Abstract
Mycobacterium smegmatis contains the low molecular weight thiols, mycothiol (MSH) and ergothioneine (ESH). Examination of transposon mutants disrupted in mshC and egtA, involved in the biosynthesis of MSH and ESH respectively, demonstrated that both mutants were sensitive to oxidative, alkylating, and metal stress. However, the mshC mutant exhibited significantly more protein carbonylation and lipid peroxidation than wildtype, while the egtA mutant had less protein and lipid damage than wildtype. We further show that Ohr, KatN, and AhpC, involved in protection against oxidative stress, are upregulated in the egtA mutant. In the mshC mutant, an Usp and a putative thiol peroxidase are upregulated. In addition, mutants lacking MSH also contained higher levels of Coenzyme F420 as compared to wildtype and two Coenzyme F420 dependent enzymes were found to be upregulated. These results indicate that lack of MSH and ESH result in induction of different mechanisms for protecting against oxidative stress.
Keywords: Mycothiol, Ergothioneine, Mycobacteria, Oxidative stress, Coenzyme F420, Thiol peroxidase
Graphical abstract
Highlights
-
•
MSH and ESH are involved in protection against metal stress in Mycobacterium smegmatis.
-
•
MSH mutants have greater macromolecular damage while ESH mutants have lesser damage than wildtype.
-
•
Different proteins associated with protection against oxidative stress are upregulated in mshC and egtA mutants.
1. Introduction
Mycothiol (MSH), the major low-molecular weight (LMW) thiol produced by Actinomycetes, acts as a glutathione (GSH) analog to maintain redox homeostasis, and protects the cell against xenobiotic agents, notably in pathogens such as Mycobacterium tuberculosis [1], [2]. MSH is present in mycobacteria at millimolar levels [1] and is synthesized in a five step pathway from glucose-6-phosphate with the enzymes MshA, A2, B, C, D [2]. A second LMW thiol, l-ergothioneine (ESH), is also synthesized by Actinomycetes but is present in lesser amounts [3]. In M. smegmatis, ESH biosynthesis is initiated by a methyltransferase (EgtD), and proceeds through an additional five-step pathway involving the enzymes EgtA-E [4]. Unlike other LMW thiols such as MSH, ESH is tautomeric [5] and exists predominantly in the thione form in neutral aqueous solutions. In addition, the redox potential of ESH is −60 mV as compared to −250 mV of GSH which may account for ESH's resistance to auto-oxidation [5].
While the role and function of MSH has been extensively studied, the role of ESH in bacteria is not as well-described. MSH mutants are more susceptible than wildtype to a number of antibiotics, oxidative stress, alkylating agents, and other stresses but resistant to the pro-drugs, isoniazid and ethioniamide [6]. An M. smegmatis mutant disrupted in egtD, which lacks ESH, and is more susceptible to the lipid peroxides, cumene hydroperoxide (CHP) and tert-butyl hydroperoxide (tBOH), but not sensitive to the antibiotics, rifampin, isoniazid, ethionamide, and ethambutol [3]. A double mutant disrupted in mshA and egtD and lacking both ESH and MSH is significantly more sensitive to the peroxides than either of the single mutants lacking either ESH or MSH. Interestingly, levels of ESH are significantly higher in the mshA mutant indicating that ESH may be able to compensate for the loss of MSH [7]. In addition, Ohr, an organic hydroperoxide reductase, is substantially upregulated in both mshA and egtD mutants. Whether this upregulation of Ohr is particular to mshA and egtD mutants or is common in mutants disrupted in other genes in the biosynthetic pathway of MSH and ESH is unknown.
In a screen for genes involved in disulfide stress, a transposon mutant library was screened for sensitivity to diamide, a thiol-oxidizing agent. Five mutants lacking MSH and disrupted in mshC, S24, D43, D48, D41, R41, and a mutant disrupted in egtA, R119, were identified as having increased sensitivity to diamide. We demonstrate that the mshC mutant, S24, has significantly more protein carbonylation and lipid peroxidation, measures of oxidative damage, while the egtA mutant has less protein and lipid damage than wildtype. We further show that Ohr and other enzymes involved in protection against oxidative stress are upregulated in the egtA mutant, while in the mshC mutant, the levels of coenzyme F420, a coenzyme involved in redox reactions, are increased.
2. Materials and methods
2.1. Bacterial strains, culture conditions, and disk sensitivity assays
M. smegmatis mc2155 and mutant strains were grown in Middlebrook 7H9 broth (Difco) with 10% OADC and then diluted in Middlebrook 7H9 broth with 0.05% Tween 80% and 1% glucose. The strains were also grown on 7H11 solid medium with 0.5% glycerol with 1% glucose for disk sensitivity assays, which were performed as previously described [6]. Antibiotics were added to the media for the transposon mutant (Kanamycin 25 μg/ml) and complemented strains (Hygromycin 75 μg/ml). Broth cultures were grown in a shaking incubator and all cultures were grown at 37 °C unless otherwise indicated.
2.2. Creation and screening of a transposon mutant library and identification of the disrupted gene
An M. smegmatis transposon mutant library (EZ::TN <kan-2> Tnp transposase and Tn5 kanamycin resistance marker) was constructed and screened for diamide sensitive mutants as described by Rawat et al. [8]. To identify the site of insertion, genomic DNA was digested with restriction enzymes Sa1I and PstI followed by self-ligation and PCR amplification with primers complementary to the transposon. The sequence of the amplified PCR product was compared to the M. smegmatis genome sequence [8].
2.3. Determination of thiol levels and Coenzyme F420
Thiols in cell extracts were labeled with monobromobimane (mBBr) as previously described [6]. High-pressure liquid chromatographic (HPLC) analysis was carried out on a HiChrom Ultrasphere Ion Pair column (5 µm; 250×4.6 mm) with the following gradient at 1 ml/min of Buffer A, 0.25% glacial acetic acid, pH 4.0; and Buffer B, methanol (0–5 min, 10% A; 35 min, 18% A; 45 min 27% A; 47 min, 100% A; 49 min, 0%). HPLC analysis for F420 content was performed as previously described [9] with the following modifications: Shimadzu Prominence system (LC 20 CE dual pumps, SIL-20A autosampler and SPD-M20A Diodearray detector) was used for chromatographic separation and quantitative analysis. Cell extracts were separated with a Vydac 218TP54 column (C18, 5 µm, 4.6 mm i.d.×250 mm) at a flow rate of 0.6 ml/min. A linear gradient of sodium acetate buffer-acetonitrile solution was used and elution was monitored at 400 nm. Identities of F420 peaks were confirmed by their UV–visible spectrum collected by an in-line diode array detector. Samples in quadruplicates were analyzed at least three times and reported as averages±SD. Student t test was performed with p≤0.05 regarded as significant.
2.4. Complementation of S24 (mshC mutant), R119 (egtA mutant)
M. tuberculosis mshC cloned into pSODIT [10] was introduced into S24 in order to obtain the complemented strain, S24C. Primers, TEshAF, with a HindIII restriction site (CTTAAGCTTCACCGACGAGGCCTGGAC) and TEshAR with a XhoI restriction site (TGCCTCGAGTGAAGTCACGACGCCCCG) were used to amplify M. tuberculosis egtA (Rv3704c) and 297 base pairs upstream of the start codon, which contained the promoter region of the gene. The 1609 bp amplicon and complementation vector pHINT were digested with the HindIII and XhoI, ligated and transformed into E. coli. After confirmation of cloning, the resulting vector, pHINTEgtA, was electroporated into R119 competent cells. Kanamycin and hygromycin resistant transformants were screened by PCR for M. tuberculosis egtA and one colony, R119C, was characterized further.
2.5. Measurement of protein carbonylation, lipid peroxidation, and peroxidase activity
Protein carbonylation and lipid peroxidation was measured using the Protein Oxidation Detection Kit and TBARS Assay Kit, following the manufacturer's instructions (Cayman Chemical). To assess protein carbonylation, cells were grown until OD600 equaled one. Then one set of 10 ml cultures in triplicate were treated with 5 mM H2O2 and another set was left untreated for one hour. The cultures were harvested and lysed using bead beating (Fast Prep FP120, Thermo Electron company). The cell lysates were centrifuged at 13,000 rpm for 30 min at 4 °C to get rid of cell debris and the protein concentration of the supernatant was determined by Bio-Rad DC assay. Two mg/ml of protein was incubated with 2,4-dinitrophenylhydrazine at room temperature in the dark for one hour with frequent vortexing. The derivatized proteins were TCA precipitated and the pellet was further extracted with ethanol/ethyl acetate (1:1) solution three times. The resulting pellet was resuspended in guanidine hydrochloride. After centrifugation, the supernatant containing the protein hydrazones were analyzed spectrophotometrically at 360 nm.
Lipid peroxidation was assessed by treating 1 ml of cells in triplicate at 1.0 OD600 in log phase with 30 mM FeSO4 and 5 mM H2O2 for two hours. Cell lysates were prepared similarly to the samples for protein carbonylation assay and to 100 μl of the cell lysate, 100 μl of 10% SDS was added. A color reagent was prepared by adding thiobarbituric acid (TBA) to acetic acid followed by the addition of sodium hydroxide to the solution. Four ml of the color reagent was added to each sample and the samples were boiled for 1 h and then transferred to ice for 10 min to stop the reaction. After centrifugation at 1600×g for 10 min at 4 °C, the supernatants were transferred to an ELISA plate and A532 was determined in duplicate. The amount of lipid peroxidation was calculated with reference to a standard curve of malondialdehyde.
Peroxidase activity was determined for each strain in triplicate using the Pierce Quantitative Peroxide Assay Kit, which detects levels of peroxides based on the oxidation of ferrous to ferric ion at acidic pH. The cell free lysate was prepared as described for the protein carbonylation assay. 300 μg of protein was incubated with 45 μM H2O2, CHP, and TbOH separately for 30 min. The amount of remaining peroxides was determined by addition of sulfuric acid solution containing ferrous ion and xylenol orange and incubation at room temperature for 20 min. The ferric ion-xylenol orange purple complex was measured at 595 nm. The amount of peroxides was calculated with reference to a standard curve of the relevant peroxide.
All the experiments were performed at least three times.
2.6. Proteomic evaluation of wildtype and mutants, S24 and R119
As described in Ta et al. [7], M. smegmatis strains were grown for 11 days on Middlebrook 7H9 agar and 1.0% glucose at 23 °C. Cells were collected by centrifugation and resuspended in lysis buffer (50 mM Tris containing 7 M urea, 2 M thiourea, and 4% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS)). Two-dimensional difference gel electrophoresis (2D-DIGE) analysis of the supernatant was performed at Applied Biomics (Hayward, CA). Briefly, samples were fluorescently labeled and subjected to isoelectric focusing (IEF) on a 13 cm pH gradient strip (pH 4–6). The changes in abundance of proteins between the wildtype and the mutants were identified using the ImageQuant TL and DeCyder softwares. Spots were excised from the gel and identified by MALDI TOF/TOF (tandem mass spectrometry MS/MS) on a 5800 mass spectrometer (AB Sciex). Resulting peptide masses and associated fragmentation spectra were submitted to GPS Explorer version 3.5 with the MASCOT search algorithm (Matrix science) and searched against the National Center for Biotechnology Information non-redundant database. Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage allowed in the search parameters. MASCOT searches were also performed to identify phosphorylation in the proteins. Candidates with score of either protein C.I.% or Ion C.I.% greater than 95 were considered significant.
3. Results
3.1. Mycothiol, ergothioneine and coenzyme F420 levels in transposon mutants lacking MSH and ESH
Inverse PCR was performed to identify the disrupted genes in diamide sensitive mutants, S24, D41, D43, D48, R41, and R119. Five of the mutants, S24, D41, D43, D48, R41, are disrupted in mshC (MSMEG_4189) and did not contain any MSH. In four of the five mutants disrupted in mshC, the transposon was inserted at 589 nucleotides from the start codon. In the D48 mutant, the insertion site was 599 nucleotides from the start codon. As the majority of the mutants had the same transposon insertion site, we decided to focus on only one of these mutants, S24, further. The amount of ESH in S24, 0.37±0.04 µmol/g dry cell weight, was higher than in wildtype (0.20±0.01) and significantly different. The increase in ESH was not as high as in the mshA mutant (1.02±0.24) grown under the same growth conditions [7]. The complemented strain, S24C, did not show a significant difference from wildtype in ESH levels (0.19±0.01) and MSH levels (wildtype: 6.21±0.17, S24C: 5.90±0.66).
In the transposon mutant, R119, where the transposon is inserted 972 nucleotides after the start codon in egtA (MSMEG_6250), there was no ESH present and MSH levels (6.10±0.37) were not statistically significantly different than wildtype. The complemented strain, R119C, did not have statistically significant different levels of MSH (5.88±0.27) and ESH from wildtype (0.24±0.04).
Previously, we had demonstrated that mutants disrupted in fbiC, which is involved in the biosynthesis of Coenzyme F420 (F420), a deazaflavin derivative of FMN found in methanogenic Archaea and actinobacteria [11], are sensitive to oxidative stress [12], [13]. We measured F420 levels to determine if F420 can compensate for the lack of MSH or ESH. F420 levels were statistically significantly higher in mshA (8.47±0.52 nmol/g wet cell weight) and mshC (S24) mutants (8.05±0.24) as compared to wildtype (6.12±0.87). Complementation of the mshA mutant showed partial restoration of wildtype F420 levels (7.16±0.57) while complementation of S24 resulted in lower F420 levels (4.81±0.32) as compared to wildtype. Interestingly, R119 had statistically significantly less F420 (4.21±0.1) than the wildtype and complementation (6.51±0.38) restored F420 to wildtype levels.
3.2. Sensitivity of S24 and R119
In liquid culture, there is no difference in growth between R119 and wildtype under normal growth conditions (data not shown). However, R119, like S24 is more susceptible to alkylating agents and the sensitivity is reversed by complementation with the M. tuberculosis homolog (Table 1). In the case of oxidants, both mutants show sensitivity to diamide and CHP, but only S24 demonstrates sensitivity to H2O2.
Table 1.
Susceptibility of Mycobacterium smegmatis ergotheionine mutants to antibiotics, toxins, metals and oxidants as determined by disk assays. All values represent averages and SD (n=4). Student t-test was performed and the p values of mutants were compared with the wildtype.
| Reagent | Zone of clearing (mm) | ||||
|---|---|---|---|---|---|
|
M. smegmatis strains | |||||
| Wt | S24 | S24C | R119 | R119C | |
| Oxidants | |||||
| H2O2 (0.5 µmol) | 20±1 | 26±2⁎⁎⁎ | 22±2⁎ | 22±1 | 19±1 |
| Diamide (15 µmol) | 19±1 | 27±1⁎⁎⁎ | 25±1⁎⁎ | 29±1⁎⁎⁎ | 20±2 |
| CHP (0.5 µmol) | 23±2 | 31±2⁎⁎⁎ | 27±1⁎ | 29±1⁎⁎ | 27±2⁎ |
| Alkylating agents | |||||
| N-ethylmalemide (0.25 µmol) | 14±2 | 23±1⁎⁎ | 16±1 | 22±1⁎⁎ | 15±1 |
| Iodoacetamide (0.0125 µmol) | 16±3 | 21±2⁎ | 18±2 | 22±2⁎ | 19±2 |
| Chlorodinitrobenzene (0.0125 µmol) | 17±2 | 22±1⁎⁎ | 18±1 | 22±1⁎ | 18±2 |
| Metal | |||||
| Cd2+ (0.5 µmol) | 22±2 | 31±1⁎⁎ | 27±1⁎ | 27±1⁎ | 23±1 |
| Cu2+ (0.5 µmol) | 19±1 | 22±2⁎ | 20±1 | 22±2⁎ | 18±2 |
| SeO32− (0.25 µmol) | 20±1 | 25±1⁎⁎⁎ | 19±1⁎ | 23±1⁎⁎ | 22±2 |
| TeO32− (0.25 µmol) | 15±2 | 22±1⁎⁎⁎ | 18±1⁎ | 19±1⁎⁎ | 16±1 |
| Fe2+ (5 µmol) | 16±1 | 21±3⁎ | 17±3 | 19±4 | 17±3 |
| CrO42− (0.5 µmol) | 20±1 | 27±2⁎⁎⁎ | 24±1⁎ | 24±1⁎⁎ | 22±1 |
P≤0.0005 using student's t-test.
P≤0.05 using student's t-test.
P≤0.005 using student's t-test.
As metal stress results in oxidative stress through either the Fenton reaction or sequesteration. of LMW thiols, we also examined metal sensitivity. Both mutants were sensitive to Cd2+, Cu2+, SeO32−, TeO32−, and CrO42−, but only S24 was more sensitive to Fe2+, as compared to wildtype (Table 1). The sensitivity to oxidants, alkylating agents, and metals was reversed in R119C and partially reversed in S24C.
3.3. Protein carbonylation and lipid oxidation in mutants lacking MSH and ESH
Protein carbonylation, as an indicator for protein damage, was measured in S24 and R119, with and without hydrogen peroxide. S24 demonstrated significantly higher levels of protein carbonylation which was partially reversed in the complemented strain, S24C. In contrast, R119 had lower levels of protein carbonylation and the complemented strain, R119C, was not significantly different from the wildtype. S24 also had significantly higher levels of lipid peroxidation and R119 had lower lipid oxidation levels. The complemented strain, R119C, was not significantly different from wildtype (Table 2). Thus, lack of MSH results in increased damage while lack of ESH results in lesser damage to cellular proteins and lipids.
Table 2.
Protein carbonylation, lipid peroxidation, and peroxidase activity in M. smegmatis mutants and complemented strains. All values represent averages and SD (n=3). Student t-test was performed and the p values of mutants were compared with the wildtype.
| Strains | Protein carbonylation (μM L−1) |
Lipid oxidation (μM) |
Peroxidase activity (μM) |
||||
|---|---|---|---|---|---|---|---|
| Untreated | 5 mM H2O2 | Untreated | 5 mM H2O2 | CHP | H2O2 | TbOH | |
| WT | 15.4±2.3 | 26.9±4.6 | 50.4±4.3 | 75.0±2.9 | 20.1±0.4 | 16.1±0.3 | 17.3±1.1 |
| S24 | 36.2±3.9⁎⁎⁎ | 55.8±2.1⁎⁎⁎ | 90.0±1.9⁎⁎ | 102.4±2.4⁎⁎ | 35.0±1.9⁎⁎ | 33.0±0.7⁎⁎⁎ | 24.5±0.8⁎⁎ |
| S24C | 29.1±2.4⁎ | 40±3.1⁎ | 64.0±1.7 | 79.1±3.8 | 19.0±0.6 | 17.5±2.0 | 19.0±0.9 |
| R119 | 8.9±1.9⁎⁎ | 21.2±4.1⁎ | 43.3±1.3⁎ | 57.1±5.1⁎⁎ | 18.1±1.6 | 16.8±1.1 | 17.6±1.0 |
| R119C | 15.0±3.3 | 26.0±4.9 | 39.0±4.9 | 62.6±5.0 | 19.0±1.2 | 18.5±1.0 | 19.0±1.9 |
|
M. smegmatis strains |
|||||
|---|---|---|---|---|---|
| Wt | S24 | S24C | R119 | R119C | |
| Protein carbonylation (μM L−1) | |||||
| Untreated | 15.4±2.3 | 36.2±3.9⁎⁎⁎ | 29.1±2.4⁎ | 8.9±1.9⁎⁎ | 15.0±3.3 |
| 5 mM H2O | 26.9±4.6 | 55.8±2.1⁎⁎⁎ | 40±3.1⁎ | 21.2±4.1⁎ | 26.0±4.9 |
| Lipid peroxidation (μM) | |||||
| Untreated | 50.4±4.3 | 90.0±1.9⁎⁎ | 64.0±1.7 | 43.3±1.3⁎ | 39.0±4.9 |
| 5mM H2O2 | 75.0±2.9 | 102.4±2.4⁎⁎ | 79.1±3.8 | 57.1±5.1⁎⁎ | 62.6±5.0 |
| Peroxidase activity (μM) | |||||
| CHP | 20.1±0.4 | 35.0±1.9⁎⁎ | 19.0±0.6 | 18.1±1.6 | 19.0±1.2 |
| H2O2 | 16.1±0.3 | 33.0±0.7⁎⁎⁎ | 17.5±2.0 | 16.8±1.1 | 18.5±1.0 |
| TbOH | 17.3±1.1 | 24.5±0.8⁎⁎ | 19.0±0.9 | 17.6±1.0 | 19.0±1.9 |
P≤0.0005 using student's t-test.
P≤0.005 using student's t-test.
P≤0.05 using student's t-test.
3.4. Peroxidase activity in mutants lacking in MSH and ESH
To determine if MSH or ESH are involved as electron donors to a peroxidase(s), the decomposition of H2O2, CHP, and tBOH was measured using xylenol orange assay. The amount of peroxides was higher in S24 than wildtype (Table 2) indicating that MSH is able to either directly reduce the peroxides or act as an electron donor to a peroxidase. In contrast, the amount of peroxides in R119 was the same as wildtype (Table 2) indicating that ESH is not directly involved in the reduction of peroxides or another mechanism is induced in R119 that detoxifies peroxides. The peroxide levels in S24C were similar to wildtype.
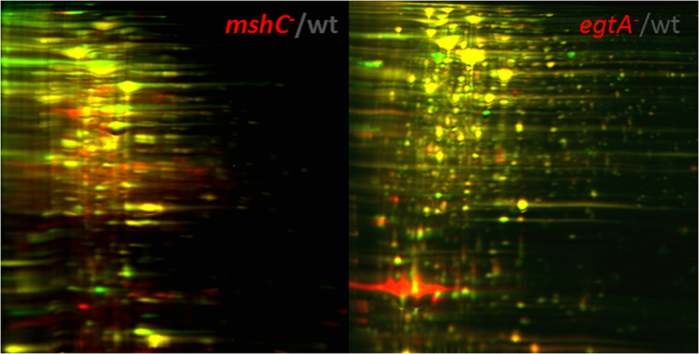
3.5. 2D-DIGE analysis of S24
Since standard SDS-PAGE analysis of transposon mutants disrupted in mshA and egtD demonstrated the overproduction of a band identified as Ohr [3], [7], 2D-DIGE was performed on wildtype and S24. In S24, 36 spots were clearly different between S24 and wildtype and of these 19 were conclusively identified (Table 3, Supp Fig. 1). A number of proteins associated with protection against oxidative stress are up-regulated in S24. One of these upregulated proteins is MSMEG_4283 (Spot 18), dihydrolipoamide acyltransferase (DlaT), which is the E2 component of pyruvate dehydrogenase complex, and contains a lipoyl binding site [14]. M. tuberculosis dlat mutant has severe growth defect in vitro, has reduced survival in bone marrow-derived macrophages from c57bl/6 and iNOS −/− mice, and is less able to grow in c57bl/6 mouse lungs, spleen and liver in vivo [15]. Together with lipoamide dehydrogenase (LpdC), AhpD, and AhpC, DlaT acts as an NADH dependent peroxidase and peroxynitrite reductase to provide protection against oxidative stress [14]. The importance of this dihydrolipoamide system is highlighted by the discovery of suppressor mutations in lipoamide dehydrogenase (lpdA) in E. coli. An E. coli mutant lacking both glutathione reductase and thioredoxin reductase is not viable but suppressor mutations in lpdA restore growth to this redox-defective mutant by causing the accumulation of dihydrolipoamide, which reduces oxidized glutaredoxins and circumvents the need for thioredoxin and glutathione in E. coli [16].
Table 3.
Differentially expressed proteins in wildtype and mshC mutant, S24.
| Spot | S24/wt | M. smegmatis | M. tuberculosis | Description |
|---|---|---|---|---|
| 18 | 8.12 | MSMEG_4283 | Rv2215 | SucB/DlaT dihydrolipoamide acyltransferase |
| 14 | 4.5 | MSMEG_4688 | Rv2466c | Thiol peroxidase |
| 11 | 8.8 | MSMEG_3084 | Rv1436 | Glyceraldehyde-3-phosphate dehydrogenase |
| 12 | −4.68 | |||
| 24 | 7.63 | MSMEG_2027 | Rv1558 | Coenzyme F420 dependent quinone reductase |
| 16 | 5.15 | MSMEG_1996 | Rv3079c | Putative Coenzyme F420 reductase |
| 23 | 5.98 | MSMEG_0415 MSMEG_2027 | Rv0245/ | NADH-FMN oxidoreductase |
| Rv1558 | ||||
| 19 | −8.36 | MSMEG_4362 | Rv2026c | Usp |
| 22 | −6.76 | MSMEG_3811 | Rv1636 | Usp |
| 27 | 42.55 | MSMEG_3811 | Rv1636 | |
| 28 | −8.08 | MSMEG_3811 | Rv1636 | |
| 29 | 5.28 | MSMEG_3811 | Rv1636 | |
| 13 | 5.13 | MSMEG_0880 | Rv0440 | GroEL chaperonin |
| 26 | 37.25 | MSMEG_6431 | Rv3849 | EspR |
| 30 | 4.83 | MSMEG_2426 | Rv2919c | GlnB nitrogen regulatory protein |
| 3 | −3.12 | MSMEG_4290 | Rv2220 | GlnA Glutamine synthetase. |
| 9 | 2.59 | MSMEG_4290 | ||
| 33 | 13.29 | MSMEI_2460 | Rv2890c | RpsB |
| 35 | 12.67 | MSMEG_1401 | Rv0685 | Tuf |
| 8 | 3.03 | MSMEG_1401 | ||
| 15 | 3.69 | MSMEG_5050 | No homolog | Map |
MSMEG_4688, also upregulated, codes for a putative thiol peroxidase and its M. tuberculosis ortholog, Rv2466c, has been shown to be involved in activation of TP053, a thienopyrimidine compound considered to be a leading drug target against tuberculosis [17]. Rv2466c is a member of the sigH regulon and is induced in the Wayne model of dormancy [18]. The physiological electron donor for this thiol peroxidase is unknown although it is able to reduce peroxides using DTT as an electron donor [17]. Interestingly, two of the proteins up-regulated include coenzyme F420 dependent proteins: MSMEG_1996 (Spot 16), a methyltetrahydromethanepterin reductase that reduces oxidized coenzyme F420, and MSMEG_2027 (Spot 24), a quinone reductase, which uses F420 as an electron donor or receptor to activate another new tuberculosis drug, PAS-824 [13]. Another indication that S24 is under oxidative stress is the upregulation of GAPDH, a marker of oxidative stress, which is present in two spots (11 and 12) that represent different post-translational modifications in the form of oxidation cysteine residues to sulfenic, sulfinic and sulfonic acid.
Several other proteins that are upregulated in S24 are associated with a general stress response. Two Usps (universal stress proteins), MSMEG_4362 (spot 19) and MSMEG_3811 (Spots 22, 27, 28, 29), are differentially expressed. MSMEG_3811 codes for an iron regulated single domain Usp and is the only Usp which has a homolog in M. leprae. The homolog in M. tuberculosis, Rv1636, is the only Usp along with kdpD, which is not up-regulated during macrophage infection [19]. Of the spots representing MSMEG_3811, spots 22 and 28 are up-regulated in the wildtype, and spots 27 and 29 are up-regulated in S24. A MASCOT search for phosphorylations revealed that the wildtype MSMEG_3811 isoforms are phosphorylated while the mshC isoforms are not.
Spot 13 is the 65 kDa heat shock protein, GroEl (MSMEG_0880), which prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions like high temperatures. Spot 26 is MSMEG_6431, the homolog of M. tuberculosis EspR (Rv3849), which regulates ESX-1, required for secretion and virulence in mice [20]. RpsB (Spot 33), 30s ribosomal protein S2, is involved in formation of the translation initiation complex and involved in the metabolism of ppGpp, the mediator of the stringent response that coordinates a variety of cellular activities in response to changes in nutritional abundance. EspR, Tuf (Spot 8, 35) and the Usp, MSMEG_3811, are regulated by iron levels, suggesting an interaction among iron levels, oxidative stress, and LMW thiols.
Also upregulated in the mutant is nitrogen regulatory protein PII (Spot 30), which becomes uridylylated to pII-UMP by GlnD when the ratio of glutamine to 2-ketoglutarate decreases. The pII-UMP allows the deadenylylation of glutamine synthetase activating the enzyme. During nitrogen excess, p-II is deuridylated and promotes the adenylation of glutamine synthetase, which was identified in two spots, 3 and 9.
3.6. 2D-DIGE analysis of R119
The DIGE protein profile of wildtype and R119 demonstrated that 98 proteins were differentially regulated (Table 4, Supp Fig. 2). Of these 17 spots had a egtA-/wt ratio greater than two and 18 spots had a egtA-/wt ratio less than −2. Ohr is the most upregulated protein and was present in four spots (Spots 88, 89, 90, 91). Oxidation of specific cysteines and perhaps mycothiolation of other cysteines may be responsible for these multiple spots of Ohr. In addition to Ohr, other proteins associated with protection against oxidative stress, AhpC (Spot 72), another thiol peroxidase, and KatN (Spot 48), a manganese containing catalase, are also up-regulated in R119. Another up-regulated protein, MSMEG_4272 (Spot 95), is a member of an iron-sulfur cluster biosynthesis protein family, which have been shown to be dependent on LMW thiols such as bacillithiol for assembly [21]. In addition, a MarR type regulator (Spot 82), upstream of a quinone reductase, is up-regulated.
Table 4.
Differentially expressed proteins in wildtype, egtA mutant, R119, and complemented strain, R119C.
| Spot | R119/WT | R119C/WT | M. smegmatis | M. tuberculosis | Description |
|---|---|---|---|---|---|
| 89 | 254.25 | 1.48 | MSMEG_0447 | – | Ohr, Organic hydroperoxide protein |
| 88 | 75.47 | 1.19 | MSMEG_0447 | ||
| 90 | 18.4 | 1.18 | MSMEG_0447 | ||
| 91 | 8.6 | −2.8 | MSMEG_0447 | ||
| 95 | 21.51 | 14.85 | MSMEG_4272 | Rv2204c | HesB/YadR/YfhF family protein/ iron sulfur proteins |
| 36 | 10.46 | 26.52 | MSMEG_3962 | – | Lactate-2-monooxygenase |
| 72 | 8.58 | 6.61 | MSMEG_4891 | Rv2428 | AhpC alkylhydroperoxide reductase |
| 82 | 5.36 | 6.82 | MSMEG_6361 | – | MarR, transcriptional regulator, upstream of quinone reductase |
| 78 | 3.62 | 6.47 | MSMEG_0965 | – | MspA, Chain A, Rim Domain Of Main Porin |
| 48 | 3.45 | 2.75 | MSMEG_6213 | – | KatN, manganese containing catalase |
| 52 | 3.34 | 1.81 | Aminoglycoside phosphotransferase-kanamycin resistance gene | ||
| 100 | 2.34 | 2.11 | MSMEG_1680 | – | Conserved hypothetical protein |
| 101 | −1.07 | −1.39 | MSMEG_3811 | Rv1636 | Universal stress protein family protein |
| 102 | −1.8 | −1.45 | MSMEG_4935 | Rv1311 | AtpC, F0F1 ATP synthase subunit epsilon |
| 10 | −1.87 | 1.19 | MSMEG_0880 | Rv0440 | GroEl |
| 59 | −3.24 | −1.45 | MSMEG_2601 (pcaH)/MSMEG_2398 (tesB) | – | Protocatechuate 3,4-dioxygenase/acyl-CoA thioesterase II |
| 40 | −4.66 | −2.12 | MSMEG_2374 | Rv3001c | IlvC, Ketol-acid isomerase |
| 15 | −5.44 | −3.85 | MSMEG_4303 | – | Methyltransferase |
| 19 | −5.7 | −2.71 | MSMEG_5412 | Rv1926c | Immunogenic protein MPT63 |
| 33 | −6.33 | −4.28 | MSMEG_1682 | – | Flavin-containing monooxygenase FMO |
| 44 | −7.76 | −6.65 | MSMEG_2079 | – | Alcohol dehydrogenase, Yhd/YhfP family |
| 22 | −8.24 | −9.5 | MSMEG_6759 | Rv3696c | GlpK, Glycerol kinase |
Proteins associated with metabolism are differentially expressed in R119 and wildtype. Lactate monooxygenase (Spot 36), which catalyzes the conversion of lactate to acetate, is upregulated while glycerol kinase (Spot 22), which catalyzes the transfer of a phosphate from ATP to glycerol dihydroxyacetone, l-glyceraldehyde and d-glyceraldehyde during carbohydrate metabolism, is the most downregulated protein. IlvC (Spot 40) associated with isoleucine and valine metabolism is also downregulated. Other downregulated proteins include an alcohol dehydrogenase (Spot 44), belonging to family of quinone reductases, a flavin-containing monooxygenase (Spot 33), and MPT63 (Spot 19), an immunogenic protein which has been implicated in virulence.
In contrast to S24, the Usp, MSMEG_3811, is down-regulated in R119. GroEl, another general stress protein, is also down-regulated in R119 but up-regulated in S24. Other protein markers for stress seen in S24 are not up-regulated in R119 and R119, thus, does not appear to be in a “stressed” state. 2D-DIGE of R119C demonstrated partial complementation by the M. tuberculosis egtA gene. In particular, the R119C/WT ratios for spots representing Ohr are significantly lower as compared to R119/WT. Although other proteins are differentially expressed in both R119 and R119C, the R119C/WT ratios are less than R119/WT ratios. As other phenotypic data for S24C demonstrated that this strain was partially complemented by the M. tuberculosis mshC, 2D-DIGE was not performed.
4. Discussion
Herein, we confirm that both MSH and ESH protect mycobacterial cells against a number of stresses, including oxidative stress and alkylating stress, and demonstrate that these two LMW thiols are also involved in protection against metal stress. S24 is extremely sensitive to oxidants and alkylating agents, as previously reported for other mutants lacking MSH [6], while R119 is less sensitive (Table 1). Similar to the M. smegmatis mutant disrupted in egtD, which is more susceptible to the lipid peroxides, CHP and tert-butyl hydroperoxide (tBOH), R119 is sensitive to CHP [3]. In contrast to the Streptomyces coelicolor egtA mutant, R119 is not sensitive to hydrogen peroxide [22].
Both mutants are also sensitive to metal stress. In vitro studies had previously demonstrated that ESH is able to scavenge free radicals and metal ions. In particular, ESH is known to form complexes with copper [23] in vitro but in vivo, copper sulfate had no effect on conidial germination or hyphal growth in a mutant of Neurospora crassa lacking ESH [24]. Other LMW thiols like GSH are known to protect against cadmium toxicity in E. coli [25] and mutants disrupted in bacillithiol, which is structurally similar to MSH, are sensitive to metal stress [26].
Recently, Servillo et al. [27] reported that oxidation of ESH with hypochlorite, peroxynitrite and hydrogen peroxide resulted in the sulfonated form (ESO3H), and hercynine, the desulfurated form of ESH, indicating that ESH, like cysteine and MSH, is oxidized irreversibly during oxidative stress. Moreover, Sao-Emani et al. (2013) reported that a double mutant lacking both ESH and MSH is significantly more sensitive to the peroxides than either of the single mutants lacking either ESH or MSH. Taken together, these results suggest that MSH and ESH may be able to partly compensate for the loss of the other thiol. However, as the levels of ESH are 25–50 fold lower than MSH in M. smegmatis, ESH is less likely to be able to compensate for MSH. In the mshA mutant, which lacks MSH, ESH levels are substantially higher but that is not the case in the mshC mutant. Instead, the levels of Coenzyme F420 are higher in S24. As disruption of mshA and eshD results in a dramatic overproduction of organic hydroperoxide resistance protein, Ohr [3], [7], we sought to establish if the same was true for mutants disrupted in mshC (S24) and eshA (R119). In S24, there is no upregulation of Ohr; however, the upregulation of DlaT which is part of the system that provides reducing equivalents to the thiol peroxidases, AhpC and Ohr, and the upregulation of the M. smegmatis homolog of the putative thiol peroxidase, Rv2466c, implies that there is a major increase in oxidative stress in the mshC mutant. Upregulation of an enzyme that likely serves as a F420-dependent quinone reductase, preventing the formation of cytotoxic semiquinones, and upregulation of a putative F420-dependent reductase implies that coenzyme F420, which has been shown to be involved in protection against oxidative stress [12], may be able to partially substitute for MSH in the mycobacterial cell.
Interestingly, multiple spots representing Usp (MSMEG_3811) are present in the 2D- DIGE. These multiple spots are a result of different amounts of phosphorylation of MSMEG_3811. Intriguingly, Usp is not phosphorylated in S24, implying a cross-talk between MSH and universal stress proteins. Previously, we observed phosphorylation of another Usp, MSMEG_3940, during stationary phase in wildtype M. smegmatis using a stain for phosphoproteins (data not shown). Phosphorylation of Usps has also been demonstrated in Arabidopsis challenged with biotic stress [28] although how this signal is translated into a protective response is not clear. Lack of MSH alters the redox environment of the cell which may cause oxidative modifications of kinases, either inhibiting or activating them [29]. The resulting phosphorylation/dephosphorylation events would positively regulate some and negatively regulate other signal transduction pathways [29]. In M. smegmatis wildtype, there would be presumably be less mycothiolation of proteins, including that of a kinase that phosphorylates Usp or perhaps less mycothiolation of MSMEG_3811 directly. This could result in phosphorylation or autophosphorylation of MSMEG_3811, as in E. coli UspG [31]. In S24, mycothiolation of the kinase would reduce activity and less phosphorylation or auto-phosphorylation. It is not clear what the exact functions of Usps are but it is likely that post-translation modifications of MSMEG_3811 in S24 has far-reaching consequences involving changes in metabolism and growth arrest.
Despite upregulation of genes involved in the stress response, S24 has increased protein carbonylation and lipid peroxidation. Paradoxically, R119 has less protein carbonylation and lipid oxidation than wildtype when cell lysates are treated with inorganic and organic peroxides. This is in contrast to HeLA cells, where disruption of the ESH transporter, ETT, results in an increase in protein and lipid damage [32] and ETT knockout fish, where there is an increase in levels of lipid peroxidation markers after incubation with Pb2+ or Cu2+ and an approximately four fold increase in DNA lesions in the skin [33]. The decreases in protein carbonylation, lipid peroxidation, and wildtype levels of peroxides in R119 can be explained by the dramatic upregulation of Ohr and other enzymes involved in reduction of oxidants, such as the manganese dependent catalase, KatN, and AhpC. Intriguingly, neither Ohr nor KatN are present in M. tuberculosis although there is a paralog of Ohr, OsmC (Rv2923c). Recombinant OsmC from both M. smegmatis (MSMEG_2421) and M. tuberculosis are able to reduce H2O2, CHP, and tBOH [34]. Multiple species of Ohr are present in R119, probably as a result of mycothiolation or oxidation of exposed cysteine residues resulting in the formation of sulfenic/sulfinic/sulfonic acids. Chi et al., 2013 [30] demonstrated that a number of proteins are mycothiolated, including the antioxidant enzymes Tpx, Gpx, and MsrA, but not Ohr, in MSH containing Corynebacterium glutamicum [30], although OhrR, the Ohr regulator, is known to be bacillithiolated in Bacillus subtilis [35]. Whether “ergothiolation” serves a similar protective role in mycobacteria remains to be seen.
5. Conclusions
Disruption of genes involved in biosynthesis of either MSH and ESH leads to up-regulation of genes involved in oxidative stress, albeit the genes that are induced are different in the two strains. The M. smegmatis MSH mutant displays protein and lipid damage despite the up-regulation of enzymes involved in protection against oxidative stress, and elevation of CoenzymeF420 levels. In contrast, the ESH mutant demonstrates less protein carbonylation and lipid peroxidation than wildtype, likely due to the up-regulation of proteins that specifically protect against oxidative stress, such as Ohr and KatN. Another key difference is result is the up-regulation of general stress proteins, GroEl and an Usp, which appears to be unphosphorylated in the MSH mutant. The lack of phosphorylation of the Usp protein in the MSH mutant suggests crosstalk between redox signaling and phosphorylation/dephosphorylation pathways in mycobacteria. Future studies are needed to determine the interplay of these two post-translational modifications in the signaling networks of mycobacteria.
Acknowledgments
This study was supported by National Institutes of Health award 5SC3GM100855-03 and CSU Program for Education and Research in Biotechnology Faculty-Student Collaborative Research award to M.R. We thank Alisha Ramlan Hussain, Kathryn Barretto, and Jason Thomas for technical assistance. E.P. was supported by award 1R21AI100039 from the National Institutes of Health.
Footnotes
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.08.006.
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.08.006.
Appendix A. Transparency document
Supplementary material
.
Appendix B. Transparency document
Supplementary material
.
Supplementary material
.
References
- 1.R.C. Fahey, Glutathione analogs in prokaryotes, Biochimica et Biophysica Acta (BBA)-General Subjects, 1830, 2013, pp. 3182–3198. [DOI] [PubMed]
- 2.Newton G.L., Buchmeier N., Fahey R.C. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sao Emani C., Williams M.J., Wiid I.J., Hiten N.F., Viljoen A.J., Pietersen R.-D.D., van Helden P.D., Baker B. Ergothioneine is a secreted antioxidant in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 2013;57:3202–3207. doi: 10.1128/AAC.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seebeck F.P. In vitro reconstitution of mycobacterial ergothioneine biosynthesis. J. Am. Chem. Soc. 2010;132:6632–6633. doi: 10.1021/ja101721e. [DOI] [PubMed] [Google Scholar]
- 5.Hand C.E., Taylor N.J., Honek J.F. Ab initio studies of the properties of intracellular thiols ergothioneine and ovothiol. Bioorg. Med. Chem. Lett. 2005;15:1357–1360. doi: 10.1016/j.bmcl.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Rawat M., Johnson C., Cadiz V., Av-Gay Y. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 2007;363:71–76. doi: 10.1016/j.bbrc.2007.08.142. [DOI] [PubMed] [Google Scholar]
- 7.Ta P., Buchmeier N., Newton G.L., Rawat M., Fahey R.C. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J. Bacteriol. 2011;193:1981–1990. doi: 10.1128/JB.01402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawat M., Heys J., Av-Gay Y. Identification and characterization of a diamide sensitive mutant of Mycobacterium smegmatis FEMS. Microbiology. 2002;220:161–169. doi: 10.1016/S0378-1097(03)00127-7. [DOI] [PubMed] [Google Scholar]
- 9.Choi K.P., Kendrick N., Daniels L. Demonstration that fbiC is required by Mycobacterium bovis BCG for Coenzyme F420 and FO Biosynthesis. J. Bacteriol. 2002;184:2420–2428. doi: 10.1128/JB.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawat M., Newton G.L., Ko M., Martinez G.J., Fahey R.C., Av-Gay Y. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chromother. 2002;46:3348–3355. doi: 10.1128/AAC.46.11.3348-3355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selengut J.D., Haft D.H. Unexpected abundance of coenzyme F420-dependent enzymes in Mycobacterium tuberculosis and other actinobacteria. J. Bacteriol. 2010;192:5788–5798. doi: 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra-Lopez D., Daniels L., Rawat M. Mycobacterium smegmatis mc2 155 fbiC and MSMEG_2392 are involved in triphenylmethane dye decolorization and coenzyme F420 biosynthesis. Microbiology. 2007;153:2724–2732. doi: 10.1099/mic.0.2006/009241-0. [DOI] [PubMed] [Google Scholar]
- 13.Gurumurthy M., Rao M., Mukherjee T., Rao S.P.S., Boshoff H.I., Dick T., Barry C.E., Manjunatha U.H. A novel F420–dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol. Microbiol. 2012 doi: 10.1111/mmi.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryk R., Lima C.D., Erdjument-Bromage H., Tempst P., Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002 doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 15.Shi S., Ehrt S. Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect. Immun. 2006;74:56–63. doi: 10.1128/IAI.74.1.56-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feeney M.A., Veeravalli K., Boyd D., Gon S., Faulkner M.J., Georgiou G., Beckwith J. Repurposing lipoic acid changes electron flow in two important metabolic pathways of Escherichia coli. Proc. Natl. Acad. Sci. 2011;108:7991–7996. doi: 10.1073/pnas.1105429108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albesa-Jove D., Chiarelli L.R., Makarov V., Pasca M.R., Urresti S., Mori G., Salina E., Vocat A., Comino N., Mohorko E. Rv2466c mediates the activation of TP053 To kill replicating and non-replicating Mycobacterium tuberculosis. ACS Chem. Biol. 2014;9:1567–1575. doi: 10.1021/cb500149m. [DOI] [PubMed] [Google Scholar]
- 18.Rao S.P.S., Camacho L., Huat Tan B., Boon C., Russel D.G., Dick T., Pethe K. Recombinase-based reporter system and antisense technology to study gene expression and essentiality in hypoxic nonreplicating mycobacteria. FEMS Microbiol. Lett. 2008;284:68–75. doi: 10.1111/j.1574-6968.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 19.Schnappinger D., Ehrt S., Voskuil M.I., Liu Y., Mangan J.A., Monahan I.M., Dolganov G., Efron B., Butcher P.D., Nathan C., Schoolnik G.K. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal Environment. J. Exp. Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghavan S., Manzanillo P., Chan K., Dovey C., Cox J.S. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosario-Cruz Z., Chahal H.K., Mike L.A., Skaar E.P., Boyd J.M. Bacillithiol has a role in Fe–S cluster biogenesis in Staphylococcus aureus. Mol. Microbiol. 2015;98:218–242. doi: 10.1111/mmi.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Nakajima, Y. Satoh, K. Yanashima, T. Matsui, T. Dairi, Ergothioneine protects Streptomyces coelicolor A3(2) from oxidative stresses, J. Biosci. Bioeng. [DOI] [PubMed]
- 23.Zhu B.-Z., Mao L., Fan R.-M., Zhu J.-G., Zhang Y.-N., Wang J., Kalyanaraman B., Frei B. Ergothioneine prevents copper-induced oxidative damage to DNA and protein by forming a redox-inactive ergothioneine–copper complex. Chem. Res. Toxicol. 2010;24:30–34. doi: 10.1021/tx100214t. [DOI] [PubMed] [Google Scholar]
- 24.Bello M.H., Barrera-Perez V., Morin D., Epstein L. The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 2011 doi: 10.1016/j.fgb.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Helbig K., Grosse C., Nies D.H. Cadmium toxicity in glutathione mutants of Escherichia coli. J. Bacteriol. 2008;190:5439–5454. doi: 10.1128/JB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajkarnikar A., Strankman A., Duran S., Vargas D., Roberts A.A., Barretto K., Upton H., Hamilton C.J., Rawat M. Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2013;436:128–133. doi: 10.1016/j.bbrc.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servillo L., Castaldo D., Casale R., D’Onofrio N., Giovane A., Cautela D., Balestrieri M.L. An uncommon redox behavior sheds light on the cellular antioxidant properties of ergothioneine. Free Radic. Biol. Med. 2015;79:228–236. doi: 10.1016/j.freeradbiomed.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Lenman M., Sörensson C., Andreasson E. Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated arabidopsis. Mol. Plant-Microbe Interact. 2008;21:1275–1284. doi: 10.1094/MPMI-21-10-1275. [DOI] [PubMed] [Google Scholar]
- 29.Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi B.K., Busche T., Van Laer K., Bäsell K., Becher D., Clermont L., Seibold G.M., Persicke M., Kalinowski J., Messens J. Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid. Redox Signal. 2014;20:589–605. doi: 10.1089/ars.2013.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber A., Jung K. Biochemical properties of UspG, a universal stress protein of Escherichia coli. Biochemistry. 2006;45:1620–1628. doi: 10.1021/bi051301u. [DOI] [PubMed] [Google Scholar]
- 32.Paul B.D., Snyder S.H. The unusual amino acid l-ergothioneine is a physiologic cytoprotectant. Cell Death Differ. 2010;17:1134–1140. doi: 10.1038/cdd.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer C., Bach M., Bauer T., da Ponte J.C., Schömig E., Gründemann D. Knockout of the ergothioneine transporter ETT in zebrafish results in increased 8-oxoguanine levels. Free Radic. Biol. Med. 2015 doi: 10.1016/j.freeradbiomed.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 34.Saikolappan S., Das K., Sasindran S.J., Jagannath C., Dhandayuthapani S. OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis. 2011;91:S119–S127. doi: 10.1016/j.tube.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J.-W., Soonsanga S., Helmann J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material