Abstract
As cardiomyocytes mature, their sarcomeres and Z-band widths increase in length in order for their myofibrils to produce stronger twitch forces during a contraction. In this study, we tested the hypothesis that tensional homeostasis is affected by altering myofibril forces. To assess this hypothesis, neonatal rat cardiomyocytes were cultured on arrays of microposts to measure cellular contractility. An optical line scanning technique was used to measure the deflections in the microposts with high temporal resolution, enabling the analysis of twitch force, twitch velocity, and twitch power. Myofibril force production was elevated by vector-mediated overexpression of ribonucleotide reductase (RR) to increase cellular dATP content or by adding the inotropic agent EMD 57033 (EMD). We found that RR and EMD treatment did not affect cardiomyocyte twitch force, but it did lead to reduced twitch velocity and twitch power. Immunofluorescent analysis of α-actinin revealed that RR-over-expressing cardiomyocytes and EMD-treated cardiomyocytes had lower spread area, sarcomere length, and Z-band width as compared to control cells. These results indicate a correlation between myofibril structure and cardiac power. This correlation was confirmed by exposing the cells to the myosin II inhibitor blebbistatin, and then subsequently washing it out. After wash-out, cardiomyocytes exhibited a reduction in twitch force, velocity, and power due to shorter sarcomere length and Z-band widths. Our results suggest that cardiac myofibril structure is regulated by tensional homeostasis. If myofibril-generated forces in cardiomyocytes are elevated, a state of tensional homeostasis is maintained by producing sufficient twitch forces with a lower degree myofibril structure.
Introduction
During maturation, neonatal cardiomyocytes undergo a hypertrophic growth phase in order to increase their capacity to perfuse the body with a sufficient supply of blood.1–7 This hypertrophic growth is associated with increased diastolic strain and systolic stress, leading to a greater density, coupling, and structural organization of myofibrils in the cardiomyocytes.1, 2, 4, 6, 7 In contrast, neonatal cardiomyocytes that are unable to contract have suppressed hypertrophic growth and reduced myofibril development, suggesting that a mechanotransduction mechanism may underlie this response.8, 9 Moreover, the stiffness of the cardiac extracellular environment, which provides a mechanical resistance to twitch contractions, can stimulate cardiomyocytes to improve their myofibril structure and amplify calcium transients in order to produce a more powerful twitch contraction.10–13 Examples such as these suggest that a feedback mechanism exists in cardiomyocytes, enabling adaptation to their mechanical environment through remodeling of their myofibril structure.
In non-muscle cells, tensional homeostasis has been suggested as a feedback mechanism by which cells respond to their mechanical environment.14–18 However, it is unclear whether or not tensional homeostasis is also a governing mechanism for cardiomyocytes. Specifically, forces that act on cardiomyocytes, whether they stem from internal or external sources, may serve as mechanical cues that stimulate hypertrophic growth and myofibril development. External forces applied to cardiomyocytes by stretching them on flexible membranes can promote their hypertrophic growth,19, 20 but the effect of these applied forces on the spontaneous contractility of the cardiomyocytes is unclear. Additionally, since external forces alter the tensional state within cardiomyocytes, it is plausible that cardiomyocytes with enhanced internal forces i.e. twitch forces, could achieve a state of homeostasis without the need for further myofibril production or organization. Thus, enhancing the force produced by myofibrils in cardiomyocytes could directly test the tensional homeostasis hypothesis. However, previous studies have only explored how cells respond to reductions in myofibril force.21
The goal of this study was to determine how tensional homeostasis – a feedback mechanism governed by cellular forces – is affected by modulating the amount of force produced by myosin heads in spontaneously beating cardiomyocytes. We utilized two strategies to increase myofibril force generation in cardiomyocytes: adenoviral overexpression of ribonucleotide reductase (RR) and treatment with EMD 57033 (EMD). RR promotes the generation of 2-deoxy-ATP (dATP) from ATP, which we have previously shown leads to a higher magnitude of force and a faster rate of force production in cardiac muscle by increasing the number of cross-bridges that bind and cycle.22–27 Alternatively, EMD 57033 (EMD) is an inotropic agent that can be used to increase the average force per actomyosin cross-bridge.28–31
To determine the effect of RR-overexpression and EMD-treatment on cardiac tensional homeostasis, we cultured cardiomyocytes on arrays of microposts and measured their twitch force, velocity and power in relation to their myofibril structure. We observed that both RR-overexpressing and EMD-treated cardiomyocytes produced twitch forces that were similar to those produced by cardiomyocytes cultured under control conditions, which indicates that the equilibrium state for cardiac tensional homeostasis was maintained. Twitch velocity and twitch power in the treated cells, however, were significantly lower than control cells. Immunofluorescent analysis of spread area, sarcomere length, and Z-band width revealed that RR-overexpression and EMD treatment both caused cardiomyocytes to have lower spreading and poor myofibril structure. These results suggest that twitch forces act as an internal cue for tensional homeostasis, which is maintained by altering myofibril structure and, subsequently, cardiac power. To the best of our knowledge, this is the first study to examine how altering the internal load produced by spontaneously-beating cardiomyocytes affects the contractile and structural properties of these cells.
Methods
Cell Culture
Cardiomyocytes were isolated from 2–4 day old newborn Fischer 344 rats as previously described,10 under approval of the University of Washington Animal Care Committee and in accordance with federal guidelines. Micropost arrays were submerged in 2 mL of plating media (4:1 ratio of DMEM to M199 Hyclone, 10% horse serum, 5% fetal bovine serum, 20 mM HEPES, 100 U/mL penicillin G, 100 µg/mL streptomycin and 4 mM glutamine) and placed in an incubator. Isolated cardiomyocytes were suspended at a density of 1000 cells/µL in plating media and gently pipetted on top of the arrays of microposts in 200 µL aliquots. The culture was then left undisturbed for 15 minutes before it was moved into the incubator. All reagents were purchased from Sigma-Aldrich unless specifically noted. Culture conditions for each treatment were the same. After treatment, the media was changed every day and was supplemented with 10 µM arabinosylcytosine to remove proliferating cells (such as fibroblasts).
Micropost Preparation
Arrays of polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) microposts were fabricated on top of glass cover slips using a replica-molding process.10 The spring constant of an individual micropost, k, was calculated by:
| (1) |
where E is the Young’s modulus of PDMS, D is the diameter of the post, and L is the post height. For these experiments, a uniform array of microposts was used. Each micropost had an approximate height of 5.7 µm, diameter of 2.9 µm, and spring constant of k = 142 ± 30 nN/µm, while the array had an effective shear modulus of 15 ± 2 kPa, which matches closely with the stiffness of the native tissue.10 To enable cardiomyocyte attachment on top of the microposts, fibronectin (50 µg/mL BD Biosciences) was deposited on the tips of the microposts using microcontact printing. The arrays were then incubated overnight in a 0.2% solution of Pluronic F127 (Sigma-Aldrich) at 4 °C to block the adsorption of additional proteins to the surface of the microposts.
Cell Force Modulation
Viral-mediated enhancement of myofibril forces was achieved by increasing dATP production in cells via transduction with an adenoviral vector for ribonucleotide reductase subunits Rrm1 and Rrm2, in addition to green fluorescent protein as a reporter protein to confirm expression (RR + GFP). Control cells were treated with an adenoviral vector for expression of GFP only.
To increase the average force per actomyosin crossbridge through pharmacological means, cardiomyocytes were exposed to10 mM EMD in dimethyl sulfoxide (DMSO) at 0.05% (v/v). Here, control cells were treated with DMSO. EMD was obtained from a generous gift by Merck. After measurements were made in the presence of EMD, it was washed out to determine the twitch capacity of these cardiomyocytes without the enhancement of cross-bridge force. All of the above treatments were performed 16 hours after seeding the cells onto the posts.
Blebbistatin (in DMSO), which binds to myosin and inhibits cell contraction, was used as a negative pharmacological control. Cells were treated with 10 µM blebbistatin after 64 hours of culture on the microposts. At this point, the cardiomyocytes were attached and had adapted to their micropost environment by increasing their contractility and myofibril structure.10 After 6 hours, blebbistatin was washed out by replacing the media with blebbistatin-free DMSO.
Measurement of Force, Velocity, and Power
Micropost deflections were measured 70 hours after cardiomyocyte seeding. These deflections were measured with IonOptix© line scanning software and IonOptix© optical equipment, as previously described.10 Briefly, optical line scanning was used to track the motion of a single micropost as it moved along a line of pixels in an optical microscope’s CCD camera.32 To determine the magnitude of the twitch force, the distance between the position of a micropost in each video frame and its initial position, i.e. its deflection, was multiplied by the spring constant (142 nN/µm) of the micropost as described by equation 1. Twitch velocity was determined by dividing the change in position of a micropost by the period of time between frames, while twitch power was calculated by multiplying the twitch force and twitch velocity together.
Post deflections were measured at the perimeter of the cell, and at least two post measurements were analyzed from each cell. We restricted our analysis to posts on the edge of the cell as these locations are associated with the terminal end of at least one myofibril, as well as producing the highest twitch forces.10 Cells that did not deflect the posts were not measured, and of the measured cells, only those posts which demonstrated steady contractions that fell within the 95% confidence interval of all contractions were analyzed. Calculations of average maximum twitch force, velocity, and power were then based on the analysis of all contractions that met this criterion, across multiple different posts, cells, and experiments.
To conduct these measurements, cardiomyocytes on microposts were placed into an IonOptix© field stimulation chamber immersed in Tyrode’s buffer (10 mM HEPES, 138 mM NaCl, 5.5 mM glucose, 12 mM NaHCO3, 0.36 mM Na2HPO4, 2.9 mM KCl, 0.5 mM MgCl2 and 1 mM CaCl2 at pH 7.4). For the study with EMD and its control group, 10 mM EMD in DMSO or only DMSO was added to the Tyrode’s buffer at 0.05% (v/v). All experiments were conducted at room temperature. The measurement time point was selected in order to give the cells sufficient time to reach a steady-state in their growth, as previously reported.9 We observed that twitch force, velocity, and power in the cells did not change significantly when cultured between 70 to 118 hours on the arrays of microposts: 69.7 ± 14.3 versus 70.8 ± 11.5 nN for twitch forces; 3.27 ± 0.52 versus 3.35 ± 0.79 µm/s for twitch velocity; and 243 ± 100 versus 203 ± 42 fW for twitch power (p > 0.05, Z-test). Thus for this study, all contractile measurements were made after 70 hours.
Immunofluorescence Analysis
Myofibril structure was assessed by using a Triton extraction and immunofluorescence staining protocol. Briefly, cells were treated with 0.5% Triton X-100 with protease inhibitors, fixed in 4% paraformaldehyde, and blocked with 10% goat serum. Z-bands were stained by incubating cells with monoclonal mouse antibodies against sarcomeric α-actinin (Sigma-Aldrich) and Alexa-Fluor 647 goat anti-mouse IgG antibodies (Invitrogen). Nuclear staining was conducted using Hoechst 33342 (Invitrogen). Image analysis of spread area, sarcomere length, and Z-band width was performed with NIS-Elements software. Spread area was measured by counting the number of pixels contained inside the perimeter of a cell. Sarcomere lengths were quantified by measuring the distance between the peak fluorescence intensities of two, linearly-oriented, parallel Z-bands, while Z-band width was quantified by applying an edge-detection algorithm in order to measure their end-to-end length.10 At least 9 measurements of sarcomere length and 15 measurements of Z-band width were taken per cell, and at least four cells were analyzed per experimental group. The approximate error in these measurements is 0.05 µm, which was determined by taking measurements of the same structure in different viewing planes.
Statistical Analysis
Due to the non-parametric nature of the data, statistical differences between GFP and RR groups were determined by a Mann-Whitney Rank Sum Test,10 while differences between EMD, blebbistatin, and DMSO conditions were determined with a Kruskal-Wallis ANOVA on Ranks. To compare multiple treatments, a Mann-Whitney post hoc test with a Bonferroni correction was performed. Normality of the data distributions was determined by the Ryan Joiner test. Statistical significance was assessed at p < 0.05, and is denoted by an asterisk. Data in text is reported as mean ± 95% confidence intervals. Error bars in graphs, as well as error intervals reported in figure captions, represent standard error of the mean.
Results
Neonate cardiomyocytes were cultured on fibronectin-stamped micropost arrays and treated with 1) adenoviruses for expressing green fluorescent protein (GFP), 2) adenoviruses for expressing muscle ribonucleotide reductase subunits M1 and M2 and GFP (RR+GFP), 3) DMSO, or 4) EMD in DMSO. At the time when the vectors or reagents were added to the culture, cardiomyocytes had attached onto the tips of the microposts, but not yet fully spread and did not exhibit spontaneous contractions. After 2 days in culture, cells from all four test groups began to spontaneously beat. From these observations, we determined that the adenoviruses and reagents were added at a time during which twitch contractions are able to influence cardiomyocyte growth and/or remodeling in vitro.8, 9 Once spontaneous contractions began, the average beat frequency was 1.0 ± 0.1 s−1 at room temperature and was not significantly different between the four groups.
RR Expression Reduces Twitch Power and Myofibril Structure
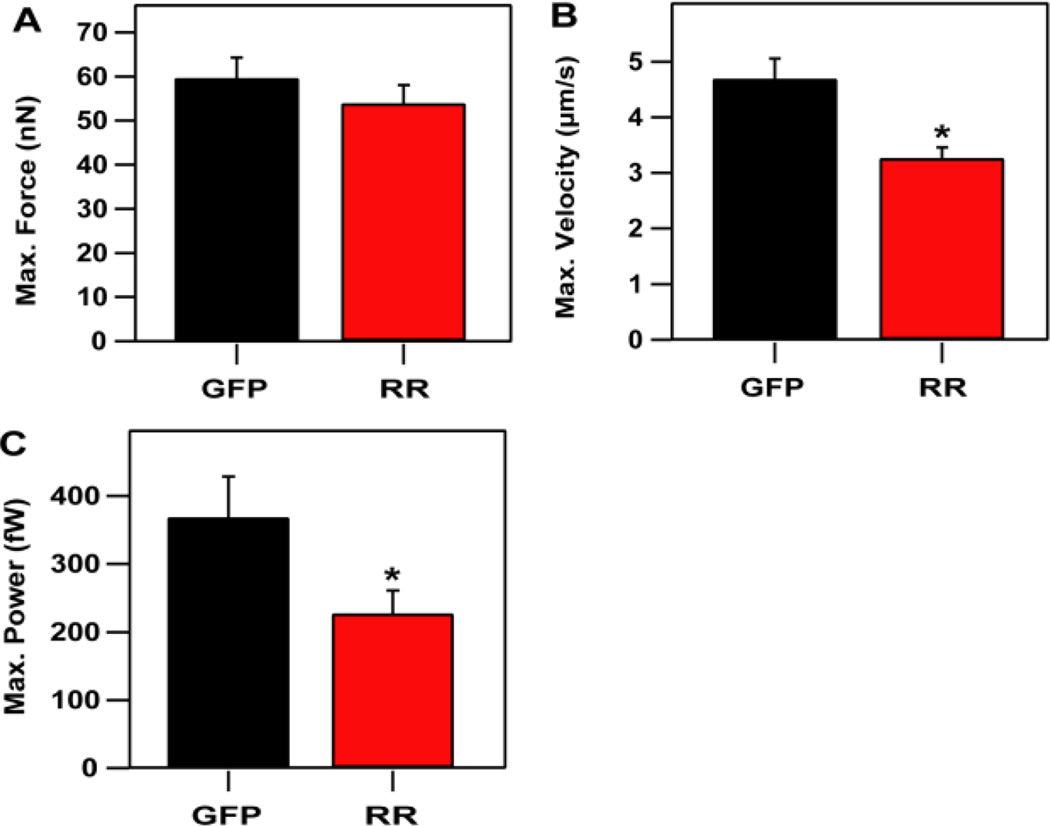
The average maximum twitch force produced by posts at the perimeter of the cell for RR + GFP-expressing cardiomyocytes was found to be statistically similar to the average produced by GFP-expressing cardiomyocytes (Fig. 1A). At first examination, the similarity between the maximal twitch forces for RR+GFP and GFP-expressing cells suggested that elevated levels of dATP do not affect contractility. This differed from our previous findings using isolated muscle fibers or adult cells.22–26 However, it was plausible that the difference may have been due to difference in experimental conditions between the two studies. In the current study, neonatal cardiomyocytes with nascent contractile machinery were used instead of fully developed adult cells. Moreover, cardiomyocytes used in this study were cultured on arrays of flexible microposts, which enabled a larger degree of shortening during a twitch contraction than would be possible for cardiomyocytes adhered to a rigid tissue culture dish. As a consequence, they would be able to equilibrate their twitch forces in order to compensate for the higher myofibril forces caused by elevated dATP levels in RR + GFP overexpressing cardiomyocytes.
Figure 1.
Twitch velocity and power decrease with increased dATP production in RR + GFP-expressing cells (RR), but twitch forces are unaffected. (A) Maximum twitch forces were not significantly altered by over-expressing RR (53.9 ± 4.1 nN) when compared to control cells expressing GFP (59.7 ± 4.6 nN). (B) Maximum twitch velocity was significantly lower in RR + GFP-expressing cells (3.27 ± 0.19 µm/s) than in GFP-expressing control cells (4.69 ± 0.36 µm/s). (C) Maximum twitch power was lower in RR + GFP-expressing cells (227 ±34 fW) than in GFP-expressing cells (369 ±60 fW). Data shown for RR+GFP-expressing cells is from 29 micropost measurements taken from 15 cells. Data shown for GFP-expressing cells is from 27 micropost measurements taken from 13 cells. Three replicate experiments were conducted for each condition.
Even though the average maximum twitch forces were similar in RR + GFP and GFP-expressing cardiomyocytes, twitch velocity and twitch power were not maintained. The average maximum velocity reached during a twitch contraction was significantly lower for RR + GFP-expressing cardiomyocytes as compared to GFP-expressing cells (Fig. 1B). Likewise, the average maximum power achieved during a twitch contraction was lower in RR + GFP-expressing cells than GFP-expressing cells (Fig. 1C). These differences in twitch velocity and twitch power indicate that elevated cellular levels of dATP can adversely affect twitch power in neonatal cardiomyocytes, which, to the best of our knowledge, has not been seen previously in adult cardiomyocytes.22–26
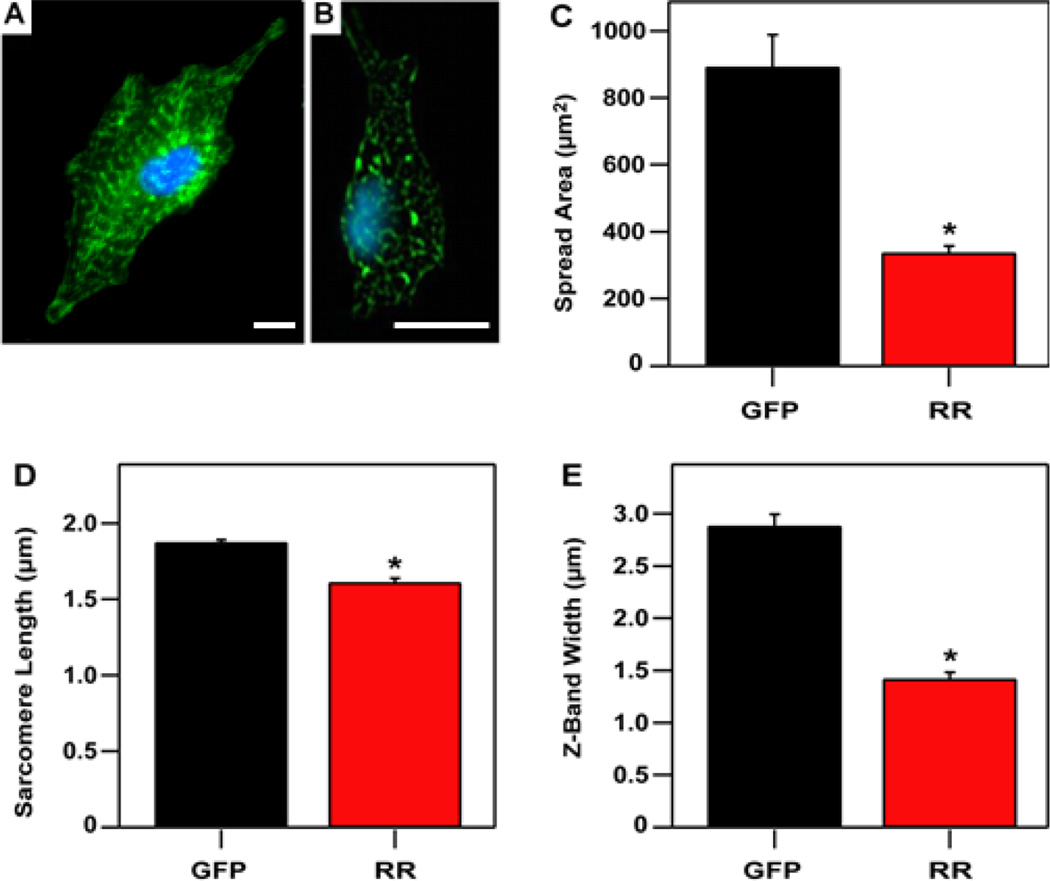
Immunofluorescent antibody staining of the treated cardiomyocytes revealed structural and morphological differences between GFP-expressing (Fig. 2A) and RR+GFP-expressing cells (Fig. 2B). For example, RR + GFP-expressing cells had spread areas that were significantly lower than the spread areas of GFP-expressing cells, indicating growth related to tension production may have been suppressed (Fig. 2C). Myofibril structure was also influenced by RR + GFP-overexpression, as evidenced by shorter sarcomere lengths and reduced Z-band width as compared to GFP-expressing cells (Fig. 2D–E).
Figure 2.
Cytosolic dATP reduces hypertrophic growth and myofibril structure. Fluorescent images showing nuclei (cyan) and α-actinin (green) for representative cells expressing (A) RR + GFP or (B) GFP. (C) Spread area was significantly less for RR + GFP-expressing cells (338 ± 19 µm2) than for GFP-expressing cells (893 ± 95 µm2). (D) Sarcomere lengths (1.61 ± 0.03 µm for RR + GFP cells and 1.88 ± 0.02 µm for GFP cells) and (E) Z-band widths (1.42 ± 0.06 µm for RR + GFP cells and 2.89 ± 0.11 µm for GFP cells) are also significantly reduced with the expression of RR. Scale bar represents 10 µm. n = 4 cells (from each group).
Sarcomere length determines the degree of thin and thick filament overlap and influences the lattice spacing between the filaments, both of which affect the probability of actomyosin cross-bridge formation and the force generating capacity of myofibril bundles.33–36 Since the GFP-expressing cardiomyocytes have a sarcomere length (~1.88 µm) that is near optimal for filament overlap, the similar twitch force for RR+GFP-expressing cells with shorter sarcomere lengths (~1.61 µm) suggests that elevated cellular dATP may increase the number of cross-bridges per unit length of filament overlap, or increase the force produced per cross-bridge, or both.
An additional factor in the structure of myofibrils is Z-band width (Fig. 2E). A Z-band adjoins neighboring sarcomeres and improves the synchronicity of their shortening during contraction.37 Additionally, increasing numbers of myofibrils in parallel increases Z-band width, providing more contractile capacity to bend individual micro-posts. Previously, we have shown that Z-band width correlates with the degree of twitch power produced by neonatal cardiomyocytes.10 The width of the Z-bands in RR + GFP-expressing cells was significantly lower than for GFP-expressing cells, indicating a lower degree of synchronicity in their myofibrils. The width of a single myofibril is approximately ~1 µm,38 so the data suggests that RR + GFP-expressing cardiomyocytes are primarily populated by single myofibrils as opposed to bundles. In contrast, Z-disk widths for GFP-expressing cells were ~ 3 µm, suggesting either two or three myofibrils per post on average. Taken together, the shorter sarcomere length and narrower Z-bands in RR + GFP-expressing cardiomyocytes suggests that these structural differences may result in lower maximum twitch velocity and twitch power.
EMD Reduces Twitch Power and Myofibril Structure
The inotropic agent EMD was used to further test the hypothesis of tensional homeostasis in cardiomyocytes. EMD increases contraction by enhancing the force produced by individual cross-bridges.28–31 A comparison of cardiomyocytes treated with EMD or those expressing RR + GFP should determine whether myofibril force per se and/or the relative number of force producing cross-bridges is regulated by tensional homeostasis. Prior to adding EMD, initial twitch behavior was assessed. After this baseline was established, EMD in DMSO was added to culture bath, and contraction measurements were repeated as the concentration of EMD was increased. After treatment with EMD, twitch forces of neonatal cardiomyocytes were increased by 20–25% over a 20-fold range of EMD concentrations in media, (Fig. 3). Furthermore, unlike over expression of RR + GFP where elevated cellular levels of dATP are not easily reversed, a series of cell washes removes EMD and restores twitch forces to pre-treatment levels.
Figure 3.
EMD 57033 increases neonatal cardiomyocyte twitch forces. Maximum twitch force was tracked at individual posts for a range of EMD concentrations (n = 6 cells).
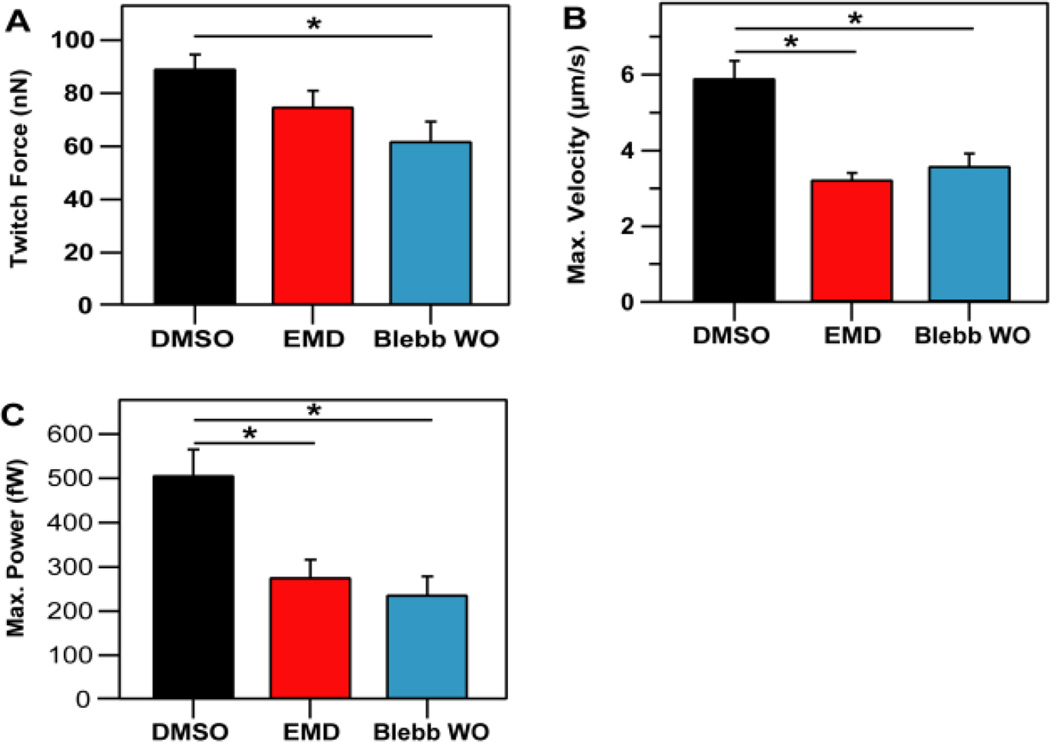
Similar to previous results, no significant difference in the average maximum twitch force per post was observed between cells cultured with or without EMD (Fig. 4A). EMD cells did have a reduced maximal twitch velocity (Fig. 4B), which resulted in lower maximum twitch power (Fig. 4C). After EMD wash-out, twitch force was reduced by 18%, and was not significantly different that pre-EMD force levels.
Figure 4.
Twitch velocities and power decreased for cells cultured with EMD and cells with an acute application of blebbistatin. (A) Maximum force per post was not significantly altered with the presence of EMD (75.1 ± 6.0 nN) as compared to DMSO control cells (89.3 ± 5.2 nN) but was significantly reduced for blebbistatin (62.0 ± 7.2 nN). (B) Maximum velocity was significantly reduced for cells treated with EMD (3.25 ± 0.17 µm/s) or blebbistatin (3.6 ± 0.32 µm/s) with respect to the DMSO group (5.91 ± 0.45 µm/s). (C) Maximum power was also significantly reduced by EMD (277 ± 39 fW) and blebbistatin (238 ± 41 fW), when compared to the DMSO control (507 ± 57 fW). Data shown for DMSO-expressing cells is from 29 micropost measurements taken from 14 cells. Data shown for cells treated with EMD is from 25 micropost measurements taken from 13 cells. Data shown for cells treated with blebbistatin is from 21 measurements taken from 9 cells. Three replicate experiments were conducted for each condition.
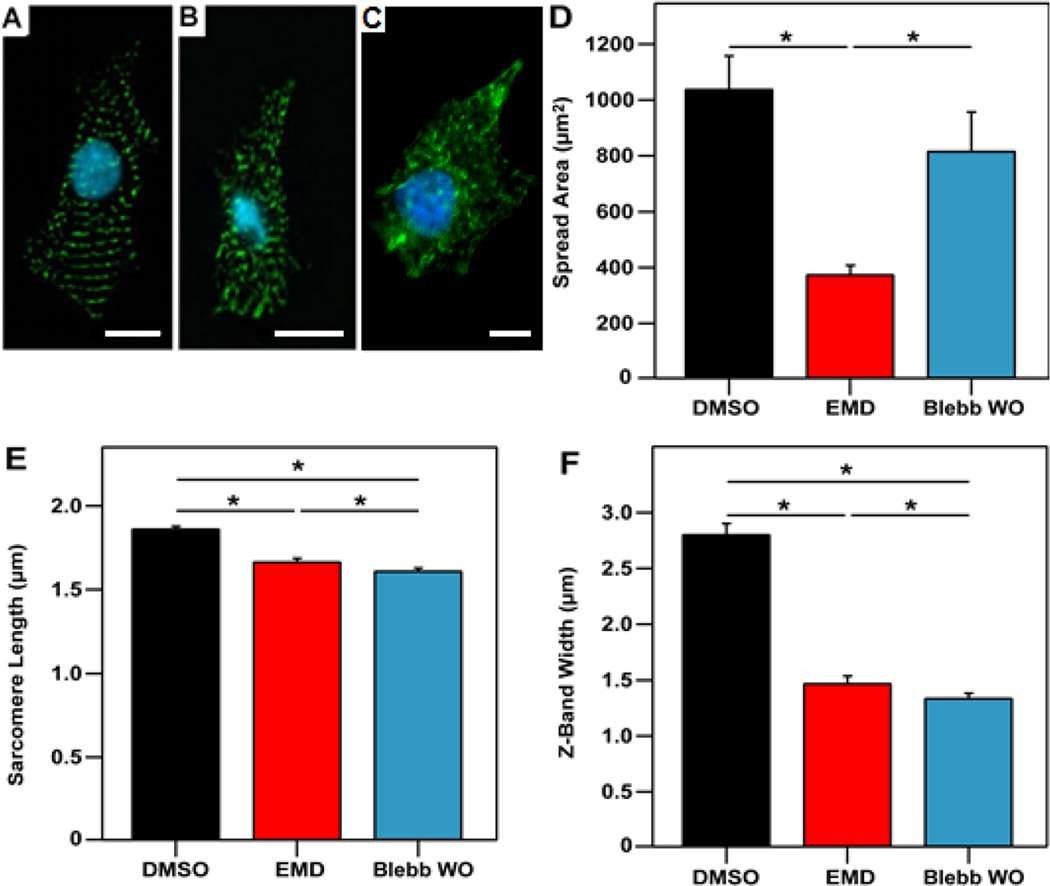
To determine if EMD affected cell spread area or myofibrillar structure, cardiomyocytes were fixed and stained against α-actinin. Cells treated with EMD had similar myofibril structure as RR + GFP cells, including significantly smaller spread areas (Fig. 5D), sarcomere lengths (Fig. 5E), and Z-band widths (Fig. 5F). Taken together, these results suggest that the effect of EMD treatment on myofibril development was similar to that of RR + GFP overexpression, and both likely result from increased myofibril force producing ability. Furthermore, both treatments alter myofibril structure and bundling (Fig. 5B, E, and F) in a similar manner, suggesting cells compensate for increases in actomyosin force by suppressing myofibril structure and organization. The compensation for increased myofilament force supports the idea of existence of mechanical feedback controls during neonatal cardiomyocyte growth to maintain tensional homeostasis.
Figure 5.
EMD and blebbistatin impair myofibril structure. Fluorescent images showing nuclei (cyan) and α-actinin (green) for representative cardiomyocytes from the (A) DMSO-treated group, (B) EMD-treated group and (C) blebbistatin-treated. (D) Spread area was found to be significantly less for the EMD (377 ± 31 µm2) as compared to the DMSO (1041 ± 117 µm2) and blebbistatin (819 ± 138 µm2) groups. Measurement of (E) sarcomere lengths (1.87 ± 0.01 µm for DMSO, 1.67 ± 0.02 µm for EMD, and 1.61 ± 0.01 µm for blebbistatin) and (F) Z-band widths (2.81 ± 0.09 µm for DMSO, 1.48 ± 0.06 µm for EMD, and 1.34 ± 0.04 µm for blebbistatin) also show a significant reduction with the addition of EMD or blebbistatin. Scale bar represents 10 µm. Data is from at least 11 cells per group.
Reduced Twitch Power is Associated with Structural Changes
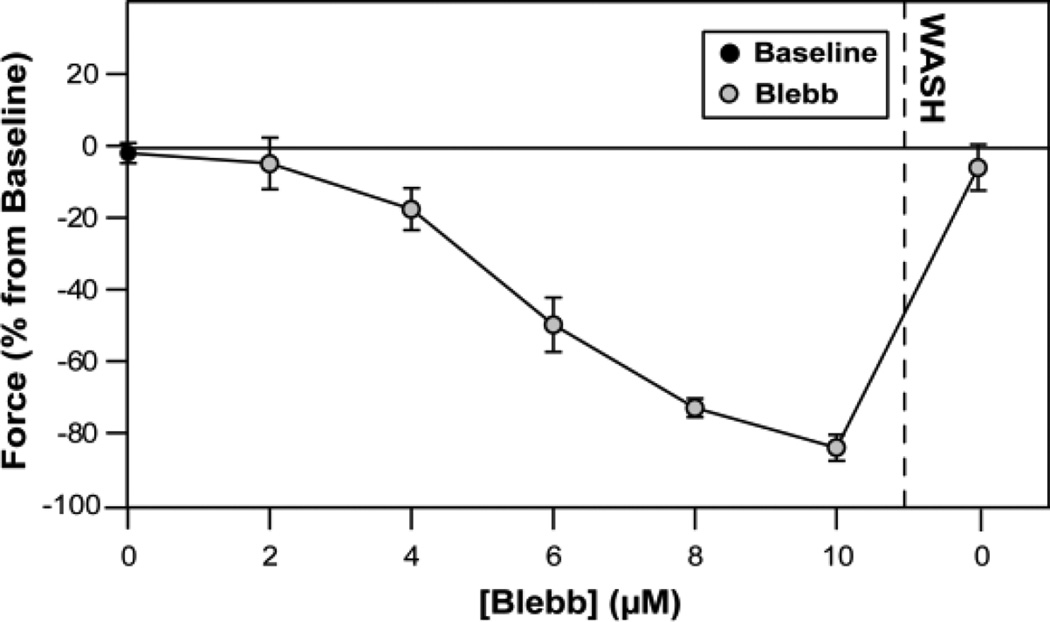
Blebbistatin was used to inhibit myosin interaction with actin and thus reduce twitch forces. Before adding blebbistatin to the culture media, twitch forces were measured to obtain a baseline measurement. Incubation with increasing concentrations of blebbistatin progressively inhibited twitch force, and at 10 µM blebbistatin twitch forces were reduced to less than 20% of the baseline force. For short term treatments, after wash out, twitch forces returned to near baseline levels (Fig. 6).
Figure 6.
Blebbistatin decreased neonatal cardiomyocyte twitch forces. Maximum twitch force was tracked at individual posts for a range of Blebbistatin concentrations (n = 5 cells).
However, following longer blebbistatin treatment (6 hrs.) and wash-out, twitch force (Fig. 4A), velocity (Fig. 4B), and power (Fig. 4C) were significantly reduced when compared to the DMSO control group. Interestingly, the decreases in force, velocity, and power were statistically similar to those seen in the cells treated with EMD. Longer incubations of cardiomyocytes with blebbistatin (4 or 20 hours) have previously been shown to inhibit spontaneous contraction and result in deleterious reductions to myofilament structure and cell morphology39.
We found similar results in this study. The effect of 6 hours of blebbistatin incubation on sarcomere length and degree of myofibril coupling was similar to that for EMD-treated cardiomyocytes (Fig. 5E & 5F). However, blebbistatin did not significantly affect spread area (Fig. 5D). Since blebbistatin has no significant effect on the calcium transient,39 and there is similarity in spread area between the blebbistatin and EMD groups, the reduced force in both cases is likely a result of the altered myofibril structure.
Discussion
The major conclusion of this work is that: increasing the ability of individual myofibrils to produce force (by either increased crossbridge binding or force per cross-bridge), appears to lead to changes in cardiomyocyte cell spread area and sarcomeric organization which likely results in no change in twitch force but a reduction in twitch power and twitch velocity. These results suggest that mechanical feedback can influence the structural development of the cell as it tries to maintain tensional homeostasis.
Tensional Homeostasis Maintained by Mechanical Feedback
Although RR-overexpression and EMD-treatment potentiate myofibril forces via different mechanisms, cardiomyocytes cultured with either approach yield similar alterations in myofibril structure (Fig. 2B & 5B), and produce a similar twitch force as control cells (Fig. 1A & 4A). This corroborating evidence suggests that enhanced actin-myosin interactions were critical in determining the degree of cytoskeletal organization, and not the specific agent. This result is indicative of cardiomyocytes reaching tensional homeostasis with the mechanical stiffness of their substrate. It is likely that once this point of stasis is reached, myofibril development is suppressed, as reflected in myofibril structure and spread area.
Tensional homeostasis would help explain mechanisms that induce controlled growth during maturation. It is known that changes in the demands of the heart lead to increased mechanical stress on developing cardiomyocytes, and eventually results in hypertrophy.1–7 This increased demand is associated with extracellular environmental changes, such as increased myocardium stiffness and diastolic stretching.1, 2, 4, 6, 7, 40, 41 In non-muscle cells, mechanical stiffness of the extracellular environment sets the degree of tension needed to reach homeostasis.15, 42 If a similar mechanism translates to cardiomyocytes, increasing stiffness will increase the necessary degree of force generation by individual twitch contractions in order to reach tensional homeostasis. A correlation between stiffness and twitch force was demonstrated previously on both microposts10 and flat polyacrylamide gels.11 These studies found that twitch force increases with substrate stiffness. Additionally, increases in force were associated with altered structure and increased calcium transient amplitude, both of which represent intracellular modifications that could be the result of mechanical feedback controls to generate additional tension. In terms of stretch, increased mechanical strain leads to better myofibril alignment and amplification of hypertrophic signaling.19, 20 Thus, external mechanical cues can control growth and development by altering the level of tension necessary for the cell to reach homeostasis.
In this study, we observed that structural modifications in response to increased myofibril force occurred on deformable microposts, but this does not occur on non-deformable substrates.26 Deformation is associated with the degree of strain sensed by the cell, and strain is an important element in initiating mechanotransduction.43–45 The feedback control associated with tensional homeostasis is likely mechanotransductive in nature,46 and is likely linked to proteins at the Z-disc and/or adhesion complexes typical of cardiomyocytes.44, 47 When cells are overstretched, hypertrophic signaling pathways are suppressed, suggesting the potential for a negative feedback loop associated with large degrees of strain.20 If a cell cannot deform its environment, it might not generate enough strain to initiate the negative feedback controls associated with tensional homeostasis. Therefore, the degree to which a cell can deform its environment is important to maintaining tensional homeostasis and preventing the development of pathophysiological phenotypes.
Twitch Power is Dependant on Myofibril Organization
Actomyosin cross-bridges within sarcomeres generate the twitch force and shortening at the myofibril level that produce the power needed for ventricular blood ejection.34, 48, 49 Here, we showed that increasing myofibril force in a developing cardiomyocyte appears to result in cells capable of producing the normal levels of twitch force with less myofibril organization. This reduction in structure is associated with lower twitch velocity, independent of the level of twitch force (Fig. 2B & 4B), and a lower capacity to generate twitch power (Fig. 2C & 4C).
It is important to note, however, that these results are specific to force-modulated neonatal cardiomyocytes. When adult rat cardiomyocytes were made to overexpress RR, Korte et al. found no significant structural differences between treated and non-treated cells.26 Furthermore, RR treated cells exhibited enhanced fractional shortening, shortening velocity, and relaxation velocity, as compared to non-treated control cells.26 Isolated cardiomyocytes from transgenic mice that overexpress RR were also seen to demonstrate similar increases in contractile properties27. These results imply that, while cardiomyocytes are in their developing state, they have greater plasticity to find ways to reach the same level contractile force. However, when cells are already in a structurally developed state, such is the case with adult cardiomyocytes, methods that produce force enhancement may have no delirious effect on myofibril development.
Velocity is a product of synchronization of multiple cross-bridges undergoing their power stroke.34 Synchronization to affect shortening velocity in a particular direction is achieved when myofibrils are tightly grouped and aligned in register with one another.37 Maximum force is somewhat dependent on synchronization of cross-bridges during contraction, but is primarily determined by the number of cross-bridges formed at the peak of contraction, and the amount of force produced by each of these cross-bridges.50 Therefore, similar levels of force may be produced independent of myofibril coupling. Thus, for equivalent force production, velocity can be lower in cardiomyocytes with reduced myofibril coupling. As such, the degree of myofibril coupling is positively correlated with the capacity to generate power due to increased contractile synchronization associated with the lateral transmission of mechanical stress.
Conclusions
With this work, we demonstrated for the first time, that increases in myofibril force can abrogate hypertrophic development and myofibril coupling of spontaneously-beating cardiomyocytes in vitro. This mechanobiological response is associated with reduced myofibril coupling and sarcomere length, which appears sufficient to cause reduced twitch velocity and power. Evidence from others investigating cellular strain suggests that mechanotransductive signaling is a potential mechanism for feedback control,43, 45 but further work is needed to directly connect this hypothesis with the results presented here. The results of this study demonstrate the complexity of cardiomyocyte response to mechanical stress and the potential role of tensional homeostasis in the development of cardiac muscle.
Supplementary Material
Notes and references
- 1.Hopkins SF, Jr, McCutcheon EP, Wekstein DR. Circ Res. 1973;32:685–691. doi: 10.1161/01.res.32.6.685. [DOI] [PubMed] [Google Scholar]
- 2.Anversa P, Olivetti G, Bracchi PG, Loud AV. Circ Res. 1981;48:561–568. doi: 10.1161/01.res.48.4.561. [DOI] [PubMed] [Google Scholar]
- 3.Morgan HE, Baker KM. Circulation. 1991;83:13–25. doi: 10.1161/01.cir.83.1.13. [DOI] [PubMed] [Google Scholar]
- 4.Anversa P, Capasso JM, Olivetti G, Sonnenblick EH. Acta Paediatrica. 1992;81:29–31. [PubMed] [Google Scholar]
- 5.Beinlich CJ, Morgan HE. Molecular and Cellular Biochemistry. 1993;119:3–9. doi: 10.1007/BF00926846. [DOI] [PubMed] [Google Scholar]
- 6.Russell B, Motlagh D, Ashley WW. J Appl Physiol. 2000;88:1127–1132. doi: 10.1152/jappl.2000.88.3.1127. [DOI] [PubMed] [Google Scholar]
- 7.Russell B, Curtis MW, Koshman YE, Samarel AM. Journal of Molecular and Cellular Cardiology. 2010;48:817–823. doi: 10.1016/j.yjmcc.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein I, Daood M, Whiteside T. J Cell Physiol. 1985;124:49–53. doi: 10.1002/jcp.1041240109. [DOI] [PubMed] [Google Scholar]
- 9.McDermott PJ, Morgan HE. Circ Res. 1989;64:542–553. doi: 10.1161/01.res.64.3.542. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez AG, Han SJ, Regnier M, Sniadecki NJ. Biophys J. 2011;101:2455–2464. doi: 10.1016/j.bpj.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacot JG, McCulloch AD, Omens JH. Biophys J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majkut SF, Discher DE. Biomech Model Mechanobiol. 2012;11:1219–1225. doi: 10.1007/s10237-012-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Clark K, Langeslag M, Figdor CG, van Leeuwen FN. Trends Cell Biol. 2007;17:178–186. doi: 10.1016/j.tcb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Curr Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provenzano PP, Keely PJ. J Cell Sci. 2011;124:1195–1205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson DG, Sharp WW, Borg TK, Price RL, Terracio L, Samarel AM. Am J Physiol. 1996;270:C1075–C1087. doi: 10.1152/ajpcell.1996.270.4.C1075. [DOI] [PubMed] [Google Scholar]
- 20.Simpson DG, Majeski M, Borg TK, Terracio L. Circ Res. 1999;85:e59–e69. doi: 10.1161/01.res.85.10.e59. [DOI] [PubMed] [Google Scholar]
- 21.Farman GP, Tachampa K, Mateja R, Cazorla O, Lacampagne A, de Tombe PP. Pflug Arch Eur J Phy. 2008;455:995–1005. doi: 10.1007/s00424-007-0375-3. [DOI] [PubMed] [Google Scholar]
- 22.Regnier M, Homsher E. Biophys J. 1998;74:3059–3071. doi: 10.1016/S0006-3495(98)78013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regnier M, Lee DM, Homsher E. Biophys J. 1998;74:3044–3058. doi: 10.1016/S0006-3495(98)78012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regnier M, Martyn DA, Chase PB. Biophys J. 1998;74:2005–2015. doi: 10.1016/S0006-3495(98)77907-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regnier M, Rivera AJ, Chen Y, Chase PB. Circ Res. 2000;86:1211–1217. doi: 10.1161/01.res.86.12.1211. [DOI] [PubMed] [Google Scholar]
- 26.Korte FS, Dai J, Buckley K, Feest ER, Adamek N, Geeves MA, Murry CE, Regnier M. Journal of molecular and cellular cardiology. 2011;51:894–901. doi: 10.1016/j.yjmcc.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowakowski SG, Kolwicz SC, Korte FS, Luo Z, Robinson-Hamm JN, Page JL, Brozovich F, Weiss RS, Tian R, Murry CE, Regnier M. Proc Natl Acad Sci U S A. 2013;110:6187–6192. doi: 10.1073/pnas.1220693110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solaro RJ, Gambassi G, Warshaw DM, Keller MR, Spurgeon HA, Beier N, Lakatta EG. Circ Res. 1993;73:981–990. doi: 10.1161/01.res.73.6.981. [DOI] [PubMed] [Google Scholar]
- 29.Strauss JD, Bletz C, Ruegg JC. Eur J Pharmacol. 1994;252:219–224. doi: 10.1016/0014-2999(94)90600-9. [DOI] [PubMed] [Google Scholar]
- 30.Kraft T, Brenner B. Biophys J. 1997;72:272–281. doi: 10.1016/S0006-3495(97)78666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipscomb-Allhouse S, Mulligan IP, Ashley CC. Pflugers Arch. 2001;442:171–177. doi: 10.1007/s004240000500. [DOI] [PubMed] [Google Scholar]
- 32.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woledge RC, Curtin NA, Homsher E. Monographs of the Physiological Society. 1985;41:1–357. [PubMed] [Google Scholar]
- 34.Gordon AM, Homsher E, Regnier M. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 35.Williams CD, Regnier M, Daniel TL. PLoS computational biology. 2010;6:e1001018. doi: 10.1371/journal.pcbi.1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leyton RA, Spotnitz HM, Sonnenblick EH. Am J Physiol. 1971;221:902–910. doi: 10.1152/ajplegacy.1971.221.3.902. [DOI] [PubMed] [Google Scholar]
- 37.Campbell KS. PLoS computational biology. 2009;5:e1000560. doi: 10.1371/journal.pcbi.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linke WA, Popov VI, Pollack GH. Biophys J. 1994;67:782–792. doi: 10.1016/S0006-3495(94)80538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- 40.Prakash YS, Cody MJ, Housmans PR, Hannon JD, Sieck GC. J Muscle Res Cell Motil. 1999;20:717–723. doi: 10.1023/a:1005585807179. [DOI] [PubMed] [Google Scholar]
- 41.Jacot JG, Martin JC, Hunt DL. J Biomech. 2010;43:93–98. doi: 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provenzano PP, Keely PJ. J Cell Sci. 124:1195–1205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 44.Janmey PA, McCulloch CA. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 45.Janmey PA, Winer JP, Murray ME, Wen Q. Cell Motil Cytoskeleton. 2009;66:597–605. doi: 10.1002/cm.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wozniak MA, Chen CS. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borg TK, Goldsmith EC, Price R, Carver W, Terracio L, Samarel AM. Cardiovasc Res. 2000;46:277–285. doi: 10.1016/s0008-6363(99)00433-2. [DOI] [PubMed] [Google Scholar]
- 48.Korte FS, Herron TJ, Rovetto MJ, McDonald KS. Am J Physiol Heart Circ Physiol. 2005;289:H801–H812. doi: 10.1152/ajpheart.01227.2004. [DOI] [PubMed] [Google Scholar]
- 49.Korte FS, McDonald KS. The Journal of physiology. 2007;581:725–739. doi: 10.1113/jphysiol.2007.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Randall DJ, Burggren WW, French K, Eckert R. Eckert animal physiology : mechanisms and adaptations. 5. New York: W.H. Freeman and Co.; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.