Abstract
Intrinsically disordered proteins (IDPs) are an emerging concept. IDPs have high flexibility in their polypeptide chains, lacking a stable 3-dimensional structure. Because of the difficulty in performing X-ray crystallography for IDPs, nuclear magnetic resonance (NMR) spectroscopy is the first choice for atomic-level investigation of their nature. Given that isotopically labeled IDP samples are necessary for NMR study, a robust and cost-effective protocol for bacterial expression and purification of IDP is also needed. We employed the Npro (EDDIE)-autoprotease fusion protein system. Although IDPs are believed to be readily degraded by endogenous proteases when expressed in Escherichia coli, Npro-fused IDPs showed excellent resistance to degradation. Seven IDPs of uncharacterized function sampled from the human genome as well as 3 constructs from IDP regions derived from human FancM and Thermococcus kodakarensis Hef were prepared. We improved the protocol of refolding of Npro (EDDIE) to use dialysis, which is convenient for subsequent purification using reversed-phase (RP) HPLC. The method is robust and widely applicable to any IDP sample, promoting the acquisition of experimental data for IDPs in a high-throughput manner.
Introduction
The emerging concept of an intrinsically disordered protein (IDP) is a key for studying protein sequence–function relationships.1–3 Historically, even under physiological conditions, IDPs (and “disordered regions”) are found to lack standard secondary structures in their polypeptide chains, as deduced by X-ray and NMR methods.4–7 Recently, some IDPs were shown to be more conformationally heterogeneous, since the presence of substantial secondary structure that may behave as molten globules has been proven (For a review see refs.3,8,9). These evidences have been accumulated in a number of highly-curated databases, such as DisProt,10,11 IDEAL,12,13 MobiDB,14 MobiDB 2.0,15 D2P2,16 and pE-DB.17 The growing awareness of IDPs has led to an increasing number of reports that IDPs play indispensable roles in various biological processes such as transcription and translation, as well as in signaling cascades. Although many researchers believe that IDPs carry a high degree of flexibility and conformational polymorphism in their polypeptide chains, their physical and physicochemical properties have not been fully understood. Reflecting the substantial difficulty underlying X-ray crystallography of IDPs in accepted facts, solution NMR methodology has become more attractive for physicochemical studies of IDPs. The growing interest in structure and function of IDPs requires the development of a robust strategy for preparing multiple IDP samples for NMR, including isotopic labeling and bacterial recombinant protein production.
In a previous study, we developed a thioredoxin fusion protein-based expression plasmid, pET-TRX-PRESAT, which is competent for high-throughput vector construction using unidirectional PCR cloning technology.18 The system was carefully selected for avoiding digestion of products by endogenous proteases based on thioredoxin (TRX) fusion system.19 TRX has an internal noncanonical signal peptide sequence, leading to its autonomous secretion to the periplasmic space of Escherichia coli, where bacterial endogenous proteases are less active.19 However, when using the technic of selective expression into the periplasmic space, freshly harvested cells should be carefully subjected to a protocol called “osmotic shock treatment,” and the final yield of the product was not always high.18 Such risks should be taken into consideration when IDPs are bacterially expressed. As another approach, Livernois et al. reported the “boiling lysis” method for obtaining highly purified IDP samples from recombinant bacteria by the rapid inactivation of bacterial proteases that are harmful for IDPs of interest.20 The idea is elegant, but the method does not protect IDPs from degradation during bacterial growth.
In this study, we employed the Npro (EDDIE) fusion protein expression system for preparing isotopically labeled NMR-ready IDP samples. The Npro fusion technology was originally developed by Auer et al. for producing recombinant proteins with native amino-terminal amino acids.21 The N-terminal autoprotease Npro from classical swine fever virus was selected as a fusion partner. The Npro (EDDIE) mutant was further developed.21 The fusion protein of Npro (EDDIE) and the protein of interest is first deposited in bacterial inclusion bodies and then refolded. During the refolding process, Npro (EDDIE) recovers its autoproteolytic activity to cleave its own C-terminus, thereby releasing the protein of interest with its native N-terminal amino acid. We focused on the tendency of inclusion body formation of the Npro (EDDIE) fusion proteins, which favors the protection of IDPs during bacterial culture. An escape of fusion proteins from the cytosol to the inclusion body is a way to avoid proteolytic degradation during bacterial expression. As a result, we succeeded in preparing several NMR-ready IDP samples with reasonable time, cost, and effort.
Materials and Methods
Materials
The restriction enzymes NdeI, SpeI, XhoI, and BamHI (New England Biolabs, Ipswich, MA, USA) were used. T4 DNA ligase, Wizard plus SV Minipreps DNA purification, and Wizard SV Gel and PCR Clean-up systems (Promega, Madison, WI, USA) were used for ligation and DNA purification. Oligonucleotide primers were obtained from Hokkaido System Science Co., Ltd. (Hokkaido, Japan). pET21b was purchased from Merck KGaA/Novagen (Darmstadt, Germany). His-AcceptTM was obtained from Nacalai Tesque Inc., (Kyoto, Japan). For PCR cloning, rTaq (Takara Bio Inc., Otsu, Japan) was used. [15N]- NH4Cl was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). All other biochemical reagents were purchased from Nacalai Tesque, Inc. (Kyoto, Japan).
Vector construction
The gene encoding Hisx6-tagged Npro (EDDIE) was designed with an E. coli optimized codon table and was chemically synthesized (Hokkaido System Science Co., Ltd.) according to the original paper.21 This Hisx6-tagged Npro gene was ligated downstream of the T7 promoter of pET-21b between NdeI and BamHI sites. A SpeI site preceding the autoproteolytic cleavage site Cys168 and a short linker containing several restriction sites were engineered as a multi-cloning site, resulting in the pET-Npro vector (Supplementary Fig. 1). The genes encoding model IDPs selected from the human genome were then inserted into the pET-Npro vector by a standard PCR cloning method from plasmids carrying the corresponding genes as templates, as previously described. The genes encoding the disordered regions from human FancM and Thermococcus kodakarensis Hef were also PCR cloned into the parent pET-Npro vector from the plasmids carrying Hef genes that were previously described.22 The amino acid sequences of all IDPs used in this study are presented in Supplementary Table 1.
Protein techniques
E. coli BL21 (DE3) transformed by pET-Npro-IDP vectors was grown in either LB medium or modified M9 minimal medium, the latter of which contained 0.5 g/L [15N]-NH4Cl as the sole nitrogen source at 37°C. Expression of the recombinant proteins was induced with IPTG (1 mM) when the cell density reached an OD (600 nm) of approximately 0.7–0.8. Cells were harvested 4 h after induction, and the cell extracts were analyzed by 15% SDS-PAGE. Purification of chimeric Npro-IDP fusion proteins was performed as follows. The cells were broken by sonication in a buffer containing 20 mM sodium-phosphate, 75 mM (for H1–H4) or 300 mM (for other IDPs) NaCl, and 5 mM EDTA (pH 8.0) and centrifuged to collect the pellet fraction. The pellet (inclusion body) was washed once with the same buffer supplemented with 0.2% Triton X-100. The inclusion body was then solubilized in a buffer containing 8 M Urea, 25 mM dithiothreitol (DTT), and 50 mM Tris-HCl (pH 7.5) by stirring overnight at 4°C. Then the denatured fusion proteins were purified by immobilized metal affinity chromatography using Ni2+-loaded His-AcceptTM (Nacalai Tesque, Inc.). Before the protein solution was charged onto the His-Accept column, the solution was diluted 5 times with buffer containing 8 M urea and 50 mM Tris-HCl (pH 7.5) to lower the DTT concentration to 5 mM. The samples were eluted using stepwise increasing concentrations (100–500 mM) of imidazole buffer containing 8 M urea and 50 mM Tris-HCl (pH 7.5). The eluents (approximately 20 mL per 1 L culture) were then dialyzed using Spectra/Por “6” regenerated cellulose membrane (MWCO 1000, Spectrum Laboratories, Inc., Rancho Dominguez, CA, USA) against the buffer containing 1 M Tris-HCl, 5% glycerol, 2 mM EDTA, and 10 mM DTT (pH 7.5) twice at 4°C for 16 h each time. The dialyzed solutions were additionally incubated for 12–24 h at room temperature for completing autocleavage of Npro (EDDIE) (up to 90%). The solution was further dialyzed against 20 mM Tris-HCl (pH 7.5) at 4°C for an additional 16 h. Finally, the IDP samples of interest were purified by C18 RP-HPLC on COSMOSIL® 5C18 -AR-300 (ϕ4.6 mm × 250 mm, Nacalai Tesque, Inc., Kyoto, Japan). A standard 0.1% TFA–water–acetonitrile solvent system was used for the all samples.
NMR spectroscopy
NMR experiments were performed on a Bruker Avance III 600 MHz spectrometer equipped with a cryogenic probe. Approximately 0.2 mM of 15N-labeled IDPs was dissolved in 0.3 mL of 90%-10% H2O‒D2O containing 20 mM sodium phosphate buffer (pH 6.5) with or without 0.5 M NaCl. SOFAST-HMQC spectra23 for 0 M NaCl samples and FHSQC spectra24 for 0.5 M NaCl samples were acquired with 4–32 transients and 256 increments at 298 K, depending on the signal-to-noise ratios of observed signals and zero-filled during spectral processing. All two-dimensional spectra were processed with nmrPipe and visualized with the program nmrDraw.25
Results and Discussion
Controlling DTT concentration
To establish a robust expression and purification protocol for IDP samples based on the Npro (EDDIE) fusion protein system, we first introduced IMAC purification under denatured conditions because IMAC purification is applicable even in the presence of a high concentration of denaturant such as 8 M urea. However, for completely solubilizing the Npro-IDP fusion proteins, not only a denaturant but also a reducing agent such as 25 mM DTT was necessary. Although several commercially available IMAC supports are not compatible with DTT, some resins including His-AcceptTM may work in the presence of DTT up to 5 mM. For simplicity, we have chosen to show an example of the expression and purification of sample IDP(H1) from the T. kodakarensis Hef protein (Fig. 1). Approximately 4 mL of His-AcceptTM resin per fusion protein sample derived from 1 L of E. coli culture was sufficient to capture approximately 95% of the Hisx6-tagged Npro-IDP fusion protein (data not shown). The Npro-IDP(H1) fusion protein was purified by elution with stepwise increasing imidazole concentrations (100–500 mM, Fig. 1B, lane 3–7).
Figure 1.
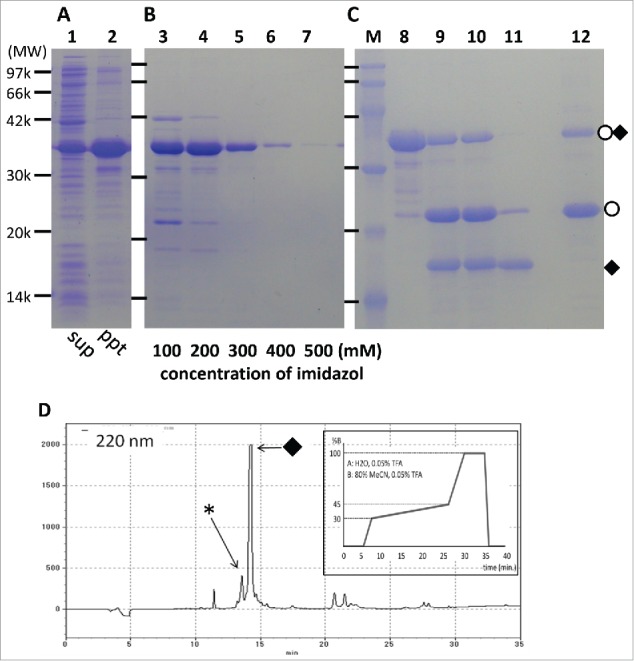
Examples from the expression–purification step of an Npro (EDDIE)-IDP(H1) fusion protein analyzed by SDS-PAGE (15% gel). (A) sup or ppt indicates soluble or pellet fraction of sonicated extract from IPTG-induced cells, respectively (lanes 1, 2). (B) Ni2+ affinity column purified Npro (EDDIE)-IDP(H1) in denatured condition (lanes 3–7) with a given imidazole concentration. (C) dialysis and autocleavage of Npro (EDDIE)-IDP(H1). MW markers (lane M), before refolding (lane 8), after the first dialysis step (lane 9), after an additional dialysis step (lane 10), supernatant after without salt/DTT buffer dialysis (lane 11), and pellet of the same dialysis (lane 12). (D) A chart of RP-HPLC purification of IDP(H1). Inset is a acetonitrile gradient profile. In (C) and (D), open circle and closed diamond represent Npro and IDP(H1), respectively. In (D), asterisk indicates a partially degraded IDP(H1).
Optimization of dialysis protocol
Next, we introduced a dialysis protocol for Npro refolding and autocleavage instead of the original dilution method, given that subsequent purification is by RP-HPLC, which may limit the working volume of the peptide solution. The buffer containing 5% glycerol with high (1 M) Tris-HCl, similar to that used in the original report, was found to be sufficient even for the refolding without any visible precipitation. During this refolding, autocleavage of Npro (EDDIE) tag and release of IDPs of interest also continued. After two sets of 16 h of dialysis, the self-cleaving reaction proceeded to roughly 70%. Examples of refolding and autocleavage of IDP(H1) are shown in Figure 1C.
Removal of cleaved Npro (EDDIE) by precipitation or second IMAC
The next step was separation of the IDP peptides from the cleaved Npro tag and the remaining uncleaved fusion protein by RP-HPLC. Given that the refolding buffer contains 5% glycerol, we further dialyzed the sample against a buffer suited for RP-HPLC. During the development of the dialysis step, we found that the cleaved Npro, but not the cleaved IDPs, tended to precipitate when dialyzed against no salt/DTT buffer, such as 25 mM Tris-HCl, pH 7.5, without salt. After centrifugation, 80–90% of the cleaved Npro was removed from the IDP sample (Fig. 1C, lanes 11, 12). Note that a similar removal effect was obtained when the cleaved mixture was applied to the second IMAC (data not shown). Thus, these 2 protocols may both contribute to improving the purification efficiency at the next RP-HPLC step. A profile of the subsequent RP-HPLC is shown in Figure 1D. A summary of the expression/purification protocol of IDPs using the Npro (EDDIE) fusion system is shown in Table 1.
Table 1.
Dialysis-based procedure for an IDP sample from inclusion bodies using the Npro (EDDIE) fusion protein expression system
| 1. Pick 3 to 5 colonies of E. coli BL21 (DE3) and add them to 1 mL LB medium, and grow the culture overnight at 37°C. |
| 2. Add 0.5 mL of the bacterial culture to 100 mL of the medium (either M9 medium containing 15NH4Cl or LB depending on the planned NMR experiments) and incubate at 37°C for 12–16 h (pre-culture medium). |
| 3. Add the pre-culture medium to 900 mL medium and incubate at 37°C until the OD (600 nm) of the culture is 0.6–0.7 (main culture medium). |
| 4. Induce protein expression with the addition of 1.0 mM IPTG (final concentration). |
| 5. Incubate the culture at 37°C for an additional 4 h. |
| 6. Harvest the cells. |
| 7. Disrupt the cells by sonication in the buffer containing 20 mM sodium-phosphate buffer (pH 8), 75 or 300 mM NaCl, 5 mM EDTA (sonication buffer). The buffer volume is 10–20 mL per 1 g wet E. coli cells. |
| 8. Centrifuge and collect the pellet fraction (IBs). |
| 9. Wash IBs with the sonication buffer supplemented with 0.1% Triton-X100 twice. |
| 10. Solubilize IBs with buffer containing 50 mM Tris-HCl (pH 7.5), 8 M urea, 300 mM NaCl and 25 mM DTT. Incubate the solution at 4°C overnight. |
| 11. Dilute the volume 5 times with the same buffer without DTT. The final concentration of DTT is now 5 mM. |
| 12. Apply the Npro fusion proteins to an immobilized Ni2+ affinity column His-AcceptTM. |
| 13. Wash the resin. |
| 14. Elute the sample with increasing concentrations (100, 200, 300, 400, and 500mM) of imidazole. |
| 15. Dialyze the protein sample against the refolding buffer (1 M Tris-HCl, pH 7.5, 5% glycerol, 10 mM DTT and 2 mM EDTA. Repeat this step twice. At the second step, keep sample at room temperature and allow autocleavage for another 16 h. |
| 16. Dialyze the protein sample against the precipitation buffer containing 25 mM Tris-HCl, pH 7.5 (at 4°C for 16 h). |
| 17. Purify the peptide by RP-HPLC. |
Technical merit
We succeeded in purifying 10 different IDPs (one of which was partly folded) from different genomes, such as mouse, human, and T. kodakarensis, which are not homologous in terms of their sequences and amino acid compositions (Supplementary Table 1). Typical yields of purified IDP samples from E. coli fermentation are summarized in Supplementary Table 2. The quality of the IDP segments after HPLC purification was assessed by MALDI-TOF mass spectrometry. The samples were further subjected to NMR experiments. Mass and NMR spectra of the selected IDPs are shown in Figure 2. We selected the IDP sequences from the mouse and human genomes essentially based on a combined Poodle-S, Poodle-W, and Poodle-I prediction.26,27 As expected, all of the purified IDPs except IDP(C9) exhibited a typical pattern of intrinsically disordered proteins in their HSQC spectra, in which the observed 1H chemical shift of NH signals showed very limited dispersion between 7.6 and 8.6 ppm. However, the line shapes and distribution of signal intensities varied depending on their sequence. In some spectra, fewer than expected HSQC signals were observed. This finding suggests a weak self-association between 2 IDP molecules that was beyond what the database search expected. Although we intended to dissolve such weak self-associations using a high molarity salt (0.5 M) solvent, no significant improvement was observed (Fig. 2G–L, cyan signals). We found IDP(C9) to be a “partial IDP” sample (C9), whose HSQC spectra showed a wider dispersion in 1H range. Indeed, IDP(C9) contains a WW domain with a flanking IDP linker, and its solution structure has already been published (PDB code: 2ysi) by another group. This (IDP(C9)) could be an example that the Npro (EDDIE) fusion system combined with dialysis refolding is also effective for NMR samples containing such the small folded protein segment.
Figure 2.
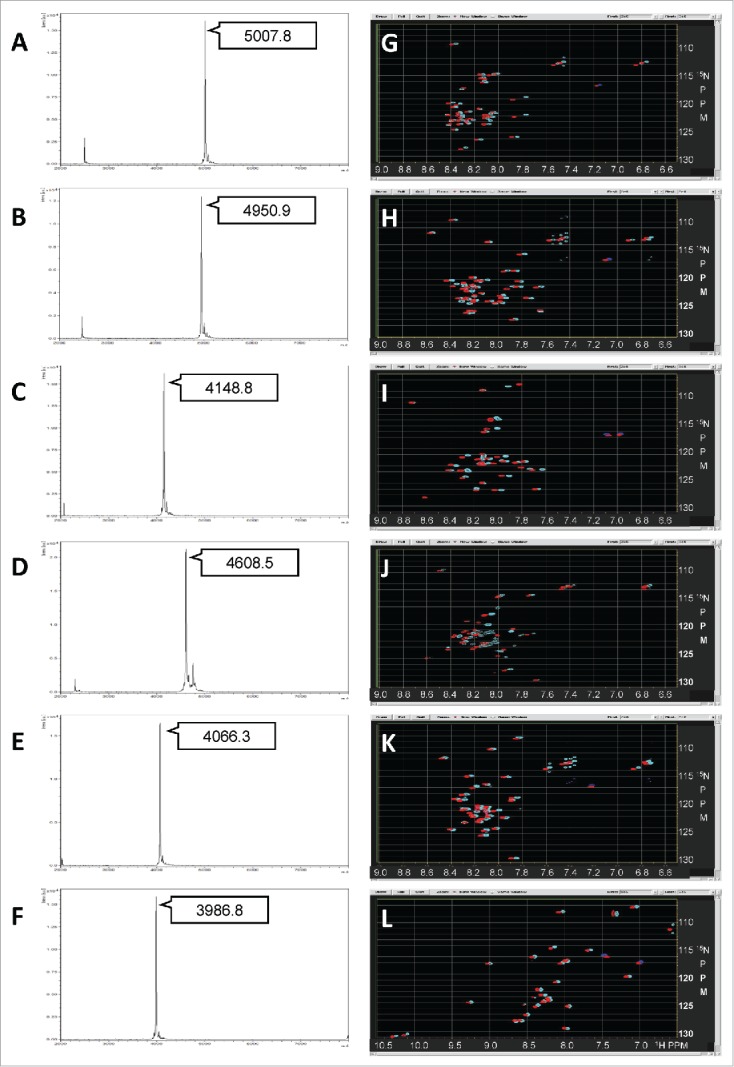
MALDI-TOF mass spectra and HSQC/SOFAST-HMQC spectra of purified IDPs. MALDI-TOF mass spectra (A–F) and NMR spectra (G–L). In the NMR spectra, the SOFAST-HMQC spectrum measured in the buffer with 0 M NaCl is shown in cyan, whereas the HSQC spectrum in 0.5 M NaCl is shown in red. (A, G) IDP(B3), (B, H) IDP(B4), (C, I) IDP(C1), (D, J) IDP(D10), (E, K) IDP(E1), (F, L) “partial”-IDP(C9).
In our previous study, we developed the PRESAT-style high-throughput construction vector for bacterial expression of biologically active peptides.18 We chose TRX as the fusion partner in view of its potential to escape from bacterial endogenous proteases.19 However, there were 2 practical disadvantages of this system: occasionally the peptides of interest were labile if the cell extract was subjected to purification, and for avoiding sample degradation, an osmotic shock treatment was necessary.18 Osmotic shock treatment is not suitable for high-throughput expression of many peptides in parallel because the treatment requires the use of fresh (not frozen) cell pellets and should be applied immediately after cell harvest. However, we also experienced an excellent yield in the bacterial expression of amyloid β 1–40 peptides from inclusion bodies, whereas the preparation from the cytosolic fraction was highly degraded (manuscript in preparation). Thus, the targeted expression of the proteins of interest in E. coli inclusion bodies is a promising candidate among the several existing strategies.28
Conclusion
We showed that the targeted expression of IDPs in E. coli inclusion bodies using Npro fusion technology is effective. Further, we optimized the Npro fusion technology to prepare multiple IDP samples suited for NMR study. We described the preparation of [15N]-labeled IDP samples. We also succeeded in preparing [13C/15N]-labeled IDP samples using the same protocol (data not shown) and propose that this method is widely applicable for other isotopically labeling methods, such as amino-acid type selective labeling,29,30 [14N]-inverse labeling,31,32 and labeling with SAIL amino acids.33 In summary, this Npro-based method can help in the acquisition of atomic-level information of IDPs via modern solution NMR techniques in a high-throughput manner.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Enago (http://www.enago.jp) for the English language review.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was partially supported by JSPS KAKENHI, Grant Number 21113007, as well as Platform for Drug Discovery, Informatics, and Structural Life Science from MEXT, Japan. This work was also partially supported by a grant from the Salt Science Research Foundation.
References
- 1. Tompa P. Structure and function of intrinsically disordered proteins. New York: Chapman and Hall/CRC; 2009. [Google Scholar]
- 2. Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 2008; 18:756-64; PMID:18952168; http://dx.doi.org/ 10.1016/j.sbi.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 3. Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci 2013; 22:693-724; PMID:23553817; http://dx.doi.org/ 10.1002/pro.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293:321-31; PMID:10550212; http://dx.doi.org/ 10.1006/jmbi.1999.3110 [DOI] [PubMed] [Google Scholar]
- 5. Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model 2001; 19:26-59; PMID:11381529; http://dx.doi.org/ 10.1016/S1093-3263(00)00138-8 [DOI] [PubMed] [Google Scholar]
- 6. Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci 2012; 37:509-16; PMID:22989858; http://dx.doi.org/ 10.1016/j.tibs.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 7. Ota M, Koike R, Amemiya T, Tenno T, Romero PR, Hiroaki H, Dunker AK, Fukuchi S. An assignment of intrinsically disordered regions of proteins based on NMR structures. J Struct Biol 2013; 181:29-36; PMID:23142703; http://dx.doi.org/ 10.1016/j.jsb.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins 2001; 42:38-48; PMID:11093259; http://dx.doi.org/ 10.1002/1097-0134(20010101)42:1%3c38::AID-PROT50%3e3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 9. Van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114:6589-631; PMID:24773235; http://dx.doi.org/ 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vucetic S, Obradovic Z, Vacic V, Radivojac P, Peng K, Iakoucheva LM, Cortese MS, Lawson JD, Brown CJ, Sikes JG, et al. DisProt: a database of protein disorder. Bioinformatics 2005; 21:137-40; PMID:15310560; http://dx.doi.org/ 10.1093/bioinformatics/bth476 [DOI] [PubMed] [Google Scholar]
- 11. Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al. DisProt: the database of disordered proteins. Nucleic Acids Res 2007; 35:D786-93; PMID:17145717; http://dx.doi.org/ 10.1093/nar/gkl893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukuchi S, Sakamoto S, Nobe Y, Murakami SD, Amemiya T, Hosoda K, Koike R, Hiroaki H, Ota M. IDEAL: Intrinsically Disordered proteins with Extensive Annotations and Literature. Nucleic Acids Res 2012; 40:D507-11; PMID:22067451; http://dx.doi.org/ 10.1093/nar/gkr884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fukuchi S, Amemiya T, Sakamoto S, Nobe Y, Hosoda K, Kado Y, Murakami SD, Koike R, Hiroaki H, Ota M. IDEAL in 2014 illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic Acids Res 2014; 42:D320-5; PMID:24178034; http://dx.doi.org/ 10.1093/nar/gkt1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di domenico T, Walsh I, Martin AJM, Tosatto SCE. MobiDB: A comprehensive database of intrinsic protein disorder annotations. Bioinformatics 2012; 28:2080-1; PMID:22661649; http://dx.doi.org/ 10.1093/bioinformatics/bts327 [DOI] [PubMed] [Google Scholar]
- 15. Potenza E, Domenico T. Di, Walsh I, Tosatto SCE. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res 2014; 43(Database issue):D315-20. PMID: 25361972; http://dx.doi.org/101093/nar/gku982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, et al. D2P2: database of disordered protein predictions. Nucleic Acids Res 2013; 41:D508-16; PMID:23203878; http://dx.doi.org/ 10.1093/nar/gks1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varadi M, Kosol S, Lebrun P, Valentini E, Blackledge M, Dunker AK, Felli IC, Forman-Kay JD, Kriwacki RW, Pierattelli R, et al. pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res 2014; 42:D326-35; PMID:24174539; http://dx.doi.org/ 10.1093/nar/gkt960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tenno T, Goda N, Tateishi Y, Tochio H, Mishima M, Hayashi H, Shirakawa M, Hiroaki H. High-throughput construction method for expression vector of peptides for NMR study suited for isotopic labeling. Protein Eng Des Sel 2004; 17:305-14; PMID:15166312; http://dx.doi.org/ 10.1093/protein/gzh044 [DOI] [PubMed] [Google Scholar]
- 19. Uegaki K, Nemoto N, Shimizu M, Wada T, Kyogoku Y, Kobayashi Y. 15N labeling method of peptides using a thioredoxin gene fusion expression system: an application to ACTH-(1-24). FEBS Lett 1996; 379:47-50; PMID:8566227; http://dx.doi.org/ 10.1016/0014-5793(95)01459-4 [DOI] [PubMed] [Google Scholar]
- 20. Livernois AM, Hnatchuk DJ, Findlater EE, Graether SP. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Anal Biochem 2009; 392:70-6; PMID:19464251; http://dx.doi.org/ 10.1016/j.ab.2009.05.023 [DOI] [PubMed] [Google Scholar]
- 21. Achmüller C, Kaar W, Ahrer K, Wechner P, Hahn R, Werther F, Schmidinger H, Cserjan-Puschmann M, Clementschitsch F, Striedner G, et al. N(pro) fusion technology to produce proteins with authentic N termini in E. coli. Nat Methods 2007; 4:1037-43; PMID:18026112; http://dx.doi.org/ 10.1038/nmeth1116 [DOI] [PubMed] [Google Scholar]
- 22. Ishino S, Yamagami T, Kitamura M, Kodera N, Mori T, Sugiyama S, Ando T, Goda N, Tenno T, Hiroaki H, et al. Multiple interactions of the intrinsically disordered region between the helicase and nuclease domains of the archaeal Hef protein. J Biol Chem 2014; 289:21627-39; PMID:24947516; http://dx.doi.org/ 10.1074/jbc.M114.554998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR 2005; 33:199-211. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16341750; PMID:16341750; http://dx.doi.org/ 10.1007/s10858-005-4425-x [DOI] [PubMed] [Google Scholar]
- 24. Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson Ser B 1995; 108:94-8; PMID:16341750; http://dx.doi.org/ 10.1006/jmrb.1995.1109 [DOI] [PubMed] [Google Scholar]
- 25. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995; 6:277-93; PMID:8520220 [DOI] [PubMed] [Google Scholar]
- 26. Shimizu K, Hirose S, Noguchi T. POODLE-S: web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics 2007; 23:2337-8; PMID:17599940; http://dx.doi.org/ 10.1093/bioinformatics/btm330 [DOI] [PubMed] [Google Scholar]
- 27. Hirose S, Shimizu K, Noguchi T. POODLE-I: Disordered region prediction by integrating POODLE series and structural information predictors based on a workflow approach. In Silico Biol 2010; 10:185-91; PMID:22430291 [DOI] [PubMed] [Google Scholar]
- 28. Hwang PM, Pan JS, Sykes BD. Targeted expression, purification, and cleavage of fusion proteins from inclusion bodies in Escherichia coli. FEBS Lett 2014; 588:247-52; PMID:24076468; http://dx.doi.org/ 10.1016/j.febslet.2013.09.028 [DOI] [PubMed] [Google Scholar]
- 29. Lin MT, Sperling LJ, Frericks Schmidt HL, Tang M, Samoilova RI, Kumasaka T, Iwasaki T, Dikanov SA, Rienstra CM, Gennis RB. A rapid and robust method for selective isotope labeling of proteins. Methods 2011; 55:370-8; PMID:21925267; http://dx.doi.org/ 10.1016/j.ymeth.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O’Grady C, Rempel BL, Sokaribo A, Nokhrin S, Dmitriev OY. One-step amino acid selective isotope labeling of proteins in prototrophic Escherichia coli strains. Anal Biochem 2012; 426:126-8; PMID:22538396; http://dx.doi.org/ 10.1016/j.ab.2012.04.019 [DOI] [PubMed] [Google Scholar]
- 31. Shortle D. Assignment of amino acid type in 1H-15N correlation spectra by labeling with 14N-amino acids. J Magn Reson B 1994; 105:88-90; PMID:7921674; http://dx.doi.org/ 10.1006/jmrb.1994.1106 [DOI] [PubMed] [Google Scholar]
- 32. Hiroaki H, Umetsu Y, Nabeshima Y, Hoshi M, Kohda D. A simplified recipe for assigning amide NMR signals using combinatorial 14N amino acid inverse-labeling. J Struct Funct Genomics 2011; 12:167-74; PMID:21866395; http://dx.doi.org/ 10.1007/s10969-011-9116-0 [DOI] [PubMed] [Google Scholar]
- 33. Miyanoiri Y, Takeda M, Okuma K, Ono AM, Terauchi T, Kainosho M. Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR 2013; 57:237-49; PMID:24057411; http://dx.doi.org/ 10.1007/s10858-013-9784-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


