Abstract
Introduction
Due to dangers associated with potential accidents from nuclear energy and terrorist threats, there is a need for high-throughput biodosimetry to rapidly assess individual doses of radiation exposure. Lipidomics and metabolomics are becoming common tools for determining global signatures after disease or other physical insult and provide a “snapshot” of potential cellular damage.
Objectives
The current study assesses changes in the nonhuman primate (NHP) serum lipidome and metabolome 7 days following exposure to ionizing radiation (IR).
Methods
Serum sample lipids and metabolites were extracted using a biphasic liquid-liquid extraction and analyzed by ultra performance liquid chromatography quadrupole time-of-flight mass spectrometry. Global radiation signatures were acquired in data-independent mode.
Results
Radiation exposure caused significant perturbations in lipid metabolism, affecting all major lipid species, including free fatty acids, glycerolipids, glycerophospholipids and esterified sterols. In particular, we observed a significant increase in the levels of polyunsaturated fatty acids (PUFA)-containing lipids in the serum of NHPs exposed to 10 Gy radiation, suggesting a primary role played by PUFAs in the physiological response to IR. Metabolomics profiling indicated an increase in the levels of amino acids, carnitine, and purine metabolites in the serum of NHPs exposed to 10 Gy radiation, suggesting perturbations to protein digestion/absorption, biological oxidations, and fatty acid β-oxidation.
Conclusions
This is the first report to determine changes in the global NHP serum lipidome and metabolome following radiation exposure and provides information for developing metabolomic biomarker panels in human-based biodosimetry.
Keywords: Lipidomics, Metabolomics, Ionizing Radiation, Nonhuman Primate
Introduction
Due to potential widespread exposures to radiation following nuclear or radiological events, tools for high-throughput biodosimetry are needed to quickly assess individual absorbed dose. Lipidomics and metabolomics (analysis of molecules < 1 kDa) represent the downstream portion of systems biology (the integration of all “omic” information from genes to proteins to metabolites) (Zhang et al. 2012; Jones et al. 2014). Global biofluid metabolite signatures provide “snapshots” of an organism’s health status and may provide biomarkers for underlying diseases (Patti et al. 2012) and radiation exposure. While global lipidomics and metabolomics offer exciting technologies with far-reaching application, they are not without limitations. Firstly, many metabolites and lipids are functionally uncharacterized or completely unknown. Characterizing unknown compound structure requires laborious methods including chromatographic fractionation, mass spectrometry (MS), nuclear magnetic resonance spectroscopy, infrared spectroscopy, among others (Reichenbächer & Popp 2012). Secondly, if a metabolite structure is known, many standards are unavailable or are prohibitively expensive to synthesize. Finally, with the vast amount of information collected from global metabolomics, robust statistical methodologies for extracting biologically relevant information are time consuming (Sugimoto et al. 2012; Mak et al. 2015). Despite these limitations, lipidomics and metabolomics have progressed rapidly in recent years leading to detection of critical disease biomarkers (Manna et al. 2010; Mapstone et al. 2014), contributed to drug development/assays (Vihervaara et al. 2014), and provided insights on natural products (Cox et al. 2014) and nutritional science (Astarita & Langridge 2013).
Lipids represent a range of compounds with varied structures and functions. Broad lipid classes encompass eight categories with varied function: 1) glycerolipids, 2) unesterified fatty acyls, 3) sterols, 4) polyketides, 5) sphingolipids, 6) saccharolipids, 7) prenols, 8) and glycerophospholipids (GPs) (Fahy et al. 2007;2009). While the major function of lipids was classically considered the formation of cellular bilayer membranes (GPs) and energy storage (triacylglycerides [TGs]), our current understanding indicates more intricate functions in cellular signaling and inflammation. Sphingosines and ceramides (CERs) have been identified as key signaling molecules involved in apoptosis and growth arrest (Hannun & Obeid 2011). Eicosanoid oxygenation by cyclooxygenase (COX), lipoxygenase (LOX), and CYP-450 pathways (or non-enzymatic pathways) lead to inflammation and are important tissue damage biomarkers (reviewed by Astarita et al. 2015). Perturbations to broad lipid classes may indicate lipoxidative stress intensity (Yin et al. 2011). Plasmalogen deficiencies are markers of peroxisomal disorders and increased oxidative stress, as the vinyl ether bond has lower dissociation energies compared to diacyl GPs (Braverman & Moser 2012). Diacyl GPs may be hydrolyzed producing higher LysoGP proportions (Subbanagounder et al. 2000; Fruhwirth et al. 2007). Monitoring shifts in lipidomic and metabolomic profiles may provide biomarkers for assessing an individual’s physical damage from xenobiotic compounds or ionizing radiation (IR).
Exposure to IR generates reactions with water to form free hydrogen and hydroxyl radicals, thus increasing oxidative stress. In addition, the response to IR involves a pro-inflammatory component, which can further contribute to oxidative stress (Mukherjee et al. 2014). Lipidomic and metabolomic radiation signatures may offer clues into the extent of physiological damage from different doses of IR and be utilized for high throughput biodosimetry. As unsaturated lipids are susceptible to oxidative damage due to IR exposure, changes in concentration of specific lipid species make them relevant for development of radiation biomarkers. Conversely, a previous study on mice exposed to IR suggested that there was a potential role of increased polyunsaturated fatty acids (PUFA) to mediate inflammation (Laiakis et al. 2014a). The roles of PUFAs in inflammation vs. degradation in a highly oxidative state remains to be elucidated.
Here, we use a nonhuman primate (NHP) model that provides a means to lower spectral “noisiness” due to genetic and pre-existing medical conditions present in human studies, while generating more human-based relevant information when compared to mouse and rat models (Tyburski et al. 2008; Johnson et al. 2011; Laiakis et al. 2014b). While previous studies have reported radiation metabolic signatures in NHP urine at various time points (Johnson et al. 2012; Pannkuk et al. 2015), information on perturbations from IR in NHP serum is lacking, as only one targeted study exists (Jones et al. 2014). These studies are needed to establish biodosimetry methodology in the event of large-scale radiological exposures (DiCarlo et al. 2010;2012).
The current study applies a biphasic liquid-liquid extraction to NHP serum to obtain global metabolomic and lipidomic radiation signatures 7 days after IR exposure using an ultra performance liquid chromatography (UPLC) quadrupole time-of-flight (QTOF) MS platform. Interestingly, we found specific polyunsaturated TGs, cholesteryl esters (ChoEs), and GPs exhibited a drastic increase in NHPs exposed to 10 Gy IR. This study provides new evidence to be used for biomarker verification and validation in high-throughput biodosimetry following large-scale radiation exposures.
Materials and Methods
Chemicals
All reagents were LC-MS grade (Fisher Scientific, Hanover Park, IL). Standards for lipidomics included phosphatidylethanolamine (PE 14:0/14:0), phosphatidylcholine (PC 14:0/14:0), lysophosphatidylcholine (LysoPC 17:1), cholesterol, cholesteryl ester (ChoE 19:0) (Avanti Polar Lipids, Inc., Alabaster, AL), TG (19:1/19:1/19:1), diacylglyceride (DG 20:1/20:1), monoacylglyceride (MG 17:1), and free fatty acid (FFA 17:1) (Nu-check Prep Inc., Elysian, MN). Metabolomic standards included debrisoquine sulfate, 4-nitrobenzoic acid, valine, hypoxanthine, carnitine, proline, tyrosine, glucose, uridine, and taurine (all Sigma-Aldrich, St. Louis, MO). All standards were of the highest purity available.
NHP System and Experimental Treatments
Study animals consisted of the cohort previously described (Pannkuk et al. 2015). Briefly, Rhesus monkeys (Macaca mulatta) (72 animals used for this study, ~ 4.75 yrs old [3.6–5.9 yrs old], ~ 4.65 kg) of Chinese origin, were acclimated to environmental conditions for at least 6 weeks prior to study initiation. Purified water was provided ad libitum to all animals and food consumption evaluated at least twice daily. Animals received a certified chow twice daily representing approximately 420 grams of food daily (Teklad Certified Hi-Fiber Primate Diet no. 7195C; Harlan Laboratories, Madison, WI). Room environment was continuously controlled (21°± 3 Celsius; 30–70% humidity) with twelve (12) air changes per hour using an HEPA system. Animals were monitored continuously (i.e., 24 h/day) by veterinarians for any clinical signs of distress or pain and the experimental protocol included strict euthanasia criteria reviewed and approved by the Institutional Animal Care and Use Committee. No animal presented clinical signs that warranted euthanasia per these pre-established criteria. Animals received a single total body irradiation (TBI) exposure at a dose rate of ~ 0.6 Gy/min with a 60Co γ source. Animals received proper medication for emesis (ondansetron, 1 mg/kg once prior to TBI and once after) and mucosal protector (sucralfate, PO, 0.5g BID). One female also received additional fruits from Day 1 to 5 post-TBI but all other animals received a standardized medical management plan (Table S1). Twelve animals (1/2 male and 1/2 female) per treatment received 2, 4, 6, 7, or 10 Gy TBI and buprenorphine (0.01 mg/kg, SC) for analgesia. These doses were administered to span both hematopoietic and minor gastrointestinal syndromes. Clinical examinations/clinical pathology were performed on all animals pre-IR and seven days post-IR. Animals were humanely euthanized on day seven followed by immediate tissue collection. Blood (2 mL each) was collected in serum separating tubes (after their morning meal) and centrifuged at 4°C, aliquoted, and stored at −70°C until shipment on dry ice to Georgetown University Medical Center (sample size; control, 3 Gy, 7 Gy n=12; 6 Gy n=11; 2 Gy, 10 Gy n=10).
Sample Preparation and Analysis
Serum samples (25 µl) were extracted with cold chloroform:methanol (100 µl, 2:1) containing internal standards, incubated for 5 min at room temperature, vortexed for 30 seconds, and centrifuged for 10 min (10,000 x g, 4°C). The lower organic phase was removed with a glass pipette, evaporated under N2, and reconstituted in 200 µl isopropanol:acetonitrile:H2O (50:25:25) (0.5 µM internal standards; see Chemicals section). The upper aqueous phase was evaporated under vacuum and reconstituted in 200 µl acetonitrile:H2O (66:34) containing internal standards for normalization (2 µM debrisoquine sulfate, 30 µM 4-nitrobenzoic acid). Samples were stored at −80°C until further LC-MS analyses.
For global metabolic profiling, samples were injected (2 µl) and analyzed by UPLC with a Waters Acquity BEH C18 1.7 µm, 2.1 × 50 mm column coupled to a Xevo® G2 QTOF-MS (Waters, Milford, MA) and analyzed in both negative and positive electrospray ionization (ESI) MSE modes as previously described (Goudarzi et al. 2015). For global lipid profiling, samples were injected (1 µl) and analyzed by UPLC with a Waters Acquity CSH C18 1.7 µm, 2.1 × 100 mm column coupled to a Synapt® G2-Si HDMS QTOF-MS (Waters, Milford, MA) and analyzed in the modes above with chromatographic conditions as previously described (Li et al. 2014; supplemental information). Select lipids were analyzed by quadrupole mass filtered tandem MS. Leucine enkephalin (556.2771 [M+H]+ or 554.2615 [M-H]−) was used to calibrate accurate mass on both instruments.
Data Processing, Statistical Analysis, and Marker Validation
Lipidomics
The total ion chromatogram (TIC) (Fig. S1) was deconvoluted and peak aligned using Progenesis QI (Nonlinear Dynamics, Newcastle, UK). Centroid raw data files were aligned to a pooled quality control (QC; 5 µl of each sample). The most suitable candidate QC file was chosen by highest similarity by Progenesis QI. Vector alignment quality was manually processed and files were aligned with sensitivity (10 ppm), retention time limits, and peak normalization (normalize to all compounds) at the default values. Putative identification was obtained by comparing ions to the LIPID MAPS database and MetaScope theoretical fragment search (Fahy et al. 2007;2009). The non-filtered dataset was normalized to an internal standard (response analyte/[response internal standard*concentration]) and analyzed with a Kruskal-Wallis test and a post-hoc Duncan test (SAS 9.4, Cary, NC). A two-way ANOVA was performed to test for significant effects between radiation dose, sex, and weight loss (>0.5 kg loss). For FFAs and ChoEs the normalized values from Progenesis QI were used for statistical analysis. The machine-learning algorithm Random Forests (RF) was applied through the programming language R producing a multi-dimensional scaling plot (MDS) and heatmap of the top 50 metabolites, as ranked through RF. A standard singular value decomposition based principal component analysis (PCA) was performed. A heatmap and volcano plot (utilizing false discovery rate [FDR] corrected P values by a classical Benjamini-Hochberg step-up correction procedure) were generated with the software MetaboLyzer on complete-presence ions, which are defined as ions that are present in at least 70% of the samples in both analysis groups (Mak et al. 2014).
Metabolomics
The metabolomic dataset was deconvoluted in a procedure identical to the lipidomic data and analyzed with univariate and multivariate statistical methods via MetaboLyzer (Mak et al. 2014). Complete-presence ions (≥70%) were analyzed with a Welch’s t-test (P<0.05) (also presented with FDR P values with Q set to 10%), while partial-presence ions (<70%) were treated as categorical data and analyzed with a Barnard’s test (P<0.05). Putative identities were assigned to ions that were found to be statistically significant via neutral mass elucidation and monoisotopic mass matching to within 20 ppm utilizing the Human Metabolome Database (HMDB), the Kyoto Encyclopedia of Genes and Genomes, BioCyc, and ChEBI (Kanehisa & Goto 2000; Degtyarenko et al. 2008; Wishart et al. 2009; Caspi et al. 2014; Croft et al. 2014; Mak et al. 2014). Putative identities were then mapped to KEGG, BioCyc, and Reactome pathways. Multivariate analysis via singular value decomposition based PCA was also conducted on the statistically significant ions. A subset of significant ions was validated to pure standards by tandem MS.
Results
Lipidomics
In order to determine the effect of radiation on the lipidome, we analyzed the organic fraction of NHP serum using LC-MS acquired in in data-independent mode and select lipids by mass filtered tandem MS. Multivariate data analysis of NHP serum lipids exposed to ionizing radiation showed that all exposed groups separated from the control group (Fig. S2,S3). IR exposed groups separated from the control with greatest separation observed in the 10 Gy group (Fig. S2A, B). Separation in the first dimension is based on differences between the in control and IR exposed groups, while dose dependent separation is seen on the second dimension. The volcano plot identified 604 putative lipid ions (Fig. S2C). Lipid molecules contributing to the highest variation on the MDS plot generated by RF analysis were identified as PCs (32:2, 32:3, 36:5, 42:6), ePC (32:2), DG (36:4), and TG (58:7) (Fig. S2B).
To identify lipid structures we searched observed adduct, retention time, and when possible fragmentation patterns from the MSE method (Fig. 1, S4,S5). As lipids can consist of multiple isomeric species, their respective carbon and double bond number represents them in this manuscript. Select lipids were further analyzed by mass filtered tandem MS to reduce fragmentation from multiple isomeric species. The response of a variety of broad lipid classes is illustrated in Figure S6. Lipids that showed significant effects between radiation dose, sex, and weight loss are listed in supplementary tables 2–5.
Fig. 1.
Low and high energy spectra for coeluting TG (52:4) and TG (54:5) species, displaying neutral losses (NL) and fragments indicative of fatty acyl chains 16:0, 18:0, 18:1, 18:2, and 18:3 acyl chains. Structure represents a potential TG (52:4) isomer.
We observed major alterations in the levels of DGs and TGs after radiation exposure (Table S2,S3 Fig. 2, S7,S8). NHP serum contained high levels of 34 and 36 C DG species, of which DG (36:3) was present in the highest abundance (Fig. S6). The levels of DG species generally decreased when exposed to IR (Table S3, Fig. S8). NHP serum contained high levels of TG species ranging from 50–62 C (Table S2, Fig. S6), and the highest abundant TG species consisted of 52 and 54 C. The levels of TGs were higher in control samples, except for select long-chain TGs (TGs [60:10, 60:11, 62:14]), whose levels were in much higher concentrations in animals exposed to 10 Gy IR (Fig. 2).
Fig. 2.
Change in lipid concentration after exposure to IR. Fatty acyl moieties for select TGs and PCs were determined by mass filtered tandem MS. A general decrease in short-chain TG, PC, and ePC is observed with high increases in their long-chain structures at 10 Gy exposure. Lysophospholipids increased after radiation exposure. (* indicates significantly different from control as determined by a Kruskal-Wallis test [P < 0.05]; [n=12; 10 Gy n=11])
The levels of selected species of ChoE (18:1, 20:4, 22:6) were significantly higher at 10 Gy (Table 1). There were significant changes in the levels of FFAs (18:3, 22:6, and 20:4) due to IR exposure (Table 1). FFAs (18:3 and 20:4) were significantly lower in animals exposed to 6, 7, and 10 Gy. Control samples had significantly lower levels of FFA (22:6) than the 10 Gy group.
Table 1.
ChoE (ESI+) and FFA (ESI−) molecules that significantly changed (trend with increasing dose) after exposure to IR exposure (Kruskal-Wallis test, P < 0.05).
| m/z_rt | Compound | P-value | H-value | MFC | LM (dose) |
HM (dose) |
HMDB ID | Trend |
|---|---|---|---|---|---|---|---|---|
| 673.5886_8.03 | ChoE (18:1) | 0.0384 | 11.751 | 5.4 | 7 | 10 | HMDB 00918 | ↑ |
| 695.5731_7.74 | ChoE (20:4) | 0.0311 | 12.280 | 3.2 | 7 | 10 | NA* | ↑ |
| 714.6178_7.64 | ChoE (22:6) | 0.0110 | 14.858 | 4.2 | 7 | 10 | NA* | ↑ |
| 277.2158_1.92 | FFA (18:3) α-Linoleic acid |
0.0059 | 16.370 | 4.6 | 10 | 4 | HMDB 00673 | ↓ |
| 303.2316_2.29 | FFA (20:4) Arachidonic acid |
<0.0001 | 26.719 | 1.7 | 7 | Con | HMDB 01043 | ↓ |
| 327.2315_2.14 | FFA (22:6) Docosahexaenoic acid |
0.0003 | 23.400 | 1.3 | Con | 10 | HMDB 02183 | ↑ |
(m/z=mass-to-charge ratio, rt=retention time, MFC=max fold change, HM=dose group with highest mean, LM=dose group with lowest mean;
indicates a HMDB bioidentifier not available, ChoE (20:4) – LMST01020014 and ChoE (22:6) – LMST01020019)
The levels of lysoGPs (LysoPC and lysophosphatidylethanolamine [LysoPE]) were significantly different among groups (Fig. 2, S9,S10, Table S3), as most were highest in animals exposed to 10 Gy IR. PEs and ePEs were significantly different among groups (Fig. S9, S11, Table S4). PCs and ePCs generally decreased in a dose dependent trend to IR exposure; however, ~33% were highest in 10 Gy exposure and the remaining highest in control or 2 Gy exposures groups (Fig. 2, S12,S13, Table S5).
To further elucidate the chemical structure of TG and GP species that were increased at 10 Gy in response to IR exposure, we analyzed their fragmentation patterns using quadrupole mass filtered tandem MS spectra. We identified PC (38:2; 18:0/20:2), PC (38:3; 18:0/20:3), PC (38:6; 16:0/22:6), PC (38:4; 18:0/20:4), TG (60:10; 18:0/20:4/22:6), TG (60:11; 18:1/20:4/22:6), and TG (62:14; 18:2/22:6/22:6 and 20:4/20:4/22:6) (Fig. 2).
Metabolomics
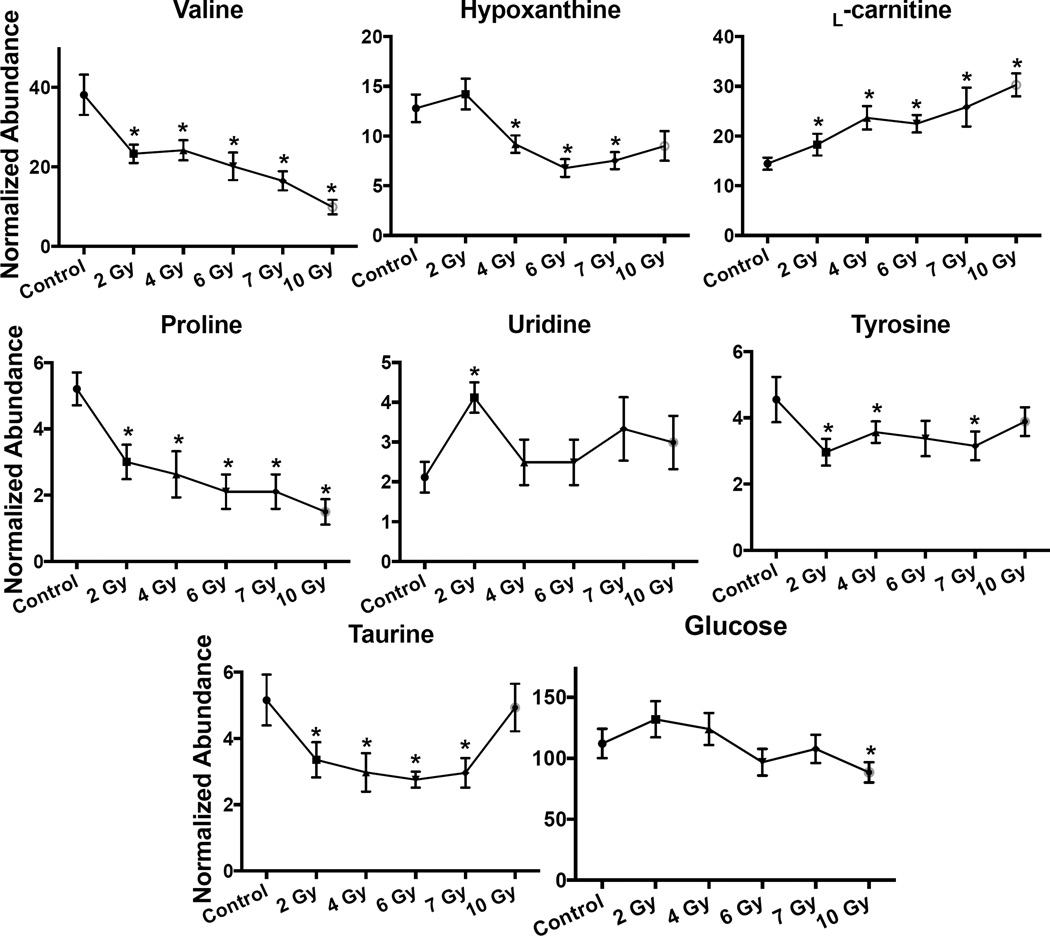
Similar to the lipidomic analysis, a PCA scores plot and heatmap of the statistically significant ions exhibits separation between control and exposed groups (Fig. S14). When the KEGG, BioCyc, and Reactome databases were searched by putative significant ions, perturbations were observed in the valine biosynthesis/degradation (2, 4, 7, 10 Gy; KEGG# ko00290), tyrosine biosynthesis/degradation (4 Gy; BioCyc# PWY-6134, TYRFUMCAT-PWY), citrulline biosynthesis (7 Gy; BioCyc# PWY-5004, CITRULBIO-PWY), protein digestion/absorption (4, 7, 10 Gy; KEGG# ko04974), biological oxidations (4, 10 Gy; Reactome# 211859), tryptophan metabolism (6, 7, 10 Gy; BioCyc# TPHAN-DEGRADATION-1) (possibly host microbiota derived, [Kurland et al. 2015; Ó Broin et al. 2015]), and proline degradation (7, 10 Gy; KEGG# ko00330). Univariate analysis detected putative ions involved in these pathways, of which taurine [M-H]−, udine [M+H]+ , glucose [M+H]+, tyrosine [M+H]+, proline [M+H]+, hypoxanthine [M+H]+, carnitine [M+H]+, and valine [M+H]+ were validated to be altered in IR exposed groups compared to the control. Valine (P=0.012 [FDR P=0.116], 0.6-fold), proline (P=0.021 [FDR P=0.160], 0.5-fold), and tyrosine (P=0.006 [FDR P=0.083], 0.7-fold) were in lower concentration at 2 Gy; however, tyrosine was not significantly different from the control at 6 and 10 Gy (Table 2, Fig. 3). Three common previously identified metabolites (taurine, carnitine, and hypoxanthine) identified in radiation metabolic profiling were detected. The levels of hypoxanthine slightly increased at 2 Gy, but were lower than controls at 4 Gy (P=0.026 [FDR P=0.162], 0.7-fold), 6 Gy (P=0.001 [FDR P=0.079], 0.5-fold), 7 Gy (P=0.003 [FDR P=0.087], 0.6-fold), and 10 Gy (P=0.052 [FDR P=0.470], 0.7-fold). The levels of uridine were significantly higher at 2 Gy from the control group (P=<0.001 [FDR P=0.007], 2.0-fold). The levels of glucose were lower at 6 Gy (P=0.060 [FDR P=0.504], 0.9-fold) and 10 Gy (P=0.005 [FDR P=0.165], 0.8-fold) than the control group. In serum, the levels of carnitine showed increases following IR exposures (10 Gy, P=<0.001 [FDR P<0.001], 2.1-fold). Taurine decreased at 2 (P=0.012 [FDR P=0.016], 0.7-fold), 4 (P=0.001 [FDR P=0.012], 0.6-fold), 6 (P=<0.001 [FDR P=0.019], 0.5-fold), and 7 (P=0.003 [FDR P=0.054], 0.6-fold) Gy; however, it increased at 10 Gy and was non-significant from the control (P=0.53 [FDR P=0.843]). Differences in these water-soluble metabolites suggest disruption to protein digestion/absorption (primarily valine, tyrosine, and tryptophan biosynthesis/degradation), biological oxidations, and fatty acid β-oxidation.
Table 2.
Validated biomarkers from global metabolomic profiling of exposure to γ radiation in NHP serum. Trend indicates change in concentration with increasing dose.
| m/z_rt | Compound | Calculated m/z |
Mass error ppm |
Formula | ESI mode | HMDB ID |
Trend |
|---|---|---|---|---|---|---|---|
| 124.0065_0.35 | Taurine | 124.0068 | 2.41 | C2H7NO3S | negative | HMDB 00251 |
↓ |
| 267.0581_0.37 | Uridine* | 267.0593 | 4.49 | C9H12N2O6 | positive | HMDB 00296 |
↑ |
| 203.0529_0.35 | Glucose* | 203.0532 | 1.48 | C6H12O6 | positive | HMDB 00122 |
↓ |
| 182.0798_0.38 | Tyrosine | 182.0817 | 10.43 | C9H11NO3 | positive | HMDB 00158 |
↓ |
| 116.0707_0.36 | Proline | 116.0712 | 4.31 | C5H9NO2 | positive | HMDB 00162 |
↓ |
| 137.0463_0.38 | Hypoxanthine | 137.0463 | 0.00 | C5H4N4O | positive | HMDB 00157 |
↓ |
| 162.1123_0.33 | L-Carnitine | 162.1130 | 4.32 | C7H15NO3 | positive | HMDB 00062 |
↑ |
| 118.0865_0.35 | Valine | 118.0868 | 2.54 | C5H11NO2 | positive | HMDB 00883 |
↓ |
Detected as Na+ adduct
Fig. 3.
Dose response of serum biomarkers from NHPs exposed to 2, 4, 6, 7, or 10 Gy γ radiation. (* indicates significantly different from control as determined by a Welch’s t-test; Mean ± SEM, considered significant at P < 0.05, [n=12; 10 Gy n=11]). A significant dose dependent increase is observed for L-carnitine. Valine, proline, tyrosine, glucose, hypoxanthine all decreased. Taurine decreased at 2, 4, 6, and 7 Gy, however the 10 Gy group levels were the same as in the control. Uridine significantly increased at 2 Gy, but not at 4, 6, 7, and 10 Gy.
Discussion
In this study we report a specific lipidomic and metabolomic serum signature from NHPs exposed to different doses of IR. We found that exposure to IR led to wide perturbations in lipid metabolism. Interestingly, the levels of three polyunsaturated TGs (TG 60:10, 60:11, 62:14) significantly increased at 10 Gy. These TGs contained primarily 20:4 (arachidonic acid) and 22:6 (docosahexaenoic acid) acyl moieties. As 20:4 and 22:6 FFAs are involved in inflammation through the COX and LOX pathways, it is possible these TGs are synthesized at an increased rate and serve as a reservoir in the 10 Gy group for possible transport to tissues (Fig. S15). It has previously been hypothesized that increased TG concentration could be attributed to lowered TG uptake due to decreased adipose LPL activity, destruction of cell membranes, or increased release from tissues (Mansour 2006). However, some of these changes may be related to changes in food intake following IR exposure or drug regimens provided. Significant decreases in body weight were reported for animals in the 7 and 10 Gy cohort (Pannkuk et al. 2015). Past reports have found decreases of TGs and GPs up to 6 days post-IR, but levels began to increase at day 12; however, when compared to a diet control group, TG concentration was higher (Feurgard et al. 1998). While the observed lipid alterations in the current study could be diet related, the observation of mixed dose dependent responses and drastic increases in select lipids in the 10 Gy group suggests that IR is responsible for altered concentrations.
The majority of significant GPs affected by IR included PEs, PCs, and their monoacyl (e.g., LysoPC) and ether-linked forms. Similar to TGs, GPs and eGPs generally decreased in concentration (some in a dose dependent manner); however, specific longer-chain polyunsaturated molecular species increased in the 10 Gy treatment. Most LysoGPs increased after exposure to IR. LysoPCs may be inflammation markers due to enzymatic release by phospholipases (e.g., PLA2) that free PUFAs that can be oxidized into eicosanoids. Further investigation by tandem MS showed the more unsaturated molecules increasing due to IR exposure were composed of 20:4 and 22:6 acyl moieties. These polyunsaturated acyls were esterified to ChoEs as well as TGs and GPs. Unlike free cholesterol that is a predominant membrane component, ChoEs may function similarly to TGs as they are used for transport in plasma or as inert storage molecules.
While 20:4 and 22:6 acyls are found in esterified form (PC, TG, and ChoE) in higher concentrations at 10 Gy, the active FFA forms increased at 10 Gy but not as high a fold change as esterified forms. FFA (20:4) (arachidonic acid) is an essential PUFA derived from FFA (18:2) (linoleic acid), usually esterified for storage, and cleaved from membrane phospholipids by phospholipase A2 (Fig. S13) (Calder 2006). Upon cleavage, FFA (20:4) is the parent molecule from which many prostaglandins, leukotrienes, and other inflammatory molecules are produced (Serhan & Savill 2005). FFA (22:6) (docosahexaenoic acid) is derived from FFA (22:5) (eicosapentaenoic acid) and inhibits angiogenesis, inflammation, apoptosis, and tumor growth (Zhang et al. 2013). FFA (22:6) forms resolvins and neuroprotectins through the LOX pathway. Increases in esterified 20:4 and 22:6 likely indicate upregulation of these compounds for transport to tissues or as signaling molecules in lethal/sublethal doses of IR exposure. Decreases of their active FFA form may be due to increased conversion in inflamed tissues. Given the importance of these molecules in signaling inflammation and inhibiting apoptosis, their roles after IR exposure warrants further investigation as PUFA mediated inflammation as been observed in murine models (Laiakis et al. 2014a).
Changes in water-soluble metabolites suggested perturbations to amino acid/protein metabolism, fatty acid β-oxidation, purine catabolism, and biological oxidations. The concentration of three amino acids (proline, valine, and tyrosine) decreased in animals exposed to IR. Perturbations of amino acids due to IR have been documented by gas chromatography (GC) mass selective detection (MSD) analysis of irradiated rat urine (Lanz et al. 2009), rat serum analyzed by GC/TOF-MS (Liu et al. 2013), and in mouse serum by UPLC-MS targeted metabolomics (Laiakis et al. 2014a) and 1H NMR (Khan et al. 2011). While many amino acids increased in mouse/rat serum 24 hours after 8 Gy IR exposure, proline, valine, and tyrosine slightly decreased, although not significantly (Laiakis et al. 2014b). Low valine and tyrosine levels may indicate kidney damage (Schrier 2006), which is a well-known symptom of radiation exposure (Dawson et al. 2010). Furthermore, biomarkers of renal failure (i.e., kynurenic acid and xanthurenic acid) were elevated in NHP urine from the same study animals (Pannkuk et al. 2015).
Three well established biomarkers of radiation exposure (L-carnitine, taurine, and hypoxanthine) were detected in the present study. Free L-carnitine and acylcarnitines exhibit high fold changes indicating radiation exposure (Laiakis et al. 2012; Goudarzi et al. 2014a; Laiakis et al. 2014b; Pannkuk et al. 2015). We found a dose dependent increase in L-carnitine, as in other studies, suggesting perturbation of fatty acid β-oxidation. The majority of L-carnitine is likely being eliminated from the body, as a 66.2-fold increase was found in urine between control and 10 Gy, but only a 2.1-fold increase is observed in serum (Pannkuk et al. 2015). Increased L-carnitine could also be a clinical sign of muscle wasting or decreased nutrition (Ringseis et al. 2013). The detection of hypoxanthine and uridine indicate perturbations to purine/pyrimidine catabolism. Hypoxanthine slightly increased at 2 Gy but decreased at higher IR exposures, which has been observed in the urine of male human patients (Laiakis et al. 2014b). In NHP urine, hypoxanthine increased suggesting elimination at day 7 (Pannkuk et al. 2015). Uridine is a glycosylated form of uracil and is a nitrogenous base found in RNA. At shorter time points after exposure to IR (e.g., 24 h) the deaminated form of uridine (2’-deoxyuridine) is found in higher concentrations compared to controls in urine. The decrease in hypoxanthine, slight increase in uridine, and the absence of 2’-deoxyuridine may indicate direct products from IR exposure are being eliminated and cellular activity is occurring at day 7. Further studies should utilize targeted methods to quantitatively determine levels of these common analytes found to be altered after IR exposure.
In addition to IR serving as a model agent to assess overall metabolomic responses to systemic injury in vivo, there is an increased need for a rapid biodosimetry “toolkit” in the event of a radiological incident and mass exposures to IR. NHPs provide excellent model organisms to elucidate potential radiation biomarkers in humans. As metabolic profiles of NHP biofluids are currently restricted to urine (Johnson et al. 2012; Pannkuk et al. 2015) or specific targeted approaches in serum (Jones et al. 2014), we have extended radiation biofluid biodosimetry by providing a global lipidomic and metabolomic analysis of NHP serum 7 days post-IR; these injury biomarkers occur at a time sufficiently later after IR exposure so as to allow triaging at local health facilities. The magnitude of lipid perturbation in serum are generally more striking than metabolic changes in urine, making serum more amenable to assess select radiation biomarkers. In particular, we found that lipids rich in polyunsaturated acyls (20:4 [arachidonic acid] and 22:6 [docosahexaenoic acid]) were increased in NHPs exposed to 10 Gy IR. About half (LD50/30) of NHP and humans show lethality without supportive care within a month after 4 to 5 Gy of whole body irradiation, while the number increases to over 7 Gy with supportive care and up 10 Gy or more with intensive therapy including bone marrow transplant (Hall and Giaccia, 2006; MacVittie et al. 2012a). A 10 Gy TBI dose in NHPs after 7 days will result in a loss of ~72% jejunum crypt cells when receiving supportive care (MacVittie et al. 2012b). Therefore, as a 10 Gy TBI dose in NHPs will bring radiation-induced lethal bone marrow damage (without transplant) and an appreciable level of gastrointestinal injury, these individuals would need immediate medical care. This may be reflected in the marked changes in some metabolites at this dose. As arachidonic acid and docosahexaenoic acid are important molecules directly involved in pro-inflammatory responses through the COX and LOX pathways, the role of these molecules should be further elucidated in NHPs exposed to high levels of IR (e.g., 10 Gy). Future studies should address other time points other than 7 days and differences in NHPs not receiving supportive care. Further use of NHP models in these lipidomic and metabolomic studies will aid in development of biodosimetry methods and assigning proper medical care in the possible event of a mass radiation exposure.
Supplementary Material
Acknowledgments
Funding: National Institutes of Health (National Institute of Allergy and Infectious Diseases) grant 1R01AI101798 (P.I. Albert J. Fornace, Jr.) and Lombardi Comprehensive Cancer Proteomics and Metabolomics Shared Resource (PMSR); partial support National Cancer Institute grant P30CA051008 (P.I. Louis Weiner)
The authors acknowledge Lombardi Comprehensive Cancer Proteomics and Metabolomics Shared Resource (PMSR) for data acquisition. Content is the responsibility of authors and does not necessarily represent official views of NCI/NIH.
Footnotes
Conflict of Interest
Evan L. Pannkuk, Evagelia C. Laiakis, Tytus D. Mak, Giuseppe Astarita, Simon Authier, Karen Wong, and Albert J. Fornace Jr. declare that they have no conflict of interest.
Compliance with ethical standards
Ethics Approval
NHP studies were conducted by CiToxLAB: North America Safety and Health Research Laboratories (Laval, Québec, Canada; study # 5013–0193) and was approved by the Institutional Animal Care and Use Committee.
References
- Astarita G, Kendall AC, Dennis EA, Nicolaou A. Targeted lipidomic strategies for oxygenated metabolites of polyunsaturated fatty acids. Biochim Biophys Acta. 2015;1851(4):456–468. doi: 10.1016/j.bbalip.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita G, Langridge J. An emerging role for metabolomics in nutrition science. J Nutrigenet Nutrigenomics. 2013;6(4–5):181–200. doi: 10.1159/000354403. [DOI] [PubMed] [Google Scholar]
- Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta. 2012;1822(9):1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Calder PC. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am J Clin Nutr. 2006;83(6 Suppl):1505S–1519S. doi: 10.1093/ajcn/83.6.1505S. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DG, Oh J, Keasling A, Colson KL, Hamann MT. The utility of metabolomics in natural product and biomarker characterization. Biochim Biophys Acta. 2014;1840(12):3460–3474. doi: 10.1016/j.bbagen.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson LA, Kavanagh BD, Paulino AC, Das SK, Miften M, Li XA, et al. Radiation-associated kidney injury. Int J Radiat Oncol Biol Phys. 2010;76(3 Suppl):S108–S115. doi: 10.1016/j.ijrobp.2009.02.089. [DOI] [PubMed] [Google Scholar]
- Degtyarenko K, De Matos P, Ennis M, Hastings J, Zbinden M, Mcnaught A, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo AL, Jackson IL, Shah JR, Czarniecki CW, Maidment BW, Williams JP. Development and licensure of medical countermeasures to treat lung damage resulting from a radiological or nuclear incident. Radiat Res. 2012;177(5):717–721. doi: 10.1667/rr2881.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo AL, Ramakrishnan N, Hatchett RJ. Radiation combined injury: overview of NIAID research. Health Phys. 2010;98(6):863–867. doi: 10.1097/HP.0b013e3181a6ee32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurgard C, Bayle D, Guezingar F, Serougne C, Mazur A, Lutton C, et al. Effects of ionizing radiation (neutrons/gamma rays) on plasma lipids and lipoproteins in rats. Radiat Res. 1998;150(1):43–51. [PubMed] [Google Scholar]
- Fruhwirth GO, Loidl A, Hermetter A. Oxidized phospholipids: from molecular properties to disease. Biochim Biophys Acta. 2007;1772(7):718–736. doi: 10.1016/j.bbadis.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Goudarzi M, Mak TD, Chen C, Smilenov LB, Brenner DJ, Fornace AJ. The effect of low dose rate on metabolomic response to radiation in mice. Radiat Environ Biophys. 2014a;53(4):645–657. doi: 10.1007/s00411-014-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Weber W, Mak TD, Chung J, Doyle-Eisele M, Melo D, et al. Development of urinary biomarkers for internal exposure by cesium-137 using a metabolomics approach in mice. Radiat Res. 2014b;181(1):54–64. doi: 10.1667/RR13479.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Weber WM, Mak TD, Chung J, Doyle-Eisele M, Melo DR, et al. Metabolomic and Lipidomic Analysis of Serum from Mice Exposed to an Internal Emitter, Cesium-137, Using a Shotgun LC-MSE Approach. J Proteome Res. 2015;14(1):374–384. doi: 10.1021/pr500913n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EC, Giaccia AJ. Radiobiology for the Radiologist. 7th. Philadelphia, PA: 2012. [Google Scholar]
- Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286(32):27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178(4):328–340. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, Luecke H, et al. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 2011;175(4):473–484. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Tudor G, Bennett A, Farese AM, Moroni M, Booth C, et al. Development and validation of a LC-MS/MS assay for quantitation of plasma citrulline for application to animal models of the acute radiation syndrome across multiple species. Anal Bioanal Chem. 2014;406(19):4663–4675. doi: 10.1007/s00216-014-7870-0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Rana P, Devi MM, Chaturvedi S, Javed S, Tripathi RP, et al. Nuclear magnetic resonance spectroscopy-based metabonomic investigation of biochemical effects in serum of gamma-irradiated mice. Int J Radiat Biol. 2011;87(1):91–97. doi: 10.3109/09553002.2010.518211. [DOI] [PubMed] [Google Scholar]
- Kurland IJ, Broin PO, Golden A, Su G, Meng F, Liu L, et al. Integrative metabolic signatures for hepatic radiation injury. PLoS One. 2015;10(6):e0124795. doi: 10.1371/journal.pone.0124795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177(2):187–199. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Strassburg K, Bogumil R, Lai S, Vreeken RJ, Hankemeier T, et al. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014a;13(9):4143–4154. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014b;181:350–361. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz C, Patterson AD, Slavik J, Krausz KW, Ledermann M, Gonzalez FJ, et al. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res. 2009;172(2):198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Tyburski JB, Wang YW, Strawn S, Moon BH, Kallakury BV, et al. Modulation of fatty acid and bile acid metabolism by peroxisome proliferator-activated receptor alpha protects against alcoholic liver disease. Alcohol Clin Exp Res. 2014;38(6):1520–1531. doi: 10.1111/acer.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Z, Zhang X, Qiao Y, Wu S, Dong F, et al. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat Prot Dosimetry. 2013;154(1):9–17. doi: 10.1093/rpd/ncs138. [DOI] [PubMed] [Google Scholar]
- Macvittie TJ, Bennett A, Booth C, Garofalo M, Tudor G, Ward A, et al. The prolonged gastrointestinal syndrome in rhesus macaques: the relationship between gastrointestinal, hematopoietic, and delayed multi-organ sequelae following acute, potentially lethal, partial-body irradiation. Health Phys. 2012a;103(4):427–453. doi: 10.1097/HP.0b013e318266eb4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macvittie TJ, Farese AM, Bennett A, Gelfond D, Shea-Donohue T, Tudor G, et al. The acute gastrointestinal subsyndrome of the acute radiation syndrome: a rhesus macaque model. Health Phys. 2012b;103(4):411–426. doi: 10.1097/HP.0b013e31826525f0. [DOI] [PubMed] [Google Scholar]
- Mak TD, Laiakis EC, Goudarzi M, Fornace AJJ. Selective paired ion contrast analysis: A novel algorithm for analyzing postprocessed LC-MS metabolomics data possessing high experimental noise. Anal Chem. 2015;87(6):3177–3186. doi: 10.1021/ac504012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TD, Laiakis EC, Goudarzi M, Fornace AJ., Jr MetaboLyzer: A novel statistical workflow for analyzing postprocessed LC-MS metabolomics data. Anal Chem. 2014;86(1):506–513. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, et al. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J Proteome Res. 2010;9:4176–4188. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour HH. Protective role of carnitine ester against radiation-induced oxidative stress in rats. Pharmacol Res. 2006;54(3):165–171. doi: 10.1016/j.phrs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, Macarthur LH, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Coates PJ, Lorimore SA, Wright EG. Responses to ionizing radiation mediated by inflammatory mechanisms. J Pathol. 2014;232(3):289–299. doi: 10.1002/path.4299. [DOI] [PubMed] [Google Scholar]
- Ó Broin P, Vaitheesvaran B, Saha S, Hartil K, Chen EI, Goldman D, et al. Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury. Int J Radiat Oncol Biol Phys. 2015;91(2):360–367. doi: 10.1016/j.ijrobp.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJ., Jr Global metabolomic identification of longer-term dose dependent urinary biomarkers in non-human primates exposed to ionizing radiation. Radiat Res. 2015;184(2):121–133. doi: 10.1667/rr14091.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbächer MJ, Popp . Challenges in Molecular Structure Determination. Springer; 2012. [Google Scholar]
- Ringseis R, Keller J, Eder K. Mechanisms underlying the anti-wasting effect of L-carnitine supplementation under pathologic conditions: evidence from experimental and clinical studies. Eur J Nutr. 2013;52(5):1421–1442. doi: 10.1007/s00394-013-0511-0. [DOI] [PubMed] [Google Scholar]
- Schrier RW. Diseases of the Kidney and Urinary Tract (Diseases of the Kidney [Schrier]) Lippincott Williams & Wilkins. 2006 [Google Scholar]
- Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6(12):1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- Subbanagounder G, Watson AD, Berliner JA. Bioactive products of phospholipid oxidation: isolation, identification, measurement and activities. Free Radic Biol Med. 2000;28(12):1751–1761. doi: 10.1016/s0891-5849(00)00233-1. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics tools for mass spectroscopy-based metabolomic data processing and analysis. Curr Bioinform. 2012;7(1):96–108. doi: 10.2174/157489312799304431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170(1):1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara T, Suoniemi M, Laaksonen R. Lipidomics in drug discovery. Drug Discov Today. 2014;19(2):164–170. doi: 10.1016/j.drudis.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xu L, Porter NA. Free radical lipid peroxidation: mechanisms and analysis. Chem Rev. 2011;111(10):5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- Zhang A, Sun H, Wang X. Serum metabolomics as a novel diagnostic approach for disease: a systematic review. Anal Bioanal Chem. 2012;404(4):1239–1245. doi: 10.1007/s00216-012-6117-1. [DOI] [PubMed] [Google Scholar]
- Zhang G, Panigrahy D, Mahakian LM, Yang J, Liu JY, Stephen Lee KS, et al. Epoxy metabolites of docosahexaenoic acid (DHA) inhibit angiogenesis, tumor growth, and metastasis. Proc Natl Acad Sci USA. 2013;110(16):6530–6535. doi: 10.1073/pnas.1304321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.