Abstract
Several ongoing clinical studies are evaluating recombinant adeno-associated virus (rAAV) vectors as gene delivery vehicles for a variety of diseases. However, the production of vectors with genomes >4.7 kb is challenging, with vector preparations frequently containing truncated genomes. To determine whether the generation of oversized rAAVs can be improved using a producer cell-line (PCL) process, HeLaS3-cell lines harboring either a 5.1 or 5.4 kb rAAV vector genome encoding codon-optimized cDNA for human B-domain deleted Factor VIII (FVIII) were isolated. High-producing “masterwells” (MWs), defined as producing >50,000 vg/cell, were identified for each oversized vector. These MWs provided stable vector production for >20 passages. The quality and potency of the AAVrh8R/FVIII-5.1 and AAVrh8R/FVIII-5.4 vectors generated by the PCL method were then compared to those prepared via transient transfection (TXN). Southern and dot blot analyses demonstrated that both production methods resulted in packaging of heterogeneously sized genomes. However, the PCL-derived rAAV vector preparations contained some genomes >4.7 kb, whereas the majority of genomes generated by the TXN method were ≤4.7 kb. The PCL process reduced packaging of non-vector DNA for both the AAVrh8R/FVIII-5.1 and the AAVrh8R/FVIII-5.4 kb vector preparations. Furthermore, more DNA-containing viral particles were obtained for the AAVrh8R/FVIII-5.1 vector. In a mouse model of hemophilia A, animals administered a PCL-derived rAAV vector exhibited twofold higher plasma FVIII activity and increased levels of vector genomes in the liver than mice treated with vector produced via TXN did. Hence, the quality of oversized vectors prepared using the PCL method is greater than that of vectors generated using the TXN process, and importantly this improvement translates to enhanced performance in vivo.
Keywords: : AAV vector production, oversized genome, producer cell line, FVIII, AAVrh8R
Introduction
Gene therapy using recombinant AAV (rAAV) vectors is promising as a therapeutic paradigm for the management of several genetic diseases. However, for some diseases, such as hemophilia A, the transgene (Factor VIII [FVIII]) expression cassette exceeds the packaging limit of the virus. Generating AAV vector preparations that harbor full-length vector genomes while limiting the number of viral particles containing incomplete genomes is challenging.1–5 The presence of smaller and fragmented genomes in oversized rAAV vector preparations can be tolerated because such vectors support the production of active FVIII protein and are efficacious in animal models of hemophilia A.6–10 However, to improve oversized vector quality, the number of incomplete genomes should be minimized. Strategies that have been proposed to overcome the limitation of oversized rAAV vectors include the use of a two-vector system with separate rAAV vectors engineered to express the FVIII heavy and light chains and the use of split (trans-splicing) AAV vectors.11,12 However, these approaches result in poor efficacy in mouse models of hemophilia A. Moreover, these strategies require the production and qualification of two rAAV vectors for clinical studies, which introduces regulatory complexity.
Currently, there are no published reports on the characterization of oversized rAAV vectors approved for use in clinical studies. Recombinant AAV vectors with predicted genome sizes of ≤4.7 kb (i.e., wild-type size) evaluated in the clinic have been produced by transient plasmid transfection of HEK293 cells,13,14 recombinant herpes simplex virus (rHSV)-mediated infection of baby hamster kidney (BHK) cells,15,16 baculovirus-mediated infection of Sf9 insect cells,17 and engineered producer cell lines.18,19 Clinical-grade vectors undergo thorough characterization, including analysis of the packaged vector DNA and quantification of the packaged non-vector DNA.20 In preparations of oversized rAAV vectors produced via transient transfection, vector genomes are heterogeneous in size and contain variable deletions at their 5′ termini.3,4,7,10,21,22 Vectors containing these fragmented genomes also exhibit slower kinetics of transgene expression and reduced gene transfer efficiency than rAAV vectors bearing a wild-type size vector genome do.22 Consequently, high vector doses are required to compensate for the lower performance of oversized rAAV vectors. Because oversized rAAV vectors typically also package proportionally more non-vector DNA,22 the need for higher doses would invariably result in administration of additional impurities. Hence, oversized rAAV vectors would benefit from alternative production platforms that enhance vector yield and quality.
Herein, the generation and characterization of slightly oversized rAAV vectors expressing FVIII using the producer cell line method in HeLaS3 cells is described. This approach is based on the use of cell lines engineered to contain an integrated vector genome encoding the transgene of interest and the AAV replication (rep) and capsid (cap) genes.18,23–25 Vector production is induced by infecting the engineered cells with a helper adenovirus. This production platform has been used successfully to manufacture GMP-grade rAAV vectors on a large scale.18,19 However, a disadvantage of this process is the time needed to generate and characterize producer cell lines (PCLs), which can be shortened using non-clonal “masterwells” (MWs).18,26 This study generated and characterized MWs producing AAVrh8R serotype vectors harboring 5.1 and 5.4 kb genomes encoding human FVIII (hFVIII). The data demonstrate the feasibility of the PCL method to produce slightly oversized rAAV vectors. These vectors were higher quality and biologically more potent than corresponding vectors generated by the transient transfection (TXN) method.
Materials and Methods
Construction and in vitro testing of plasmid expression cassettes encoding human FVIII
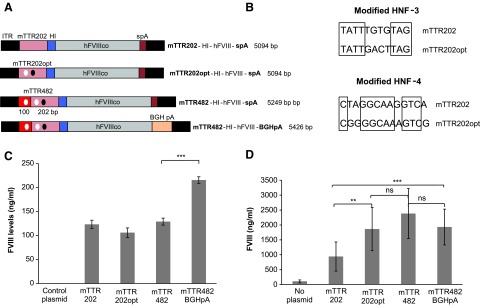
The construction of the previral plasmid encoding hFVIII (pITR-mTTR202-hFVIIIco-spA), which consists of a 5.1 kb vector genome with a mouse transthyretin (mTTR) promoter (202 bp core sequence), a hybrid intron (HI), the codon-optimized human B-domain deleted FVIII (hFVIIIco) cDNA, and a synthetic polyadenylation site (spA) flanked by the rAAV2 inverted terminal repeat sequences (ITRs), has been previously described.22 This previral plasmid was altered to contain modifications in the hepatic nuclear factor (HNF)-4 and HNF-3 binding sites within the core mTTR promoter to generate pITR-mTTR202opt-hFVIIIco-spA (Fig. 1A and B). This plasmid was further modified to include a 100 bp mTTR enhancer27 with an altered HNF-4 binding site to obtain pITR-mTTR482-hFVIIIco-spA. In another variation, the polyadenylation site in pITR-mTTR482-hFVIIIco-spA was exchanged for that of bovine growth hormone (BGH) to generate pITR-mTTR482-hFVIIIco-BGHpA with a 5.4 kb vector genome (Fig. 1A).
Figure 1.
Characterization of previral plasmids with mouse transthyretin human Factor VIII (mTTR-hFVIII) expression cassettes. (A) Diagram of hFVIII expression cassettes flanked by AAV inverted terminal repeat sequences (ITRs) and with vector genome sizes ranging from 5.1 to 5.4 kb. (B) Sequence modifications in hepatic nuclear factor (HNF)-3 and HNF-4 binding sites. Location of modified binding sites for HNF-3 (black circles) and HNF-4 (white circles) are shown in (A). (C) FVIII levels from mTTR-FVIII expression plasmids in vitro. Previral plasmids were transfected into Huh7 cells, and FVIII protein levels in the culture media were measured 72 h later by enzyme-linked immunosorbent assay (ELISA). Values were adjusted for transfection efficiency by secreted embryonic alkaline phosphatase (SEAP) activity obtained from co-transfected SEAP expressing plasmid. Two independent experiments were performed (n = 3 for each construct) with comparable results. (D) FVIII levels from mTTR-FVIII expression plasmids in vivo. The previral plasmid vectors (30 μg/mouse) were administered by hydrodynamic high-volume injections (2.4 mL/mouse) into the tail veins of C56BL/6 mice (n = 8–10 for plasmid groups; n = 5 for naïve group), and plasma FVIII protein levels were measured by ELISA. Values in each graph represent the average ± standard deviation, and significance was calculated using Student's t-test, with significance indicated as *p < 0.05, **p < 0.01, and ***p < 0.001. Abbreviations: ITR, recombinant adeno-associated virus (rAAV) inverted terminal repeat; mTTR202, mouse core transthyretin promoter (202 bp); mTTR202opt, mTTR with modified HNF-3 and -4 binding sites; mTTR482, mTTR202opt and 100 bp transthyretin enhancer; HI, hybrid intron; hFVIIIco, B-domain deleted human FVIII cDNA; spA, synthetic polyA; BGH, bovine growth hormone; pA, polyA.
In vitro analysis of previral plasmid vectors was performed by co-transfecting FVIII plasmids (0.5 μg/well) and pSEAP (0.05 μg/well; transfection control) into human Huh7 and HepG2 cells grown in 24-well plates using Lipofectamine 2000 or Lipofectamine 3000 reagents.22 Conditioned media was collected 48–72 h post transfection, and hFVIII protein levels were quantitated by enzyme-linked immunosorbent assay (ELISA; Human Coagulation Factor FVIII Total Antigen ELISA kit; Innovative Research) using pooled normal human plasma (Innovative Research) as a standard. All FVIII protein values were normalized for transfection efficiency by secreted embryonic alkaline phosphatase (SEAP) activity in the culture media.
Generation and characterization of producer cell lines
To support the production of rAAV-hFVIIIco producer cell lines, a series of pTP plasmids with hFVIIIco expression cassettes (5.1 and 5.4 kb), AAV2 replication (rep), capsid (cap; AAVrh8R or AAV8), and puromycin resistance (puroR) genes were generated as follows. The AAV2 cap in pAF-SEAP-AAV226 was changed to AAVrh8R to generate pTP-SEAP-caprh8R. The ITRs flanking the SEAP expression cassette were removed and replaced with a blunt-ended PvuI and SapI fragment containing the 5.1 kb FVIII vector genome from pITR-mTTR202-hFVIIIco-spA to generate pTP-FVIII-5.1-caprh8R. The analogous pTP-FVIII-5.4-caprh8R plasmid was created by cloning the 5.4 kb vector genome from pITR-mTTR482-hFVIIIco-BGHpA. Additionally, pTP-FVIII-5.1-cap8 was generated by replacing the AAVrh8R cap with the AAV8 cap gene.
To generate the producer cell lines, the pTP plasmids encoding the hFVIIIco, AAV2 rep, cap (AAVrh8R or AAV8), and puroR genes were transfected into HeLaS3 cells (ATCC CCL-2.2) using Lipofectamine transfection reagent, as previously described.26 Puromycin-resistant MWs were isolated and screened for relative vector production by infection of adherent cells with a multiplicity of infection (MOI) of 100/cell for wild-type adenovirus 5 (wtAd5). The highest-producing MWs were evaluated for vector production by plating 2 × 105 cells/mL in suspension cultures using serum-free production media followed by infection with wtAd5.18,26
Genomic DNA was extracted from the producer cells using a Gentra Puregene cell kit (Qiagen) according to the manufacturer's directions. The DNA was digested with SpeI (a single site in the plasmid) or with BglII and HincII (sites within the mTTR promoter, hFVIII cDNA and spA). The digested genomic DNA and the parental plasmid were electrophoresed on a 0.8% agarose gel. DNA was transferred onto a nylon membrane and probed with a DIG-labeled 2.7 kb NcoI fragment of FVIII, as described previously.22
AAV vector production and purification
Vector production by the MWs was induced as previously described.26 Briefly, the cells were maintained in shaker flask suspension cultures in EX-CELL HeLa medium (Sigma–Aldrich) with 6 mM of L-glutamine at 37°C in 10% CO2. For virus production, the cells were transferred to production media and infected with wtAd5 (MOI of 100/cell) for 3 days. The cells were lysed and purified by affinity column chromatography (AVB Sepharose High Performance medium; GE Healthcare), as previously described.26,28
Vector production according to the TXN method was performed, as previously described.28 Briefly, pITR-mTTR202-hFVIIIco-spA (5.1 kb) or pITR-mTTR482-hFVIIIco-BGHpA (5.4 kb), p5repCMVcaprh8R (AAVrh8R), and pAd helper (Stratagene Technologies) were transfected at a 1:1:1 ratio into HEK293 cells using polyethyleneimine (PEI). The virus was purified as described above.
Quantitative polymerase chain reaction analyses
The AAV vector was quantified by a real-time quantitative polymerase chain reaction (RT-qPCR) assay (7500 Real-Time PCR System; Applied Biosystems) with primers specific to FVIII (FVIII-A2,22 located in A2 region; FVIII-A3, in A3; and FVIII-C2, in C2 region of FVIII) and to BGHpA26 and a standard curve of serially diluted linearized hFVIIIco plasmid DNA (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hgtb). Vector levels were expressed as vector genomes (vg) per cell or milliliter. Packaging of the plasmid backbone sequences in vector lots was measured by quantifying the levels of ampicillin (ampR)22 or puroR genes (Supplementary Table S1). The copy numbers of vector (FVIII-A2) and rep and puroR genes in the genomic DNA were assessed by qPCR using primer/probe sets shown in Supplementary Table S1 or as previously described.22,26
Characterization of the vector and packaged vector genomes
Vector lots were characterized using 4–12% Tris-Bis sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Sypro protein staining (Invitrogen).22 The homogeneity of the vectors was assessed by analytical ultracentrifugation (AUC) using a Proteome Lab XL-1 apparatus (Beckman Coulter).22,28 The results were plotted as the normalized differential coefficient distribution value, C(S), versus the sedimentation coefficient (S).
To analyze the packaged vector genomes by Southern or DNA dot blot, DNA was first extracted from the purified virions.22 For DNA dot blot analyses, genomes were denatured in TE buffer pH 7.0 by heating at 95°C for 5 min followed by a 5 min chill on ice. DNA was applied in twofold serial dilutions onto the membrane and fixed by UV cross-linking. For Southern blot analyses, the genomes were separated by gel electrophoresis using a 1.2% alkaline gel in buffer comprising 30 mM of NaOH and 1 mM of EDTA. The samples were then transferred onto a Hybond membrane (Amersham). The dot blot or Southern blots were probed with a 4.7 kb AvrII-Spe1 fragment (containing the promoter, HI and FVIII cDNA) from pITR-mTTR202-hFVIIIco-spA labeled with a DIG High Prime DNA Labeling kit (Roche) according to the manufacturer's instructions. For strand-specific analyses by Southern and dot blot methods, various 25- to 30-mer oligonucleotide probes (Supplementary Table S1) were 3′ end-labeled using the DIG Oligo 3′-End Labeling Kit (Roche) according to the manufacturer's instructions. Briefly, hybridization was performed in “Easy Hyb” buffer for 18 h followed by washes at 68°C for fragment probes and 50°C for oligonucleotide probes. After a 30 min blocking step, detection was performed with alkaline phosphatase-conjugated anti-DIG Fab fragments for 30 min followed by further washes, reaction with CDP-Star substrate for 5 min, and exposure to X-ray film. The signal density in the Southern and dot blots was quantified using ImageJ software (http://rsb.info.nih.gov/ij).
In vivo studies and analysis
Male C57BL/6 mice (Taconic) and hemophilia A mice (C56BL/6, 129S-F8 tm1Kaz; Jackson Laboratories) were obtained at 8–12 weeks of age and housed and maintained in accordance with humane guidelines for animal care and use. All animal procedures were approved by Sanofi's Institutional Animal Care and Use Committee (IACUC). For expression plasmid analysis, plasmids (30 μg/mouse) in sterile saline were administered by hydrodynamic high-volume injections into the tail veins of C57BL/6 mice. Recombinant AAV vectors (4 × 1010 vg/mouse) were administered intravenously via the tail vein into hemophilia A mice. Blood was collected via the retro-orbital sinus into sodium citrate tubes, and the plasma was stored frozen until analysis. Livers were collected and frozen until analysis.
Plasma samples were analyzed for FVIII activity levels using the Coatest® assay (Diapharma) according to manufacturer's protocol (modified to a 96-well format). Values were measured as % FVIII activity present in normal plasma and were converted to ng/mL (100% FVIII = 150 ng FVIII/mL). Some samples were also tested for activated partial thromboplastin time (aPTT, IDEXX).
For vector genome quantification in the liver, liver samples (approximately 250 mg) in 0.9 mL of genomic digestion buffer (PureLink Genomic DNA kit; Invitrogen) and a quarter of an inch of 1 mm zirconia beads (BioSpec Product, Inc.) were homogenized with a bead beater-16. A 100-μL aliquot of Proteinase K (20 μg/mL) was added to the homogenates and incubated at 55°C for 2–4 h. A 100-μL aliquot of the homogenate was applied to DNA isolation columns (PureLink) according to the manufacturer's instructions. FVIII vector genome copies in the liver DNA were quantified by qPCR using primer/probe sets specific to FVIII shown in Supplementary Table S1 or as previously described.22
Statistical analyses
For in vitro studies, comparison was based on a minimum of three animals per group, and experiments were performed at least twice. For in vivo studies, a minimum of five animals per group were used (the number of animals in each experiment is stated in the figure legend). Data were analyzed for significance using an unpaired Student's t-test, and p-values of <0.05 were considered statistically significant.
Results
Characterization of plasmid expression cassettes encoding human FVIII
A number of previral plasmid expression cassettes (ranging in size from 5.1 to 5.4 kb) encoding human FVIII were generated (Fig. 1A). It was previously reported that the 5.1 kb mTTR-FVIII expression cassette (mTTR202-HI-hFVIII-spA) conferred expression of active FVIII.22 The current study evaluated the effects of altering the HNF-3 and HNF-4 binding sites (mTTR202opt-HI-hFVIIIco-spA; Fig. 1B), adding a mTTR enhancer (mTTR482-HI-hFVIIIco-spA) and using a BGH polyadenylation (polyA) signal sequence instead of a synthetic signal (mTTR482-HI-hFVIIIco-BGHpA). The previral plasmid vectors were evaluated in two human hepatoma cell lines: Huh7 and HepG2 cells. For both cell lines, increased FVIII levels were only detected in the culture media when the synthetic polyA was replaced with the BGH polyA (Fig. 1C). When the previral plasmids were tested in vivo by hydrodynamic injection into the tail vein of mice, the mTTR202opt-HI-FVIII-spA cassette (with HNF-3 and HNF-4 modifications) conferred a twofold higher level of plasma FVIII compared with the unmodified (mTTR202-HI-FVIII-spA) construct (Fig. 1D). Incorporation of the mTTR enhancer and BGH polyA signal sequence did not significantly enhance FVIII production compared to the HNF-3 and HNF-4 modifications (Fig. 1D). The mTTR202-HI-hFVIIIco-spA (5.1 kb) and mTTR482-HI-hFVIIIco-BGHpA (5.4 kb) vector genomes were selected for further evaluation in the producer cell line platform as examples of oversized expression cassettes of different lengths.
Generation of producer cell lines with oversized rAAV-FVIII vectors
For the comparative studies, HeLaS3-based PCLs containing the 5.1 or 5.4 kb FVIII vector genomes and the AAVrh8R cap gene were generated. To evaluate the effect of capsid serotype, PCLs containing the 5.1 kb FVIII genome and the AAV8 cap gene were also generated. The efficiency of PCL generation was compared to cell lines engineered to produce AAV8/SEAP-4.3 (4.3 kb vector genome) expressing the SEAP reporter gene. The relative production screening was performed using qPCR assay and a primer/probe set (FVIII-A2) recognizing an internal part of the genome, as terminal deletions have been reported for the oversized rAAV vectors.3,4,7,10,21,22 Although this primer/probe set may not detect all vector genomes due to the expected heterogeneity of genome sizes, nonetheless it should allow relative ranking of the candidate cell lines. Approximately 200–400 non-clonal cell lines referred to as MWs were identified in relative production screens for each vector (Supplementary Table S2). A total of 371 MWs were isolated and analyzed for production of the rAAVrh8R/FVIII-5.1 vector. Of these, one MW (MW#35) was a high producer (vector yield >1 × 1010 vg/mL), seven were medium producers (vector yield between 1 × 109 and 1 × 1010 vg/mL), and 10 were low producers (vector yield between 1 × 108 and 1 × 109 vg/mL), corresponding to frequencies of 0.3%, 1.9%, and 2.7%, respectively. Similar results were obtained for the larger rAAVrh8R/FVIII-5.4 vector. In comparison, the frequency of high-producing MWs for rAAV8/FVIII-5.1 and AAV8/SEAP-4.3 vectors was slightly higher at 1.2% and 3.3%, respectively (Supplementary Table S2). The specific productivity for each of the highest-producing MWs was confirmed following scale-up to shake flasks (high producers defined as yielding ≥50,000 vg/cell; Supplementary Table S3). Hence, the results show that it is possible to isolate high-producing MWs for oversized rAAV-FVIII vectors.
The integrity of the integrated FVIII expression cassette, puroR, and rep and cap genes in the high-producing MWs was analyzed. Southern blot analysis of genomic DNA isolated from three MWs (#272, #418, and #35) producing the AAVrh8R/5.1 vector is shown in Supplementary Fig. S1A. Restriction enzyme digestion with an enzyme (SpeI) that cleaves only once in the pTP-FVIII plasmid resulted in the generation of a band of the predicted unit length (approximately 13 kb) and that co-migrated with the linearized parental plasmid. This result suggests that in these cell lines, the entire plasmid was integrated into the HeLaS3 genome as head-to-tail tandem repeats. All MWs also exhibited evidence of single-copy fragments of varying sizes that likely represent junction fragments containing genomic DNA flanking the integration sites. MW#272 (medium producer) and MW#35 (high producer) displayed a single integration site (two flanking fragments observed), while MW#418 (high producer) had multiple flanking fragments, which suggests that this MW comprised a mixture of clones or a single clone with multiple integration sites (Supplementary Fig. S1A). To characterize the 5.1 kb vector in the MWs further, the genomic DNA was digested with the restriction enzymes HincII and BglII, which cleave at sites within the FVIII expression cassette.22 Fragments of the expected size were observed that co-migrated with bands derived via digestion of the parental pTP-FVIII plasmid (Supplementary Fig. S1B). Similar analysis of the AAV8/FVIII-5.1 and AAVrh8R/FVIII-5.4 MWs produced comparable results (Supplementary Fig. S1C and D). Thus, all the MWs contained intact integrated vector genomes with no evidence of rearrangements or deletions in the expression cassette.
The integrated plasmid copy numbers for selected MWs was quantitated by qPCR analysis (Supplementary Table S3). While the copy numbers were variable among the MWs (ranging from 15 to approximately 1,000 copies per cell), each MW had comparable copy numbers for the FVIII and rep and puroR genes, as would be expected if intact, tandemly repeated copies of the pTP-FVIII plasmid were integrated into the genome (Supplementary Table S3). In general, there was little correlation between the integrated plasmid copy number and vector yield.
The specific vector production levels from multiple MWs were assessed using small-scale shaker flask (20 mL) cultures, and the data are summarized in Supplementary Table S3. The stability of vector production was evaluated for the high-producing AAVrh8R/FVIII-5.1 (MW#35), AAVrh8R/FVIII-5.4 (MW#163), and AAV8/FVIII-5.1 (MW#287) MWs. Sustained high levels of vector yield (≥1 × 1010 vg/mL) was observed for all clones cultured for a minimum of 20 passages (Supplementary Fig. S2A). Further characterization of the high-producing MW (#35) for AAVrh8R/FVIII-5.1 showed peak rAAV production on days 3 and 4 (Supplementary Fig. S2B). This MW also provided similar production levels at the 250 mL and 1 L scale (Supplementary Fig. S2C). In each scale, the vector was detected in equal amounts in the cell pellets and culture media (Supplementary Fig. S2C).
Characterization of oversized rAAV vectors produced by producer cell lines and triple transfection
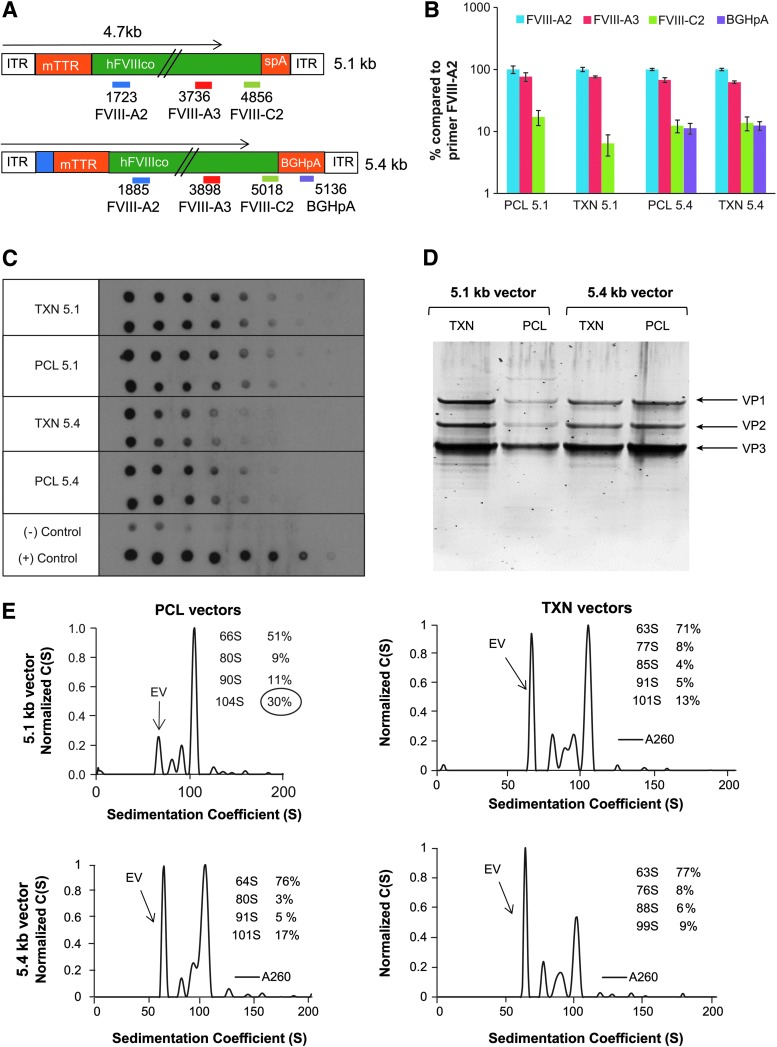
For comparative studies, the vectors AAVrh8R/FVIII-5.1 (MW#35) and AAVrh8R/FVIII-5.4 (MW#163) generated by PCL process were also produced via the TXN production process. Both PCL and TXN derived vectors were titered by qPCR using multiple primer/probes sets detecting various parts of the genome (Fig. 2A). Primers complementary to the internal regions (FVIII-A2, FVIII-A3; within 4.7 kb from either terminus) resulted in comparable titers, while primers complementary to regions beyond 4.7 kb (FVIII-A3, BGHpA) showed an average of eightfold lower titers (Fig. 2B). A DNA dot blot analysis was then performed for the genomes isolated from the 5.1 and 5.4 kb vectors applied at equal amounts (based on FVIII-A2 titers) and probed using a full vector genome probe to account for all genomes. This resulted in comparable signal detection for vectors derived from the two production platforms (however, slightly lower titers were observed for the 5.4 kb vector compared with the 5.1 kb vector). Since this result validated the titer values from the FVIII-A2 primer/probe set, all subsequent vector quantitations were performed using this primer/probe set unless otherwise indicated.
Figure 2.
Characterization of the AAVrh8R/FVIII-5.1 and AAVrh8R/FVIII-5.4 vector preparations. rAAV vectors were generated via producer cell line (PCL; MWs #35 and #163) or transient transfection (TXN) followed by purification via column chromatography. (A) Location of primer/probe sets used to measure titers for 5.1 and 5.4 kb FVIII vector genomes. Primer/probe sets used detect regions located internally (FVIII-A2, FVIII-A3) or toward the terminal ends (outside 4.7 kb from the 3′ terminus; FVIII-C2, BGHpA) of the vector genomes. (B) Vector quantitation with multiple primer/probe sets. The purified vectors used in vivo were titered with multiple primer/probe sets shown in (A). The results are calculated as % of titer compared to the internal primer/probe FVIII-A2 used for most analyses. (C) Evaluation of vector titers by DNA dot blot analysis. Equal amounts of vector genomes (1.5e9 vg) based on quantitative polymerase chain reaction (qPCR) with FVIII-A2 primer/probe were loaded in duplicates onto membrane and serially diluted twofold, followed by probing with fragment consisting of the majority of the vector genome (4.7 kb fragment containing promoter and FVIII cDNA). Plasmids containing non-FVIII (negative control) or FVIII (positive control) cDNA were used as controls for signal specificity. (D) Purity of vector lots. Equal amounts of vector (1 × 1010 vg/lane) were separated by SDS-PAGE, followed by protein staining with SYPRO stain. The locations of the capsid proteins (VP1, VP2, and VP3) are shown on the right. (E) Quality analysis of vector preparations by analytical ultracentrifugation analysis (AUC). The insert shows the vector sedimentation value (S) and percentage (%) for each peak. The empty virion (EV) peak has an S value of 63–64, while virions with 4.7 kb vector genomes are typically 100S–103S. Additional AAV/FVIII-5.1 lots analyzed by AUC are shown in Supplementary Fig. S3.
SDS-PAGE analysis of capsid proteins was performed by loading an equal amount of vector genomes. The results showed that the AAVrh8R/FVIII-5.1 vector produced by the TXN method contained an approximately fivefold higher level of capsid proteins (VP1, VP2, and VP3) than that generated via the PCL process (Fig. 2D). This result implies that a higher level of empty virions were present in the TXN-derived preparation. However, the larger AAVrh8R/FVIII-5.4 vectors generated by both platforms exhibited comparable levels of capsids, indicating that the preparations contained similar amounts of empty and full virions.
The proportion of empty and genome-containing particles of the rAAV vectors was further evaluated by AUC. This method can accurately determine the fractional content of capsids harboring the full-length vector genomes, capsids harboring no vector genomes (empty virions), or capsids harboring fragmented vector genomes. The size of the encapsidated vector genome correlates to the sedimentation coefficient (S) of each peak, and hence the fractional content of each capsid species in the vector preparation can be calculated.28 Analysis of three independent preparations of AAVrh8R/FVIII-5.1 vectors derived from the PCL process demonstrated that the empty virions (63 to 64S species) accounted for approximately 51% of the vector preparation (Fig. 2E and Supplementary Fig. S3). By contrast, approximately 71% of the virions were empty when the same vector was generated using the TXN method (Fig. 2E and Supplementary Fig. S3). Consistent with the SDS-PAGE data above, for the larger AAVrh8R/FVIII-5.4 vectors, the proportion of empty virions (76–77%) generated by both processes was comparable. Multiple peaks (76 to 101S species) representing genome-containing particles ranging in size from 1.3 to 4.7 kb were also observed for both 5.4 kb vectors, indicating that the packaged genomes generated by both production methods were heterogeneous. Neither production method yielded preparations containing a substantial population of sedimentation values expected for an AAV particle with 5.1 or 5.4 kb vector genomes (106S and 108S, respectively). The majority of the oversized vectors were packaged at the wild-type size for AAV (4.7 kb), as represented by the populations from 101 to 103S. However, the proportion of these capsids was approximately twofold higher in the PCL-derived 5.1 kb vector preparations (Fig. 2E and Supplementary Fig. S3). Although the same trend was observed for the 5.4 kb vectors, it should be noted that only one 5.4 kb vector lot generated by each process was analyzed.
The study also determined whether the method used to produce the AAV vectors affected the extent of non-vector DNA packaging. Measurement of the amount of encapsidated plasmid backbone sequences (ampR or puroR genes) revealed that a low level of puromycin DNA (<1%) was present in the PCL-produced material (Supplementary Table S4). However, approximately 10-fold higher levels of non-vector DNA were observed in vectors generated by the TXN process. These results suggest that the production method can influence the level of impurities and consequently the quality of the vector preparations.
Characterization of packaged genomes in oversized rAAV vectors
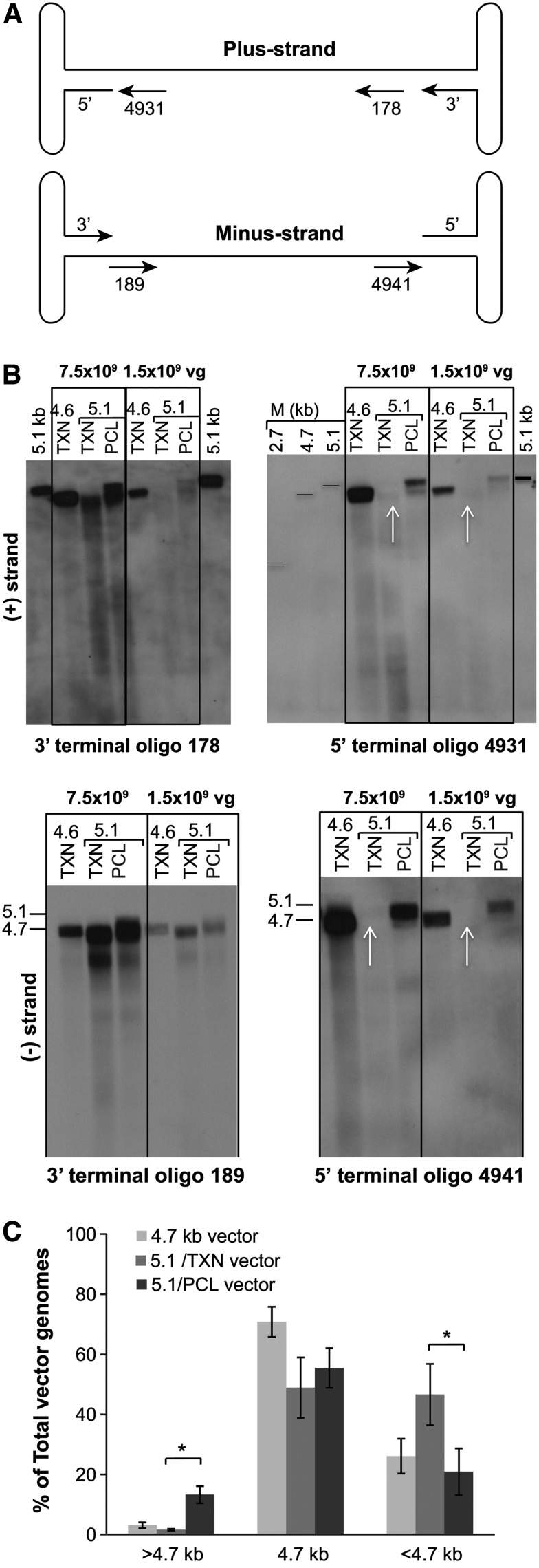
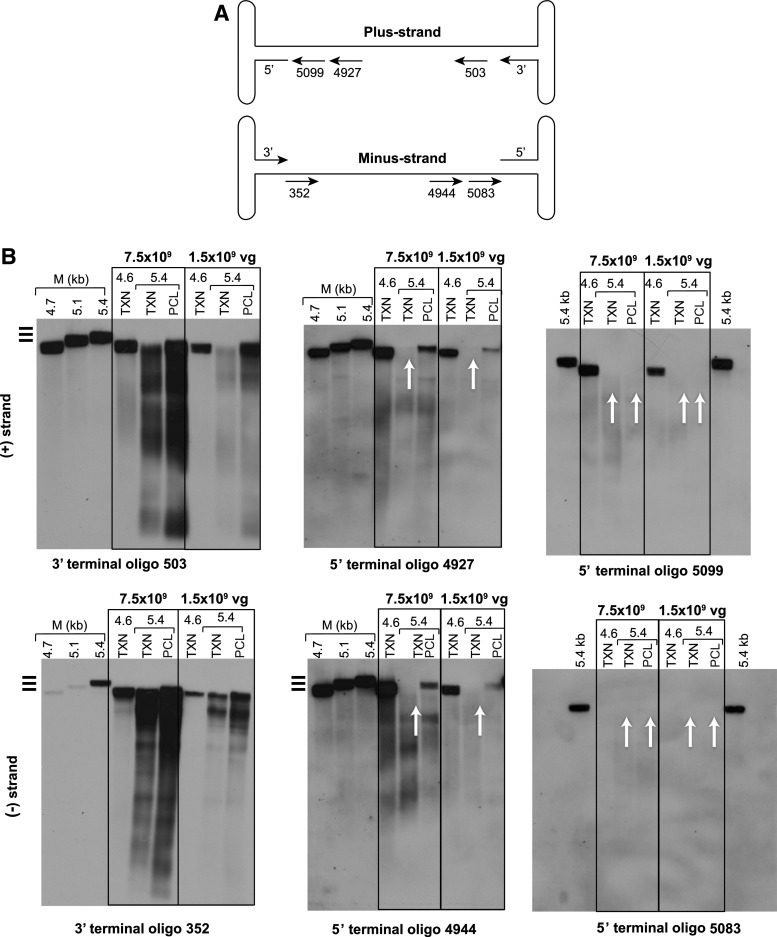
Southern blot analyses were performed to evaluate the size of the packaged genomes and the presence of terminal deletions. Equal amounts of encapsidated vector genomes isolated from the AAVrh8R/FVIII-5.1 and AAVrh8R/FVIII-5.4 vectors prepared using TXN or the PCL processes were applied to agarose gels. The genomes were probed with various strand-specific oligonucleotides detecting either the 5′ or 3′ terminal ends (Figs. 3A and 4A and Table S1). Because packaging of the genomes initiates from the 3′ termini of the ITRs,5,29 all vectors should harbor 3′ termini while the detection of 5′ terminal ends may be reduced or absent (as confirmed by the vector titer results using the terminal primer/probe sets; Fig 2B).
Figure 3.
Characterization of the 5′ and 3′ ends of the packaged vector genomes in PCL- and TXN-generated AAVrh8R/FVIII-5.1 vector preparations. (A) Diagram showing the location of strand-specific oligonucleotide probes complementary to the 5′ and 3′ termini of the plus (+) and minus (−) strands of the 5.1 kb vector. The numbering of the probes indicates the distance from the 3′ end of the 5.1 kb vector genomes. Sequences for these probes (Supplementary Table S1) are also present in the control AAVrh8R/FVIII-4.6 control vector.22 (B) Southern blot analysis using 3′ and 5′ terminus probes for the AAVrh8R/FVIII-5.1 vector. The vectors were loaded at 1.5 and 7.5 × 109 vg/lane and run as described in the Materials and Methods. Top panels, (+) strand analysis; bottom panels, (−) strand analysis. The probes and size markers (2.7, 4.7, and 5.1 kb) used for each panel are shown. White arrows indicate missing signals in the TXN-derived vector. (C) Quantitation of genome sizes in each vector. The signal intensity in the panels probed with the 3′ terminal probes (detects all packaged genomes) was quantified by ImageJ software. The intensity of each distinct genome size (>4.7 kb, 4.7 kb, and <4.7 kb) was quantified for the (+) and (−) strands (both concentrations), and the average of the four measurements was graphed as the % of the total signal.
Figure 4.
Characterization of the 5′ and 3′ ends of the packaged vector genomes in PCL- or TXN-generated rAAVrh8R/FVIII-5.4 vectors. (A) Diagram showing the location of the oligonucleotide probes complementary to the 5′ and 3′ termini of the (+) and (−) strands of the vector genomes. The numbering of the probes indicates the distance from the 3′ end of the 5.4 kb vector genomes. Complementary sequences for all probes except oligo #5083 (located in the BGHpA region of [−] strand) are present in the rAAVrh8R/FVIII-4.6 control vector.22 (B) Southern blot analysis of rAAVrh8R/FVIII-5.4 vectors using 3′ and 5′ terminal oligonucleotide probes. The experiment was performed as described in Fig. 3. The vector genome was not detected for the AAVrh8R/FVIII-4.6 control vector (TXN 4.6) with oligo probe #5083 because the complementary sequence is not present in this vector (the 4.6 kb vector contains spA, whereas the probe targets BGHpA). The white arrows indicate missing signals in the vectors.
Probing the vector preparations produced by the TXN or PCL processes with oligonucleotides complementary to the 3′ termini of the genomes (both plus and minus strands) demonstrated that the AAVrh8R/FVIII-5.1 vectors contained more heterogeneously sized genomes than the AAVrh8R/FVIII-4.6 vector, regardless of the production method (Fig. 3B; oligo probes 178 and 189). A similar vector genome size pattern was observed when probing a Southern blot with a full-length genome probe (data not shown). However, the PCL-derived AAVrh8R/FVIII-5.1 vector preparations contained more genomes approximately ≥4.6 kb compared to the TXN-derived material.
Previous reports have indicated that oversized vectors may lack complete sequences at the 5′ ends of the minus and plus strands.3,4,7,10,21,22 Southern blot analysis using oligonucleotide probes specific to the 5′ termini (oligo probes 4931 and 4941) detected vector genomes that co-migrated with the 5.1 kb marker in the PCL vector, whereas these fragments were not detected in the TXN-generated AAVrh8R/FVIII-5.1 vector (Fig. 3B, white arrows). Analysis of the signal intensities suggested that between 10% and 20% of the vector genomes in the PCL-derived AAVrh8R/FVIII-5.1 vector were ≥4.7 kb and potentially full-length 5.1 kb in size (Fig. 3C). In summary, although both production platforms generated AAVrh8R/FVIII-5.1 viral preparations that contained more fragmented genomes than observed in the AAVrh8R/FVIII-4.6 vector, the PCL-derived vector had more genomes with an approximate size of 5.1 kb than the TXN-generated preparations. Thus, the data indicate that the methods for producing oversized AAV vectors can result in qualitative differences, with the PCL process generating less heterogeneous genome sizes and packaging more intact genomes.
Southern blot analysis was also performed on the larger AAVrh8R/FVIII-5.4 vector produced using the PCL and TXN methods (Fig. 4). Probing with oligonucleotides complementary to the 3′ ends of the strands (oligo probes 503 and 352; Fig. 4B, left panel) revealed a higher level of heterogeneity in vector genome sizes than noted for the AAVrh8R/FVIII-5.1 vector, irrespective of the method used for production. Moreover, little to no signals for genomes in the 5.1 to 5.4 kb size range were detected. This result was confirmed using probes complementary to the 5′ terminal end of the strands (at 4.9 kb from the 3′ ends; oligo probes 4927, 4944; middle panel). These probes detected genomes of approximately 4.9 kb in the PCL-derived vector but not the TXN material (white arrows). Additional testing using an oligonucleotide that was closer to the 5′ terminus (at 5.1 kb from the 3′ ends; oligo probes 5099, 5083; right panel) did not reveal any signal for 5.4 kb vectors produced by either process (white arrows). Thus, similar to the AAVrh8R/FVIII-5.1 vector, the AAVrh8R/FVIII-5.4 vector generated via the PCL process contained a higher proportion of larger-sized genomes (although none were full-length) than the vector produced by the TXN method did.
Efficacy of the oversized rAAV vectors in the mouse model of hemophilia A
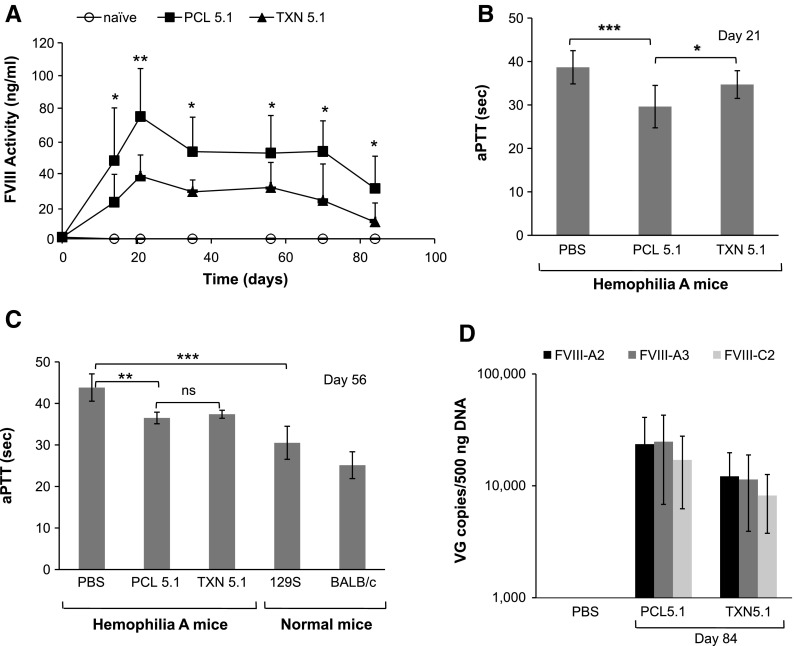
The transduction activity of AAVrh8R/FVIII-5.1 vectors produced using either the PCL or TXN process was compared in a mouse model of hemophilia A. Equal doses of vectors (4 × 1010 vg/mouse [2 × 1012 vg/kg]) generated by PCL and TXN were delivered via intravenous injection, and FVIII activity was monitored for 84 days post administration. Mice treated with the PCL-produced AAVrh8R/FVIII-5.1 vector displayed approximately twofold higher plasma FVIII activity than mice treated with vector produced via TXN did, which translated to significantly shorter clotting times on day 21 (Fig. 5A and B). However, the clotting times at day 56 were similar for the PCL- and TXN-generated viruses, suggesting that the difference noted on day 21 may be due to more rapid expression kinetics from the PCL-derived virus (Fig. 5C). Analysis of the livers from these mice on day 84 revealed that the number of vector genomes was on average twofold higher in mice treated with the PCL-generated virus than it was in those treated with the vector produced via TXN (Fig. 5D). Vector genome quantitation with internal and terminal primer/probe sets showed comparable amounts of vector genomes in liver reflecting the presence of full-length genomes (in contrast to the qPCR results for the administered vector; Fig. 2B), as would be expected based on the plasma FVIII detection. There was no significant difference in vector genome copies among the two treatment groups at day 84, though a significant difference in plasma FVIII levels was observed at all time points.
Figure 5.
Comparison of PCL- and TXN-produced AAVrh8R/FVIII-5.1 vectors in vivo using hemophilia A mice. Vectors were administered via the tail vein at 4 × 1010 vg/mouse. (A) Plasma FVIII protein activity. Activity was measured in day 21, 35, 56, 70, and 84 samples using the Coatest® assay. (B) Plasma clotting times on day 21 measured by the aPTT assay. (C) Plasma clotting times on day 56. The clotting times for mouse strains 129S and BALB/c are shown for comparison. (D) Vector genome levels in the liver. Vector genome copies on day 84 were quantitated by qPCR using three primer/probe sets (FVIII-A2, FVIII-A3, and FVIII-C2). For all analyses, the method of virus production is indicated (PCL or TXN). All treatment groups contained eight mice per group. Values in each graph represent the average ± standard deviation, and significance was calculated using Student's t-test, with significance indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.
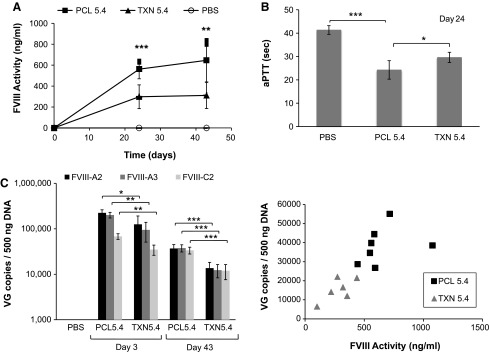
Hemophilia A mice were also administered comparable doses of TXN- or PCL-derived AAVrh8R/FVIII-5.4 vector (4 × 1010 vg/mouse), and plasma FVIII levels were measured for 43 days. Similar to the AAVrh8R/FVIII-5.1 vector, the AAVrh8R/FVIII-5.4 vectors prepared by the PCL platform produced twofold higher plasma FVIII activity than those generated by the TXN method did (Fig. 6A). The levels of FVIII activity were higher in the AAVrh8R/FVIII-5.4-treated mice than they were in the AAVrh8R/FVIII-5.1-treated mice, presumably because of the use of a stronger expression cassette in the AAVrh8R/FVIII-5.4 kb vector (Fig. 1C and D). Interestingly, while only a twofold increase in FVIII activity was observed with the previral plasmids with the optimized larger promoter in vivo, a 5- to 10-fold increase in FVIII levels was noted for the rAAV vector in vivo (Fig. 1D vs. Fig. 6A). Similar to the results observed for the AAVrh8R/FVIII-5.1 vector above, the PCL-derived AAVrh8R/FVIII-5.4 vector resulted in shorter clotting times on day 24 (Fig. 6B) but not day 43 (not shown).
Figure 6.
Comparison of PCL- and TXN-produced rAAVrh8R/FVIII-5.4 vectors in vivo using hemophilia A mice. Vectors were administered via the tail vein at 4 × 1010 vg/mouse. (A) Plasma FVIII protein activity. Activity was measured on days 24 and 43 by a Coatest® assay. (B) Day 24 plasma clotting times measured by an aPTT assay. (C) Vector genome levels in the liver. Vector genome copies on days 3 and 43 were quantified by qPCR using three primer/probe sets (FVIII-A2, FVIII-A3, and FVIII-C2). (D) Correlation of plasma FVIII activity and vector genomes in the liver on day 43. Vector genome levels quantitated by FVIII-A2 primer/probe set are shown. Each point represents one mouse. For all analyses, the method of virus production is indicated (PCL or TXN). Each virus treatment group contained four (day 3) or six to eight (day 43) mice per group (five for the control, PBS, group). Values in each graph represent the average ± standard deviation, and significance was calculated using Student's t-test, with significance indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.
Quantitation of vector genome levels in the liver demonstrated two- to threefold higher copies on days 3 and 43, respectively, in mice treated with the PCL-derived vector compared with the mice treated with the TXN-generated vectors (Fig. 6C). This result is supported by the correlation between the liver vector genomes and plasma FVIII activity in individual animals (Fig. 6D). Both PCL- and TXN-derived vectors had only partial detection of the terminal regions (detected with FVIII-C2 primer/probe set) on day 3, indicating that the correction of truncated terminal ends continued beyond day 3. However, similar to results with the 5.1 kb vectors, all qPCR primer/probe sets showed comparable levels of vector genomes in the liver on day 43. Note that vector genomes were reduced six- to eightfold from day 3 to day 43, indicating that only a portion of the input genomes was processed into a persistent form.
Discussion
Oversized rAAV vectors encoding FVIII are efficacious at correcting disease manifestations in animal models of hemophilia A.6–10 However, the methods used to generate the vectors in these studies have generally produced vectors poor in yield and quality, delaying the deployment of these vectors in clinical studies. Comparisons of the yield and quality attributes of oversized rAAV vectors generated using various production platforms are scarce. This study evaluated and characterized the properties and activity of slightly oversized rAAV vectors encoding FVIII produced by either a PCL or TXN process.
Various production platforms have been developed to manufacture rAAV vectors for clinical trials.19 These include methods based on the use of transient transfection of HEK 293 cells with plasmids containing the vector genomes and helper functions (e.g., to support hemophilia B trials14,30), BHK cells infected with rHSV (e.g., to support α1-antitrypsin deficiency trials15,16), HeLa cell-based PCLs infected with adenovirus (e.g., to support a heart failure trial19), and Sf9 cells infected with baculovirus (e.g., to support a lipoprotein lipase trial31). Both the PCL- and baculovirus-based systems are easily scalable serum-free production processes; such scalability is necessary for the treatment of diseases that require large vector doses or which have large patient populations.17–19 The present study evaluated the PCL/adenovirus-based approach for preparing oversized rAAV vectors because of its reported ability to generate high-quality vectors in high yield (≥50,000 vg/cell).18,23,26 The system uses a HeLaS3 cell line engineered to contain an integrated vector genome and AAV2 rep and cap genes; these genes enable rapid scale-up of production upon infection with wild-type adenovirus. Previous analyses of this system focused on generating rAAV vectors containing vector genome sizes of ≤4.7 kb to confirm the merits of this platform.26 However, factors such as an oversized genome, capsid serotype, and the characteristics of the transgene and expression cassette are likely to impact the success of obtaining high-producing cell lines. The present report is the first to evaluate the merits of the adenovirus-based PCL process for generating slightly oversized rAAV vectors. This study demonstrates that it is feasible to generate high-yielding cell lines for rAAV vectors that harbor either a 5.1- or 5.4-kb vector genome in the context of the AAVrh8R serotype. Though the frequency of high-producing cell lines for oversized rAAV vectors was reduced compared to vectors with genomes at ≤4.7 kb, it is not clear if this frequency was significantly impacted due to the limited number of oversized vector cell lines generated.
The PCLs produced oversized rAAV vectors at high yield over multiple passages (tested up to 60 passages), an important characteristic for manufacturing on a large scale (e.g., to ≥250 L bioreactors). Analysis of the integration patterns of the plasmids containing the transgene and cap and rep genes in high-producing cell lines revealed no evidence of deletions or rearrangements of these genes. The copy number of the integrated plasmids was comparable to that observed for the wild-type size rAAV vector, indicating that generation of PCLs for oversized vectors is feasible.26 There was little correlation between the observed copy number of integrated plasmids and vector production levels, suggesting that other factors, such as the site of integration and perhaps the extent of amplification of rep–cap sequences, may play a role in influencing production yields. This incomplete concordance between genome copies and vector production has been previously reported for MWs producing wild-type size rAAV vectors and does not appear to be a specific property of oversized vectors.24–26
Typically, rAAV vectors are designed not to exceed the size of the wild-type size AAV genome (4.7 kb) because efforts to produce oversized rAAV vectors have invariably led to the generation of virions with heterogeneously sized genomes, with the majority approximately ≤4.7 kb. This outcome is likely caused by partial degradation of genomes by cellular exonucleases too large to fit into capsids during packaging.3,4 In the analysis of the 5′ ends of the packaged genomes, methods that extended or amplified the sequences were avoided, and detection of the native 5′ ends using strand-specific oligonucleotide probes was relied on instead. Comparison of oversized rAAV vectors generated by the PCL or TXN platforms demonstrated that both processes packaged genomes with heterogeneous sizes and contained genomes deleted in the 5′ termini. However, differences in the quality of the vector preparations were observed. For example, approximately 10–20% of AAVrh8R/FVIII-5.1 vectors produced by the PCL process contained full-length genomes (5.1 kb), while no full-length genomes were detected in vector packaged by the TXN method. Approximately half of the viral particles from both platforms comprised fragmented genomes of 4.7 kb, with the remainder consisting of smaller encapsidated genomes that may be due to premature termination of packaging.
As many of the aspects of AAV viral genome packaging are not understood, the mechanism by which the PCL process might enhance packaging of oversized genomes is unclear. However, the PCL method does closely mimic the natural process in which AAV replication is initiated by adenovirus infection, resulting in amplification and coordinated expression of the rep and cap genes.24–26 Undefined attributes of this process (e.g., the temporal and spatial organization of AAV gene expression, the contributions of specific adenoviral gene products, and the alterations in the cellular milieu by adenovirus infection) may result in more efficient assembly of rAAV particles and vector DNA packaging. Accordingly, this more efficient packaging may be responsible for the improved encapsidation of oversized genomes and the reduction in packaging of fragmented genomes. This overall efficiency is also consistent with the generally higher proportion of virions containing vector genomes in the PCL-derived vector (approximately 70–90%) compared with the vector produced via TXN (approximately 20%).26 Similarly, the PCL process produced oversized AAVrh8R/FVIII-5.1 vector preparations containing a greater proportion of genome-containing particles (50%) than the TXN method did (30%). Taken together, these results indicate that upstream production processes can have a significant impact on vector quality.26,28
The results for the larger AAVrh8R/FVIII-5.4 vector demonstrated that some of the benefits of PCL platform did not extend to larger vector genomes. Although the AAVrh8R/FVIII-5.4 vector yield in high-producing cell lines was comparable to AAVrh8R/FVIII-5.1 vector, the percentage of vector genome-containing particles was lower (24%) than that of the TXN-derived vector. These results suggest that a larger vector genome size reduced the efficiency of vector assembly. The packaged genomes were also more heterogeneous than the AAVrh8R/FVIII-5.1 vector was, presumably due to an increase in premature termination of packaging. This phenomenon has been suggested to occur more frequently at the initiating 3′ITR when the 5′ITR is not within the normal packaging limit.5 Although the PCL-derived AAVrh8R/FVIII-5.4 vector preparations contained a higher portion of larger genomes than the TXN-derived vector did, none was the full-length size (5.4 kb), with the largest genomes ranging from 4.6 to 4.9 kb. These results suggest that genome sizes of 5.4 kb exceed the constraints of the normal packaging process, whereas some 5.1 kb genomes may be packaged as full-size genomes. These data are consistent with previous reports indicating that the maximum packaging capacity of AAV vectors is approximately 5.0–5.1 kb.2,4 However, as only one vector lot was generated by each method, the reproducibility of the quality differences between the PCL- and TXN-derived 5.4 kb vector is unknown. Furthermore, whether the quality differences observed between the 5.1 and 5.4 kb vectors were affected by any sequence-based packaging bias is unclear, since similar analyses with comparable sized alternate vector sequences were not performed.
The study also determined whether the method of producing rAAV vectors affected the extent of packaging of non-vector DNA. Non-vector DNA is an impurity that cannot be easily removed by downstream processing.19,20 It was previously observed that increasing amounts of plasmid backbone are packaged into rAAV8 vectors prepared by the TXN method as the size of the genome increases.22 The present study demonstrated that rAAV vectors produced via the PCL process contain reduced amounts of non-vector DNA (e.g., puroR gene) compared with those generated by the TXN process. This result may be due to the absence of plasmid DNA in the PCL process for vector production. Modifications such as the use of large vector plasmid backbones and reduced rep expression can reduce packaging of plasmid-derived DNA (0.02%) in TXN-prepared vectors.14,28,32 Although packaging of rep and cap sequences in the PCL vector preparations were not measured, only very low levels (0.4–0.6%) of these sequences have been reported, despite the 100-fold amplification of rep–cap sequences upon infection with adenovirus.24–26 The TXN and PCL processes both typically result in very low levels of encapsidation of host genomic DNA.14,18,26
The goal of the studies performed in hemophilia A mice was to evaluate the impact of the vector quality differences on the efficiency of hepatic transduction. The PCL-derived AAVrh8R/FVIII-5.1 and AAVrh8R/FVIII-5.4 vectors both generated twofold higher levels of FVIII activity than the corresponding TXN-derived vectors did. These higher levels of FVIII activity translated to faster clotting times on days 21 and 24, although these differences were reduced at later time points. The PCL-generated AAVrh8R/FVIII-5.1 and AAVrh8R/FVIII-5.4 vectors also resulted in a twofold higher number of vector genomes in the liver than their TXN-derived counterparts. The comparable levels of vector DNA in the liver for the 5.1 and 5.4 kb vectors were unexpected because the latter vector was significantly more heterogeneous. However, this is consistent with previous observations of AAV8/FVIII vector genomes ranging from 4.9 to 5.4 kb all resulting in comparable vector genome copies in the liver.22 Because the AAVrh8R/FVIII-5.4 vectors generated by the two production methods contained similar amounts of empty capsids, differences in the quality of the packaged genomes rather than the ratio of full to empty capsids were likely responsible for the higher efficacy of the PCL-generated vector. The presence of more intact genomes in the PCL-derived vectors may have permitted more rapid and perhaps more efficient conversion of incomplete genomes into full-length transcriptionally active genomes and persistent circular forms. This rapid rate of conversion is supported by the observation of twofold more vector genomes at 3 days post administration in the livers of animals treated with the PCL-derived vector compared with that of the TXN vector. Vector genomes released after capsid uncoating are rapidly degraded within the first few days, unless they are converted into more stable forms, such as double-stranded and/or circular forms, by host DNA repair proteins.12,33–35 The presence of intact ITR and D sequences plays an important role in the process, and hence any full-length genomes may be preferentially converted to the more stable forms.36,37 However, this may only apply to the 5.1 kb vector, as no full-length ITR containing genomes could be detected for the 5.4 kb vector. The eightfold decrease in the amount of 5.4 kb vector genomes between day 3 and 43 demonstrated that only a portion of these genomes were processed into persistent genomes. This inefficiency has also been demonstrated for wild-type sized genomes and thus is not restricted to oversized vector genomes.34 Lastly, in vivo evaluation of multiple 5.1 and 5.4 kb lots generated by the two production processes would be required to confirm the consistency of these observations and whether the significant differences in FVIII production levels between the PCL and TXN lots were simply due to differences in the number of genome copies or were due to qualitative differences in the persistent forms of vector-derived genomes. Furthermore, analysis of input vector genomes shortly after administration (i.e., 6 h) would be helpful to examine the kinetics and nature of genome processing that is required to generate full-length genomes from the predominantly truncated vector genomes.
Lastly, a critical aspect of these experiments was an accurate quantitation of the vector preparations containing heterogeneously sized genomes. Given that most of the genomes in the 5.1 kb AAVrh8R/FVIII vector were approximately 4.7 kb in length and thus contained terminal deletions, the use of an internal primer was adopted to capture the large majority of the genomes for the estimation of production yields during screening, MW ranking, and characterization. For the 5.4 kb vector that showed more variability in packaged genome sizes, titering with a single primer/probe set could be more challenging. However, the titer analysis of both 5.1 and 5.4 kb vectors resulted in comparable values when lots were quantitated with a primer/probe set specific to sequences within 4.7 kb of the vector termini while detection of terminal regions was consistently reduced. DNA dot blot analysis using the vector genome as a probe confirmed the titer values as signal intensities were comparable for vectors of the same, size though the 5.1 and 5.4 kb vectors showed a slight difference. Hence, these data support the accurate vector quantitation, particularly when comparing vectors of the same size from the two production platforms. However, oversized vectors with heterologous genomes add more complexity with respect to the ability to quantitate them accurately.
In summary, this study has demonstrated that the PCL platform is feasible for generating oversized rAAV vectors. While the quality of the vector preparations was lower than that of wild-type sized rAAV vectors, the PCL-derived vectors typically contained a greater proportion of ≥4.7 kb genomes than those generated via the TXN method did. Importantly, this study demonstrated that this improvement in quality translates into greater efficacy in a mouse model of hemophilia A.
Supplementary Material
Acknowledgments
We appreciate the support from Abraham Scaria. We thank John Martin, Amy Frederick, Yuxia Luo, and Chris Renzi for their help with the producer cell line work. We also thank the members of Virus Production and Comparative Medicine for producing the vectors and supporting the animal studies, respectively. The hydrodynamic injection technique was conducted under a research license grant from Arrowhead Madison, Inc.
Author Disclosure
All authors were employees of Sanofi at the time of the study.
References
- 1.Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther 1996;7:2101–2112 [DOI] [PubMed] [Google Scholar]
- 2.Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol 2005:79:9933–9944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther 2010;18:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther 2010;18:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, Chen L, Dederich D, et al. Molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum Gene Ther 2012;23:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Lillicrap D, Patarroyo-White S, et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 2006;108:107–115 [DOI] [PubMed] [Google Scholar]
- 7.Lu H, Chen L, Wang J, et al. Complete correction of hemophilia A with adeno-associated viral vectors containing a full-size expression cassette. Hum Gene Ther 2008;19:648–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monihan PE, Lotrop CD, Sun J, et al. Proteosome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol Ther 2010;18:1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatino DE, Lange AM, Altynova ES, et al. Efficacy and safety of long-term prophylaxis in severe hemophilia A dogs following liver gene therapy using AAV vectors. Mol Ther 2011;19:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIntosh J, Lenting PJ, Rosales C, et al. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood 2013;121:3335–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scallan CD, Liu T, Parker AE, et al. Phenotypic correction of a mouse model of hemophilia A using AAV2 vectors encoding the heavy and light chains of FVIII. Blood 2003;102:3919–3926 [DOI] [PubMed] [Google Scholar]
- 12.Hirch ML, Li C, Bellon I, et al. Oversized AAV transduction is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol Ther 2013;21:2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright JF. Transient transfection methods for clinical adeno-associated viral vector production. Hum Gene Ther 2008,15:840–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allay JA, Sleep S, Long S, et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated virus vector for a hemophilia B clinical trial. Hum Gene Ther 2011:22:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas DL, Wang L, Niamke J, et al. Scalable recombinant adeno-associated virus production using recombinant herpes simplex virus type 1 co-infection of suspension adapted mammalian cells. Hum Gene Ther 2009;20:861–870 [DOI] [PubMed] [Google Scholar]
- 16.Flotte TR, Trapnell BC, Humphries M, et al. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing a1-antitrypsin: interim results. Hum Gene Ther 2011;22:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotin RM. Large-scale recombinant adeno-associated virus production. Hum Mol Genet 2011;20:R2–R6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorne BA, Takeya RK, Peluso RW. Manufacturing recombinant adeno-associated viral vectors from producer cell clones. Hum Gene Ther 2009:20:707–714 [DOI] [PubMed] [Google Scholar]
- 19.Clement N, Grieger JC. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol Ther Methods Clin Dev 2016;3:16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright JF. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther 2008;15:840–848 [DOI] [PubMed] [Google Scholar]
- 21.Grose WE, Clark KR, Griffin D, et al. Homologous recombination mediates functional recovery of dysferlin deficiency following AAV5 gene transfer. PLos One 2012;7:e39233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyostio-Moore S, Berthelette P, Piraino S, et al. The impact of minimally oversized adeno-associated viral vectors encoding human Factor VIII on vector potency in vivo. Mol Ther Methods Clin Dev 2016;24:16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark KR, Voulgaropoulou P, Fraley DM, et al. Cell lines for the production of recombinant adeno-associated virus. Hum Gene Ther 1995,6:1329–1341 [DOI] [PubMed] [Google Scholar]
- 24.Chadeuf G, Favre D, Tessier J, et al. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J Gene Med 2000;2:260–268 [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Voulgaropoulou F, Chen R, et al. Selective rep-cap gene amplification as a mechanism for high titer recombinant AAV production from stable cell lines. Mol Ther 2000;2:394–403 [DOI] [PubMed] [Google Scholar]
- 26.Martin J, Frederick A, Luo Y, et al. Generation and characterization of adeno-associated virus cell lines for research and preclinical vector production. Hum Gene Ther Methods 2013;24:253–269 [DOI] [PubMed] [Google Scholar]
- 27.Costa RH, Lai E, Darnell JE., Jr. Transcriptional control of the mouse prealbumin (transthyreting) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol 1986;6:4697–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnham B, Nass S, Kong E, et al. Analytical ultracentrifugation as an approach to characterize recombinant AAV vectors. Hum Gene Ther Methods 2015;26:228–242 [DOI] [PubMed] [Google Scholar]
- 29.King JA, Dubielzig R, Grimm D, et al. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsid. EMBO J 2001;20:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathwani AC, Tuddenham EGD, Rangarajan S, et al. Adeno-associated virus vector mediated gene transfer in hemophilia B. New Engl J Med 2011;22:2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaudet D, Methot J, Dery S, et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther 2013;20:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauck B, Murphy SL, Smith PH, et al. Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol Ther 2009;17:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai H, Storm TA, Kay MA. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J Virol 2000;74:9451–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas CE, Storm TA, Huang Z, et al. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol 2004;78:3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi VW, McCarthy DM, Samulski RJ. Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol 2006;80:10346–10356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling C, Wang YL, Jayndharan GR, et al. The adeno-associated virus genome packaging puzzle. J Mol Genet Med 2015;9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Z, Zak R, Engelhart JF. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J Virol 2005;79:364–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.