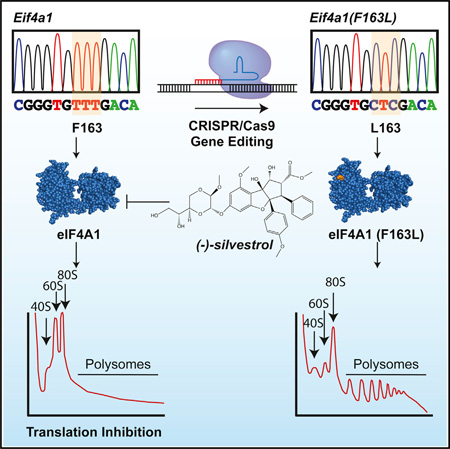
SUMMARY
Targeting translation initiation is an emerging anti-neoplastic strategy that capitalizes on de-regulated upstream MAPK and PI3K-mTOR signaling pathways in cancers. A key regulator of translation that controls ribosome recruitment flux is eukaryotic initiation factor (eIF) 4F, a hetero-trimeric complex composed of the cap binding protein eIF4E, the scaffolding protein eIF4G, and the RNA helicase eIF4A. Small molecule inhibitors targeting eIF4F display promising anti-neoplastic activity in preclinical settings. Among these are some rocaglate family members that are well tolerated in vivo, deplete eIF4F of its eIF4A helicase subunit, have shown activity as single agents in several xenograft models, and can reverse acquired resistance to MAPK and PI3K-mTOR targeted therapies. Herein, we highlight the power of using genetic complementation approaches and CRISPR/Cas9-mediated editing for drug-target validation ex vivo and in vivo, linking the anti-tumor properties of rocaglates to eIF4A inhibition.
Graphical abstract
INTRODUCTION
Protein synthesis is a tightly controlled process that is deregulated in many human cancers and is required to sustain several cancer hallmarks (Bhat et al., 2015). In part, this is attributed to hyper-activation of the MAPK and PI3K-mTOR pathways, both of which impact on the activity of eukaryotic initiation factor (eIF) 4F. As well, resistance to targeted therapies aimed at inhibiting the PI3K-mTOR and MAPK signaling pathways in various cancers has been linked to elevated eIF4F activity (Bhat et al., 2015). Therefore, there is significant interest in developing eIF4F inhibitors as anti-neoplastic compounds (Bhat et al., 2015).
The eIF4F heterotrimeric complex binds to m7GpppN mRNA cap structures through its eIF4E subunit, remodels proximal secondary structure via its eIF4A RNA helicase subunit, and recruits 40S ribosomes (with associated initiation factors) through its eIF4G subunit. The mammalian genome encodes two highly related (>90% identity) eIF4A isoforms, eIF4A1 and eIF4A2. These two isoforms were initially thought to be functionally redundant, but there is evidence suggesting they may also possess distinct biological properties (Galicia-Vázquez et al., 2012).
Strategies aimed at inhibiting eIF4F include blocking eIF4E:eIF4G and eIF4E-cap interaction, interfering with eIF4A1/2 activity, and suppressing eIF4E expression with antisense oligonucleotides (ASOs) (Bhat et al., 2015). The development of eIF4E ASOs has provided proof-of-concept validation for targeting eIF4F in xenograft models, as well as generating safety data profiling from phase I clinical trials (Graff et al., 2007; Hong et al., 2011). Transient inhibition of eIF4E (and hence eIF4F) is tolerated at the organismal level (Lin et al., 2012), despite its essential nature (Truitt et al., 2015). The most potent small molecule inhibitors of the eIF4F complex derive from a family of compounds referred to as rocaglates, which are characterized by a common cyclopenta[b]benzofuran skeleton. Extensive structure-activity relationship into the biological activity of these compounds has been obtained (Pan et al., 2014), with a few compounds capable of potently inhibiting translation (Bordeleau et al., 2008; Rodrigo et al., 2012).
Rocaglates decrease eIF4A1/2 levels present in the eIF4F complex (Bordeleau et al., 2008), exhibit anti-tumor activity in a number of pre-clinical models (Bordeleau et al., 2008; Cencic et al., 2009; Wolfe et al., 2014), and are thought to exert their effects by preferentially inhibiting the translation of key oncogenic mRNAs (e.g., MYC) (Cencic et al., 2009; Rubio et al., 2014; Wolfe et al., 2014). Chemogenomic profiling in yeast have identified the eIF4A orthologs, TIF1 and TIF2, as targets of rocaglates (Sadlish et al., 2013). However, prohibitins (PHB) 1 and 2 have also been proposed as potential rocaglate targets in mammals (Polier et al., 2012). PHB1/2 are involved in a variety of processes, including activation of MAPK signaling (Rajalingam et al., 2005). Given the possibility of alternative targets (PHB1/2 or others), it is critical to validate the rocaglate-eIF4A1/2 drug target relationship in vivo since poor drug-target characterization is a frequent cause of drug development failures (Smurnyy et al., 2014). Here, we validate the rocaglate-eIF4A1 drug target relationship by identifying a drug-resistant and functional mammalian eIF4A1 allele that is capable of rescuing rocaglate anti-neoplastic activity upon introduction into cells either by genetic complementation or genome editing.
RESULTS
eIF4A1 F163L Is Unresponsive to Rocaglates In Vitro
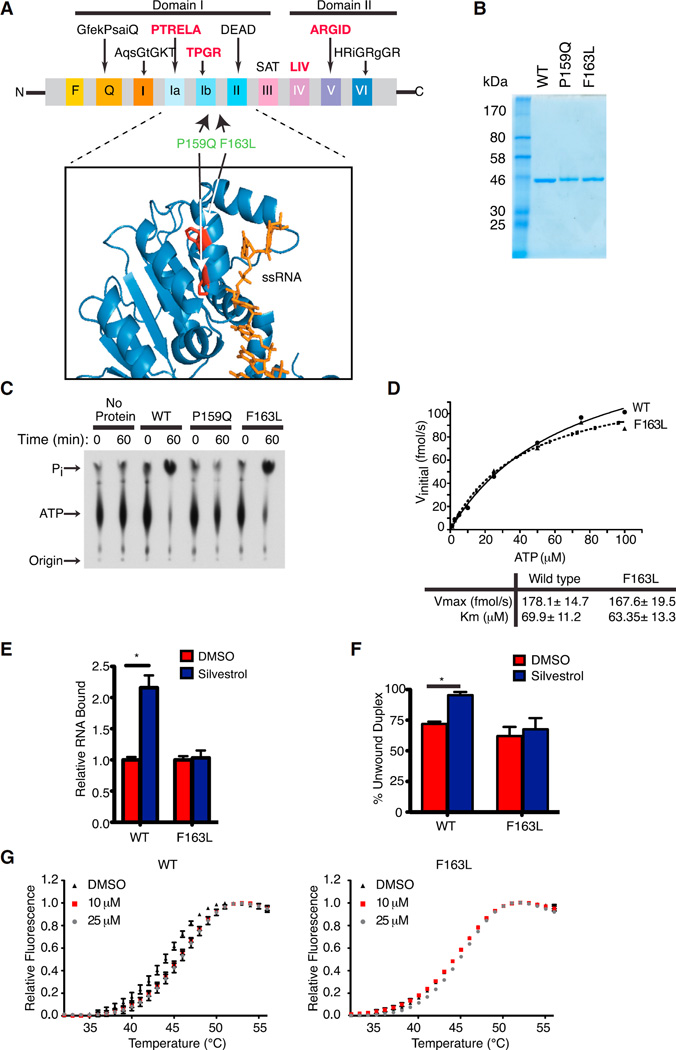
We previously identified mutations in the yeast eIF4A orthologs, TIF1 and TIF2, that conferred resistance to rocaglates, which mapped to the TIF-RNA interface (P147Q, F151L, and Q183E) (Sadlish et al., 2013). We sought to determine whether analogous mutations in the mammalian setting (P159Q and F163L) could inform on the eIF4A:rocaglate relationship. P159Q and F163L map within, or adjacent of, the conserved TPGR Ib motif, which along with the PTRELA motif, is implicated in RNA binding (Figure 1A). Following purification of recombinant proteins (Figure 1B), we performed ATPase assays and noted that eIF4A1(F163L), but not eIF4A1(P159Q), retained robust ATPase activity (Figure 1C). The kinetics of ATP hydrolysis by eIF4A1(F163L) were similar to wild-type (WT) eIF4A and to published values (Figure 1D) (Lorsch and Herschlag, 1998). Since rocaglates increase the RNA binding of eIF4A and lead to its depletion from the eIF4F complex (Bordeleau et al., 2008; Rodrigo et al., 2012), we monitored binding of eIF4A1 to 32P-labeled RNA in vitro. Whereas silvestrol stimulated WT eIF4A1:RNA binding, eIF4A1(F163L):RNA complex formation was unaffected (Figure 1E). As well, eIF4A1(F163L) helicase activity was comparable to WT eIF4A1 but remained unaffected by silvestrol (Figure 1F).
Figure 1. Generation and Characterization of a Rocaglate-Resistant eIF4A1 Allele.
(A) Schematic illustrating conserved motifs of the DEAD-box helicase family. Sequences of the conserved motifs are denoted with motifs involved in RNA binding highlighted in bold red. The structure of eIF4A indicates it to be dumbbell in shape with two domains (I and II) linked via a flexible linker sequence. The inset shows a ribbon diagram of eIF4A1 (PDB 2ZU6) aligned to eIF4A3 (not shown; PDB 2HYI). The residues targeted for mutagenesis are highlighted and the single-stranded RNA substrate (positioned relative to the eIF4A3 crystal structure) is shown in orange.
(B) Coomassie stain of purified recombinant eIF4A1 proteins.
(C) Assessment of ATP hydrolysis by recombinant proteins via thin layer chromatography.
(D) Kinetics of ATP hydrolysis by eIF4A1 and eIF4A1(F163L). ATPase assays were performed with 1 µg protein and varying ATP concentrations. Graph represents the Michaelis-Menten fit from two independent experiments.
(E) RNA binding activity of eIF4A1 and eIF4A1(F163L) using [32P]-labeled RNA generated from pSP/CAT (see Supplemental Experimental Procedures). Assays were performed in the presence of 0.5% DMSO or 1 µM silvestrol and the retained eIF4A:RNA complexes are set relative to DMSO controls. n = 3 biological replicates performed in triplicate ±SEM; *p < 0.001.
(F) Quantitation of eIF4A1 and eIF4A1(F163L) helicase activity performed with 0.5 µg recombinant eIF4A1 and an 11-nt radiolabeled RNA duplex in the presence of DMSO or 50 µM silvestrol. n = 3 biological replicates ±SEM; *p < 0.05.
(G) DSF analysis of eIF4A1 or eIF4A1(F163L) in the presence of DMSO or (−)-SDS-1-021.
Rocaglate resistance of eIF4A1(F163L) was also demonstrated using differential scanning fluorimetry (DSF) (Figure 1G). This approach monitors temperature-dependent protein unfolding with an increase in fluorescence arising from dye binding to newly exposed hydrophobic domains. Protein/ligand interactions are expected to promote protein stability, leading to a shift (increase) in denaturation temperature (Niesen et al., 2007). As this approach consumes large quantities of compounds, we used the related synthetic rocaglate (−)-SDS-1-021 instead of the scarcer natural product silvestrol (Figure S1A). In our hands, (−)-SDS-1-021 is more active than silvestrol with respect to stimulating eIF4A RNA binding activity (Chu et al., 2016), inhibiting translation (Chu et al., 2016), and affecting cell viability (Figure S1B). Only WT eIF4A1 showed a transition midpoint temperature shift of ~+2–3°C (Figure 1G), which is consistent with the ability of rocaglates to interact with eIF4A1, but not as robustly (if at all) with eIF4A1(F163L). Taken together, these experiments indicate that eIF4A1(F163L) ATPase activity and RNA binding are resistant to rocaglates in vitro while displaying Vmax and Km values for ATP hydrolysis that are similar to WT eIF4A1.
Introduction of eIF4A1(F163L) Confers Cellular Resistance to Silvestrol
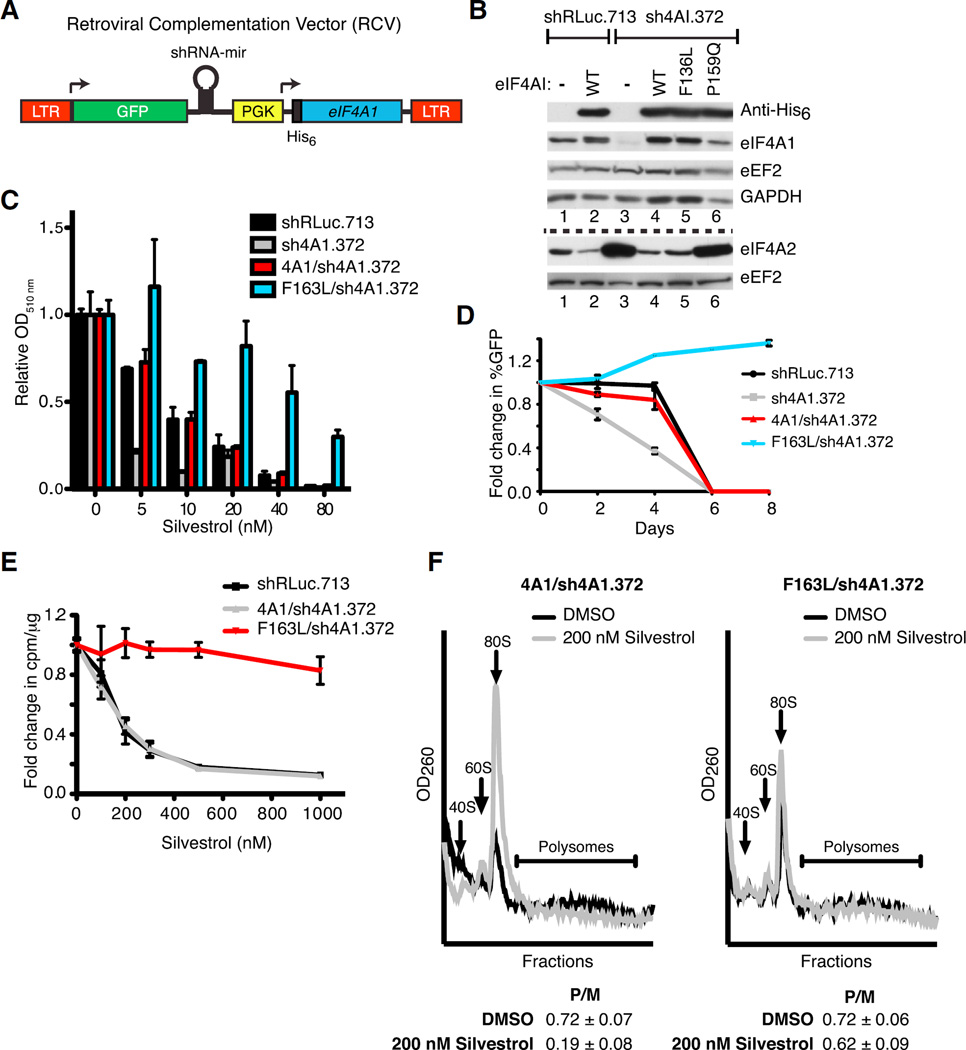
To determine whether eIF4A1(F163L) could complement for loss of WT eIF4A1 in cellulo, we engineered a retroviral complementation vector (RCV) that affords simultaneous small hairpin RNA (shRNA)-mediated suppression of endogenous eIF4A1 while co-expressing exogenous His-tagged eIF4A1 (Figure 2A). As previously reported, knockdown of eIF4A1 led to increased expression of eIF4A2 (Figure 2B) (Galicia-Vázquez et al., 2012). Ectopic expression of WT eIF4A1 or eIF4A1(F163L), but not the inactive eIF4A1(P159Q) mutant, rescued this response (Figure 2B). Cells expressing eIF4A1(F163L) showed resistance to silvestrol as assessed by monitoring cell viability (Figure 2C), growth competition assays (Figure 2D), 35S-methionine metabolic labeling (Figure 2E), and polysome profiling (Figure 2F). The resistance phenotype observed upon ectopic expression of the eIF4A1(F163L) allele was not pleiotropic since these cells were still sensitive to the structurally unrelated eIF4A inhibitor, hippuristanol (Figures S1A and S1C). Taken together, these results demonstrate that the eIF4A1(F163L) allele can functionally compensate for suppression of eIF4A1 and is sufficient to confer cellular resistance to silvestrol.
Figure 2. Ectopic Expression of eIF4A1(F163L) Confers Resistance to Rocaglates in Mammalian Cells.
(A) Schematic diagram of RCV designed to simultaneously express an shRNA-resistant His6-eIF4A1 allele while suppressing endogenous eIF4A1.
(B) Representative western blot of NIH/3T3 cells transduced with RCVs. The dashed line separates the two sets of western blots.
(C) Viability assay of RCV-transduced NIH/3T3 cells. Cells were exposed to the indicated concentrations of silvestrol and relative viability was assessed 6 days later by sulforhodamine B (SRB). n = 2 biological replicates performed in duplicates ±SEM.
(D) Competition assay of transduced NIH/3T3 cells. Transduced cells (GFP+) were mixed with parental cells (GFP−) and cultured in the presence of 20 nM silvestrol. The percentage of GFP+ cells was determined on the indicated days. n = 2 biological replicates performed in triplicate ±SEM.
(E) Cells expressing eIF4A1(F163L) are resistant to translation inhibition by silvestrol. Transduced cells were incubated with the indicated concentrations of silvestrol for 1 hr and labeled with [35S]-methionine/cysteine during the last 15 min. n = 4 biological replicates ±SEM.
(F) Polysome profiles of transduced NIH/3T3 cells following exposure to 200 nM silvestrol for 30 min. P/M represents the polysome/monosome ratio. n = 3 biological replicates ±SEM.
See also Figure S1.
Cas9-Mediated Editing of the Eif4a1 Locus Rescues Cells from the Inhibitory Effects of Rocaglates
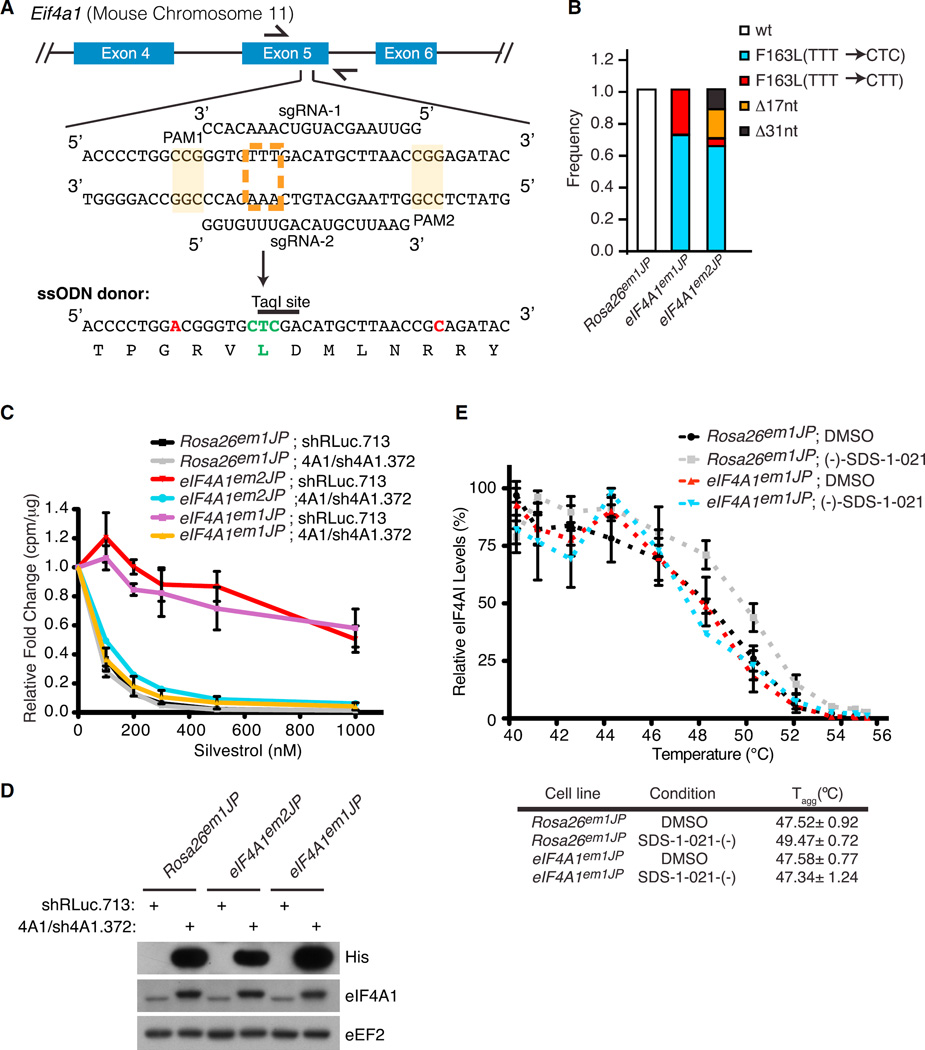
To strengthen these results, we utilized CRISPR/Cas9 gene editing to introduce the F163L mutation into the endogenous Eif4a1 locus. To this end, two sgRNAs were designed to target Eif4a1 exon 5 and co-transfected with a single-stranded oligonucleotide (ssODN) donor template (Figure 3A). In addition to harboring the desired F163L change, two silent mutations were present in the ssODN that altered the protospacer adjacent motifs (PAMs) to prevent re-cleavage (Figure 3A, indicated in red). Control cells received Cas9 and sgRNAs targeting the neutral Rosa26 locus (Rosa26em1JP). Two cell populations, eIF4A1em1JP and eIF4A1em2JP, derived from sgRNA2 and sgRNA1, respectively, were characterized by sequencing exon 5 PCR products (Figure 3B). The results indicate that the eIF4A1em1JP population contains Eif4a1 alleles that harbor CTT or CTC codons encoding for leucine at position 163, whereas the eIF4A1em2JP population also has Eif4a1 alleles with deletions within exon 5 (Figures 3B and S2A). No silvestrol-resistant colonies arose from Rosa26 targeted cells, and we did not detect mutant Eif4a1 alleles in Rosa26em1JP cells. The growth of eIF4A1em1JP cells showed increased resistance (~10-fold) to silvestrol and (−)-SDS-1-021 (Figure S2B). To ensure that the observed resistance was not due to off-target alterations by CRISPR/Cas9, we suppressed the mutated Eif4a1 alleles in eIF4A1em1JP and eIF4A1em2JP using the RCV system (Figures 3C and 3D). Resensitization was monitored using 35S-methionine/cysteine protein labeling. As expected eIF4A1em1JP showed increased resistance (~10- to 20-fold) to silvestrol compared to control Rosa26em1JP cells (Figure 3C). Importantly, suppressing endogenous eIF4A1(F163L) using sh4A1.372 and co-expressing WT eIF4A1 resensitized eIF4A1em1JP cells to silvestrol (Figure 3C). Similar results were also obtained with eIF4A1em2JP cells (Figure 3C).
Figure 3. Cas9-Mediated Editing of Eif4a1.
(A) Strategy for introducing the Eif4a1(F163L) mutant allele. The sequence of two sgRNAs targeting exon 5 and the partial sequence of the ssODN donor are shown. The PAMs are shaded, and the nucleotide changes in the ssODN donor that abolishes their presence are indicated in red. The targeted TTT (F) codon is indicated by a dashed orange box, and engineered CTC (L) change in the ssODN donor is indicated in green.
(B) Sequence analysis of the PCR products from eIF4A1em1JP and eIF4A1em2JP cells indicating loss of the wild-type Eif4a1 allele and composition of mutant alleles.
(C) Relative translation rates in Rosa26em1JP, eIF4A1em1JP, eIF4A1em2JP cells transduced with the indicated retroviruses.
(D) Western blot assessing His6-eIF4A1 and total eIF4A1 in the cell lines used in (C).
(E) CETSA of Rosa26em1JP and eIF4A1em1JP cells. Cells were incubated with 1 µM (−)-SDS-1-021 or DMSO for 1 hr at 37°C and heated at the indicated temperatures for 3 min. Soluble lysates were prepared and used for western blotting. n = 4 biological replicates ±SEM.
See also Figure S2.
To assess whether eIF4A1(F163L) showed altered rocaglate binding in cellulo, we implemented a cellular thermal shift assay (CETSA) by measuring the thermal stability of WT eIF4A1 or eIF4A1(F163L) from Rosa26em1JP and eIF4A1em1JP cells, respectively, that had been exposed to vehicle or (−)-SDS-1-021 (Figure 3E). In this assay, thermal stability is assessed by heating cells over a range of temperatures, followed by separation of insoluble (i.e., denatured/aggregated) from soluble proteins (Jafari et al., 2014). Levels of the protein of interest remaining in the soluble fractions are then determined by immunoblotting. Similar to DSF, CETSA is based on the principle that thermal stability of a protein is increased upon binding to a ligand (Jafari et al., 2014). For Rosa26em1JP cells, WT eIF4A1 displayed a 2°C increase in thermal stability when cells were exposed to (−)-SDS-1-021 (Figure 3E). For eIF4A1em1JP cells, we observed no differences in the denaturation profile of eIF4A1(F163L), suggesting reduced target engagement for eIF4A1(F163L) (Figure 3E).
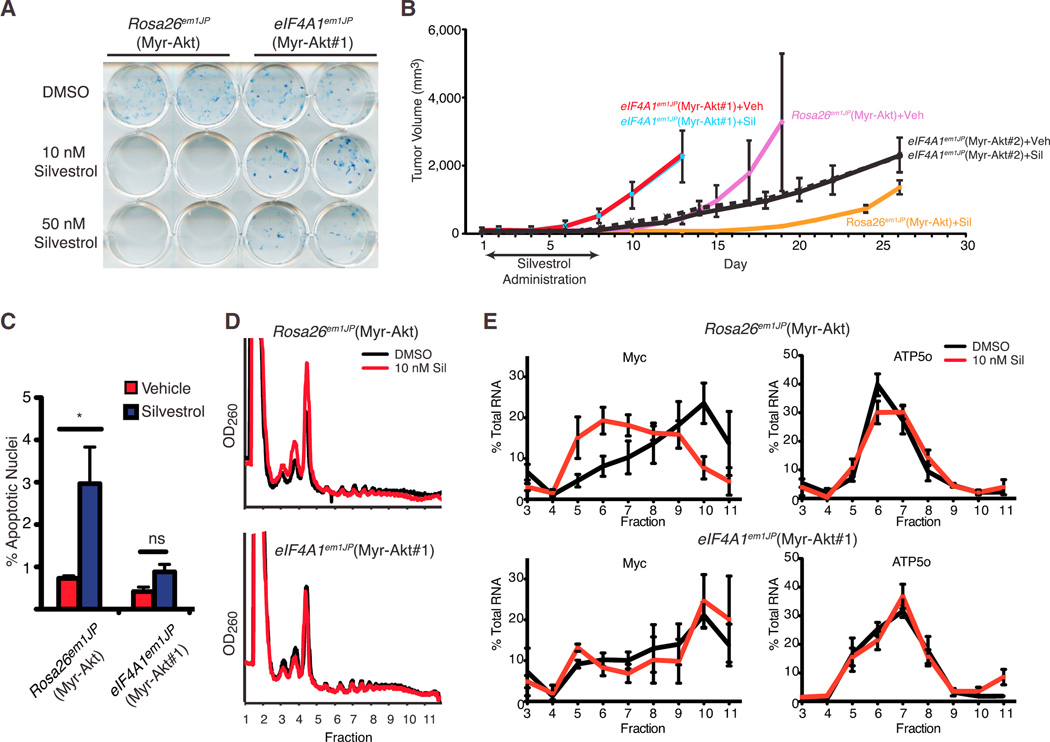
We next investigated the relationship between eIF4A1 status and the in vivo anti-neoplastic activity of rocaglates. To this end, Rosa26em1JP and eIF4A1em1JP cells were transduced with a retrovirus expressing Myr-Akt and single clones from these populations were isolated (referred to as Rosa26em1JP(Myr-Akt), eIF4A1em1JP(Myr-Akt#1), and eIF4A1em1JP(Myr-Akt#2)). The relative distribution of the different Eif4a1 alleles in eIF4A1em1JP(Myr-Akt#1) was assessed by cloning and sequencing exon 5 PCR products and revealed the presence of four different Eif4a1(F163L) alleles in approximately equimolar ratios, with no intragenic deletions (Figures S3A and S3B). These results are consistent with NIH/3T3 cells being tetraploid for chromosome 11 (the location of murine Eif4a1) (Leibiger et al., 2013). Direct sequencing of the exon 5 PCR products independently demonstrated that a mixture of CTC and CTT alleles were present (Figure S3B). Colony formation assays demonstrated that eIF4A1em1JP(Myr-Akt#1) cells were resistant to rocaglates ex vivo (Figure 4A). In vivo, eIF4A1em1JP(Myr-Akt#1) cells formed tumors faster than Rosa26em1JP(Myr-Akt) cells (Figure 4B), despite displaying similar doubling rates ex vivo (Figure S3C). Nevertheless, an 8-day treatment course with silvestrol significantly curtailed tumor outgrowth in mice transplanted with Rosa26em1JP(Myr-Akt) cells while having no effect on eIF4A1em1JP(Myr-Akt#1) tumors (Figure 4B). Independently generated tumors from the second cell line, eIF4A1em1JP(Myr-Akt#2), formed tumors slower than eIF4A1em1JP(Myr-Akt#1) in mice but nonetheless remained unresponsive to silvestrol (Figure 4B). The failure to respond to silvestrol in vivo was associated with an ~3-fold reduction in apoptosis (Figure 4C). Previous studies have identified several rocaglate-responsive mRNAs, of which c-myc is a representative anti-cancer target (Robert et al., 2014; Wolfe et al., 2014). Polysome analysis revealed that c-myc mRNA distribution shifts from heavy polysome fractions to light polysome fractions when Rosa26em1JP(Myr-Akt) cells were exposed to silvestrol but remains unaffected in eIF4A1em1JP(Myr-Akt#1) cells (Figures 4D and 4E). Translation of Atp5o, a prototypical mRNA harboring a translation initiator of short 5′ UTR (TISU) element, which confers eIF4A independence (Elfakess et al., 2011), was unaffected when either cell line was exposed to silvestrol (Figure 4E). Immunoblotting and immunoprecipitation of metabolically labeled proteins were consistent with the ability of silvestrol to inhibit MYC protein production in Rosa26em1JP(Myr-Akt), but not eIF4A1em1JP(Myr-Akt#1), cells (Figure S4). These results indicate that the antineoplastic activity of silvestrol is a consequence of eIF4A1 suppression, which subsequently is associated with diminished expression of Myc, a quintessential oncogene.
Figure 4. Cas9-Mediated Editing of the Eif4a1 Locus Confirms the Drug-Target Hypothesis.
(A) Colony formation assay of Myr-Akt-transformed Rosa26em1JP or eIF4A1em1JP cells in the presence of silvestrol.
(B) Response of Myr-Akt-transformed Rosa26em1JP or eIF4A1em1JP xenografts in vivo to silvestrol. On the indicated days, mice were treated with silvestrol (0.2 mg/kg) following tumor appearance. n = 6–7 mice/cohort ±SEM.
(C) Bar graph of the percentage of apoptotic nuclei from tumor sections. Three hours before harvesting of tumors, mice were treated with vehicle or 0.2 mg/kg silvestrol. n = 2 biological replicates (with ~7,000 nuclei analyzed per tumor) ±SD; *p < 0.05; ns, not significant.
(D) Polysome profiles of Myr-Akt-transformed Rosa26em1JP or eIF4A1em1JP cells exposed to 10 nM silvestrol for 1 hr.
(E) Distribution of mRNAs in polysome fractions shown in (D).
See also Figures S3 and S4.
DISCUSSION
Using genetic complementation and CRISPR/Cas9-mediated gene editing, we validate eIF4A1 as the primary molecular target of rocaglates in mammalian cells responsible for the inhibition of translation observed with this class of compounds. These results significantly strengthen the drug-target link between rocaglates and eIF4A1 in vivo and were critical to undertake since rocaglates have also been reported to bind to prohibitins (PHB) 1 and 2 and block their interaction with cRaf (Polier et al., 2012). Our results indicate that this is not the mechanism by which rocaglates inhibit translation (Chu et al., 2016) and in our hands cannot be responsible for the ex vivo or in vivo activity of rocaglates. However, rocaglates may have other unsuspected biological targets. Although both eIF4A1 and eIF4A2 can cycle through the eIF4F complex in vitro (Yoder-Hill et al., 1993), there is emerging evidence suggesting that their activities are not interchangeable (Galicia-Vázquez et al., 2012). Whether eIF4A2 plays a significant role in response to rocaglate-mediated inhibition and could be responsible for the partial sensitivity of eIF4A1em1JP to high rocaglate concentration (Figure S2B) remains to be elucidated. Nevertheless, these results consolidate the position of eIF4A1 as an anti-cancer target.
We were surprised to find several alleles of Eif4a1(F136L) in our silvestrol-resistant population, both at the codon encoding leucine and at the PAM motifs (Figures 3B, S3A, and S3B). This may be a consequence of strand-switching during homology directed repair (HDR) occurring before incorporation of the second mutation present on the ODN. This has previously been documented when simultaneously introducing two mutations at a given locus, with as much as a 17% drop in efficiency occurring with mutations positioned 8–10 bp apart (Elliott et al., 1998; Yang et al., 2013). These results highlight the importance of thorough characterization of cell lines generated by CRISPR/Cas9.
In silico analysis of the yeast TIF1/2 proteins predicted a rocaglate- binding pocket at the TIF1-RNA interface (Sadlish et al., 2013). Our results from DSF and CETSA assays are consistent with rocaglates directly interacting with eIF4A in vitro and in cellulo. Although structural studies of eIF4A1-rocaglate interaction are necessary to unequivocally identify the rocaglate binding site and understand how the F163L mutation affects rocaglate binding, our results suggest that eIF4A1(F136L) is likely resistant to rocaglates due to reduced compound engagement (Figures 1G and 3E).
Precise genome editing of change-of-function alleles coupled with drug selection has been shown to be a powerful approach to generate tailored cell lines and validate drug:target interactions (Smurnyy et al., 2014). Our results extend this paradigm to gain-of-function alleles coupled with activity screening to validate drug-target relationships in vivo. Although our previous work had identified several TIF1 alleles as capable of conferring rocaglate-resistance in yeast (Sadlish et al., 2013), we found that not all corresponding Eif4a1 alleles encoded for functional proteins in the mammalian setting. This highlights the importance of undertaking detailed biochemical studies coupled with an approach, such as the RCV system (Figure 2), to demonstrate complementation before undertaking the more labor- and time-intensive task of engineering the alleles into the cellular genome. Our genetic complementation and genome editing approaches, in combination with in cellulo and in vivo data converge to position eIF4A1 as a critical anti-neoplastic target.
EXPERIMENTAL PROCEDURES
Cell Lines and Retrovirus Generation
All cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin/streptomycin, and 2 mM L-glutamine at 37°C and 5% CO2. For retroviral transductions, 25 µg of plasmid was transfected into ecotropic Phoenix cells using calcium phosphate in the presence of 25 µM chloroquine, and the media was changed the following day. Starting 48 hr after transfection, viral supernatant was filtered and added to NIH/3T3 cells in the presence of 4 µg/ml polybrene. Cells were spinoculated at 1,000 × g for 1 hr at 30°C. Infections were performed every 8 hr, for a total of six infections. Cas9-modified cell lines are named as suggested (http://www.informatics.jax.org/mgihome/nomen/gene.shtml#endim); for example, eIF4A1em1JP indicates the first endonuclease-induced mutation (em1) of the Eif4a1 gene produced in J.P.’s lab).
Xenograft Models
Four million cells were injected with Matrigel subcutaneously into 4- to 6-weekold female Balb/c-nu/nu mice. Tumor growth was monitored every second day using calipers. Treatments began when tumors had reached 25–50 mm3 with silvestrol (0.2 mg/kg) delivered by intraperitoneal (IP) injection daily for 8 consecutive days. Tumor growth was monitored for the remainder of the experiment, and no further drug treatments were performed. For tumor analysis, mice were treated with compound or vehicle 3 hr prior to harvesting of the tumors. Tumors were collected into 10% formalin, embedded in paraffin, and sectioned. TUNEL staining was performed using the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer’s instructions. The percentage of cells undergoing apoptosis was determined by counting the number of TUNEL positive nuclei on Fiji (ImageJ, NIH) and dividing it by the total number of nuclei in the field. All animal studies were approved by the McGill University Faculty of Medicine Animal Care Committee.
Additional details regarding methodology are presented in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
CRISPR/Cas9 gene editing is a powerful approach for in vivo drug-target validation
Rocaglates interact with eIF4A1 in vitro and in vivo
Anti-neoplastic activity of rocaglates is a consequence of eIF4A1 inhibition
In Brief.
Rocaglates are anti-neoplastic agents that are thought to inhibit the RNA helicase eIF4A, although alternative targets have also been proposed. Using a series of biochemical assays and CRISPR/Cas9 genome editing, Chu et al. provide genetic evidence that the antineoplastic activities of rocaglates are a consequence of eIF4A1 inhibition.
Acknowledgments
We thank Mr. Steven Stone and Dr. Neil Lajkiewicz (Boston University) for the synthesis of (−)-SDS-1-021. This work was supported by grants from the Richard and Edith Strauss Canada Foundation and the Canadian Institutes of Health Research (MOP-106530 and MOP-115126, to J.P.) and the NIH (R01 GM073855, to J.A.P.). Research in the BU-CMD is supported by NIH grant GM111625.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.05.005.
AUTHOR CONTRIBUTIONS
J.C. and J.P. conceived and designed the study. J.C. performed all experiments with the following exceptions: G.G.-V. generated the CRISPR-modified cell lines and performed experiments in Figures 3B, S2, and S3. R.C. performed experiments in Figure 4B, J.R.M. generated the RCV constructs. A.K. undertook analysis of tumors derived from Rosa26em1JP(Myr-Akt) and eIF4A1em1JP(Myr-Akt#1) cells. J.A.P. provided unique rocaglate derivatives. J.P. supervised the study. J.C. and J.P. wrote the manuscript, and all authors contributed to editing the manuscript.
REFERENCES
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA, Jr, Pelletier J. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vázquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Cencic R, Wang W, Porco JA, Jr, Pelletier J. Translation Inhibition by Rocaglates Is Independent of eIF4E Phosphorylation Status. Mol. Cancer Ther. 2016;15:136–141. doi: 10.1158/1535-7163.MCT-15-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39:7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Vázquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 2012;18:1373–1384. doi: 10.1261/rna.033209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, Callies S, André V, Kadam SK, Nasir A, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin. Cancer Res. 2011;17:6582–6591. doi: 10.1158/1078-0432.CCR-11-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundbäck T, Nordlund P, Martinez Molina D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- Leibiger C, Kosyakova N, Mkrtchyan H, Glei M, Trifonov V, Liehr T. First molecular cytogenetic high resolution characterization of the NIH 3T3 cell line by murine multicolor banding. J. Histochem. Cytochem. 2013;61:306–312. doi: 10.1369/0022155413476868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Nasr Z, Premsrirut PK, Porco JA, Jr, Hippo Y, Lowe SW, Pelletier J. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 2012;1:325–333. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Pan L, Woodard JL, Lucas DM, Fuchs JR, Kinghorn AD. Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat. Prod. Rep. 2014;31:924–939. doi: 10.1039/c4np00006d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier G, Neumann J, Thuaud F, Ribeiro N, Gelhaus C, Schmidt H, Giaisi M, Köhler R, Müller WW, Proksch P, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012;19:1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- Robert F, Roman W, Bramoullé A, Fellmann C, Roulston A, Shustik C, Porco JA, Jr, Shore GC, Sebag M, Pelletier J. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc. Natl. Acad. Sci. USA. 2014;111:13421–13426. doi: 10.1073/pnas.1402650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J. Med. Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, Helliwell SB, Hoepfner D, Knapp B, Riedl R, et al. Evidence for functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem. Biol. 2013;8:1519–1527. doi: 10.1021/cb400158t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurnyy Y, Cai M, Wu H, McWhinnie E, Tallarico JA, Yang Y, Feng Y. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat. Chem. Biol. 2014;10:623–625. doi: 10.1038/nchembio.1550. [DOI] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J. Biol. Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.