Abstract
There is little published data on the performance of hand-portable polymerase chain reaction (PCR) systems that can be used by first responders to determine if a suspicious powder contains a potential biothreat agent. We evaluated 5 commercially available hand-portable PCR instruments for detection of Bacillus anthracis. We used a cost-effective, statistically based test plan to evaluate systems at performance levels ranging from 0.85-0.95 lower confidence bound (LCB) of the probability of detection (POD) at confidence levels of 80% to 95%. We assessed specificity using purified genomic DNA from 13 B. anthracis strains and 18 Bacillus near neighbors, potential interference with 22 suspicious powders that are commonly encountered in the field by first responders during suspected biothreat incidents, and the potential for PCR inhibition when B. anthracis spores were spiked into these powders. Our results indicate that 3 of the 5 systems achieved 0.95 LCB of the probability of detection with 95% confidence levels at test concentrations of 2,000 genome equivalents/mL (GE/mL), which is comparable to 2,000 spores/mL. This is more than sufficient sensitivity for screening visible suspicious powders. These systems exhibited no false-positive results or PCR inhibition with common suspicious powders and reliably detected B. anthracis spores spiked into these powders, though some issues with assay controls were observed. Our testing approach enables efficient performance testing using a statistically rigorous and cost-effective test plan to generate performance data that allow users to make informed decisions regarding the purchase and use of field biodetection equipment.
Keywords: : PCR, Anthrax, Bacillus anthracis, Ricin, First responders, Field screening
The authors evaluated 5 commercially available hand-portable PCR instruments for detecting Bacillus anthracis. These systems exhibited no false-positive results or PCR inhibition with common suspicious powders and reliably detected B. anthracis spores spiked into these powders, though some issues with assay controls were observed.
The anthrax attacks of 2001 and other bioterror plots highlight the need for first responders to have field biodetection tools that can rapidly and accurately identify potential biothreats to support short-term tactical decision making.1,2 Early identification of and rapid response to biological threats can save lives, minimize the potential spread of contamination, and reduce costs associated with additional exposures, response, and remediation.
First responders to a “suspicious powder” event may perform initial presumptive screening tests to inform public safety actions and incident management. To enable effective short-term tactical decisions, equipment that can be used in the field to quickly screen suspicious samples for the presence of potential biothreat agents is needed. Market research to identify commercially available biodetection instruments and assays designed for use by first responders has been carried out and the results compiled in a product guide titled Biodetection Technologies for First Responders, which can be downloaded at no cost (http://biodetectionresource.pnnl.gov).3 The technologies identified in this product guide include general biological indicator tests such as protein or adenosine triphosphate tests, biothreat agent–specific immunoassays, and polymerase chain reaction (PCR) systems.4 Immunoassays and PCR assays provide more specific identification of biological threats than do biological indicator tests.5,6 PCR provides the highest level of sensitivity and specificity, although immunoassays are generally more sensitive for toxin detection.7-12 Unfortunately, scant statistically based performance data are available for these biodetection technologies to allow first responders and other end users to make informed procurement decisions.
Standards exist for evaluation of biodetection systems,13-15 including a protocol for PCR evaluation for B. anthracis detection that was developed by the Association of Official Analytical Communities (AOAC) in cooperation with the US Department of Homeland Security–sponsored Stakeholder Panel on Agent Detection Assays (SPADA).16,17 However, these protocols typically require extensive effort and expenditures to evaluate a single instrument. For example, BioFire Diagnostics reported testing 2,479 samples on a RAZOR® EX system to gain certification as an AOAC Performance-Tested Method following the test plan outlined by AOAC Standard Method Performance Requirement (SMPR) 2010.003.12,17 The assay evaluated for this previous certification is not commercially available; however, using the RAZOR® EX Ten™10 Target Screen Kit, this testing would have cost $495,800 for the test kits alone. Biodetection technologies are rapidly advancing—for example, one of the instruments we tested (Tetracore T-COR 4™) has already been replaced by a newer model (T-COR 8™), while another has been discontinued (Smith Detection Bio-Seeq PLUS™). Thus, testing approaches are needed that balance the need for a validated instrument/assay with the need for a process that is neither cost- nor time-prohibitive to allow for timely testing of new technologies and improved assays.
Therefore, we developed a statistically based test plan that provides a more time- and cost-effective approach. This plan allows users to test to a given standard, knowing in advance the maximum number of tests that will be required. We conducted a side-by-side comparison of 5 different hand-portable PCR instruments that had commercially available assays for B. anthracis. The PCR systems are approximately the size of a car battery or smaller, and, with the exception of the BioFire Diagnostics FilmArray® and GeneReach USA Pockit™, the systems can all run off of self-contained battery power. These systems range in weight from 5 to 20 pounds with assay times of 30 to 60 minutes. We used DNA from 13 B. anthracis strains for inclusivity testing (to assess the potential for false-negative results) and 18 near-neighbor strains for exclusivity testing (to assess the potential for false-positive results). We also tested whether 22 commonly encountered suspicious powders generated false-positive results or caused other problems with the assay, and we tested the powders spiked with B. anthracis spores to determine if the powders interfered with the instruments' performance (eg, caused a false-negative result).
Our instrument evaluation metrics were guided by 2 published standards. The highest performance metric is described in AOAC SMPR 2010.003 for laboratory PCR detection of B. anthracis,17 which specifies a 5% lower confidence limit on the probability of detection of 0.95. The lower performance metric we used was guided by American Society for Testing and Materials (ASTM) standard E2885-13 for handheld chemical vapor detectors,19 which specifies a probability of detection of ≥0.85 with an approximate 80% confidence level. Because the probability of detection needed for evaluation of instrument performance represents a minimum level, we used a one-sided confidence interval on the probability of detection parameter, where only the lower confidence bound (LCB) is calculated, rather than the traditional 2-sided confidence interval. Imposing minimum requirements on the lower confidence bound of 0.85-0.95 with a stated level of confidence ensures that the estimate of the probability of detection is at least as good as the lower confidence bound. In this way, requirements on the lower confidence bound with a prescribed level of confidence can help ensure that a sufficiently large number of replicates are used for testing and the performance of the system is adequately described. We designed a test plan to assess performance of the 5 instruments over a similar range of criteria in these published standards (0.85 LCB/80% CL to 0.95 LCB/95% CL), but using a slightly different statistical approach. The performance metrics applied to the inclusivity (target), exclusivity (nontarget), and suspicious powder testing and establish the lower confidence bound on the probability of obtaining the correct answer, whether it is a true positive (inclusivity) or a true negative (exclusivity and powder samples).
Four of the instruments we evaluated also offered assays for the protein toxin ricin by indirectly targeting DNA from Ricinus communis. We conducted limited testing of these assays, and those experimental results are presented in the supplemental information that accompanies this article (http://online.liebertpub.com/doi/suppl/10.1089/hs.2017.0031).
Materials and Methods
Instruments and Assays
The 5 instruments and associated B. anthracis assays that we evaluated are listed in Table 1. Although some previous testing of the FilmArray® has occurred for B. anthracis, Yersinia pestis, and Francisella tularensis, DNA detection11 and some RAZOR™ EX system testing has been done for the analysis of aerosol filter extracts,12 the previous studies were not intended to evaluate the detection of biothreats in suspicious powders. Therefore, these instruments were included in this study to provide a side-by-side comparison using the same statistically based test plan for evaluating the performance of the hand-portable PCR systems for screening powders for B. anthracis. Some manufacturers offered individual assays for B. anthracis, and others offered multiplexed assays designed to detect more than one biothreat agent in a single test. All instruments and assay kits were purchased from the manufacturer.
Table 1.
PCR Systems and Assays Evaluated in this Study
| Instrument | Manufacturer | B. anthracis Assay Tested |
|---|---|---|
| FilmArray® | BioFire, Salt Lake City, UT | BioThreat Panel with internal controls |
| RAZOR® EX | BioFire, Salt Lake City, UT | Ten™ Target Screen Kit with internal controls |
| T-COR 4™ | Tetracore, Rockville, MD | T-COR 4™ pXO2 assay with internal control |
| POCKIT™ | GeneReach USA, Lexington, MA | pXO2 assay, no internal control |
| Bio-Seeq™ PLUS | Smiths Detection, Edgewood, MD | pXO2 assay with internal control |
FilmArray® Assays
The FilmArray® instrument analyzes a single sample at a time for 16 biothreats (the “BioThreat Panel,” which includes 27 targets for the 16 biothreat agents).3 The pouch includes 3 B. anthracis targets (pXO1, pXO2, and B. anthracis chromosome) and requires detection of all 3 targets before a B. anthracis positive result is generated; with fewer than 3 positive targets, the sample is identified as Bacillus species. The pouch includes inhibition and positive controls. Samples were loaded and analyzed according to the manufacturer's instructions.
RAZOR®
The RAZOR® instrument can analyze a single sample at a time using an individual 10-threat pouch (Ten™ Target Screen). The pouch includes a pXO2 assay for B. anthracis along with analyses for 9 other biothreats.3 The pouch incorporates a PCR inhibition control and a positive control. Samples were loaded and analyzed according to the manufacturer's instructions, with minor modifications to ensure a known concentration of DNA or powder was used.
T-COR 4™
The T-COR 4™ is a 4-channel instrument that can analyze up to 4 samples at a time. The pXO2 assay includes an internal positive control. Samples were loaded and analyzed according to the manufacturer's instructions.
POCKIT™
The POCKIT™ is an 8-channel instrument that can analyze up to 8 samples at a time (not including external positive controls). The pXO2 assay does not include internal controls, so a sample position must be used for an external positive control, which is provided as a separate solution. Including a blank (negative) and external positive control with each analytical run allows the analysis of up to 6 analytical samples per run. Samples were loaded and analyzed according to the manufacturer's instructions.
Bio-Seeq™
The Bio-Seeq™ PLUS is a 6-channel instrument capable of programming the temperature and cycle time of each channel independently. Each assay includes an internal positive control. Samples were loaded and analyzed according to the manufacturer's instructions. The Bio-Seeq™ PLUS generates a positive, negative, or indeterminate result. Indeterminate results were re-read once per manufacturer instructions, and, if still indeterminate, the result was considered an assay failure and not a false positive or negative (ie, it was not considered in the statistical performance assessment).
Each day prior to all testing, the laboratory space was disinfected by thoroughly wiping down the working area with the following solutions in sequence: (1) 10% bleach, (2) ultrapure water, (3) DNA AWAY™ (Thermo Scientific, Carlsbad, CA), and (4) 70% ethanol. If blanks (no template/negative controls) analyzed at any time during testing were positive, the space was cleaned again and the analysis was repeated. The analyst wore safety glasses, nitrile gloves, and a lab coat during all work.
The instruments underwent the following tests:
• Inclusivity: DNA from 13 different B. anthracis strains
• Exclusivity: DNA from 18 different near-neighbor strains
• Suspicious powders: 22 commonly encountered suspicious powders
• Biothreat-spiked suspicious powders: 22 common powders, each spiked with B. anthracis spores
Samples
Inclusivity and Exclusivity DNA Panels
The AOAC SMPR 2010.00317 was used to guide the selection of inclusivity and exclusivity Bacillus strains for DNA-based testing. (See Supplemental Information and Tables S1 and S2 for further information.)
All inclusivity strain DNA was obtained from the Critical Reagents Program (CRP) through the Biodefense and Emerging Infections Research Resources Repository (BEI Resources, Manassas, VA) with the exception of PAK-1. The apparent PAK-1 strain obtained from CRP was found to be lacking the pXO2 plasmid; therefore, we obtained a PAK-1 strain from Midwest Research Institute (Kansas City, MO), which did test positive for pXO2 (see discussion below on DNA stock screening).
Exclusivity strains and sources are listed in Table S2 in the supplemental information. We obtained DNA from BEI Resources, and for those DNA strains not available from BEI, we obtained cultures from the American Type Culture Collection (ATCC, Rockville, MD) and from Dr. Karen Hill of Los Alamos National Laboratory. Exclusivity DNA that was prepared on-site was isolated from freshly cultured stocks using the Qiagen DNeasy Blood and Tissue kit (Valencia, CA) as per the manufacturer's protocol.
All DNA stocks were quantified using the Invitrogen Quant-iT™ PicoGreen® assay kit (Grand Island, NY), using either the lambda phage DNA provided in the kit or lambda phage DNA from New England Biolabs (Ipswitch, MA). To confirm that the DNA stocks were free of PCR inhibitors, DNA from the inclusivity (2,000 GE/mL) and exclusivity (20,000 GE/mL) panel strains were analyzed by PCR in triplicate using an ABI 7500 Fast Real-Time PCR System (Life Technologies, Grand Island, NY), with primer and probe sets designed to detect the respective DNA.20 The purified genomic stocks did not exhibit any PCR inhibition.
B. anthracis Spores
Spore stocks were cultured, prepared, loaded into assay pouches or cartridges, and analyzed inside of a biological safety cabinet in a biosafety level 3 (BSL-3) laboratory space following all health and safety requirements for BSL-3 operations. All BSL-3 work was conducted at the Battelle Eastern Science and Technology Center in Aberdeen, MD. B. anthracis Ames was grown on nutrient agar plates with 5 μg/mL manganese sulfate, and after approximately 14 days the spores were removed from the plates and washed twice with sterile water. The concentration of the spores was determined via colony counting on tryptic soy agar plates in triplicate. Shaeffer-Fulton staining of the prepared spores was used to screen for the presence of vegetative cells. Phase contrast microscopy (bright and dark-field conditions) was also used to assess spore quality, where the stock was demonstrated to have nearly 100% “phase bright” spores. Based on these assessments, spores were deemed acceptable for use in further testing. The mean concentration from all enumerations of the working stock was calculated and used as the final concentration of the stock for use in spore sample preparations. All spore samples were prepared from this working stock.
Suspicious Powders
Suspicious powders (see Supplemental Information Table S3 for details) were selected from those listed in AOAC SMPR 2010.005,15 which is a method developed for testing immunoassays for screening visible powders. Additional powders were selected based on feedback from more than 2 dozen first responders. Powders were acquired from Amazon.com. Epson salt and Dipel dust were purchased from local home and garden stores.
Stock solutions/suspensions of the powders were prepared in microcentrifuge tubes at a concentration of 10 mg/mL using Tris-EDTA (TE) buffer and vortex mixing. The stock solution/suspension of each powder was prepared fresh daily. The stock powder solution was diluted using the manufacturer-recommended assay buffer to a final concentration of 0.1 mg/mL for testing. The PCR systems were tested using pure powder suspensions and powder suspensions that had been spiked with B. anthracis Ames spores. The powder suspensions were spiked with spores at the lowest concentration of spores that was consistently detected (ie, detection of all replicates at a given concentration) by the instrument (see Tables 2 and 3). Sample tests resulting in assay control failures were not repeated, as control failures can be indicative of PCR inhibition by the powder. Assay control failures did not count toward a “statistical” failure in the performance metrics.
Table 2.
Estimated Sensitivity of the PCR Systems When Challenged with Specific Numbers of Spores or Spore Concentrations. Data are reported as number of positive results/total tests performed.
| Spores/Assay: | 20 | 200 | 2,000 | * | * |
|---|---|---|---|---|---|
| Spores/mL: | * | * | * | 2,000 | 20,000 |
| FilmArray® | 0/3 | 5/5 | Not tested | 3/3 | Not tested |
| RAZOR® EX | Not tested | 2/5 | 5/5 | 3/3 | Not tested |
| T-COR 4™ | 1/3 | 5/5 | Not tested | 3/3 | Not tested |
| Bio-Seeq™ PLUS | Not tested | 1/5 | 5/5 | Not tested | 5/5 |
| POCKIT™ | 1/3 | 5/5 | Not tested | 3/3 | Not tested |
Varies depending on sample volume required for each assay (see Table 3).
Table 3.
Concentration of spores in the various assays when specific numbers of spores were added, and the lowest concentration of spores that was consistently detected in our tests
| PCR Platform | 20 Spores/Assay (spores/mL in test sample) | 200 Spores/Assay (spores/mL in test sample) | 2,000 Spores/Assay (spores/mL in test sample) | LCCDaSpores/mL |
|---|---|---|---|---|
| FilmArray® | 67 | 675 | Not tested | 675 |
| RAZOR® EX | Not tested | 100 | 1,000 | 1,000 |
| T-COR 4™ | 667 | 66,700 | Not tested | 2,000 |
| Bio-Seeq™ PLUS | Not tested | 4,000 | 40,000 | 20,000 |
| POCKIT™ | 400 | 4,000 | Not tested | 2,000 |
Lowest concentration consistently detected; see Table 2.
Results
Sample Concentrations for Testing
AOAC SMPR 2010.00317 was written as guidance for laboratory analysis of aerosol collection filters and/or liquids, rather than powders. The AOAC standard establishes a required probability of detection of ≥0.95 as demonstrated by an estimated 5% lower confidence limit at a defined acceptable minimum detection level (AMDL) of 2,000 spores/mL or GE/mL. The smallest amount of a powder that can be considered a “visible powder” from the standpoint of a potential biothreat incident is between 0.01 and 0.1 mg, which represents 107 to 108 spores, because there are approximately 109 spores in 1 mg.21 Therefore, a test concentration of 2,000 spores/mL is more than sufficient for the application of screening visible suspicious powders, even considering losses for potential sample collection and extraction step inefficiencies when analyzing spores.
Initial testing established whether the various instruments could detect at least 2,000 spores/mL. In 3 out of 3 trials, all systems except the BioSeeq™ PLUS detected B. anthracis in the samples at this concentration. After consultation with the manufacturer, we performed additional testing of the BioSeeq™ PLUS with a suspension of 20,000 spores/mL, and it positively identified B. anthracis in 3 out of 3 trials.
We also tested whether the instruments could successfully identify B. anthracis when specific numbers of spores were added to the assays. All the systems were able to detect either 200 or 2,000 spores in 5 of 5 trials (Table 2). Spores, however, can have significant extracellular DNA adhered to their surface. No system successfully detected 20 spores in all trials. Because each assay used different sample volumes, when the same number of spores was deposited into each assay, the concentration of spores in each assay varied (Table 3). The lowest concentration of spores consistently detected (LCCD) by the various systems in our limited testing is shown in Table 3. The values for the FilmArray® and RAZOR® EX were in good agreement with the limits of detection reported by the manufacturer (500 and 1,000 spores/mL, respectively). The limit of detection reported by the manufacturer for the BioSeeq™ PLUS (3,750/mL) was determined using vegetative cells rather than spores and so is not directly comparable with our results. No limits of detection were reported by the manufacturers of the POCKIT™ or the T-COR 4®.
Based on the results shown in Tables 2 and 3 and the AOAC SMPR 2010.003 sample test concentration, we used 2,000 and 20,000 GE/mL when performing the inclusivity and exclusivity tests, respectively, with all the systems except the BioSeeq™ PLUS. We conducted inclusivity and exclusivity testing on the BioSeeq™ PLUS using 20,000 and 200,000 GE/mL, respectively. For the powder testing, we spiked suspensions of each powder with the lowest concentration of spores consistently detected by the different instrument systems (Table 3).
Performance Metrics and Test Plan Design
AOAC SMPRs require each strain to be tested only 1 time. If a failure occurs, an additional 96 tests are performed for the strain with the failure. Under this testing approach, assay failures will tremendously increase the number of samples (requiring up to thousands of tests if failures occur for multiple strains) and also significantly increase the time and cost to complete the evaluation for each instrument. In addition, this approach does not achieve the AOAC acceptance criteria of 0.95 probability of detection with 95% confidence level if there are no failures. With no failures for B. anthracis testing,17 20 exclusivity strains are analyzed and 15 inclusivity strains are analyzed with only 1 replicate each, which results in an estimated lower confidence bound of the probability of detection of only 0.88 with 95% confidence level and an estimated lower confidence bound of the probability of detection of only 0.85 with 95% confidence level, respectively. Furthermore, with only 1 replicate per sample when there are no failures, these performance metrics assume strain equivalency, because without assuming equivalency, obtaining a positive result for a single assay replicate would lead to nearly no information regarding the lower confidence bound of the probability of detection.
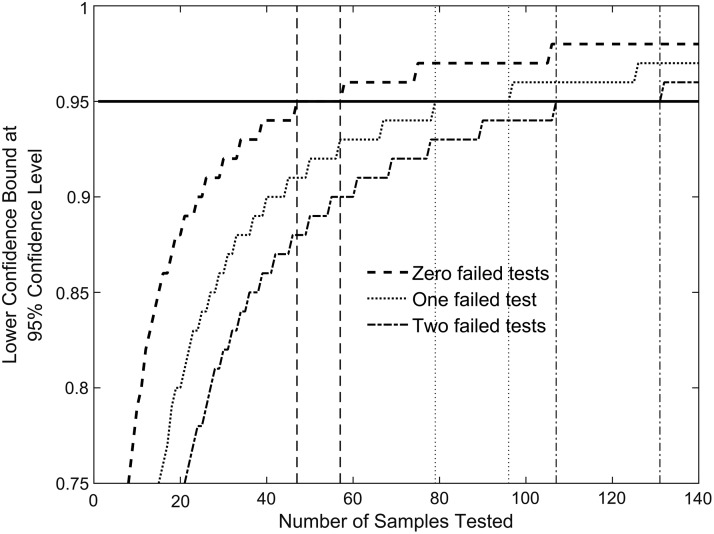
Therefore, we developed an alternative testing approach that can be used to test to a range of acceptable performance criteria while minimizing the number of samples required for testing. Using a 1-sided score confidence interval, the number of samples that need to be tested to achieve a lower confidence bound of 0.95 of the probability of detection at a 95% confidence level can be determined.22 Under this testing approach (see Figure 1):
Figure 1.
Number of independent tests required to meet the performance criterion of 0.95 LCB and 95% confidence at the sample test concentration. If the number of failed results is 0 out of 47 tested samples, 1 out of 79 samples, or 2 out of 107 samples, then a device meets the lower confidence bound of the probability of detection of 0.95 (indicated as the horizontal line) at the sample concentration with 95% confidence. The vertical lines matching the curves bound the regions, in number of samples tested, where the lower confidence bound for a 1-sided 95% confidence interval is equal to 0.95 to 2 decimal places for the cases where 0, 1, or 2 failed tests are observed. For example, the 2 dashed vertical lines indicate that a lower confidence bound estimate of 0.95 (to 2 decimal places) will be observed with 0 failed tests and between 47 and 57 samples tested.
• 47 samples must be tested without a single failed result;
• 79 samples must be tested with no more than a single failed result; or
• 107 samples must be tested with no more than 2 failed results.
The reduction per test can be as significant as going from 96 replicates (AOAC methodology) to 79 replicates with the methodology we propose for the case where 1 failed result is observed. When there are no failed tests, 47 replicate tests result in an estimated lower confidence bound of 0.95 in a one-sided 95% confidence interval, which is fairly rigorous, and, as we have mentioned, a level of performance that is not achieved by testing each of the 15 or 20 strains a single time. When failed tests were observed, we tested up to 79 replicates, because 1 failure in 79 tests provides the same lower confidence bound in a one-sided 95% confidence interval as the case with 47 tests and no failures. If more than 1 failure is observed, the number of tests needed to maintain the statistical standards of 0.95 lower confidence bound and 95% confidence level in a 1-side confidence interval quickly become cost- and time-prohibitive, which is why the testing was limited to 79 samples/test panel/instrument. If multiple failed tests are observed, a lower confidence bound with a specified confidence level can still be computed after the 79 tests are completed, providing the user with a known performance metric for the PCR system.
It is important to note that for the purposes of testing, all inclusivity strains are assumed to be equivalent and all exclusivity strains are assumed to be equivalent. This does not mean that these strains are biologically equivalent, but rather that from an inclusivity and exclusivity panel perspective, they should result in true-positive or true-negative results, respectively. In addition, while the different suspicious powders are clearly not equivalent from a chemical or physical property perspective, all powders are assumed to be equivalent for testing purposes because they should all generate true-negative results (ie, powders are a type of exclusivity or interference panel). This assumption means that it is not necessary to test each individual strain or powder 47 times (with no failures).
Our approach is practical and more reasonable than the AOAC guidance because the current AOAC guidance calls for testing each strain a single time if there are no failures, resulting in not enough replicates to have any confidence that the desired probability of detection is achieved. However, if a failure is observed for the AOAC approach, then 96 tests must be performed for each strain that fails, which is very time- and cost-intensive. If multiple failures are observed, using the AOAC guidance can result in thousands of tests being required, and the instrument under evaluation still may fail to achieve acceptable statistical performance. While assuming equivalency is not ideal and must be considered from a statistically based testing panel perspective (ie, not from a biological or chemical equivalency standpoint), it allows the development of a test plan that is not cost- and time-prohibitive. Our approach will allow the baseline performance of instruments that are currently being used or being considered for field screening by first responders to be evaluated where there would otherwise be no published performance data at all.
Our test plan was used to evaluate whether the performance of the PCR instruments met the specifications of AOAC SMPR 2010.003 (0.95 lower confidence limit of the probability of detection with 95% confidence level) or ASTM E2885-13 (≥0.85 probability of detection with an approximate 80% confidence level) (but we used a slightly different statistical approach than these standards, as discussed previously). These 2 standards are the only known published standards for field detection of chemical or biological threat agents by first responders and provided guidance for the upper and lower limits of the levels of performance we propose: 0.95 LCB/with 95% CL and 0.85 LCB/80% CL, respectively. Testing to a metric within this range of performance constitutes a reasonable and practically achievable approach that will allow producers and consumers to understand the true performance of field detection equipment and select a product with a suitable level of performance for the particular application.
We chose to conduct a maximum of 79 tests for each test module (ie, inclusivity, exclusivity, and suspicious powders) as shown in Table 4. If the assay had 47 correct results with no failures, the system achieved 0.95 lower confidence bound with 95% confidence level for that test. However, if there were 1 or more failures, up to 79 tests were performed. If there were 10 failed tests, then the results fell below our lowest performance metric (<0.85 LCB/<80% CL, shown in dark gray in Table 4), and testing was halted. This test plan resulted in conducting as few as 10 (if there are 10 initial failures) and at most 79 tests (if there are 1-9 failures) for a given DNA inclusivity, DNA exclusivity, or suspicious powder panel.
Table 4.
Lower confidence bound (LCB) and confidence level (CL) achieved with 79 tests, as a function of the number of failed tests
| Total Number of Tests = 79 | |||||
|---|---|---|---|---|---|
| Confidence Level | 95% | 90% | 85% | 80% | 75% |
| Number of Failed Tests | Lower Confidence Bound Values | ||||
| 1 | 0.95 | 0.96 | 0.97 | 0.97 | 0.98 |
| 2 | 0.93 | 0.94 | 0.95 | 0.96 | 0.96 |
| 3 | 0.91 | 0.92 | 0.93 | 0.94 | 0.94 |
| 4 | 0.89 | 0.91 | 0.92 | 0.92 | 0.93 |
| 5 | 0.88 | 0.89 | 0.90 | 0.91 | 0.92 |
| 6 | 0.86 | 0.88 | 0.89 | 0.90 | 0.90 |
| 7 | 0.84 | 0.86 | 0.87 | 0.88 | 0.89 |
| 8 | 0.83 | 0.85 | 0.86 | 0.87 | 0.87 |
| 9 | 0.81 | 0.83 | 0.84 | 0.85 | 0.86 |
| 10 | 0.80 | 0.82 | 0.83 | 0.84 | 0.85 |
White: meets 0.95 LCB/95% CL performance metric.
Light gray: meets 0.85 LCB/80% CL performance metric.
Dark gray: does not meet lowest performance metric.
Inclusivity, Exclusivity, and Suspicious Powder Testing
Inclusivity DNA testing was conducted using 3 replicates each of 13 B. anthracis strains, plus at least 8 additional tests (to obtain a total of 47 tests), where the 8 additional strains were selected in a statistically randomized fashion. Selecting samples randomly ensures that effects on the test due solely to the identity of a sample (eg, how it was prepared or collected) are mitigated, and that conclusions reached from the tests are therefore valid across the range of samples used for the evaluation. The total numbers of assays performed (47 or 79) are needed to demonstrate that the above-described statistical requirements are met (when 0 failures occur or 1 failure occurs, respectively).
Each test was conducted using 2,000 GE/mL final concentration, with the exception of the Bio-Seeq™ PLUS, which was tested at 20,000 GE/mL. All of the systems met the 0.95 LCB/95% CL performance metric with no failures in 47 tests except for the BioSeeq™ PLUS (Table 5). The BioSeeq™ PLUS had 10 failed inclusivity tests in 53 trials, and testing was halted.
Table 5.
Performance of the 5 PCR systems for inclusivity, exclusivity, and suspicious powder testing. Data are reported as number of positive results/total tests performed.
| Test Panel | FilmArray® | RAZOR®EX | T-COR 4™ | Bio-Seeq™PLUS* | POCKIT™ |
|---|---|---|---|---|---|
| Inclusivity strain DNA (2,000 GE/mL)a | 47/47 | 47/47 | 47/47 | 38/53 | 47/47 |
| Exclusivity species DNA (20,000 GE/mL)a | 1/79 | 1/79 | 0/47 | 0/47 | 3/79 |
| Common suspicious powders (0.1 mg/mL) | 0/66 | 0/66 | 0/66 | 1/79 | 15/66 |
Used 10X higher concentration for Bio-Seeq™ PLUS testing.
White: meets 0.95 LCB/95% CL performance metric.
Light gray: meets 0.85 LCB/80% CL performance metric.
Dark gray: does not meet minimum performance metric.
Exclusivity DNA testing was conducted using 2 replicates each of 18 near-neighbor Bacillus strains plus at least 11 additional tests where strains were selected at random. Exclusivity testing was conducted at 20,000 GE/mL final assay concentration, except for the Bio-Seeq™ PLUS, which was tested at 200,000 GE/mL. All of the systems met the 0.95 LCB/95% CL performance criterion except the POCKIT™, which achieved a slightly lower performance metric of 0.91 LCB/95% CL, with 3 failures in 79 tests (Table 5).
The 22 suspicious powders (see Supplementary Information Table S3) were each tested in triplicate to assess any potential impact to the assay. The FilmArray®, RAZOR EX®, and T-COR 4™ had no false-positives (Table 5). The Bio-Seeq™ PLUS had 1 false-positive (polyethylene glycol [PEG] was the sample), so we conducted an additional 13 tests using randomly selected powders with no further false-positives. The POCKIT™ had 15 false-positives and therefore failed to meet our lowest performance metric.
Assay Interference by Common Suspicious Powders
We also tested the common suspicious powders to determine if they interfered with the assays. We spiked 0.1 mg/mL suspensions/solutions of the 22 powders with B. anthracis Ames spores at the lowest concentration consistently detected by each instrument in our previous tests (Table 3). The agent concentrations were therefore different for each instrument. Each spiked powder was tested twice.
All 22 spiked powders tested positive for B. anthracis in 2 of 2 tests on the FilmArray®, the T-COR 4™, and the POCKIT™. White flour was tested 3 times with the FilmArray® because 1 assay was deemed invalid due to a software error, while with the T-COR 4™, 1 assay with PEG resulted in an expected true-positive B. anthracis detection but was negative for the internal positive control.
Of the 22 spiked powders, popcorn salt generated 1 false-negative result on the RAZOR® EX. The rest of the powders tested positive 2 of 2 times. Two of the assays had internal control errors: 1 assay with borax and 1 assay with powdered toothpaste generated positive inhibition controls, but the internal positive control was negative, suggesting a possible manufacturing defect in the RAZOR® EX pouch.
Only 13 of the 22 spiked powders tested positive for B. anthracis in 2 of 2 trials on the Bio-Seeq™ PLUS. Coffee creamer, baking powder, and road dust generated 1 true-positive and 1 false-negative result each. Baking soda and Epsom salt generated 1 true-positive and 1 indeterminate result each. PEG, chalk dust, and borax generated 2 indeterminate results each. White flour and kaolin resulted in 1 indeterminate and 1 negative result each.
Discussion
Determination of Test Sample Concentration
The PCR systems we evaluated for screening suspicious powders are hand-portable instruments and assays that are used or could be used in the field by first responders. ASTM E2458-1018 provides a protocol for field collection of powders suspected of containing a biological threat agent, such that the maximum amount of powder is collected and sent to a reference laboratory in the Centers for Disease Control Laboratory Response Network for confirmatory analysis (Method A). The standard specifies that any powder remaining after this collection procedure can be sampled with a moistened swab and subjected to field biological assessment (Method B). We conducted a limited study and determined that 1 to 10 (average 6 ± 3) mg of powder typically remained after carrying out this procedure on 22 different powders in triplicate. Published data indicate there are approximately 1012 B. anthracis spores/g;21 therefore, 1 mg of pure spores would contain approximately 109 spores. One possible scenario for an intentional biological release is the use of a common powder spiked with biothreat agent. If a trace amount of biothreat agent (eg, 10 micrograms) is spiked into a benign powder (eg, 1 mg), this sample would contain approximately 107 spores. The protocols for the assays use a moistened swab to collect and suspend the sample in buffer, typically about 1 mL. Suspending 0.1 mg of a powder spiked with 1% spores, as described above, would provide approximately 106 spores/mL, far higher than the minimum concentrations required for consistent detection for all of the PCR systems (Table 3).
Biodetection Instrument Performance
We tested all the instruments except the BioSeeq™ PLUS at 2,000 spores/mL or GE/mL. We used 20,000 GE/mL to evaluate the BioSeeq PLUS™, based on the initial results of our sensitivity screening.
Our DNA testing results indicated that the assays tested on 3 PCR instruments (FilmArray®, RAZOR® EX, and T-COR 4™) met the 0.95 LCB/95% CL for inclusivity DNA (true-positives), exclusivity DNA (true-negatives), and tests with the suspicious powders. Of these 3 platforms, the FilmArray® required the fewest manipulations by the operator to carry out a test, while the T-COR 4™ required the most.
When tested for sample matrix effects or interference by powders (Table S3), the FilmArray® tested positive for Coxiella burnetii in 2 samples, 1 each with milk powder and infant formula (the BioThreat Panel assay included targets for this pathogen as well as B. anthracis and 15 additional biothreats). Coxiella burnetii is a common pathogen found in milk, and the positive PCR results are presumed to be from residual C. burnetii DNA fragments that survived pasteurization.23 This study was focused only on B. anthracis assays, and the additional signatures were not part of this evaluation, so these were not counted as FilmArray® failures for B. anthracis (and they were likely not failures because C. burnetti DNA actually may have been present in the milk powder). Neither the RAZOR nor the T-COR 4™ exhibited any false-positive results with any of the pure powders tested; however, the T-COR 4™ had numerous internal control failures or had internal controls with low fluorescence readings when tested with pure powders, which were not counted as assay failures.
All 3 of these instruments detected B. anthracis in the spiked powders at the lowest consistently detected concentration we estimated for each instrument (Table 3), except for one RAZOR® false-negative for B. anthracis spiked into popcorn salt. This false-negative was at a concentration of 1,000 spores/mL, which is lower than our minimum test concentration of 2,000 spores/mL. The RAZOR EX™ had 2 positive control failures, and the T-COR 4™ had 1 internal control failure for the spiked powders. These control failures were not counted as assay failures. Both the FilmArray® and RAZOR have been tested in other studies, and our results are consistent with current literature.3,7-9,11,12
The Bio-Seeq™ PLUS pXO2 assay was tested at a concentration 10 times higher than other instruments based on our initial LCCD scoping studies (Table 3). Even at this concentration, the platform returned 10 false-negatives in the inclusivity testing. Consequently, this instrument failed to meet our lowest performance criterion of 0.85 LCB/80% CL for inclusivity, but it did achieve a 0.95 LCB/95% CL for exclusivity testing and with the suspicious powders (Table 5). For powder samples spiked with spores, a large number of false-negative results were obtained and the system failed to meet the lowest performance criteria.
The POCKIT™ pXO2 assay met the 0.95 LCB/95% CL metric for inclusivity testing, with no failures in 47 tests, and it correctly detected B. anthracis in 2 of 2 trials with each of the 22 spiked powders. It met the 0.85 LCB/80% CL metric for exclusivity testing with 3 sample failures in 79 tests (false-positive readings for B. cereus S2-8, B. cereus 3A, and B. cereus D17.) It failed to meet the lowest performance metric in the suspicious powder testing, where false-positive results were noted for 15 of the 22 pure suspicious powders.
Testing Framework and Guidelines
This study provides a framework and guidelines for developing test plans for field biodetection system evaluation. Importantly, the development and implementation of this test plan took into consideration prior work and also the intended application (field detection of visible suspicious powders). Our choice of B. anthracis inclusivity and exclusivity strains was guided by the existing AOAC standard for aerosol filter testing,17 the literature,12,16 and knowledge of the assays and the application. We evaluated instrument performance in the range spanned by 2 published standards: AOAC SMPR 2010.003 (0.95 lower confidence limit of POD/95% CL) and ASTM E2885-13 (0.85 lower limit of POD/80% CL), and we developed a test plan that requires a minimum of 47 samples without failure and 79 samples if there were up to 9 failures for the inclusivity, exclusivity, and powder samples. To meet the more stringent performance criteria (0.95 LCB/95% CL), only a single failure would be acceptable in 79 tests. Therefore, at most 237 replicates are required for the inclusivity, exclusivity, and powder testing. If costs are further limiting, initial screening could be done with 47 each of the inclusivity, exclusivity, and powder samples with the requirement of no failures. This is far fewer than the maximum number of tests required by existing AOAC SMPR standards, which have a similar criteria of 0.95 LCB/95% CL but require 96 replicates be tested for each strain that has an assay failure.18 This could result in testing nearly 3,000 samples in a worst-case scenario.
Conclusions
This study demonstrates the use of a statistically based approach for assessing the performance of PCR-based systems for screening visible quantities of powders for B. anthracis. These instruments can be used by first responders for screening trace amounts of suspicious powders for public safety actions and short-term tactical decision making, after the bulk of the sample is collected, transported, and analyzed by public health laboratories and other officials for confirmatory analysis. Three of the 5 PCR instruments (FilmArray®, RAZOR® EX, and T-COR 4™) achieved the specificity testing criteria of 0.95 LCB/95% CL with 2,000 GE/mL inclusivity DNA and 20,000 GE/mL exclusivity DNA. We have published a similar study that evaluated immunoassays and general biological indicator tests (eg, protein or ATP tests) performance using B. anthracis spores and suspicious powder. The indicator tests resulted in numerous false-positives and -negatives with powder samples, while approximately half of the B. anthracis immunoassays evaluated could not detect our test concentration of 107 spores/mL.24
The instruments performed well in additional screening with commonly encountered suspicious powders and B. anthracis–spiked powders: the FilmArray® had no false-negatives or false-positives, the RAZOR® EX had only 1 false-negative and 2 positive control failures, and the T-COR 4™ had no false-positive results but did have numerous problems with internal controls (either failures or low fluorescence), which would require additional time for retesting in an operational environment. The manufacturer of the T-COR 4™ has replaced it with the T-COR 8™, which has not been evaluated. We tested all of these instruments under laboratory conditions; additional testing is needed to ensure that the instruments perform to the same levels in the field and in the hands of first responders.
While this study was focused on the performance for B. anthracis detection, other types of testing could be adapted to this approach to meet the needs of different applications. It is important to note that as more equipment and assays become commercially available, it will be necessary to quickly and cost-effectively compare the performance of these emerging technologies with existing instrumentation using an objective testing framework that measures instrument performance relative to a performance standard. Our statistically based testing approach enables efficient instrument performance testing to allow users to make informed decisions regarding the purchase and use of biodetection equipment in the field.
Supplementary Material
Acknowledgments
We acknowledge Dr. Karen Hill from Los Alamos National Laboratory for providing stocks of exclusivity strain organisms that were not available through CRP/BEI or ATCC. We thank Kimberly Weber, Andrew Phipps, Bobbi Horne, Rebecca Limmer, and Jan Nevy from the Battelle Memorial Institute for their assistance in performing the tests using B. anthracis Ames spores. We also thank Cheryl Baird for technical reviews and Hayley Cardamone for conducting powder sampling measurements when they were at Pacific Northwest National Laboratory (PNNL). The Department of Homeland Security, Science and Technology Directorate, provided funding for this research through contracts HSHQDC-08-X00843/6 and HSHQPM-12-X-00169/6 to PNNL. PNNL is operated by Battelle Memorial Institute for the US Department of Energy under contract DE-AC06-76RLO. This evaluation does not indicate an endorsement or adoption of any of the technologies by PNNL or the US Department of Homeland Security. The authors declare no conflicts of interest related to this work.
References
- 1.Kman NE, Bachmann DJ. Biosurveillance: a review and update. Adv Prev Med 2012;2012:301408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt E, Shanker T. Qaeda trying to harness toxin for bombs, U.S. officials fear. New York Times August 12, 2011. http://www.nytimes.com/2011/08/13/world/middleeast/13terror.html Accessed November30, 2016
- 3.Ozanich RM, Bartholomew RA, Bruckner-Lea CJ, Hess BM, Arce JS, Cardamone HC. Biodetection Technologies for First Responders: 2015 Edition. Richland, WA: Pacific Northwest National Laboratory; 2015. http://biodetectionresource.pnnl.gov Accessed November30, 2016 [Google Scholar]
- 4.Poore C, Clark P, Emanuel PA. An evaluation of suspicious powder screening tools for first responders. J Hazard Mater 2009;172(2-3):559-565 [DOI] [PubMed] [Google Scholar]
- 5.Gessler F, Pagel-Wieder S, Avondet MA, Böhnel H. Evaluation of lateral flow assays for the detection of botulinum neurotoxin type A and their application in laboratory diagnosis of botulism. Diagn Microbiol Infect Dis 2007;57(3):243-249 [DOI] [PubMed] [Google Scholar]
- 6.King D, Luna V, Cannons A, Cattani J, Amuso P. Performance assessment of three commercial assays for direct detection of Bacillus anthracis spores. J Clin Microbiol 2003;41(7):3454-3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slotved HC, Sparding N, Tanassi JT, Steenhard NR, Heegaard NH. Evaluating 6 ricin field detection assays. Biosecur Bioterror 2014;12(4):186-189 [DOI] [PubMed] [Google Scholar]
- 8.Slotved HC, Tanassi JT, Sparding N, Lindqvist A, Steenhard NR, Heegaard NH. Botulinum toxin field assays evaluated using cosmetic botox preparations. Biosecur Bioterror 2013;11(4):280-286 [DOI] [PubMed] [Google Scholar]
- 9.Peckham GD, Hew BD, Waller DF, Holdaway C, Jen M. Amperometric detection of Bacillus anthracis spores: A portable, low-cost approach to the ELISA. Int J Electrochem 2013;(Article 803485):1-6 [Google Scholar]
- 10.Zasada AA, Formińska K, Zacharczuk K, Jacob D, Grunow R. Comparison of eleven commercially available rapid tests for detection of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. Lett Appl Microbiol 2015;60(5):409-413 [DOI] [PubMed] [Google Scholar]
- 11.Seiner DR, Colburn HA, Baird C, et al. Evaluation of the FilmArray® system for detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis. J Appl Microbiol 2013;114(4):992-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spaulding UK, Christensen CJ, Crisp RJ, et al. RAZOR® EX anthrax air detection system. J AOAC Int 2012;95(3):860-891 [DOI] [PubMed] [Google Scholar]
- 13.AOAC SMPR 2010.001. Standard method performance requirements for polymerase chain reaction (PCR) methods for detection of Francisella tularensis in aerosol collection filters and/or liquids. J AOAC Int 2011;94(4):1338-1341 [PubMed] [Google Scholar]
- 14.AOAC SMPR 2010.002. Standard method performance requirements for polymerase chain reaction (PCR) methods for detection of Yersinia pestis in aerosol collection filters and/or liquids. J AOAC Int 2011;94(4):1342-1346 [PubMed] [Google Scholar]
- 15.AOAC SMPR 2010.004. Standard method performance requirements for immunological-based handheld assays (HHAs) for detection of Bacillus anthracis spores in visible powders. J AOAC Int 2011;94(4):1352-1355 [PubMed] [Google Scholar]
- 16.Coates SG, Brunelle SL, Davenport MG. Development of standard method performance requirements for biological threat agent detection methods. J AOAC Int 2011;94(4):1328-1337 [PubMed] [Google Scholar]
- 17.AOAC SMPR 2010.003. Standard method performance requirements for polymerase chain reaction (PCR) methods for detection of Bacillus anthracis in aerosol collection filters and/or liquids. J AOAC Int 2011;94(4):1347-1351 [PubMed] [Google Scholar]
- 18.ASTM E2458-10: Standard practices for bulk sample collection and swab sample collection of visible powders suspected of being biological agents from nonporous surfaces. In: E2458-10. West Conshohoken, PA: ASTM International; 2010 [PubMed] [Google Scholar]
- 19.ASTM. E2885-13: Standard specification for handheld point chemical vapor detectors (HPCVD) for homeland security applications. In: E2885-13. West Conshohocken, PA: ASTM International; 2013 [Google Scholar]
- 20.Wielinga PR, Hamidjaja RA, Agren J, et al. A multiplex real-time PCR for identifying and differentiating B. anthracis virulent types. Int J Food Microbiol 2011;145(Suppl 1):S137-S144 [DOI] [PubMed] [Google Scholar]
- 21.St. Amant DC, Carey LF, Guelta BA. An improved method for the production of Bacillus subtilis var. niger spores for use as a simulant for biological warfare agents—quality analysis. Aberdeen Proving Ground, MD: Edgewood Chemical Biological Center; 2003. Accession No.: ADA483822 [Google Scholar]
- 22.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat 1998;52(2):119-126 [Google Scholar]
- 23.Tilburg JJ, Roest HJ, Nabuurs-Franssen MH, Horrevorts AM, Klaassen CH. Genotyping reveals the presence of a predominant genotype of Coxiella burnetii in consumer milk products. J Clin Microbiol 2012;50(6):2156-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartholomew RA, Ozanich R, Arce J, et al. Evaluation of immunoassays and general biological indicator tests for Bacillus anthracis and ricin detection. Health Secur 2016; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.