pausing is typically regulated by proximal sequences within the RNA or DNA, as well as by protein factors (19). RNA secondary structures >20 nt upstream can influence RNAP pausing (20,21), although it is not clear how they operate.
The complex sequence of events required for Rho to trigger transcription termination provides many opportunities for intracellular conditions to impact the efficiency of this process, thereby making Rho-dependent terminators useful platforms for gene control (22). Regulatory mechanisms that employ Rho-dependent terminators generally function via modulation of rut site accessibility (23,24). However, the Salmonella mgtA leader mRNA conformation modulates Rho-dependent termination efficiency by regulating both rut site availability (25) and RNAP pausing (20). In addition, the Salmonella mgtCBR leader mRNA can adopt alternative conformations that expose or sequester a Rho-antagonizing RNA element that traps Rho in an inactive complex without affecting rut site accessibility (26). Thus, regulation of Rho-RNA-RNAP interactions independent of rut site recognition may be widespread. This notion is supported by the finding that the primary binding function of Rho is largely dispensable in vivo; Rho SBS mutants, but not PBS mutants, were found to be defective for genome-wide Rho-dependent transcription termination (27).
Here, we report a singular example of an mRNA leader that can regulate three distinct aspects of Rho-dependent termination. We previously established that the corA leader mRNA in Salmonella controls accessibility of a rut site via selective secondary structure formation (3). We now demonstrate that the corA leader also regulates two additional aspects of the interaction between itself, Rho and RNAP. Specifically, the same RNA conformation that sequesters the rut site suppresses RNAP pausing at a single site over 100 nt downstream, which may further reduce the efficiency of Rho-dependent termination when the corA leader adopts this structure. This interaction occurs over a significantly longer distance than that at which RNA structures typically influence pausing. In addition, formation of an additional, conserved RNA hairpin promotes Rho activity and prevents Rho from triggering transcription termination at a major RNAP pause site. Together, these findings reveal multiple points at which intracellular signals may regulate expression of corA.
MATERIALS AND METHODS
Bacterial strains and growth conditions
In vivo experiments were carried out with wild-type Salmonella enterica serovar Typhimurium strain 14028s (28) harboring plasmid pYS1040, pYS1040ext or mutant derivatives (Supplementary Table S1). Escherichia coli DH5α (29) or XL-1 blue (Agilent) strains were used for cloning and plasmid DNA preparation. Bacteria were grown at 37°C in LB broth (Difco) or SOB medium (Difco). For plasmid maintenance, 20 μg/ml chloramphenicol (cm) or 50 μg/ml ampicillin (amp) was used.
Plasmid construction
Nucleotide substitutions in plasmids pMK100 and pYS1040 (3) were generated using the QuikChange II site-directed mutagenesis kit (Agilent) and primer pairs listed in Supplementary Table S2.
In vitro transcription termination assays
Transcription templates were prepared by performing polymerase chain reaction with plasmid pMK100 or mutant derivatives as template and primers IA17 and IA256. Purified Salmonella RNAP core enzyme and σ70 were mixed at a 1:3 molar ratio in RNAP storage buffer (50% v/v glycerol, 10 mM Tris pH 7.9, 0.1 M NaCl, 0.1 mM ethylenediaminetetracetic acid (EDTA), 0.1 mM DTT) and allowed to associate by incubating at 30°C for 30 min.
Single round, synchronized in vitro transcription reactions were carried out as described (3,30). Reactions contained 100 mM Tris HCl (pH 8.0), 100 mM KCl, 1 mM MgCl2, 5 μM each of ATP and UTP, 1 μM GTP, 100 μM ApU (TriLink), 1.5 μl [α-32P]-GTP (3000 Ci/mmol, Perkin Elmer) per 100 μl reaction volume, 50 nM DNA template and 56 nM σ70+RNAP. This mixture was incubated at 37°C for 15 min to generate halted transcription elongation complexes (hTEC). hTEC were divided into aliquots and incubated at 37°C for 3 min. To initiate synchronized transcription elongation, a 5X chase ±Rho was added (100 mM Tris HCl (pH 8.0), 100 mM KCl, 0.1mg/ml rifampicin, 1 mM NTPs, with 600 nM Rho or an equal volume of Rho buffer). Reactions were incubated at 37°C for 5 min, terminated by addition of 2X stop solution (80% v/v formamide, 50 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and placed on ice. After heating at 95°C for 3 min, transcription products were resolved on denaturing 6% polyacrylamide sequencing gels (Sequagel system, National Diagnostics) and visualized using a Typhoon FLA 9000 laser scanner (GE Healthcare).
To map transcription termination sites, a portion of hTEC were used for RNA sequencing reactions as described (30). Band intensities were measured using ImageQuant software (GE Healthcare). Termination efficiency was calculated using the formula: 100 – 100* (runoff band intensity/sum of all band intensities).
In vitro pausing assays
Pausing assays and gel electrophoresis were performed as described for in vitro transcription termination assays, except that the reactions contained 5 mM MgCl2 and the 5X chase solution contained 250 μM NTPs and no Rho or Rho buffer. Samples were removed at 10–20 s intervals starting 10 s after synchronization of transcription elongation and added to 2X stop solution that had been pre-aliquoted on ice.
Intensities of each lane and pause bands were measured using ImageQuant software (GE Healthcare) and normalized by subtracting the intensity of a blank region of the same area. Pause bands were further normalized by subtracting the band intensity of the last time point. Pause efficiency and duration were calculated as described (31) by plotting ln(p/t) over time, where p is the normalized pause intensity and t is the normalized lane intensity. Pause half-life was extrapolated from the slope of the linear portion of the graph according to the formula half-life = -ln(2)/slope. Pause efficiency was extrapolated from the y-intercept of the linear portion according to the formula % eff = 100 × e∧yint. The position of major pause sites was mapped by comparison with an RNA sequencing ladder generated as described (30).
Enzymatic and filter binding assays
Accumulation of free phosphate was measured using the EnzChek Phosphate Assay kit (Invitrogen) as described (3). All experiments shown were performed in 0.75 mM MgCl2.
β-galactosidase assays and filter binding assays were performed as described (3).
RESULTS
The Salmonella corA leader can adopt two mutually exclusive conformations that control accessibility of a rut site
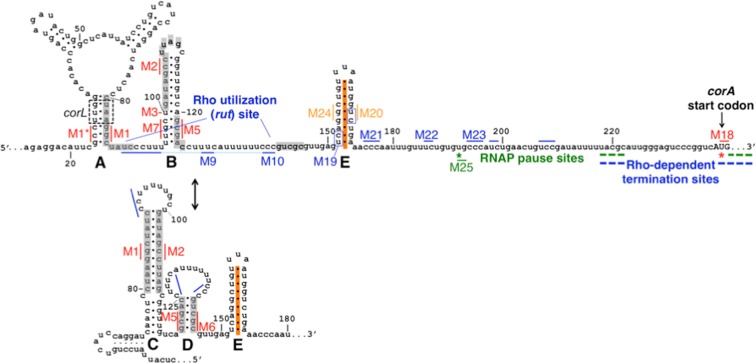
In vitro structure probing assays previously demonstrated that the corA leader mRNA is capable of adopting mutually exclusive secondary structures termed stem-loops A+B and stem-loops C+D (Figure 1) (3). However, stem-loop A is unlikely to form in vivo because a short open reading frame, termed corL, overlaps stem-loop A such that ribosomes translating corL should prevent base-pairing (Figure 1) (3). Our previous work demonstrated that the stem-loop B conformation promotes Rho-dependent transcription termination within the corA leader by exposing nucleotides comprising a rut site (Figure 1) (3). By contrast, the stem-loop C+D conformation hinders Rho-dependent termination within the corA leader by sequestering the rut site (Figure 1) (3). Efficient translation of corL promotes formation of stem-loop B, whereas impaired corL translation favors stem-loops C+D; thus, transcription of the corA coding region is inversely coupled to the efficiency of corL translation (3).
Figure 1.
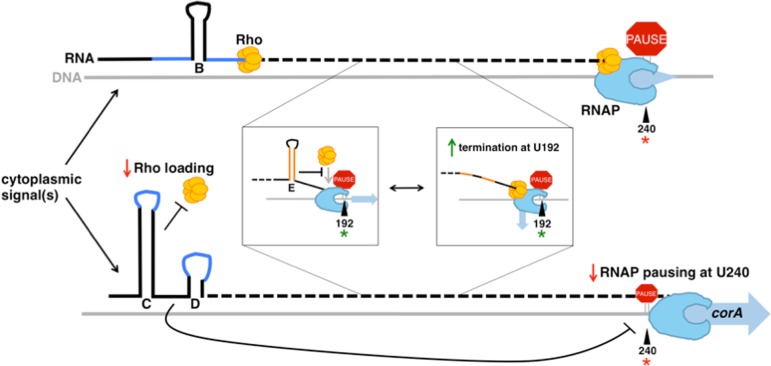
Schematic of the corA leader mRNA. The Salmonella corA leader can adopt two mutually exclusive conformations, stem-loops A+B or C+D, that control transcription elongation into the corA coding region by modulating accessibility of a rut site (3). Stem-loop A may not form in vivo due to translation of the overlapping ORF corL (dashed black box denotes corL start and stop codons) (3). Major sites of in vitro RNAP pausing (green) and Rho-dependent termination (blue) are denoted. In this work, we identify an additional RNA structure, stem-loop E (orange), whose formation may be independent of stem-loops B versus C+D. Stem-loops C+D suppress RNAP pausing at U240 (red star). Stem-loop E prevents Rho from triggering transcription termination at U192 (green star).
Because the corA leader extends ∼100 nt downstream of the RNA structures that regulate Rho's ability to access the rut site (Figure 1) and the nucleotide sequence of this region is highly conserved (Supplementary Figure S1), we reasoned that it might play additional role(s) in regulating Rho-dependent transcription termination. For example, the Salmonella mgtA leader RNA conformation controls Rho-dependent termination by modulating rut site accessibility and RNAP pausing, and regulation of both aspects is required for gene control (20).
RNAP pauses frequently within the corA leader
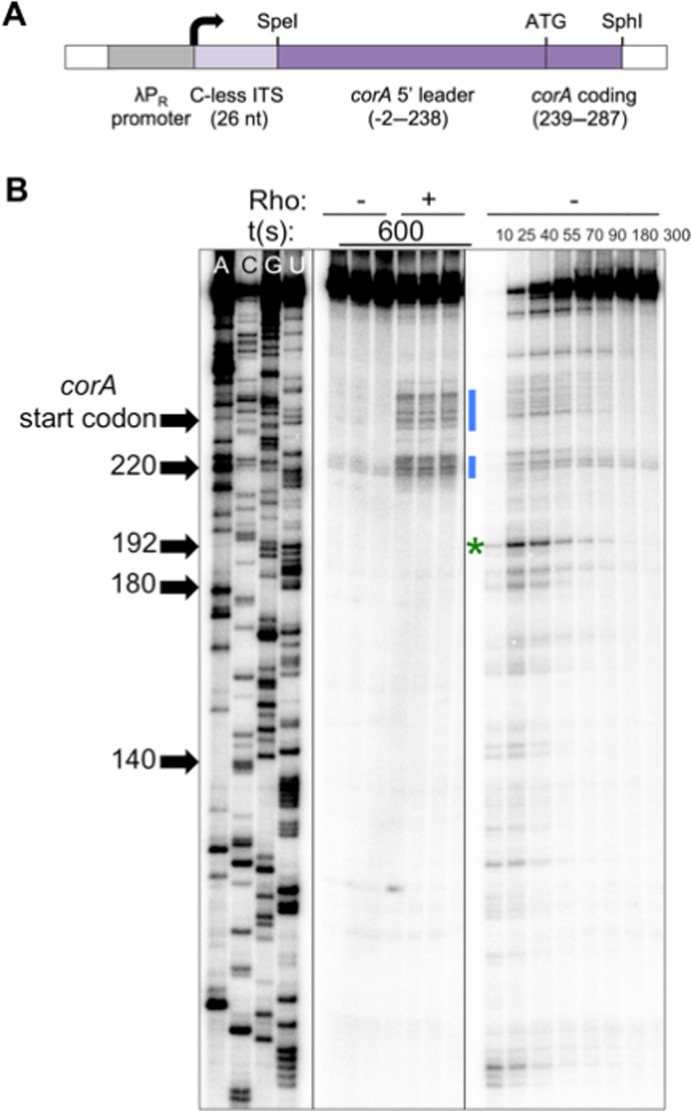
To investigate the possibility that the corA leader influences RNAP pausing, we first mapped RNAP pause sites within this region by performing single-round, synchronized in vitro transcription reactions. The DNA template used in these experiments encodes a 26 nt C-less initial transcribed sequence, to facilitate synchronization of transcription elongation, followed by the complete corA leader and first 49 nt of the corA coding region, all under the control of a constitutive promoter (Figure 2A) (20,30). Samples were collected over time on the order of seconds; bands that appear and then disappear throughout the time course correspond to positions at which some fraction of RNAP molecules pauses before continuing transcription.
Figure 2.
Mapping of RNAP pause sites in the corA leader. (A) Schematic of DNA templates used for in vitro transcription assays, modified from (20). Inclusion of a 26 nt C-less initial transcribed sequence (ITS) facilitates synchronization of transcription (30). (B) A 6% denaturing gel showing RNA sequencing (left), transcription termination (center) and pausing assays (right) performed using a wild-type corA leader template generated from pMK100 as described in Materials and Methods. All panels are from the same gel and were re-ordered for clarity. RNAP pauses frequently within the corA leader, including at the sites of Rho-dependent termination (blue bars). The major pause site, U192 (green star) is not a site of Rho-dependent termination. The results are representative of at least three independent experiments and only the relevant portion of the gel is shown.
RNAP pauses frequently within the corA leader in vitro (Figure 2B, right), including at the expected clusters of sites that correspond to the Rho-dependent termination sites observed in vitro (Figure 2B, blue bars). Interestingly, the most efficient site of pausing, which is located at position U192 (Figure 2B, green star) and conforms to the recently identified consensus pause sequence G -10Y-1G+1 (32,33), is not a Rho-dependent termination site (Figure 2B).
Stem-loops C and D reduce the efficiency of RNAP pausing at a site over 100 nt downstream
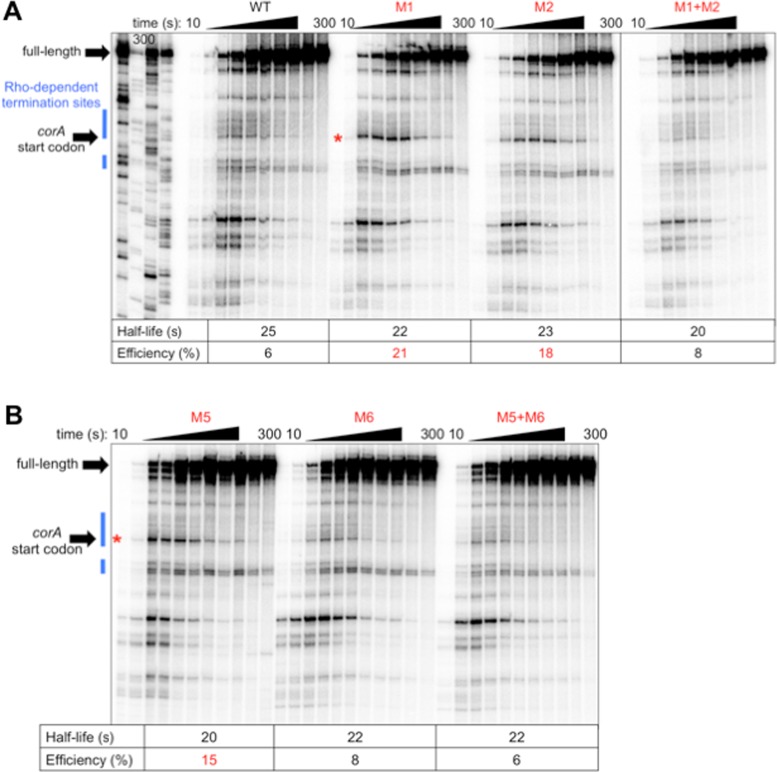
We next tested whether the two known conformations of the corA leader, stem-loop B and stem-loops C+D, differentially affect RNAP pausing. Stem-loops C+D sequester the rut site (3). Disruption of this conformation by mutation M1 (G84C G85C C86G, Figure 1) significantly increased pausing at U240, a site of Rho-dependent termination (Figure 3A). This result was unexpected considering that the affected pause site is 100–150 nt downstream of M1 and stem-loops B-D (Figure 1). M1 specifically increases the efficiency of the pause, rather than its half-life (Figure 3A), indicating modulation of RNAP entry into the paused state.
Figure 3.
The same RNA conformation that sequesters the rut site (stem-loops C+D) reduces efficiency of RNAP pausing at U240. (A and B) A 6% denaturing gel of RNAP pausing assays performed using DNA templates generated from plasmid pMK100 or derivatives containing mutations in (A) stem-loop C or (B) stem-loop D (Figure 1). A red star indicates the position of the U240 pause, which is within the corA start codon. Pause efficiency and half-life were calculated as described in Materials and Methods. The results are representative of at least three independent experiments and only the relevant portion of the gel is shown. Data shown in (A) are from the same gel; solid line denotes where intervening lanes were omitted for clarity.
The M1 mutation theoretically disrupts both stem-loops A and C, but stem-loop A is not likely to form in vivo due to the presence of ribosomes translating corL, an open reading frame that overlaps stem-loop A (Figure 1) (3). Indeed, the effect of M1 on RNAP pausing is specifically due to disruption of stem-loop C rather than disruption of stem-loop A because restoration of stem-loop C base-pairing potential by combining M1 with M2 (G105C C106G C107G, Figure 1) returned pausing to wild-type efficiency (Figure 3A), whereas restoration of stem-loop A base-pairing potential by combining M1 with M1* (G24C C25G C26G, Figure 1) did not reverse the effect of M1 (Supplementary Figure S2). It is important to note that mutation M2, which disrupts both stem-loops B and C, elicits the same pause behavior as M1 (Figure 3A), indicating that the effect at U240 is due to suppression of pausing by stem-loop C rather than promotion of pausing by stem-loop B.
Because stem-loop C is expected to facilitate formation of stem-loop D (by virtue of precluding stem-loop B formation) (Figure 1), we tested whether disruption of stem-loop D also leads to efficient RNAP pausing at U240. Indeed, a mutation that disrupts stem-loop D (M5 [G121T G123T], Figure 1) displayed the same increase in U240 pause efficiency as a mutation that disrupts stem-loop C (M1) (Figure 3B). Although M5 is predicted to disrupt both stem-loops B and D, the effect of this mutation on pausing is specifically due to disruption of stem-loop D because combining M5 with M6 (C143A C145A, Figure 1) to restore stem-loop D base-pairing potential reinstated wild-type pause efficiency (Figure 3B), whereas combining M5 with M7 (T97G C99G, Figure 1) to restore stem-loop B base-pairing potential retained elevated pause efficiency at U240 (Supplementary Figure S3). Unexpectedly, M6 alone did not increase U240 pause efficiency like M5, despite the fact that both mutations disrupt stem-loop D (Figure 3B).
In addition to disrupting stem-loop C, mutation M1 should also prevent stem-loop D formation because stem-loop B will have a kinetic advantage over stem-loop D when stem-loop C is unable to form (Figure 1). Thus, it seemed possible that the ability of M1 to enhance pausing at U240 is due solely to disruption of stem-loop D. However, this does not appear to be the case because M1+M5+M6, which is expected to adopt stem-loop D despite disruption of stem-loop C, displayed enhanced pausing at U240 (Supplementary Figure S4). Therefore, stem-loops C and D are both required for suppression of pausing at U240. Complete elimination of pausing at U240 by mutation M18 (T240G G241A, Figure 1) (Supplementary Figure S5A) further de-repressed expression of a corA leader-lacZ transcriptional fusion carried on plasmid pYS1040 that contains a mutation favoring adoption of stem-loops C+D (Supplementary Figure S5D), suggesting that suppression of pausing at U240 enhances the ability of stem-loops C+D to promote corA expression.
YC dinucleotides downstream of stem-loop D can influence Rho activity
U192 is not a site of Rho-dependent transcription termination despite being the most efficient RNAP pause site in the corA leader (Figure 2B). The rut site was previously reported to encompass positions 89–140 of the corA leader and thus lie well upstream of U192 (Figure 1) (3). However, because this region contains only three appropriately spaced, single-stranded YC dinucleotides (positions 89–91, 129–130, 138–140, Figure 1) and the Rho PBS can accommodate up to six YC dinucleotides, we explored the possibility that YC dinucleotides downstream of position 140 might be involved in Rho loading. The corA leader sequence contains seven YC dinucleotides between position 140 and the first site of significant Rho-dependent termination (∼220), four of which would not be available to Rho when RNAP is paused at U192 (Figure 1). Thus, it is possible that Rho does not terminate transcription at U192 because it has not completed loading by the time RNAP reaches this position.
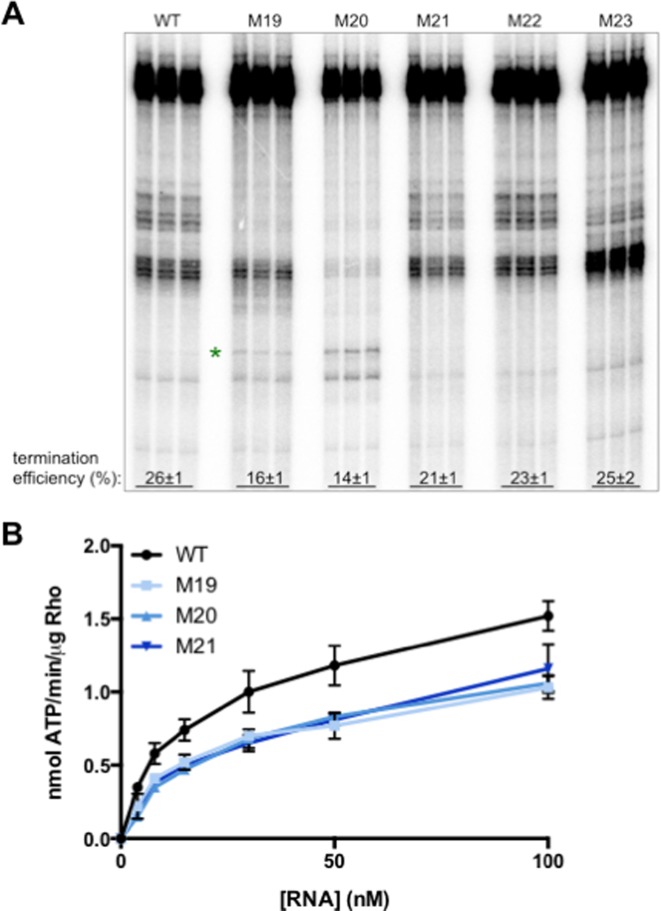
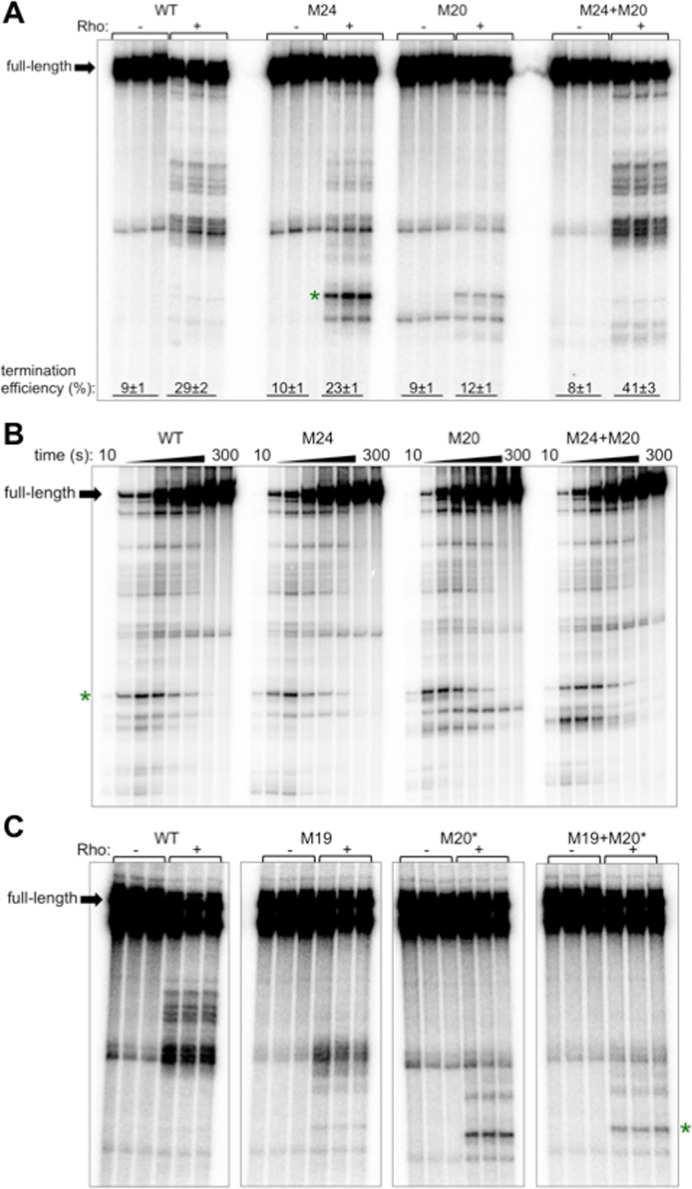
We first tested whether removal of a given YC dinucleotide decreased the efficiency of Rho-dependent transcription termination in vitro. Mutation of each of the first three YC dinucleotides downstream of position 140 (M19 [T152C C153A], M20 [T168G C169G] and M21 [C176G C177T]) resulted in reduced efficiency of Rho-dependent termination (Figure 4A), revealing that these YC dinucleotides may be part of the rut site. By contrast, mutation of the fourth and fifth YC dinucleotides (M22 [T186A C187G T188A], M23 [C194A C195G C196T]) did not significantly affect termination efficiency (Figure 4A), indicating that they are not required for Rho-dependent termination in the corA leader.
Figure 4.
Rho can utilize YC dinucleotides downstream of C140. (A) A 6% denaturing gel of transcription termination assays performed using DNA templates generated from plasmid pMK100 or derivatives harboring mutations in YC dinucleotides between positions 152–196 (Figure 1), in the presence of purified Rho protein. Reactions were performed in triplicate and only the relevant portion of the gel is shown. The results are representative of at least three independent experiments. Green star indicates the position of the U192 pause. (B) The accumulation of free phosphate was monitored to quantify Rho ATPase activity. Purified Rho protein was incubated with RNA corresponding to the wild-type corA leader mRNA or variants harboring mutations YC dinucleotides between positions 152–196 (Figure 1). Data shown correspond to mean values and standard deviation of technical duplicates and are representative of three independent experiments.
If the YC dinucleotides disrupted by M19, M20 and M21 reduce termination efficiency by serving as components of the rut site, then these mutations should weaken the interaction between Rho and corA leader mRNA, irrespective of the transcription process. To test this notion, we measured the ability of corA leader mRNA harboring mutations M19, M20 and M21 to stimulate Rho ATPase activity in vitro. All three mutant RNAs exhibited a reduced ability to stimulate Rho ATPase activity (Figure 4B), indicating that the YC dinucleotides at positions 152–153, 168–169 and 176–177 promote Rho activity and thus may be directly involved in Rho PBS–RNA recognition. As a result, we conclude that the rut site encompasses positions 89–177.
Stem-loop E prevents Rho from accessing RNAP complexes paused at U192 in a rut site-independent manner
Paradoxically, mutations M19 and M20 caused some Rho-dependent termination to take place at an upstream site (Figure 4A, green star) that we determined to be U192, the major RNAP pause site (Supplementary Figure S6). This result indicates that nucleotides 152–153 and 168–169 somehow inhibit Rho-dependent termination at U192. We noticed a potential RNA hairpin located at position 152–172 (Figure 1) that is conserved among analyzed corA leader sequences (Supplementary Figure S1); we designated this hairpin stem-loop E. We tested whether the ability of M20 to promote Rho-dependent termination at U192 is a consequence of disrupting stem-loop E. Indeed, mutation of the nucleotides predicted to base pair with those disrupted by mutation M20 (M24 [G155C G156C], Figure 1) also allowed Rho-dependent termination to take place at U192 (Figure 5A), whereas combining M24+M20 to restore stem-loop E base-pairing potential prevented termination at U192 and restored the wild-type pattern of termination locations (Figure 5A). Taken together, these results suggest that stem-loop E formation prevents Rho-dependent termination at U192.
Figure 5.
Stem-loop E prevents Rho from terminating transcription at a major RNAP pause site, U192, in a rut site-independent manner. (A) A 6% denaturing gel of transcription termination assays performed using templates generated from plasmid pMK100 or derivatives containing mutations in stem-loop E (Figure 1). Green star indicates the position of the U192 pause. Calculation of termination efficiency was performed as described in Materials and Methods. Reactions were performed in triplicate and only the relevant portion of the gel is shown. The results are representative of at least three independent experiments. (B) A 6% denaturing gel of RNAP pausing assays performed using DNA templates generated from plasmid pMK100 or derivatives containing mutations in stem-loop E (Figure 1). The results are representative of at least three independent experiments and only the relevant portion of the gel is shown. Green star indicates the position of the U192 pause. (C) A 6% denaturing gel of transcription termination assays performed using templates generated from plasmid pMK100 or derivatives containing mutations that disrupt stem-loop E and/or remove YC nucleotides predicted to participate in stem-loop E base-pairing (Figure 1). Reactions were performed in triplicate and only the relevant portion of the gel is shown. The results are representative of at least three independent experiments. Green star indicates the position of the U192 pause. The data shown are from the same gel; the panels were re-ordered and intervening lanes were omitted for clarity.
The M24 and M20 mutations respectively introduce and eliminate a YC dinucleotide from the corA leader RNA sequence. To decouple the effects of M24 and M20 on stem-loop E structure from rut site composition, we tested the alternative mutations M24∧ (G155T G156T C157G T158A) and M20∧ (G166T G167C T168A C169A). M24∧ disrupts the left arm of stem-loop E without introducing a YC dinucleotide, and M20∧ disrupts the right arm of stem-loop E but retains a YC two nucleotides upstream of its wild-type position (Figure 1). As with the original mutations, M24∧ and M20∧ both promote Rho-dependent termination at U192, and the combination of M24∧+M20∧ restored the wild-type termination locations (Supplementary Figure S7A). In addition, combination of M24∧ with M20* (T168A C169T), which does not restore stem-loop E base-pairing potential, did not restore the wild-type termination pattern (Supplementary Figure S7B). Together, these results indicate that stem-loop E itself, rather than the nucleotide sequences disrupted by the M24 and M20 mutations, prevents Rho from accessing transcription complexes paused at U192.
Importantly, templates harboring the M24 and/or M20 mutations retain wild-type RNAP pausing at U192 (Figure 5B), indicating that stem-loop E prevents termination at U192 by a mechanism other than inhibition of RNAP pausing at this site. This result suggests that stem-loop E instead influences the Rho-RNA interaction. One possibility is that stem-loop E prevents the Rho PBS from accessing the YC dinucleotides at position 152–153 and 168–169 by sequestering them within the RNA hairpin (Figure 1), thus forcing Rho to utilize downstream YC dinucleotides such as C175-177 and resulting in termination at downstream sites. However, this explanation is not sufficient because Rho-dependent termination at U192 was observed in the presence of mutations that both disrupt stem-loop E and remove the YC dinucleotides from position 152–153 and 168–169 (M20*+M19, Figure 5C). This result indicates that termination at U192 resulting from disruption of stem-loop E is not a consequence of improved Rho access to the YC dinucleotides within stem-loop E.
Stem-loop E promotes Rho activity in a rut site-independent manner
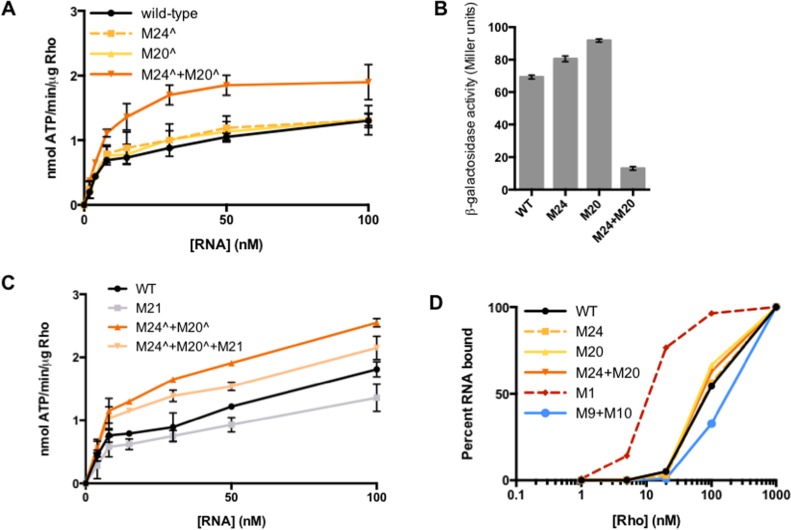
To further investigate how stem-loop E affects the Rho-RNA interaction, we tested the ability of corA leader mRNA harboring mutations M24∧ and/or M20∧ to stimulate Rho ATPase activity in vitro. M24∧ or M20∧ alone, which prevent stem-loop E formation, did not significantly affect ATPase activity (Figure 6A). By contrast, combining M24∧+M20∧ to restore stem-loop E base-pairing potential significantly increased Rho ATPase activity (Figure 6A). A possible interpretation of these results is that stem-loop E is not the predominate conformation under the conditions tested (thus, disrupting base-pairing would have little effect), and that M24∧+M20∧ forces adoption of stem-loop E, which, in turn, promotes Rho activity. This result is consistent with the ability of M24+M20 to significantly increase the efficiency of Rho-dependent termination in vitro (Figure 5A) and significantly repress expression in vivo (Figure 6B). Together, these results indicate that stem-loop E specifically enhances the Rho-RNA interaction.
Figure 6.
Stem-loop E promotes Rho activity in a rut site-independent manner. (A) The accumulation of free phosphate was monitored to quantify Rho ATPase activity. Purified Rho protein was incubated with RNA corresponding to the wild-type corA leader mRNA or variants harboring mutations in stem-loop E (Figure 1). Data shown correspond to mean values and standard deviation of technical duplicates and are representative of three independent experiments. (B) β-galactosidase activity (Miller units) from wild-type Salmonella (14028s) harboring plasmid pYS1040 or derivatives harboring mutations that disrupt or restore stem-loop E base-pairing potential (Figure 1). Data shown indicate mean values and standard deviations from an experiment performed in biological duplicate that is representative of three independent experiments. (C) The accumulation of free phosphate was monitored to quantify Rho ATPase activity. Purified Rho protein was incubated with RNA corresponding to the wild-type corA leader mRNA or variants harboring mutations in stem-loop E and/or the C-rich region at position 175–177 (Figure 1). Data shown correspond to mean values and standard deviation of technical duplicates and are representative of three independent experiments. (D) Percentage of 5′ radiolabeled RNA bound by Rho and retained following vacuum filtration. RNAs correspond to the wild-type corA leader (black) or mutant derivatives containing mutations in stem-loop E (yellow, orange), stem-loop C (red, positive control) or the rut site (blue, negative control). Data shown are from a single experiment that is representative of three independent experiments.
We next sought to determine how stem-loop E promotes Rho activity. A possible function of stem-loop E could be to bring disparate rut site elements into a favorable spatial orientation. For example, if stem-loop E enhances the Rho-RNA interaction by positioning the C175-177 YC dinucleotide near the rest of the rut site (Figure 1), then mutation of this YC dinucleotide should reverse the ability of a mutant RNA locked in stem-loop E to promote Rho ATPase activity in vitro. However, mutation of C175-177 (M21) in a stem-loop E-locked background (M24∧+M20∧) only slightly decreased Rho ATPase activity relative to M24∧+M20∧ (Figure 6C). That disruption of C175-177 is additive with rather than epistatic over locking the RNA in stem-loop E suggests that the mutations affect independent aspects of Rho activity. Thus, the ability of stem-loop E to promote Rho activity cannot be attributed to its effect on the orientation of C175-C177.
The effect of stem-loop E on Rho-dependent termination is independent of the RNA structures that regulate accessibility of upstream rut site components (stem-loops B versus C+D) because in vitro, formation of stem-loops B versus C+D influences the efficiency, rather than the location, of Rho-dependent termination (3), whereas stem-loop E affects the location and only modestly impacts termination efficiency (Figure 5A). Moreover, the effects of combining mutations that disrupt stem-loop E with those that favor stem-loop B (M1) or stem-loops C+D (M3) are additive (Supplementary Figure S8), thus indicating that these mutations operate in different pathways. Since disruption of stem-loop E has effects in both stem-loop B-locked and stem-loop C+D-locked backgrounds (Supplementary Figure S8), formation of stem-loop E appears to be independent of whether the upstream RNA adopts stem-loop B or stem-loops C+D.
Because the abilities of stem-loop E to promote Rho activity and influence the location of termination appear to be rut site-independent, we hypothesized that stem-loop E influences step(s) of the Rho-RNA interaction after initial Rho PBS-rut site binding has taken place. If this is the case, then alteration of stem-loop E should have no impact on the affinity of Rho for the corA leader mRNA. To test this possibility, we measured the ability of purified Rho protein to retain radiolabeled corA leader mRNA, or derivatives containing mutations in stem-loop E, on a filter. RNAs harboring M24, M20 and M24+M20 all yielded binding curves very similar to that of the wild-type RNA (Figure 6D), indicating that these mutations do not affect the ability of Rho to bind to corA leader mRNA. By contrast, Rho displayed a significantly higher affinity for M1 RNA, which constitutively exposes the rut site, and a lower affinity for M9+M10 RNA, in which key rut site nucleotides have been mutated (Figure 6D) (3). Therefore, we conclude that stem-loop E promotes Rho activity by influencing step(s) subsequent to rut site recognition.
DISCUSSION
We have now established that the corA leader controls at least three aspects of Rho-dependent transcription termination. We previously reported that formation of stem-loop B facilitates the initial Rho-RNA interaction by exposing sequences comprising a rut site, whereas adoption of the alternative conformation, stem-loops C+D, reduces accessibility of the rut site (3). Our new findings demonstrate that stem-loops C and D also reduce the efficiency of RNAP pausing at a single site over 100 nt downstream. To our knowledge, this is the first description of RNA structures acting over such a long distance to modulate efficiency of a specific pause. In addition, stem-loop E affects the location and overall efficiency of Rho-dependent termination. Specifically, formation of stem-loop E prevents Rho from accessing RNAP complexes stalled at a major pause site, leading to termination farther downstream. Together, the results show how a single leader RNA can regulate three independent features of a Rho-dependent terminator, revealing multiple points at which physiological signals may regulate corA expression.
The same RNA structures that modulate rut site accessibility also regulate efficiency of RNAP pausing at a single site over 100 nt downstream
The corA leader conformation that sequesters the rut site (stem-loops C+D) suppresses RNAP pausing at U240, which is over 100 nt downstream (Figure 3). In general, pause regulation by RNA structures occurs over much shorter distances. For example, short RNA hairpins can stabilize pauses as they emerge from the RNAP exit channel, but only within a strict spacing window (∼11 nt) due to the requirement for interaction of the nascent RNA stem-loop with the flap domain of RNAP (19). Because RNA oligonucleotides can stabilize pausing in trans by base pairing with nascent RNA near the RNAP exit channel (34), it is feasible that an upstream portion of the corA leader could base pair with sequences emerging from RNAP as it approaches T240 to control pausing. Alternatively, suppression of pausing by stem-loops C and D could be due to propagation of structural information along the RNA via unknown elements. The latter possibility may be less likely because the only known intervening RNA structure (i.e. stem-loop E) does not influence RNAP pausing at U240 (Figure 5B), and its formation is independent of whether the RNA adopts stem-loops B versus C+D (Supplementary Figure S8).
An additional possibility is that the corA leader regulates RNAP pausing by interacting directly with the transcription elongation complex. For example, a structured region of bacteriophage HK022 putL RNA appears to bind the β′ subunit of RNAP to suppress pausing 21–22 nt downstream by limiting the extent of RNAP backtracking (21,35). In addition, the EAR RNA element in Bacillus subtilis can inhibit intrinsic transcription termination over 1 kb downstream and was predicted to interact with RNAP (36), although regulation by the EAR element could not be recapitulated in vitro and thus requires factor(s) in addition to the RNA structure. Whereas putL and EAR both modify the transcription elongation complex to cause processive antitermination, the corA leader conformation alters RNAP pausing only at a single site (Figure 3A). As a result, it is unlikely that the mechanism by which stem-loops C+D suppress pausing is analogous to the EAR or put elements.
Like corA, the mgtA leader in Salmonella regulates both rut site accessibility and RNAP pausing at a site of Rho-dependent termination (U218) (20). In the case of mgtA, the DNA sequence directly downstream of T218 is required for pause entry, and the pause is stabilized by an RNA stem-loop that is similar to canonical pause hairpins in terms of size and distance from the pause site (20). Intriguingly, mgtA's stem-loop B (which presents an accessible rut site analogously to corA's stem-loop B) further stabilizes the U218 pause, even though it is located 57 nt upstream (20). Thus, the corA and mgtA leaders both utilize features of a single RNA conformation to influence rut site accessibility and RNAP pausing at a site unusually far downstream. In both cases, the RNA structures that present an accessible rut site are associated with enhanced RNAP pausing. However, these pauses differ in that the mgtA leader specifically affects pause duration (i.e. exit from the pause state), while the corA leader controls pause efficiency (i.e. entry into the pause state).
What is the physiological relevance of regulated pause efficiency at U240 in the corA leader? Pause suppression at this site should naturally enhance the ability of stem-loops C and D to promote transcription elongation into the corA coding region by reducing the number of RNAP complexes accessible to Rho. In accordance with this notion, elimination of pausing at U240 further de-repressed expression of a corA-lacZ fusion harboring a mutation favoring formation of stem-loops C+D (Supplementary Figure S5D). Efficient RNAP pausing at U240 is not required for stem-loop B to fully promote Rho-dependent termination within the corA leader (Supplementary Figure S5B and C). By contrast, modulation of RNAP pausing at U218 in the mgtA leader is required for regulation of transcription elongation into the coding region (20). This is likely because U218 is the only site in the mgtA leader at which Rho can trigger transcription termination, whereas for corA, Rho loading leads to termination at many sites besides U240 (Figure 2B).
U240 lies within the corA start codon (Figure 1). RNAP pausing within start codons is widespread due to the similarity of typical pause sequences (G-10 Y-1 G+1) to translation initiation signals (32). Such pausing was proposed to promote translation by preventing formation of secondary structures that sequester the RBS (32). However, RNAP occupancy of the start codon precludes ribosome loading until RNAP resumes transcription. As a result, suppression of pausing with the corA start codon by stem-loops C+D might favor corA translation by preventing RNAP from occluding the RBS. Therefore, formation of stem-loops C+D would favor both transcription of the corA coding region and translation of the corA ORF.
Regulation of late step(s) of Rho loading by stem-loop E dictates the efficiency and location of transcription termination
Formation of stem-loop E prevents Rho from accessing RNAP complexes paused at U192, resulting in Rho-dependent termination taking place at downstream sites (Figure 5A). Stem-loop E appears to control step(s) after rut site recognition (e.g. RNA-Rho SBS interactions or Rho translocation) because it had no effect on RNAP pausing (Figure 5B) or the affinity of Rho for corA leader mRNA (Figure 6D). This notion is consistent with a previous report that amino acid substitutions in the Rho SBS can alter the location of transcription termination (37).
RNA structures within other Rho-dependent terminators have been shown to influence the location of transcription termination. For example, introduction of an RNA hairpin (boxB) into the rut site of the trp t′’ terminator shifted the sites of Rho-dependent termination downstream (38). In this case, the position of Rho-dependent termination was not dictated by kinetic competition between Rho translocation and transcription elongation because the sites of termination did not shift upstream when RNAP was slowed down by reducing the concentration of NTPs (14). Instead, it was proposed that Rho may ‘loop out’ structured regions, effectively increasing the length of RNA that must be synthesized before loading can be completed (14). Crystallographic data supports this notion: double-stranded RNA is too bulky to pass into Rho's central channel (7). Such a model is consistent with the possibility that stem-loop E prevents Rho from completing the loading process until downstream of U192.
An alternative possibility is that Rho does complete loading upstream of stem-loop E, but must ‘step around’ the structured region in a manner that prevents termination of transcription at U192. For example, translocating Rho molecules bypassed an artificial RNA roadblock located between a rut site and Rho-dependent termination sites without unwinding it, suggesting that Rho accommodates RNA structures in a ‘composite’ primary binding site and re-establishes translocation downstream (15). Thus, even if Rho completely loads upstream of stem-loop E, it may be that stem-loop E forces the Rho-RNA complex to rearrange in such a way that the protein does not regain termination competence until after U192.
That the corA leader mRNA controls its interaction with Rho after initial binding has taken place (Figure 6D) is reminiscent of how the mgtCBR leader operates (26), suggesting that regulation of steps subsequent to rut site recognition may be a general phenomenon (22). The Salmonella mgtCBR leader can adopt two mutually exclusive conformations that expose or sequester an RNA motif with the capacity to antagonize Rho activity, without affecting rut site recognition (26). Such RNA motifs may enhance the specificity of the Rho-RNA interaction beyond that provided by the rut site (26). This notion agrees with the observation that Rho is associated with most nascent RNAs but only terminates transcription of a fraction of them (39).
What is the physiological relevance of preventing Rho-dependent termination at U192? One possibility is that longer corA transcription termination products generated by prevention of transcription termination at U192 possess regulatory function(s). For example, such products might act in trans as sRNAs, as has been observed for the truncated RNA generated by S-adenosyl methionine riboswitch action in Listeria monocytogenes (40), and this function might require sequences between position 192 and the normal termination sites at positions ∼220–250.
Stem-loops B-E regulate three independent steps of Rho-dependent termination
We previously demonstrated that adoption of stem-loop B increases accessibility of the rut site in the corA leader, whereas stem-loops C+D sequester nucleotides required for Rho loading (3). Because YC dinucleotides downstream of stem-loop D were found to promote Rho activity (Figure 4), we now conclude that stem-loop E is also located within the rut site. Interestingly, stem-loop E promotes Rho activity (Figure 6A) despite its ability to prevent Rho-dependent termination at a particular site (Figure 5A) and despite the fact that it sequesters YC dinucleotides that the Rho PBS appears to recognize (Figures 1 and 4). Its location notwithstanding, the effects of stem-loop E were determined to be rut site-independent (Figures 5C and 6C and D). Stem-loops B versus C+D are the major determinants of rut site accessibility because these upstream structures more strongly affect Rho-dependent termination efficiency in vitro (Supplementary Figure S8A) and affect the Rho-RNA interaction at an earlier step (Figure 6D) compared to stem-loop E.
Stem-loop E has the potential to form in both the stem-loop B and stem-loops C+D conformations (Figure 1). Moreover, mutations that hinder stem-loop E base pairing have effects in both stem-loop B-locked and stem-loop C+D-locked backgrounds (Supplementary Figure S8), suggesting that formation of stem-loop E is independent of the upstream RNA conformation. Thus, it remains unclear whether and how formation of stem-loop E is regulated. We noticed that stem-loop E might compete with potential long-distance base pairing between C47-A54 and U148-G155, especially in the C+D conformation (Figure 1, Supplementary Figure S1). Such competition could explain why wild-type corA leader RNA appears to adopt stem-loop E in the kinetically driven transcription termination assay (Figure 5A) but not in the thermodynamically driven Rho ATPase assay (Figure 6A).
In sum, the corA leader mRNA regulates three distinct steps of Rho-dependent transcription termination, namely rut site recognition, Rho translocation dynamics and RNAP pausing, that determine whether transcription proceeds into the corA coding region (Figure 7). Specifically, formation of stem-loops B versus C+D initially determines the accessibility of upstream elements within the rut site to the Rho PBS. Next, stem-loop E influences later step(s) of Rho loading and/or translocation to influence the location and overall efficiency of Rho-dependent termination. Lastly, stem-loops B versus C+D also dictate the efficiency of RNAP pausing within the corA start codon, which may enhance the ability of these structures to modulate Rho-dependent termination efficiency and/or affect the efficiency of corA translation initiation.
Figure 7.
The corA leader RNA controls three steps of Rho-dependent transcription termination. Once RNAP (light blue) begins transcribing the corA gene, cytoplasmic signal(s) can influence whether the corA leader RNA adopts the stem-loop B conformation or the stem-loops C+D conformatiion. The stem-loop B conformation presents an accessible rut site (dark blue), whereas stem-loops C+D sequester nucleotides specifically recognized by Rho (yellow). Stem-loops C+D also reduce the efficiency of RNAP pausing at U240 (red star). Stem-loop E (orange), whose formation is independent of stem-loops B versus C+D, prevents Rho from accessing RNAP complexes paused at U192 (green star), thus promoting termination at downstream sites. Black lines represent RNA and grey lines represent DNA.
Regulation of multiple steps may facilitate fine-tuning of Rho-dependent termination
Beyond contributing to understanding of how structured regions influence interactions of RNA with Rho and RNAP, our findings reveal multiple steps at which distinct physiological signals could influence the efficiency of Rho-dependent termination. Regulation of RNAP pausing efficiency and Rho's ability to access RNAP complexes paused at particular sites is likely to have more subtle effects compared to modulation of rut site accessibility, thus allowing for tight control of termination efficiency. These features may be general properties of Rho-dependent terminators involved in gene regulation, whose efficiencies must be responsive to changing cellular conditions (22).
Acknowledgments
The authors thank Anastasia Sevostyanova for insightful discussions, technical advice and providing purified Salmonella Rho, RNA polymerase and σ70 proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [5T32AI007640-10, 5T32GM007223-38 and AI49561]; Yale University [E.A.G.]. Funding for open access charge: Yale University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Peters J.M., Mooney R.A., Kuan P.F., Rowland J.L., Keles S., Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters J.M., Mooney R.A., Grass J.A., Jessen E.D., Tran F., Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriner M.A., Groisman E.A. The Bacterial Transcription Termination Factor Rho Coordinates Mg(2+) Homeostasis with Translational Signals. J. Mol. Biol. 2015;427:3834–3849. doi: 10.1016/j.jmb.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dombroski A.J., Platt T. Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2538–2542. doi: 10.1073/pnas.85.8.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briercheck D.M., Wood T.C., Allison T.J., Richardson J.P., Rule G.S. The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions. Nat. Struct. Biol. 1998;5:393–399. doi: 10.1038/nsb0598-393. [DOI] [PubMed] [Google Scholar]
- 6.Ray-Soni A., Bellecourt M.J., Landick R. Mechanisms of bacterial transcription termination: All good things must end. Annu. Rev. Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- 7.Skordalakes E., Berger J.M. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 8.Koslover D.J., Fazal F.M., Mooney R.A., Landick R., Block S.M. Binding and translocation of termination factor rho studied at the single-molecule level. J. Mol. Biol. 2012;423:664–676. doi: 10.1016/j.jmb.2012.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan W.D., Bear D.G., von Hippel P.H. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J. Biol. Chem. 1983;258:9565–9574. [PubMed] [Google Scholar]
- 10.Richardson J.P. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J. Biol. Chem. 1982;257:5760–5766. [PubMed] [Google Scholar]
- 11.Wei R.R., Richardson J.P. Identification of an RNA-binding Site in the ATP binding domain of Escherichia coli Rho by H2O2/Fe-EDTA cleavage protection studies. J. Biol. Chem. 2001;276:28380–28387. doi: 10.1074/jbc.M102444200. [DOI] [PubMed] [Google Scholar]
- 12.Gocheva V., Le Gall A., Boudvillain M., Margeat E., Nollmann M. Direct observation of the translocation mechanism of transcription termination factor Rho. Nucleic Acids Res. 2015;43:2367–2377. doi: 10.1093/nar/gkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinmetz E.J., Brennan C.A., Platt T. A short intervening structure can block rho factor helicase action at a distance. J. Biol. Chem. 1990;265:18408–18413. [PubMed] [Google Scholar]
- 14.Zhu A.Q., von Hippel P.H. Rho-dependent termination within the trp t' terminator. II. Effects of kinetic competition and rho processivity. Biochemistry. 1998;37:11215–11222. doi: 10.1021/bi972912s. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz A., Walmacq C., Rahmouni A.R., Boudvillain M. Noncanonical interactions in the management of RNA structural blocks by the transcription termination rho helicase. Biochemistry. 2007;46:9366–9379. doi: 10.1021/bi700493m. [DOI] [PubMed] [Google Scholar]
- 16.Epshtein V., Dutta D., Wade J., Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudvillain M., Figueroa-Bossi N., Bossi L. Terminator still moving forward: Expanding roles for Rho factor. Curr. Opin. Microbiol. 2013;16:118–124. doi: 10.1016/j.mib.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee S., Chalissery J., Bandey I., Sen R. Rho-dependent transcription termination: more questions than answers. J. Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 19.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 20.Hollands K., Sevostiyanova A., Groisman E.A. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1999–E2007. doi: 10.1073/pnas.1319193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komissarova N., Velikodvorskaya T., Sen R., King R.A., Banik-Maiti S., Weisberg R.A. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol. Cell. 2008;31:683–694. doi: 10.1016/j.molcel.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kriner M.A., Sevostyanova A., Groisman E.A. Learning from the Leaders: Gene regulation by the transcription termination factor Rho. Trends Biochem. Sci. 2016;41:690–699. doi: 10.1016/j.tibs.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konan K.V., Yanofsky C. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J. Bacteriol. 2000;182:3981–3988. doi: 10.1128/jb.182.14.3981-3988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Bossi N., Schwartz A., Guillemardet B., D'Heygère F., Bossi L., Boudvillain M. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 2014;28:1239–1251. doi: 10.1101/gad.240192.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollands K., Proshkin S., Sklyarova S., Epshtein V., Mironov A., Nudler E., Groisman E.A. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevostyanova A., Groisman E.A. An RNA motif advances transcription by preventing Rho-dependent termination. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6835–E6843. doi: 10.1073/pnas.1515383112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shashni R., Qayyum M.Z., Vishalini V., Dey D., Sen R. Redundancy of primary RNA-binding functions of the bacterial transcription terminator Rho. Nucleic Acids Res. 2014;42:9677–9690. doi: 10.1093/nar/gku690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields P.I., Swanson R.V., Haidaris C.G., Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 30.Artsimovitch I., Henkin T.M. In vitro approaches to analysis of transcription termination. Methods. 2009;47:37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landick R., Wang D., Chan C.L. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 32.Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vvedenskaya I.O., Vahedian-Movahed H., Bird J.G., Knoblauch J.G., Goldman S.R., Zhang Y., Ebright R.H., Nickels B.E. Transcription. Interactions between RNA polymerase and the ‘core recognition element’ counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolb K.E., Hein P.P., Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen R., King R.A., Weisberg R.A. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol. Cell. 2001;7:993–1001. doi: 10.1016/s1097-2765(01)00243-x. [DOI] [PubMed] [Google Scholar]
- 36.Irnov I., Winkler W.C. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol. Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- 37.Tsurushita N., Shigesada K., Imai M. Mutant rho factors with increased transcription termination activities. I. Functional correlations of the primary and secondary polynucleotide binding sites with the efficiency and site-selectivity of rho-dependent termination. J. Mol. Biol. 1989;210:23–37. doi: 10.1016/0022-2836(89)90288-x. [DOI] [PubMed] [Google Scholar]
- 38.Zhu A.Q., von Hippel P.H. Rho-dependent termination within the trp t' terminator. I. Effects of rho loading and template sequence. Biochemistry. 1998;37:11202–11214. doi: 10.1021/bi9729110. [DOI] [PubMed] [Google Scholar]
- 39.Mooney R.A., Davis S.E., Peters J.M., Rowland J.L., Ansari A.Z., Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh E., Dussurget O., Gripenland J., Vaitkevicius K., Tiensuu T., Mandin P., Repoila F., Buchrieser C., Cossart P., Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]