Abstract
Pancreatic Cancer (PC) is a devastating lethal disease. Therefore, there is an urgent need to develop new intervention strategies. The mammalian Target of Rapamycin (mTOR) is a conserved kinase and master regulator of metabolism and cell growth. mTOR is dysregulated in chronic diseases including diabetes and pancreatic cancer. Recent reports indicate that 50% of Pancreatic Ductal Adenocarcinoma (PDAC) patients are diabetic at the time of diagnosis. Furthermore, the anti-diabetic drug, metformin, which indirectly inhibits mTOR, has emerged as a potential therapeutic target for PC.
The objective of this study is to determine the targeted-metabolomics profile in PDAC cell line (HPAF-II) with mTOR inhibition and the interaction between mTOR ATP-competitive inhibitor (Torin 2) and metformin as potential combined therapy in PC.
HPAF-II cell lines were cultured in the presence of either Torin 2, metformin, both, or control vehicle. We utilized targeted LC/MS/MS to characterize the alterations in glycolytic and tricarboxylic acid cycle metabolomics, and employed Western Blot analysis for cell signaling activation by phosphorylation. Comparisons between groups were analyzed using one-way Analysis of Variance followed by secondary post-hoc analysis.
After 1 h incubation with metformin, AMP concentration was significantly increased compared to other groups (p<0.03). After 24 h, Torin-2 significantly decreased glycolysis intermediates (fructose 1,6-bisphosphate (FBP), and 2-phosphoglycerate/3-phosphoglycerate), TCA intermediate metabolites (citrate/isocitrate, and malate), as well as Nicotinamide Adenine Dinucleotide (NAD+) and Flavin Adenine Dinucleotide (FAD), and ATP levels. When HPAF-II cells were incubated with both Torin-2 and metformin, there was a significant reduction in NAD+ and FAD, suggesting decreased levels of the energy equivalents that are available to the electron transport chain.
Targeted metabolomics data indicate that mTOR complexes inhibition by Torin 2 reduced glycolytic intermediates and TCA metabolites in HPAF- II and may synergize with metformin to decrease the electron acceptors NAD+ and FAD which may lead to reduced energy production.
Keywords: mTORC1, mTORC2, Metabolomics, Glycolysis, TCA cycle
Introduction
Pancreatic Cancer (PC) is a devastating disease with an estimated 53,070 new cases and an estimated 41,780 deaths respectively in 2016 [1]. PC is a lethal disease attributable to the late diagnosis and PC is the 3th leading cause of cancer-related death with the 5-year survival rate 8% as reported between years 2005-2011 [1]. The most common type of pancreatic cancer is Pancreatic Ductal Adenocarcinoma (PDAC). PDAC is the also most aggressive type of PC due to late diagnosis, high metastatic capacity, aggressive infiltrating nature of the disease and the current in effective treatment [2]. Since pancreatic cancer is diagnosed at an advanced stage, the early events that trigger cancer development and metastasis are largely unknown. Therefore, there is an urgent need to develop new strategies for prevention, early detection, and therapeutic interventions for this aggressive cancer. Recent reports indicate that 50% of PDAC patients are diabetic at the time of diagnosis. To that end, tumor-related diabetes is now considered as Type 3c Diabetes Mellitus (3cDM). Mounting evidence links the glycolytic metabolic disturbances to the adverse pancreatic cancer prognosis. Furthermore, the anti-diabetic drug, metformin, which indirectly inhibits the mammalian Target of Rapamycin (mTOR) via activation of AMPK (5’ adenosine monophosphate-activated protein kinase), has emerged as a potential therapeutic target in the treatment of PC, particularly when associated with type 3c Diabetes Mellitus
The best chances of survival are while the disease is still localized in the pancreas and surgical dissection is still an option. Thus using a traceback approach, which allows us to investigate the underlying metabolite platform associated with effective therapy, may provide cues to understanding the early events that preceded distant metastasis.
mTOR is a 289 kDa serine/threonine conserved protein kinase and is ubiquitously expressed in eukaryotes [3]. mTOR cellular signaling cascade serves as a master regulator of metabolism, cell growth and proliferation. mTOR is dysregulated in several diseases of metabolism including diabetes and pancreatic cancer [4,5]. mTOR kinase nucleates two functionally and structurally distinct complexes namely; mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). The first complex, mTORC1 selectively binds Raptor (Regulatory associated protein of mammalian Target of Rapamycin), and other proteins including mLST8 (mammalian Lethal with Sac 13 protein 8), Deptor, and PRAS40. mTORC1 integrates signals from the insulin pathway, growth hormones, amino acids, AMP/ATP energy levels, and mitogens at the cellular level to effectuate downstream targets [6]. mTORC1 is activated by nutrients, glucose, amino acid, growth factors, mitogens, and cellular energy and is inhibited by the prototype drug, rapamycin [7]. As such, mTORC1 activates anabolism including lipid and protein synthesis, ribosome biogenesis, beta cell mass expansion, and nucleotides biosynthesis and inhibits catabolism and autophagy [4,8,9]. On the contrary, mTORC2 binds exclusively to Rictor (Rapamycin-Insensitive companion of TOR), in addition to other proteins including mSIN1 (mammalian stress-activated map kinase-interacting protein 1), MLST8, Deptor and Protor 1, 2. Unlike mTORC1, mTORC2 complex is only activated by growth factors and is insensitive to rapamycin treatment, at least during acute treatment [10]. It is worth noting that in some cell lines, mTORC2 was reported to be responsive to rapamycin with chronic and prolonged long-term treatment due to inhibition of the assembly of mTORC2 [11]. mTORC2 activates actin cytoskeleton rearrangement. Importantly, mTORC2 phosphorylates PKC isoforms including, Akt regulatory site, serine 473, which in turn regulates cellular processes and plays a significant role in tumor growth and proliferation, angiogenesis and tissue invasion. Thus both mTORC1 and mTORC2 pathways play a fundamental role in cancer cells including growth and proliferation; cell cycle, genomic instability and cellular and tumor metabolism [12].
Metformin, an antihyperglycemic drug, is the first-line of treatment of type II diabetes and is a widely prescribed anti-diabetes drug [13-15]. Additionally, metformin inhibits mTORC1 by activation of AMPK, and has emerged as a potential therapeutic target in the treatment of cancer [16]. Further, Sah and Colleagues reported that the majority of pancreatic cancer patients have diabetes mellitus or hyperglycaemia which may manifest even prior to the time of diagnosis [17]. Epidemiological studies indicated that the use of metformin in type II diabetes patients was associated with reduced cancer incidence and cancer-related death [18-20]. While the mechanisms of action of metformin that confer anticancer and chemopreventive properties are not entirely elucidated; one proposed action is serving as indirect inhibitor of mTOR via AMPK activation. Studies have shown that metformin may mediate some of the anticancer properties via mTOR inhibition in mice and human liver cancer cell lines [21-23]. It has been suggested that mTOR inhibition by AMPK activators may lead to cancer metabolic reprogramming, which is the hallmark of cancer [24,25]. Mounting evidence links glycolytic metabolic disturbances and the adverse pancreatic cancer prognosis. Therefore, we determined the targeted-metabolomic profile in human PDAC cell line (HPAF-II); to address the possible synergism between mTOR inhibition by ATP competitors (which inhibits both mTORC1 and mTORC2), and Metformin administration (AMPK activator which indirectly inhibits mTORC1) as potential targets for therapeutic intervention in pancreatic cancer [26].
In this study we used HPAF II which is a well-differentiated human pancreatic adenocarcinoma cell line. This differentiated epithelial cell line is well characterized and proved to be a useful model to investigate the molecular mechanism of pancreatic tissue disease state [27]. This cell line displays the ductal structure including secretory granules and mucin production [28]. Therefore, we utilized HPAF II cell lines to investigate the impact of mTORC1 and mTORC2 inhibition and AMPK activation on the profile of glycolysis and TCA cycle and as a tool to further investigate biomarkers of pancreatic cancer response to treatment. The metabolomic outputs, as small-molecule metabolites, are sensitive to the pathophysiological changes, and thus can be used to detect novel biomarkers in pancreatic cancer, as well as to monitor therapeutic efficacy and response. Modeling such interactions can both validate assumptions and help uncover additional pathways or regulatory steps not currently understood, to delineate more fully insulin insensitivity.
Materials and Methods
Reagents
Reagents were obtained from the following sources: Metformin hydrochloride (N,N-Dimethyllimidodicarbonimidic diamide hydrochloride) (cat # 2864) and Torin 2 (9-(6-Aminopyridin-3-yl)-1-(3-trifluromethyl)-phenyl)benozo[h][1,6]naphthyridin-2(1H) (Cat # 4248) were obtained from from Tocris Bioscience (R & D Systems, USA). Rapamycin was obtained from Cell Signaling (Cat # 9904). Other chemicals were obtained either from Sigma or Fisher. Immobilon-P polyvinylidene difluoride membrane (0.45 μm) was from Millipore, reagents for enhanced chemiluminescence (ECL) were from Millipore (Immobilon Western chemiluminescent horseradish peroxidase). HPAF II cells (Cat # CRL 1997), EMEM media (Cat 30-2003); Fetal Bovine Serum (Cat # 30-2020) were purchased from ATCC.
Antibodies
The following antibodies were purchased from Cell Signaling: Total mTOR (Cat # 2983); Serine P-2481 mTOR (cat # 2976); S6 (Cat # 2217); Serine 235/236 Phospho-S6 ribosomal protein (Cat # 2211); Akt (Cat # 4691); Serine P-473 Akt (Cat # 4060). Actin antibody was obtained from Sigma (Cat # A2103). Sheep anti-rabbit secondary antibodies were obtained from (GE Health Care Bioscience, Corp, Piscataway, NJ).
Cell culture
HPAF II cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) cultured in Eagle's Minimum Essential Media containing D-glucose (1 g/L), L-glutamine (292 mg/L) and sodium pyruvate (110 mg/L) supplemented with 10% fetal bovine serum (FBS) (ATCC). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cells were plated at a density 1 × 106 cells Fresh media was added to cells prior to incubation with either Torin 2 (100 nM) or metformin (1 mM) or both at 1:1 ratio, control cells were incubated with DMSO. When indicates some cells were incubated with rapamycin (100 nM).
Methods
Cell growth and quenching
Well differentiated pancreatic cancer cell line HPAF-II (ATCC CRL # 1997) was cultured in EMEM media supplemented with L-glutamine 0.292 g/L (ATCC # 30-2003, Manassas, VA), 10% fetal bovine serum (ATCC Cat # 30-2020), 25 U/ml penicillin, and 25 ug/ml streptomycin as described [27]. Briefly, Cells were grown to confluence and plated as 1 × 106 cells per 100 mm polystyrene dishes at 37°C 5% CO2 and all plates contained equal number of cells. Prior to the experiment, the cells were replenished with fresh EMEM media containing 5 mM D-glucose. Cells were incubated the presence of either Torin 2 (ATP-competitive mTOR inhibitor (TORCIS Bioscience, Cat # 4248), Metformin (Fisher Sci, Cat # 2864), both, or vehicle control (DMSO) for 1 h and 24 h in five replicate samples per group. HPAFII cells were treated with either metformin at a final concentration 1 mM or Torin 2 final concentration of 100 nM for 1 h and 24 h as described [29,30]. Some HPAFII cells were treated with rapamycin at 100 nM concentration. After the indicated incubation time, media was aspirated and cells were rinsed briefly with 6 ml deionized water. Cells were quenched by flash freezing in liquid nitrogen while placed on dry ice. Cells were immediately stored at −80°C and then shipped on dry ice until received for metabolomics analysis at the University of Michigan Metabolomics Core [31]. We utilized targeted LC/MS/MS to characterize the alterations in the glycolytic and tricarboxylic acid (TCA) cycle metabolomics, and employed Western Blot analysis for cell signaling activation by phosphorylation.
Cell lysis and immunoblotting
Cells were washed twice with ice-cold PBS (pH 7.4), and collected in ice-cold lysis Buffer A containing KPO4, 1 mM EDTA, 5 mM EGTA, 10 mM MgCl2, 50 mM β-glycerophosphate, 1 mM Sodium Orthovanadate (Na3VO4), 5 ug/ml pepstatin A, 10 ug/ml leupeptin, 40 ug/ml phenylmethylsulfonyl fluoride (PMSF). Cells were lysed in the presence of Nonidet P-40 as detergent. Cell lysates were centrifuged at 13,200 rpm for 5 min at 4°C and the supernatants were collected. Protein concentration was measured by Bradford assay to normalize the concentration for immunoblotting. Western blot was performed by blocking the polyvinylidene difluoride membranes in TBST (40 mM Tris HCL, pH 7.5), 0.9% NaCl, 0.1% Tween 20) containing 5% nonfat milk. Equal loading of samples was confirmed by Ponceau staining. The membranes were incubated in TBST with 2% bovine serum albumin containing either the primary or the secondary antibodies. The blots were developed by Enhanced Chemiluminescence (ECL).
Targeted metabolomics analysis by LC/MS/MS
Frozen cells were extracted using organic solvents-150 ml of chilled methanol: chloroform: water at a ratio of 8:1:1 13C-labeled glycolysis and TCA cycle standards respectively. Following extraction, samples were sonicated on ice (20% duty cycle and 20% maximum power for 20 seconds), cells were kept at 4°C for 5 min and centrifuged at 15,000 xg for 5 min. The supernatant was transferred to the autosampler vials and directly analyzed by LC/MC based on the method by Lorenz et al. [31].
The peak chromatographic separation was conducted for 18 targeted glycolytic and TCA intermediate metabolites as described in Lorenz and colleagues [31]. The ratio of each metabolite peak to the 13C-labeled closest match internal standard was calculated. The targeted metabolite concentrations were determined using a standard calibration curve generated from the known standard. The concentrations were normalized to the total cells in the 10 cm dish (1 × 106 cells per plate).
Data processing and statistical analysis
Quantification was done at the University of Michigan Metabolomics Core as previously described [31,32]. Briefly, quantification was performed using Agilent Technologies Mass Hunter Quantitative software. Metabolites are identified based on the retention time and m/z match to injections of known standards. Data was expressed as mean ± Standard Deviation. Statistical analysis was performed using SPSS software. Comparisons between the treatment groups were performed using one-way ANOVA for detection of the overall statistical significance. Significance was present as (p<0.05). If an overall statistical significance was detected between groups, a secondary posthoc test for multiple comparisons was performed using the Least Significant Difference (LSD).
Results
Torin 2 inhibits mTORC1 and mTORC2-associated phosphorylation
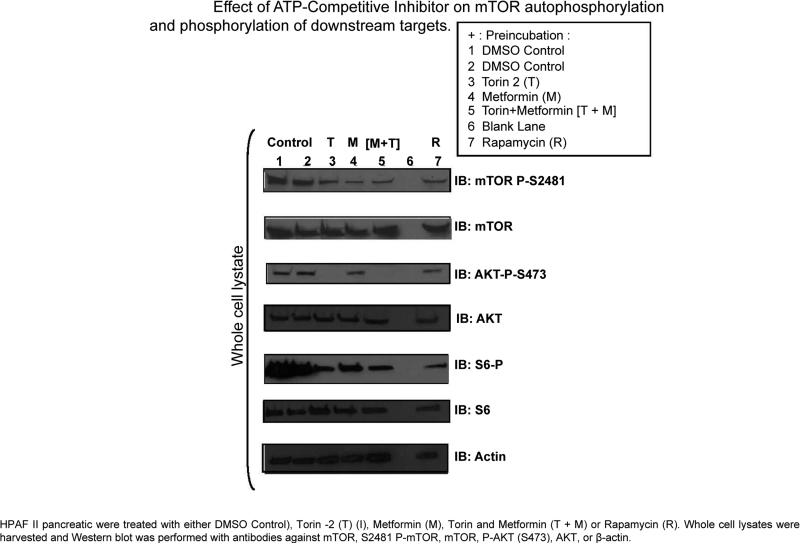
The mTORC1 pathway plays a critical role in cellular metabolism including protein synthesis and glycolysis, and its dysregulation plays a significant role in the progression of several types of cancer [33-36]. Additionally, mTORC2 has been recently shown to play an integral role in cancer metabolic reprogramming [37]. Therefore, in this study, we investigated a dual mTORC1 and mTORC2 inhibitor to determine the effect on pancreatic cancer cell line metabolite output. To confirm that Torin 2 inhibits both mTORC1 and mTORC2, we prepared whole cell lysate from HPAF II cells and incubated them for 24 h in the absence or presence of rapamycin, Torin 2, metformin or both. Our earlier work showed that mTOR autophosphorylation site at Ser 2481 serves as a readout of mTORC1 activity. Torin 2 inhibited the mTOR Ser 2481 autophosphorylation and phosphorylation of S6 as a surrogate of mTORC1 activity when immunoblotted in whole cell lysate. Under the conditions of our experiments, we found that Torin 2 also inhibited the phosphorylation of mTORC1 downstream target phospho-S6 ribosomal protein at serine 235/236. Metformin and rapamycin also decreased the phosphorylation of S6 [pS6] (mTORC1). However, Torin 2 selectively inhibited Akt phosphorylation at S473 indicating that Torin 2 inhibits_mTORC2 complex. However, neither rapamycin pretreatment nor metformin alone had an effect on Akt phosphorylation. Addition of Torin 2 to metformin led to the inhibition of Akt phosphorylation at Ser 473 as a readout of mTORC2 activity (Figure 1).
Figure 1.
Impact on mTOR inhibition and AMPK activation on cell signaling.
Torin 2 decreased malate and metformin increased AMP levels after 1 h
In most cancer cells, tumor proliferation and invasion depends on the cell glycolytic capacity to provide the needed energy [38]. Therefore, we conducted targeted metabolomics analysis for the intermediate metabolites of glycolysis and Tricarboxylic Acid (TCA) cycle in HPAFII pancreatic cancer cell lines following incubation with Torin 2 or metformin or both for 1 h (Table 1) and 24 h (Table 2). The serine/threonine kinase AMPK (AMP-activated protein kinase) is allosterically activated by AMP, and gauges the cellular fuel to regulate glucose metabolism [39]. AMPK has been shown to repress mTOR anabolic pathway and ATP-consuming metabolic pathway [40]. As an AMPK activator, metformin increased AMP levels following 1 h incubation compared to DMSO control, Torin-2, and combined metformin and Torin 2 (2.742 vs. 0.909, 0.158, and 0.837, p<0.03) (Table 1). Torin 2 significantly decreased malate, the TCA cycle intermediate formed by hydration of furmate, compared to metformin-treated group (6.906 vs. 11.453, p<0.5) (Table 1). However, the combination of metformin and Torin-2 abolished this effect compared to the DMSO control (8.445 vs 6.906, NS) (Table 1). No other metabolites were different after 1 h incubation.
Table 1.
Comparison of HP LC-MS metabolites peak areas between groups after 1 h treatment in HPAF-II well differentiated pancreatic cancer cell lines.
| Metabolite Concentration in nmol | |||||
|---|---|---|---|---|---|
| Metabolite | DMSO (Control) | Torin 2 | Metformin | Met + Torin 2 | P Value |
| NAD | 2.560 ± 1.202 | 1.810 ± 1.301 | 3.345 ± 1.149 | 3.230 ± 1.931 | 0.33 |
| SUC | 1.035 ± 0.443 | 2.579 ± 3.692 | 9.185 ± 17.319 | 1.967 ± 1.048 | 0.51 |
| FAD | 0.136 ± 0.052 | 0.119 ± 0.073 | 0.171 ± 0.042 | 0.180 ± 0.105 | 0.47 |
| MAL | 6.906 ± 2.340 | 4.779 ± 1.883a | 11.453 ± 6.257b | 8.445 ± 1.895 | 0.01 |
| F6P/G6P | 1.231 ± 0.264 | 0.923 ± 0.455 | 2.045 ± 1.402 | 1.035 ± 0.471 | 0.14 |
| AMP | 0.909 ± 0.735c | 0.158 ±0.226c | 2.742 ± 2.182d | 0.837 ± 1.023c | 0.03 |
| Citrate/iso | 4.276 ± 0.525 | 3.427 ± 1.882 | 4.793 ± 2.480 | 4.133 ± 2.283 | 0.74 |
| NADP | 0.072 ± 0.021 | 0.054 ± 0.041 | 0.075 ± 0.017 | 0.100 ± 0.031 | 0.12 |
| 6PG | 1.778 ± 0.397 | 1.065 ± 0.659 | 1.832 ± 0.919 | 1.072 ± 0.433 | 0.45 |
| 2PG/3PG | 1.148 ± 0.325 | 0.860 ± 0.433 | 3.088 ± 4.779 | 1.113 ± 0.485 | 0.12 |
| ADP | 2.621 ± 1.365 | 1.552 ± 1.508 | 9.403 ± 13.426 | 2.909 ± 1.870 | 0.29 |
| aCoA | 0.044 ± 0.047 | 0.047 ± 0.031 | 0.042 ± 0.081 | 0.028 ± 0.038 | 0.94 |
| PEP | 0.022 ± 0.019 | 0.005 ± 0.007 | 0.022 ± 0.036 | 0.016 ± 0.016 | 0.67 |
| FBP | 5.796 ± 1.996 | 4.218 ± 1.609 | 3.981 ± 2.635 | 4.507 ± 2.008 | 0.54 |
| ATP | 32.202 ± 9.829 | 31.128 ± 15.697 | 23.813 ± 14.983 | 33.489 ± 16.666 | 0.72 |
HPAF-II Well-Differentiated Pancreatic Cancer Cell lines were treated for 1 h or 24 h with either DMSO control, Torin-2 alone, Metformin, or a combination of Torin-2 and Metformin. Data is presented as mean ± Standard Error (SEM). One way Analysis of Variance (ANOVA) was conducted to compare overall differences between groups (p<0.05). Secondary Post-hoc Least Significant Difference (LSD) test was performed following ANOVA analysis to determine the significant differences between DMSO control at treatment groups after 24 h of incubation with either Torin-2, Metformin or both Torin-2 and Metformin (p<0.05). Groups with different superscripts are statistically significantly different from the other groups. NAD, Nicotinamide adenine dinucleotide; Suc, succinate, FAD Riboflavin adenine dinucleotide; G6P/F6P, glucose-6-phosphate/fructose-6-phosphate; AMP, adenosine monophosphate; Citrate/iso, citrate/isocitrate, FBP, fructose 1,6-bisphosphate, 3PGS, 2PG 3-phosphoglycerate/2-phosphoglycerate; PEP: Phosphoenolpyruvate.
Table 2.
Comparison of HP LC-MS metabolites peak areas between groups after 24 h treatment.
| Metabolite Concentration in nmol | |||||
|---|---|---|---|---|---|
| Metabolite | DMSO (Control) | Torin 2 | Metformin | Met + Torin 2 | P Value |
| NAD | 4.600 ± 0.782ab | 2.494 ± 1.663a | 4.031 ± 1.382 | 2.617 ± 1.133b | 0.05 |
| SUC | 2.170 ± 0.814 | 1.621 ± 0.911 | 2.049 ± 1.331 | 1.204± 0.745 | 0.41 |
| FAD | 0.200 ± 0.022ab | 0.107 ± 0.054ac | 0.188 ± 0.049cd | 0.117± 0.036bc | 0.004 |
| MAL | 9.147 ± 1.363a | 5.731 ± 2.125ab | 10.383 ± 3.384b | 7.577 ± 2.491 | 0.04 |
| F6P/G6P | 0.834 ± 0.095 | 0.566 ± 0.317 | 0.913 ± 0.210 | 0.652 ± 0.403 | 0.22 |
| AMP | 0.254 ± 0.212 | 0.428 ± 0.540 | 0.793 ± 0.915 | 0.460 ± 0.404 | 0.532 |
| Citrate/iso | 5.225 ± 0.448abc | 2.719 ± 0.830a | 3.110 ± 0.646bc | 2.210 ± 0.488cd | 0.0005 |
| NADP | 0.092 ± 0.028 | 0.078 ± 0.045 | 0.079 ± 0.027 | 0.092 ± 0.011 | 0.80 |
| 6PG | 0.912 ± 0.235 | 0.451 ± 0.305 | 0.623 ± 0.212 | 0.501 ± 0.333 | 0.07 |
| 2PG/3PG | 1.267 ± 0.177ab | 0.585 ± 0.198ac | 1.082 ± 0.330c | 0.888 ± 0.339b | 0.007 |
| ADP | 2.159 ± 1.034 | 1.738 ± 1.028 | 2.288 ± 1.377 | 2.563 ± 1.215 | 0.73 |
| aCoA | 0.079 ± 0.052 | 0.020 ± 0.010a | 0.109 ± 0.087ab | 0.031 ± 0.019b | 0.051 |
| PEP | 0.020 ± 0.017 | 0.012 ± 0.016 | 0.008 ± 0.013 | 0.013 ± 0.010 | 0.618 |
| FBP | 5.584 ± 1.659abc | 2.846 ± 0.857a | 3.870 ± 0.963b | 3.726 ± 1.285c | 0.02 |
| ATP | 37.521 ± 3.532ab | 18.832 ± 4.407ac | 29.509 ± 9.981c | 23.639 ± 5.832b | 0.001 |
HPAF-II Well-Differentiated Pancreatic Cancer Cell lines were treated for 1 h or 24 h with either DMSO control, Torin-2 alone, Metformin, or a combination of Torin-2 and Metformin. One way Analysis of Variance (ANOVA) was conducted to compare overall differences between groups. Data is presented as mean ± Standard Error (SEM). Secondary Post-hoc LSD test was performed following ANOVA analysis to determine the significant differences between DMSO control at treatment groups after 24 h of incubation with either Torin-2, Metformin or both Torin-2 and Metformin. Groups with different superscripts in the same line are statistically significantly different from the other groups for the same metabolite.
Torin 2 decreased the glycolytic intermediates after 24 h
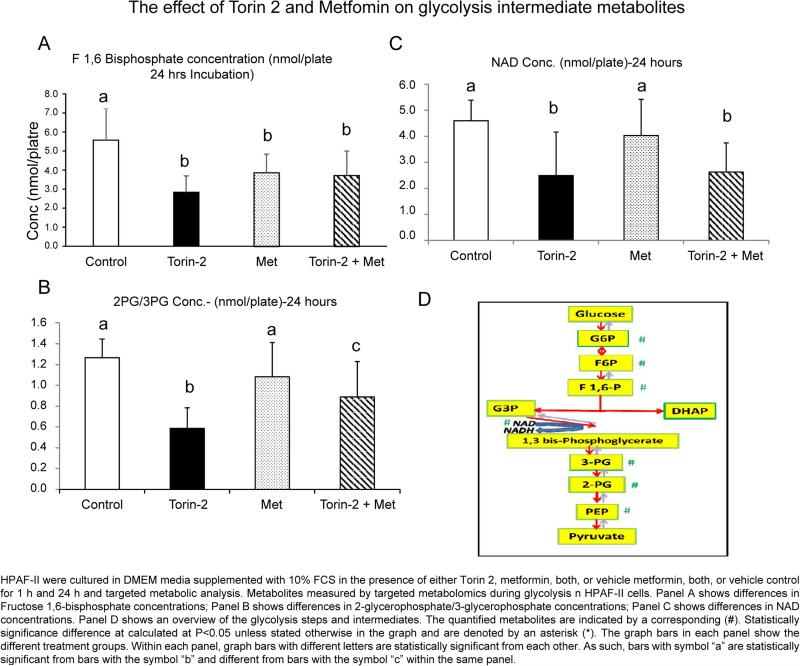
The results indicate that Torin-2 significantly decreased the glycolysis intermediates. The irreversible phosphorylation of fructose 6 phosphate to Fructose 1,6-bisphosphate (FBP) catalyzed by phosphofructokinase is the first committed and rate-limiting step of glycolysis and thereby is an important control point [41]. Since the rate of aerobic glycolysis is significantly increased in pancreatic cancer cells through a phenomenon commonly known as Warburg effect, we investigated whether mTOR inhibition would decrease the products of the committed step of glycolysis [42,43]. Compared to DMSO control, Fructose 1,6-bisphosphate (FBP) level decreased in response to incubation with Torin 2, metformin or the combination of Torin 2 and metformin (5.584 vs. 2.846, 3.870, 3.726, p<0.03) (Table 2 and Figure 2A). FBP is subsequently converted to two trioses: dihydroxyacetone phosphate and glyceraldehyde 3 phosphate. Subsequent two-step oxidation and substrate level phosphorylation of yields 3 phosphoglycerate and yields ATP. So, we further determined the ratio of 3 phosphoglycerate and 2 phosphoglycerate. Torin-2 also decreased the ratio of 2-phosphoglycerate/3-phosphoglycerate (2PG/3PG) compared to DMSO, metformin and combined Torin 2 and metformin (0.585 versus 1.267, 1.082, 0.888 respectively, p<0.007) (Table 2). Also, there was significant difference between metformin, combined Torin 2 and metformin, and Torin 2 alone (1.082 vs. 0.888 vs. 0.585, p<0.007) (Table 2 and Figure 2B). Torin 2 also decreased the ratio of 2PG/3PG compared to the control group and metformin (0.585 vs. 1.267, and 1.082, p<0.007) (Table 2 and Figure 2C); Torin 2 treatment was also significantly different from Torin 2 and metformin combined (1.267 vs. 0.888, p<0.007) (Table 2 and Figure 2C). The glycolytic metabolites measured by MS/LS/LS are summarized in Figure 2D. Taken together; the results indicate that the combination of Torin 2 and metformin has an intermediate effect on glycolysis metabolites relative to either metformin or Torin 2 alone. Overall our results demonstrate that mTOR complexes enhance the glycolytic properties of HPAFII cells and this effect is blocked by the competitive ATP inhibition of mTOR.
Figure 2.
Glycolysis targeted-metabolomics in HPAF-II cell lines.
Torin 2 decreased the TCA cycle intermediates and metformin increased acetyl-CoA after 24 h
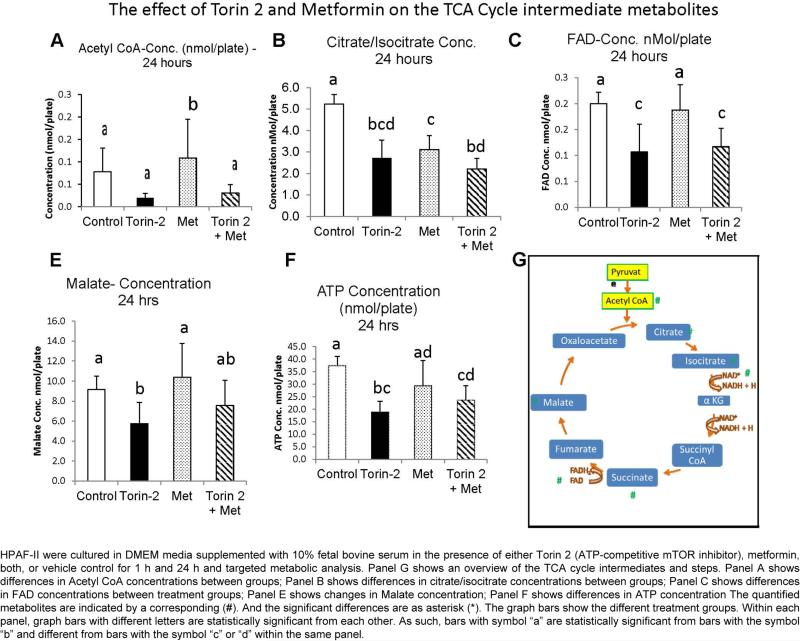
As we observed that the glycolytic intermediates were decreased with Torin 2 treatment, we sought to analyze the impact on the downstream TCA cycle. The end product of glycolysis is pyruvate, which is transported to the mitochondria to be converted to Acetyl CoA as it enters the TCA cycle. After 24 h, only metformin increased Acetyl CoA levels compared to the DMSO, Torin 2, or combined Torin 2 and metformin (0.109 vs. 0.079, 0.020, p<0.03) (Figure 3A). This effect was abolished by the combination of Torin 2 and metformin suggesting that the combination has a different effect than the single agent (0.109 vs. 0.031, p<0.03) (Figure 3A). Condensation of three of the two carbon acetyl-CoA yields the six carbon citrate, and subsequent irreversible oxidative decarboxylation of citrate generates three NADH energy equivalents. Torin 2 and metformin both individually and combined decreased citrate/isocitrate compared to the DMSO control (2.719, 3.110, 2.210 vs. 5.225, respectively, p<0.0005, Figure 3B). Torin 2 incubation decreased malate levels compared to the DMSO control and metformin (5.731 vs. 9.147, 10.383 respectively, p=0.045) (Table 2). When combined with metformin this effect was abolished (5.731 vs. 7.577).
Figure 3.
TCA cycle targeted-metabolomics profile in HPAF-II cell lines.
Torin 2 decreases the levels of NAD+ and FAD electron acceptors
As we observed decreased glycolytic and TCA cycle intermediates, we next investigated the levels of on the electron acceptors NAD+ and FAD as the precursors of the energy equivalents NADH and FADH2. The glycolytic and TCA cycle intermediates donate electrons to the electron carriers NAD+, FAD to form energy rich reduced coenzymes NADH, FADH2. We found that NAD+ levels were decreased when the HPAF II cells were incubated with Torin 2 or combined Torin 2 and metformin compared to the DMSO control and metformin alone (2.494 and 2.617 vs. 4.600 and 4.031) (Figure 2C). Similarly, FAD levels were decreased when the cells were incubated with Torin 2 or combined Torin 2 and metformin relative to the DMSO control or metformin (0.107 and 0.117 vs. 0.200 and 0.188) (Table 2, Figures 3C and 3D). Malate concentration was also significantly decreased by Torin 2 treatment compared to DMSO control, metformin, and Torin combined with metformin (5.73 vs. 9.14, 10.38, 7.57) (Table 2 and Figure 3E). Additionally, ATP concentration was significantly decreased in Torin 2 treated group compared to the DMSO control, and metformin (18.83 vs. 37.52, 29.50 and 23.63 respectively) (Table 2 and Figure 3F). The TCA cycle metabolite intermediates measured by MS/LS/LS are summarized in Figure 3G. Overall our results indicate the mTORC1 and mTORC2 inhibition led to decreased electron acceptors and energy equivalents and thus leads to decreased glycolytic flux and energy production which may have therapeutic advantage in cancer therapy.
Torin 2 and metformin alter the metabolic bioenergetics AMP, ADP, and ATP
Finally, we determined the changes in bioenergetics at baseline, 1 h and 24 h treatment. We observed that AMP levels increased with metformin treatment after 1 h, but returned to basal level after 24 h (Table 3). There were no significant differences in ADP levels between basal, 1 h and 24 h treatment. There were no differences in ATP levels in all groups after 1 h incubation. However, after 24 h, Torin 2 significantly decreased ATP levels compared to metformin and DMSO control (18.83 vs. 37.52, 29.50, p<0.01). There was no significant statistical difference between metformin and combined metformin and Torin 2 (29.509 vs. 23.639) (Table 3 and Figure 3F). As shown in Table 3, it is worth noting that Torin-2 reduced the concentration of ATP after 24 h of treatment compared to the control and metformin alone (p<0.001) suggesting that as an ATP competitor, Torin 2 led to reduction of ATP and the development of cell stress. Incubation of HPAF II with metformin alone for 24 h did not lead to significant differences in ATP concentration compared to incubation with the DMSO control (28.813 vs. 32.202, LSD p=0.066 respectively). However, incubation with metformin alone led to ATP concentration that was significantly different than incubation with Torin-2 alone (29.509 vs. 18.832, LSD p=0.018). This observation suggests that the metformin may be beneficial to reduce cell stress induced by Torin-2 treatment.
Table 3.
AMP, ADP, and ATP energy units peak areas time course.
| AMP | Baseline Conc. (nmol) ± SEM [CI] | 1 h Conc. (nmol) ± SEM [CI] | 24 h Conc. (nmol) ± SEM [CI] |
|---|---|---|---|
| DMSO (C) | 0.415 ± 0.202 [−0.145, 0.976] | 0.909 ± 0.336b [−0.025, 1.834] | 0.254 ± 0.094 [−0.010, 0.517] |
| Torin-2 | 0.777 ± 0.237 [0.117, 1.437] | 0.158 ± 0.101b [−0.122, 0.439] | 0.428 ± 0.241 [−0.242, 1.098] |
| Metformin | 0.836 ± 0.445 [−0.400, 2.072] | 2.742 ± 0.975a [−0.033, 5.451] | 0.793 ± 0.409 [−0.342, 1.929] |
| Met + Torin2 | 0.467 ± 0.290 [−0.329, 1.281] | 0.837 ± 0.457b [−0.432, 2.107] | 0.460 ± 0.180 [−0.042, 0.961] |
| P Value | 0.7 | 0.03 | 0.53 |
| ADP | |||
| DMSO (C) | 2.419 ± 0.634 [0.658, 4.179] | 2.621 ± 0.610 [0.925, 4.316] | 2.159 ± 0.463 [0.875, 3.442] |
| Torin-2 | 2.233 ± 0.524 [0.777, 3.688] | 1.552 ± 0.674 [−0.320, 3.424] | 1.738 ± 0.459 [0.461, 3.014] |
| Metformin | 1.484 ± 0.360 [0.483, 2.483] | 9.403 ± 6.004 [−7.267, 26.074] | 2.288 ± 0.615 [0.578, 3.998] |
| Met + Torin2 | 1.977 ± 0.293 [1.163, 2.790] | 2.909 ± 0.836 [0.587, 5.231] | 2.563 ± 0.543 [1.054, 4.072] |
| P Value | 0.43 | 0.29 | 0.73 |
| ATP | |||
| DMSO (C) | 33.693 ± 5.215 [19.212, 48.174] | 32.202 ± 4.395 [19.997, 44.405] | 37.521 ± 1.579a [33.136, 41.906] |
| Torin-2 | 18.924 ± 3.168 [10.127, 27.720] | 31.128 ± 7.019 [11.637, 50.618] | 18.832 ± 1.971bc [13.359, 24.348] |
| Metformin | 19.233 ± 5.874 [2.921, 35.543] | 23.813 ± 6.700 [5.209, 42.417] | 29.509 ± 4.463ad [17.115, 41.902] |
HPAF-II Well-Differentiated Pancreatic Cancer Cell lines were treated for 1 h or 24 h with either DMSO control, Torin-2 alone, Metformin, or a combination of Torin-2 and Metformin. One way Analysis of Variance (ANOVA) was conducted to compare overall differences between groups. Data is presented as the mean ± SEM and the 95% Confidence Interval for Mean [C.I Lower and Upper Bounds]. Groups with different superscript letters are statistically significantly different from the other groups (a, b, c, d). Groups with similar superscripts are not statistically significant from each other. There was no statistically significant differences between baseline data for all treatment groups for AMP (p=0.70), ADP (p=0.43), and ATP (p=0.12) as determined by ANOVA (SPSS, Statistical Software). Secondary Post-hoc LSD test was performed following ANOVA analysis to determine the significant differences between DMSO control at treatment groups after 24 h of incubation with either Torin-2, Metformin or both Torin-2 and Metformin. Groups with different superscripts are statistically significantly different from the other groups.
Both glucose and glutamine are required for anabolism and cell proliferation [4,64]. Therefore, we sought to confirm whether glutamine was depleted overtime and thus may have contributed to altered metabolic parameters. Glutamine concentration did not differ between treatment groups at 1 h (p=0.39) and 24 h (p=0.81) suggesting that the amount of glutamine in the media did not contribute to the observed changes in metabolite concentration.
Discussion
mTOR is a targetable and actionable gene and protein in the treatment of PDAC. mTOR is a nutrient-sensing pathway that plays a fundamental role in anabolic cell growth and proliferation, as well as in tumor metabolism [44]. Metabolic reprogramming is a hallmark of cancer which allows for the provision of nutrients to support the rapidly proliferating tumor biomass, fulfill bioenergetics requirements, and also enables chemoresistance. Cancer cells increase their uptake of glucose and increase aerobic glycolysis and lactate production to promote tumor energy production to support the rapid tumor growth and proliferation [45]. Hence, mTOR complexes pathways that alter glucose metabolism are important therapeutic target. Our approach is to target a highly-conversed protein kinase, (mTOR), which is required for cell growth and metabolism and is frequently dysregulated in cancer; and characterize the synergistic impact of its dual inhibition on the metabolomic readouts. Intriguingly, the biguanide drug, metformin, which activates AMP-activated Protein Kinase (AMPK) pathway, indirectly inhibits mTOR signaling, and has been recently investigated as an anti-tumor agent [26]. As anticipated as an AMPK activator, we found metformin increased AMP levels after 1 h treatment (Table 1). Rapamycin, the prototype of mTOR inhibition, was an attractive chemotherapeutic target, however recent results in clinical trials are less promising than originally anticipated. This finding is due-in-part to the rapamycin-resistant components of mTORC1 as well as rapamycin-insensitive properties of mTORC2, at least with acute treatment. Furthermore, 4EBP, a downstream effector of mTORC1, is not inhibited by rapamycin suggesting the presence of rapamycin-resistant mTORC1 functions [46,47]. Recently, a new class of mTOR inhibitors termed ATP-competitive mTOR kinase inhibitors, have been developed and are shown to inhibit both mTORC1 and mTORC2 complexes [30,48]. The mTORC1/mTORC2 inhibitor used in this study is Torin-2 [30].
Although both mTORC1 and mTORC2 are nucleated by mTOR kinase, they have similar and distinctive binding partners and therefore, these two complexes have different activities and different drug sensitivity [37]. Recent studies revealed a new role of mTORC2 in promoting carcinogenesis and increasing drug resistance to chemotherapy [49]. Furthermore, Mausi and colleagues documented that acetylation of Rictor by acetyl-CoA promotes growth factor signaling, leading to targeted chemotherapy resistance [50]. On the contrary, mTORC2 partner, DEPTOR, a negative regulator of mTORC2, has been shown to have cell growth suppressive activity in pancreatic cancer cells. Recent reports indicate the 50% of PDAC patients are diabetic at the time of diagnosis [51-53]. To that end, tumor-related diabetes is now considered diabetesT3c (T3cDM) [54]. Mounting evidence links the glycolytic metabolic disturbances to the adverse pancreatic cancer prognosis [55]. Further, Morin et al. suggested that the onco-metabolites that accumulate during tumor progression are involved in the hypoxic response and epigenetic reprogramming [56]. Additionally, Ben-Sahra et al. [55] reported that mTORC1 regulates the metabolic flux by controlling denovo synthesis of pyrimidine which can lead to increased DNA synthesis required for tumor growth. Thus it appears the both mTORC1 and mTORC2 may play a cooperative and distinctive role in carcinogenesis. Our study shows that TORIN 2 decreases the glycolytic and TCA intermediates. After 1 h of incubation, as an AMPK activator metformin increased AMP and malate levels compared to all other groups, but no other changes were detected (Table 1).
Cancer increases the metabolic reprogramming and rewiring to promote survival, tumor growth and proliferation. mTORC1 activation in cancer is associated with increased aerobic glycolysis, commonly known as the Warburg effect, with preference of conversion of pyruvate to lactate which allows survival under hypoxia. This aerobic glycolysis, even in the presence of oxygen facilitates tumor growth advantage by generating an acidic microenvironment conducive to tumor growth and proliferation. However, in our study we did not observe differences in lactate production. The “Warburg effect” is also coupled with 100 fold increase in glucose uptake [57]. Torin 2 is a potent orally available ATP-competitive inhibitor of mTOR complexes 1 & 2. The half maximal Effective Concentration (EC50) of Torin 2 for mTOR inhibition is 0.25 nM and possess 800 times more selectivity than PI3 kinase [58]. Therefore, we chose Torin 2 to determine the impact of selective mTOR inhibition on glycolysis and TCA cycle intermediates. We sought to determine whether Torin 2 may synergize with other anti-cancer agents. Metformin exhibits anti-cancer effects both as AMPK dependent and independent mechanisms. We reasoned that metformin indirectly inactivates mTOR via AMPK dependent activation of AMPK and there may synergize with Torin-2 and decrease the significant inhibition to ATP levels observed in our study.
We determined the targeted-metabolomic profile in human PDAC cell line (HPAF-II) and the possible synergism between Torin-2 and metformin administration as potential targets for therapeutic intervention in pancreatic cancer. Torin-2 significantly decreased the glycolysis intermediates (fructose 1,6-bisphosphate (FBP), and 2-phosphoglycerate/3-phosphoglycerate (2PG/3PG)) and TCA intermediate metabolites (acetyl-CoA, citrate/isocitrate, and malate). Torin-2 also reduced the electron acceptors (NAD+, and FAD). Metformin treatment alone reduced FBP and citrate/isocitrate ratio. However, when HPAF-II cells were incubated with both Torin-2 and metformin, there was significant reduction of NAD+ and FAD, suggesting that there is a decrease in the levels of energy equivalents available to the electron transport chain.
Metformin, the biguanide antihyperglycemic first-line treatment in diabetes mellitus type II, has recently emerged as an antineoplastic agent [59,60]. Metformin also exerts anticancer properties both via AMP-dependent and AMP-independent pathways [61]. Metformin increases the reactive oxygen species and reduces mitochondrial transmembrane potential leading to hampering the self-renewal capacity of Cancer Stem Cells (CSC) [62]. Epidemiological studies have shown that treatment with metformin is associated with decreased cancer risk and/or improved survival. As a member of the biguanide family, metformin causes pharmacological activation of AMPK and thus has a protective response to energy stress. As such, AMPK may serve as a tumor suppressor. Furthermore, metformin also decreases gluconeogenesis [55]. Metformin, which also indirectly inhibits mTOR via activation of AMPK, has emerged as a potential therapeutic target in the treatment of PC, particularly when associated with type 3c diabetes. Intriguingly, recent studies conducted in xenograft models revealed that metformin decreases pancreatic cancer growth in a dose-dependent manner [29,63]. The impact of metformin on inhibiting mTORC1 indirectly via AMPK activation was remarkably different than rapamycin or active-site mTOR inhibitors [29].
Combination therapy is more effective than single-agent drugs and improves cell response in cancer. Hitting mTOR pathway at multiple points may shut off the alternative pathways that promote energy metabolism in cancer. Given that metformin has anticancer properties via AMPK activation, is an indirect inhibitor of mTORC1 pathway, and that Torin 2 blocks both mTORC1 and mTORC2; it is reasonable to suggest that the combination of both drugs will have an augmented effect on decreasing glycolysis and TCA cycle and may provide therapeutic advantage in cancer management. Our results show that combination of metformin and Torin 2 may alter cancer metabolic reprogramming by decreasing glycolytic and TCA intermediates, while reducing Torin 2 side effect of significant ATP-competitive inhibition which may interfere with normal cellular functions.
Our findings indicate the mTOR pathway enhances glucose metabolism and this effect is abrogated by competitive ATP mTOR inhibitors. However, in addition to this favorable effect, competitive ATP mTOR inhibition can reduce normal metabolism cell function as a result of the marked decrease in ATP. Addition of metformin will decrease the concentration needed for Torin 2 to be effective, and by the direct anticancer effects of metformin will also decrease the side effects and dose needed. Future studies will determine the lowest effective concentration of Torin 2 and optimal synergism with metformin. The next step is to validate the cell culture data in pancreatic cancer animal models in vivo and in phase I clinical trials.
Conclusions
The targeted metabolomics data indicate that mTORC1 and mTORC2 inhibition by Torin 2 reduced glycolytic intermediates and TCA metabolites pools in HPAF- II and that Torin 2 may synergize with metformin to decrease the electron acceptors NAD+ and FAD by the TCA cycle which may lead to reduced energy production.
Understanding the molecular and metabolic mechanisms behind the low survival rate and resistance to therapy in PC will be instrumental in developing metabolic biomarkers for early detection and acquiring targets for effective therapy. Future work will determine whether interactions between mTOR inhibition by ATP competitors and AMPK activation by metformin will alter the PPP and hence nucleotides and DNA synthesis in pancreatic cancer cells. The potential applications of this work may lead to the use of glycolytic and TCA intermediates metabolites levels altered by mTOR pathway to gauge the response to Torin 2 and metformin. The implications of this work may provide mechanistic underpinning to show how mTOR signaling influences the metabolomics profile in cancer. The metabolomics profile can be developed for early detection, and targeted therapy, utilizing metabolic laboratory tests that will improve the clinical outcomes.
Acknowledgements
The work on this research utilized Metabolomics Core Services supported by grant U24 DK097153 of NIH to the University of Michigan. The authors would like to thank Daphne Baber for the excellent technical assistance.
Funding
This research is funded in-part by a grant from the College of Public Health Cancer Prevention and Control Pilot Grant. The investigator is funded by Faculty Diversity Funding from the University of Nebraska Medical Center. The work on this research utilized Michigan Regional Comprehensive Metabolomics Core Services supported by grant U24 DK097153 of NIH to the University of Michigan.
Abbreviations
- FAD
Flavin Adenine Dinucleotide
- G6P/F6P
Glucose-6-Phosphate/Fructose-6-Phosphate
- NAD+
Nicotinamide Adenine Dinucleotide
- mTOR
Mammalian Target of Rapamycin
- mTORC1
Mammalian Target of Rapamycin Complex 1
- mTORC2
Mammalian Target of Rapamycin Complex 2
- PDAC
Pancreatic Ductal Adenocarcinoma
- Suc
Succinate
- AMP
Adenosine Monophosphate
- Citrate/Iso
Citrate/Isocitrate
- FBP
Fructose 1,6-Bisphosphate
- 3PGS/2PG
3-phosphoglycerate/2-phosphoglycerate
- PEP
Phosphoenolpyruvate
Footnotes
Citation: Soliman GA, Steenson SM, Etekpo AH (2016) Effects of Metformin and a Mammalian Target of Rapamycin (mTOR) ATP-Competitive Inhibitor on Targeted Metabolomics in Pancreatic Cancer Cell Line. Metabolomics (Los Angel) 6: 183. doi: 10.4172/2153-0769.1000183
Author Contribution
GS designed the study, established the HPAF cell culture work, collected the samples for the metabolomics, obtained the results, performed the analysis, interpretations of the results and wrote than manuscript, SS and AE performed the cell signaling study and Western Blot analysis, participated in the writing and editing the final draft of the manuscript.
Disclosure Statement
The authors have nothing to disclose. There is no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute (NCI) NCI: A Snapshot of Pancreatic Cancer. US Department of Health and Human Services; 2014. [Google Scholar]
- 3.Hall MN. The TOR signalling pathway and growth control in yeast. Biochemical Society Transactions. 1996;24:234–239. doi: 10.1042/bst0240234. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Current Opinion in Cell Biology. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Tee AR, Proud CG. DNA-damaging agents cause inactivation of translational regulators linked to mTOR signalling. Oncogene. 2000;19:3021–3031. doi: 10.1038/sj.onc.1203622. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Rhodes CJ, Lawrence JC., Jr Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J Biol Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- 8.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Reviews Molecular Cell Biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends in Endocrinology and Metabolism: TEM. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochemical Society Transactions. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell IW. Metformin and glibenclamide: comparative risks. BMJ. 1984;289:289. doi: 10.1136/bmj.289.6440.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blonde L. Management of type 2 diabetes: update on new pharmacological options. Managed Care. 2000;9:11–17. [PubMed] [Google Scholar]
- 15.Melchior WR, Jaber LA. Metformin: an antihyperglycemic agent for treatment of type II diabetes. Ann Pharmacother. 1996;30:158–164. doi: 10.1177/106002809603000210. [DOI] [PubMed] [Google Scholar]
- 16.Muniraj T, Chari ST. Diabetes and pancreatic cancer. Minerva Gastroenterologica e Dietologica. 2012;58:331–345. [PMC free article] [PubMed] [Google Scholar]
- 17.Sah RP, Nagpal SJ, Mukhopadhyay D, Chari ST. New insights into pancreatic cancer-induced paraneoplastic diabetes. Nat Rev Gastroenterol Hepatol. 2013;10:423–433. doi: 10.1038/nrgastro.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libby G, Donnelly LA, Donnan PT, Alessi DR, Morris AD, et al. New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care. 2009;32:1620–1625. doi: 10.2337/dc08-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang ZJ, Zheng ZJ, Kan H, Song Y, Cui W, et al. Reduced risk of colorectal cancer with metformin therapy in patients with type 2 diabetes: a meta-analysis. Diabetes Care. 2011;34:2323–2328. doi: 10.2337/dc11-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Fang W, Xia T, Chen Y, Gao Y, et al. Metformin potentiates rapamycin and cisplatin in gastric cancer in mice. Oncotarget. 2015;6:12748–12762. doi: 10.18632/oncotarget.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Wahab Z, Mert I, Tebbe C, Chhina J, Hijaz M, et al. Metformin prevents aggressive ovarian cancer growth driven by high-energy diet: similarity with calorie restriction. Oncotarget. 2015;6:10908–10923. doi: 10.18632/oncotarget.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obara A, Fujita Y, Abudukadier A, Fukushima T, Oguri Y, et al. DEPTOR-related mTOR suppression is involved in metformin's anti-cancer action in human liver cancer cells. Biochem Biophys Res Commun. 2015;460:1047–1052. doi: 10.1016/j.bbrc.2015.03.148. [DOI] [PubMed] [Google Scholar]
- 24.Oliveras-Ferraros C, Vazquez-Martin A, Cuyas E, Corominas-Faja B, Rodriguez-Gallego E, et al. Acquired resistance to metformin in breast cancer cells triggers transcriptome reprogramming toward a degradome-related metastatic stem-like profile. Cell Cycle. 2014;13:1132–1144. doi: 10.4161/cc.27982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Chhipa RR, Pooya S, Wortman M, Yachyshin S, et al. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc Natl Acad Sci U S A. 2014;111:435–444. doi: 10.1073/pnas.1311121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soliman GA, Etekpo A. Effects of metformin and ATP-competitive inhibitor of mTOR on targeted-metabolomic profile in HPAF-II pancreatic cancer cell lines. Cancer Res. 2015;75:53. [Google Scholar]
- 27.Rajasekaran SA, Gopal J, Espineda C, Ryazantsev S, Schneeberger EE, et al. HPAF-II, a cell culture model to study pancreatic epithelial cell structure and function. Pancreas. 2004;29:77–83. doi: 10.1097/00006676-200410000-00016. [DOI] [PubMed] [Google Scholar]
- 28.Rajasekaran SA, Barwe SP, Gopal J, Ryazantsev S, Schneeberger EE, et al. Na-K-ATPase regulates tight junction permeability through occludin phosphorylation in pancreatic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:124–133. doi: 10.1152/ajpgi.00297.2006. [DOI] [PubMed] [Google Scholar]
- 29.Soares HP, Ni Y, Kisfalvi K, Sinnett-Smith J, Rozengurt E. Different patterns of Akt and ERK feedback activation in response to rapamycin, active-site mTOR inhibitors and metformin in pancreatic cancer cells. PloS One. 2013;8:57289. doi: 10.1371/journal.pone.0057289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Xu C, Kirubakaran S, Zhang X, Hur W, et al. Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 2013;73:2574–2586. doi: 10.1158/0008-5472.CAN-12-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical Chemistry. 2011;83:3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz MA, El Azzouny MA, Kennedy RT, Burant CF. Metabolome response to glucose in the beta-cell line INS-1 832/13. J Biol Chem. 2013;288:10923–10935. doi: 10.1074/jbc.M112.414961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soliman GA. The integral role of mTOR in lipid metabolism. Cell Cycle. 2011;10:861–862. doi: 10.4161/cc.10.6.14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 35.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discovery. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. TEM. 2014;25:364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 39.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metabolism. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Molecular and Cellular Biology. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey LAF. D: Lippincott's illustrated Reviews. Biochemistry. (5th edn.) 2011 [Google Scholar]
- 42.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 43.Semenza GL, Artemov D, Bedi A, Bhujwalla Z, Chiles K, et al. ‘The metabolism of tumours’: 70 years later. Novartis Foundation Symposium. 2001;240:251–260. [PubMed] [Google Scholar]
- 44.Cairns RA, Harris I, McCracken S, Mak TW. Cancer cell metabolism. Cold Spring Harbor Symposia on Quantitative Biology. 2011;76:299–311. doi: 10.1101/sqb.2011.76.012856. [DOI] [PubMed] [Google Scholar]
- 45.Cairns RA. Drivers of the Warburg phenotype. Cancer J. 2015;21:56–61. doi: 10.1097/PPO.0000000000000106. [DOI] [PubMed] [Google Scholar]
- 46.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, et al. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Ren T, Fresques T, Oppliger W, Niles BJ, et al. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem Biol. 2012;7:982–987. doi: 10.1021/cb300058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masui K, Tanaka K, Ikegami S, Villa GR, Yang H, et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci U S A. 2015;112:9406–9411. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soares HP, Ming M, Mellon M, Young SH, Han L, et al. Dual PI3K/ mTOR Inhibitors Induce Rapid Overactivation of the MEK/ERK Pathway in Human Pancreatic Cancer Cells through Suppression of mTORC2. Mol Cancer Ther. 2015;14:1014–1023. doi: 10.1158/1535-7163.MCT-14-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Sun GY, Zhao Y, Thomas D, Greenson JK, et al. DEPTOR has growth suppression activity against pancreatic cancer cells. Oncotarget. 2014;5:12811–12819. doi: 10.18632/oncotarget.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li D. Diabetes and pancreatic cancer. Molecular Carcinogenesis. 2012;51:64–74. doi: 10.1002/mc.20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ewald N, Bretzel RG. Diabetes mellitus secondary to pancreatic diseases (Type 3c)--are we neglecting an important disease? Eur J Case Rep Intern Med. 2013;24:203–206. doi: 10.1016/j.ejim.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 55.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morin A, Letouze E, Gimenez-Roqueplo AP, Favier J. Oncometabolites-driven tumorigenesis: From genetics to targeted therapy. Int J Cancer. 2014;135:2237–2248. doi: 10.1002/ijc.29080. [DOI] [PubMed] [Google Scholar]
- 57.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, et al. Discovery and optimization of potent and selective benzonaphthyridinone analogs as small molecule mTOR inhibitors with improved mouse microsome stability. Bioorg Med Chem Lett. 2011;21:4036–4040. doi: 10.1016/j.bmcl.2011.04.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheen AJ. New therapeutic approaches in type 2 diabetes. Acta Clinica Belgica. 2008;63:402–407. doi: 10.1179/acb.2008.083. [DOI] [PubMed] [Google Scholar]
- 60.Pollak MN. Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov. 2012;2:778–790. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- 61.Sinnett-Smith J, Kisfalvi K, Kui R, Rozengurt E. Metformin inhibition of mTORC1 activation, DNA synthesis and proliferation in pancreatic cancer cells: dependence on glucose concentration and role of AMPK. Biochem Biophys Res Comm. 2013;430:352–357. doi: 10.1016/j.bbrc.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lonardo E, Cioffi M, Sancho P, Sanchez-Ripoll Y, Trabulo SM, et al. Metformin targets the metabolic achilles heel of human pancreatic cancer stem cells. PloS One. 2013;8:76518. doi: 10.1371/journal.pone.0076518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kisfalvi K, Moro A, Sinnett-Smith J, Eibl G, Rozengurt E. Metformin Inhibits the Growth of Human Pancreatic Cancer Xenografts. Pancreas. 2013;42:781–785. doi: 10.1097/MPA.0b013e31827aec40. [DOI] [PMC free article] [PubMed] [Google Scholar]