Abstract
Propose of this review
The objective of this review article is to summarize the recent findings about the importance of microRNAs (miRNAs) in regulating lipoprotein metabolism. We highlight the recent findings that uncover the importance of miRNAs in controlling plasma LDL-cholesterol (LDL-C) levels.
Recent Findings
In 2013, several studies reported a number of miRNAs that regulate plasma LDL-C levels, including miR-30c. In this review article, we discuss those miRNAs that modulate LDL-cholesterol (LDL-C) levels and lipoprotein secretion. We also discuss the numerous studies that demonstrate the critical role of miRNAs in governing the many facets of high-density lipoprotein (HDL) metabolism, such as the ATP transporters, ABCA1 and ABCG1, and the scavenger receptor, SRB1.
Summary
The understanding of how these miRNAs modulate lipoprotein metabolism promises to reveal new therapeutic targets to treat dyslipidemias and related cardiovascular disorders.
Keywords: miRNAs, lipoprotein metabolism, cholesterol homeostasis
INTRODUCTION
In recent years, prominent roles for microRNAs (miRNAs) have been uncovered in regulating almost all physiological processes, including lipid metabolism [1–3]. miRNAs are small (~22 nucleotides) endogenous double-stranded non-coding RNAs that regulate gene expression at the post-transcriptional level [4–7]. These tiny RNAs bind to the 3’-untranslated region (3’UTR) of mRNAs causing RNA destabilization or translational repression [5, 6]. A single miRNA targets multiple genes, thereby controlling numerous components of complex intracellular networks. In this review article, we discuss recent advances in our understanding of the role of miRNAs in controlling cholesterol and lipid homeostasis, with particular emphasis on those miRNAs that regulate lipoprotein metabolism. Specifically we highlight those miRNAs that influence LDL-cholesterol (LDL-C) levels, such as the well-characterized liver-specific miRNA, miR-122, and the role of miR-30c in controlling lipoprotein secretion [8–10]. Several miRNAs (i.e. miR-96/182/183 and miR-185) that affect LDL metabolism by controlling the sterol response element binding protein-2 (SREBP2) expression are also discussed [11, 12]. In addition, we review those miRNAs that influence HDL-cholesterol (HDL-C) levels by targeting the ATP transporter, ABCA1, such as miR-33a/b and miR-144 [13–17].
MIRNAS AND LDL METABOLISM
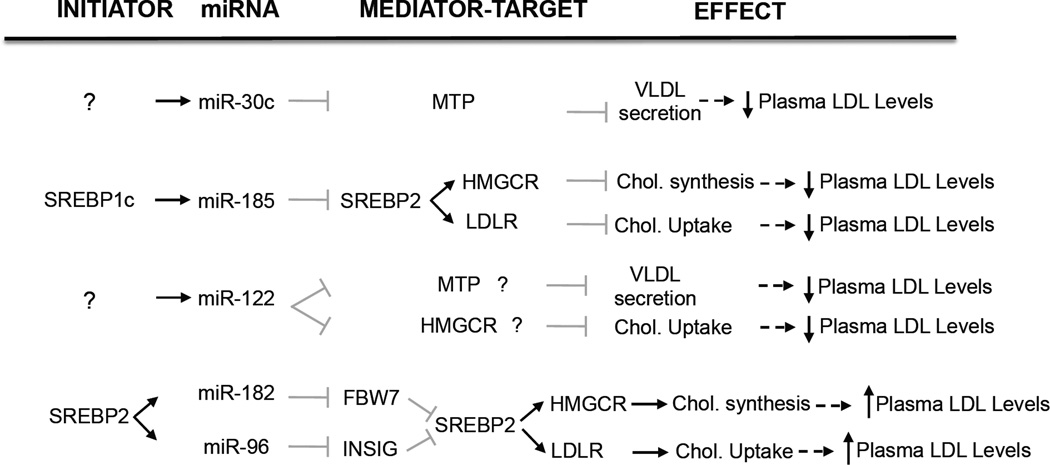
A number of studies have recently reported the importance of miRNAs such as miR-122, miR-30c, miR-185, miR-96 and miR-182 in controlling LDL metabolism by regulating very-low density lipoprotein (VLDL) secretion, cholesterol biosynthesis and LDL receptor (LDLR) activity (Figure 1).
Figure 1. miRNAs that regulate LDL metabolism.
The figure represents the initiator, miRNA target genes and the overall effect of miRNAs that control LDL metabolism. ? indicates unknown initiator or miRNA target gene.
miR-122
Relatively little is known about how miRNAs modulate LDL-C levels. To date, only miR-122 has been shown to play a direct role in LDL cholesterol metabolism [8, 9, 18]. miR-122 is the most abundantly expressed miRNA in the liver and one of the first miRNAs to be linked to hepatic function. Specifically, several reports have highlighted the role of miR-122 in regulating cholesterol and fatty acid metabolism, namely through loss-of-function experiments. Early studies showed that antisense targeting of hepatic miR-122 in mice resulted in significant reductions in total plasma cholesterol, which reflected decreases in both HDL-C and LDL-C [19]. In a separate study, antisense silencing of miR-122 led to decreased hepatic cholesterol and fatty acid biosynthesis and increased fatty-acid oxidation. This was associated with reduced circulating total cholesterol and triglycerides, as well as decreased hepatic steatosis in mice fed a high-fat diet [9]. In subsequent studies, efficient silencing of miR-122 was successfully achieved in the liver of non-human primates, which resulted in reduced plasma cholesterol without any signs of toxicity [8]. However, unlike in mice, these changes were primarily associated with LDL-C. Given the success of anti-miR-122 therapy in non-human primate models, miR-122 has become a promising therapeutic target in humans. Despite this, the development of anti-miR-122 therapies to treat hypercholesteremia has diminished, as further studies have associated anti-miR-122 therapy with adverse effects, such as decreased HDL-C and hepatocellular carcinoma [9, 19]. Additionally, the mechanism by which miR-122 regulates lipid metabolism remains unclear, as direct targets of miR-122 associated with cholesterol homeostasis have yet to be identified (Figure 1). Further studies in mice and non-human primates are needed to gain a better mechanistic understanding of the role of miR-122 in directly regulating cholesterol and lipid metabolism.
miR-30c
In addition to miR-122, miR-30c has also recently been associated with lipid metabolism [10]. miR-30c was the first miRNA shown to play a role in lipoprotein assembly. It is expressed primarily in the heart, skeletal muscle, and kidney and resides in intron 5 of the human nuclear transcription factor Y subunit C (NFYC) gene. Interestingly, NFYC cooperates with SREBP transcription factors in regulating the expression of genes associated with lipid metabolism [20, 21]. Using computational approaches, Soh et al. identified miR-30c as a miRNA that affects lipid metabolism by regulating microsomal triglyeride transfer protein (MTP), a gene crucial for the assembly of ApoB-containing lipoproteins. Overexpression and inhibition of miR-30c in human hepatic cells reduced and increased MTP levels, activity, and ApoB secretion respectively [10]. Accordingly, lentiviral delivery of miR-30c to Western-diet fed mice reduced plasma cholesterol levels by decreasing lipid synthesis and secretion of triglyceride-rich ApoB lipoproteins with no effect on hepatic steatosis [10]. Moreover, inhibition of miR-30c increased hyperlipidemia and atherosclerosis in mice, consistent with a physiological role for miR-30c in controlling lipoprotein assembly. Taken together, this study suggests that miR-30c mimetics may be useful to treat hyperlipidemia and other related cardiovascular disorders in humans.
miRNA regulation of cholesterol homeostasis
The hepatic low-density lipoprotein receptor (LDLR) is essential for clearing circulating LDL and is an important therapeutic target for treating cardiovascular disease [22]. As such, the intracellular and membrane levels of the LDLR and other lipid regulators are highly controlled by a series of feedback mechanisms that operate at the transcriptional and post-transcriptional level. One of the best-characterized transcriptional regulators of the LDLR is the ER-bound SREBPs. SREBPs bind to the SREBP cleavage-activating protein (SCAP), a transmembrane protein that serves as an escort protein and sterol sensor. In mammals, the SCAP/SREBP exit from the ER to the Golgi for activation is regulated by the interaction between SCAP and the insulin-induced gene (INSIG) [20, 21, 23–25]. When sterol concentrations are low, the INSIG-SCAP/SREBP interaction is disrupted, allowing the trafficking of the SCAP/SREBP complex to the Golgi, where SREBP is cleaved and activated [24, 25]. The N-terminal fragment translocates to the nucleus and activates the transcription of genes involved in cholesterol biosynthesis and uptake including 3-hydroxy-3methylglutaryl coenzyme A reductase (HMGCR) and LDLR, respectively [21]. SREBP stability is regulated in the nucleus by the E3 ubiquitin ligase, FBXW7 that targets nuclear SREBPs for proteosomal degradation, thus inhibiting LDLR transcriptional activation [26].
Additionally, the LDLR is also subject to post-transcriptional regulation such as its proprotein convertase sutilisin/kexin type 9 (PCSK9)- dependent degradation and inducible degrader of idol (IDOL)-dependent ubiquitination [27–29]. Despite these advances, to date, no studies have shown a role for miRNAs in regulating LDLR expression directly. The miRNA-dependent regulation of SREBP2, however, has recently been explored and represents another layer of regulatory control of LDLR gene expression. In particular, two recent studies have highlighted the role of miR-185 and the miR-96/182/183 locus in post-transcriptionally controlling SREBP2 expression [11, 12].
miR-96/182/183 locus
To identify hepatic miRNAs that are regulated by cholesterol levels, Osborne and colleagues treated mice with statins (to inhibit cholesterol biosynthesis) and ezetimibe (to inhibit dietary cholesterol absorption). They found that the miR-96/182/183 miRNA cluster was significantly upregulated in the livers of mice treated with statins plus ezetimibe compared with untreated mice [11]. The miR-96/182/183 primary transcript is directly regulated by SREBP2 and its processing yields three mature miRNAs (miR-96, miR-182 and miR-183). Interestingly, miR-96 inhibits INSIG2 expression and miR-182 targets FBXW7 [11]. Both proteins regulate the levels and activation of SREBP2 by controlling the processing (INSIG2) and stability (FBXW7) of this transcription factor. Overexpression of miR-96 and miR-182 increases SREBP2 transcriptional activity and lipid synthesis in culture cells [11]. However, antagonism of miR-182 in mice did not influence hepatic cholesterol and triglyceride content or plasma cholesterol and triglyceride levels. These findings suggest that under cholesterol depletion conditions SREBP2 activates its own transcription and at the same time regulates the expression of miRNAs that inhibits negative regulators of its activity. This elegant feedback loop of SREBP2/miR-96/miR-182 regulation illustrates a novel mechanism by which cholesterol metabolism is tightly regulated at the organismal and cellular level.
miR-185
In addition to the miR-96/182/183 cluster, it has recently been shown that miR-185 is also regulated by SREBP transcription factors. The miR-185 primary transcript is regulated by SREBP1c and its expression is elevated in the livers of mice fed a high fat diet (HFD) compared with mice fed a chow diet [12]. miR-185 targets SREBP2, thereby inhibiting de novo cholesterol biosynthesis and LDL uptake. miR-185 overexpression in human hepatic cell lines (HepG2) results in significant decreases in HMGCR activity and LDL internalization. Notably, mice infected with lentiviral constructs encoding the primary miR-185 sequence showed markedly reduced expression of SREBP2 regulated genes including LDLR, HMGCR and PCSK9 compared with mice injected with a control lentivirus [12]. Even though these data strongly suggest that miR-185 regulates lipid metabolism in vivo, additional experiments using miR-185 inhibitors will be necessary to define the role of hepatic miR-185 in controlling cholesterol metabolism. These results uncover a novel feedback loop by which the SREBP1c-induced miR-185 regulates SREBP2 expression, thus controlling cellular cholesterol metabolism.
MIRNAS AND HDL METABOLISM
Whereas the contribution of miRNA regulation to LDL metabolism has been less well-studied, the body of literature describing the role of miRNAs in governing HDL homeostasis is quite extensive. Indeed, work over the past years has demonstrated that miRNAs control the expression of many genes associated with HDL metabolism, including those that regulate HDL biogenesis, cellular cholesterol efflux, HDL-C uptake in the liver, and bile acid transport [1–3] (Figure 2). In particular, these findings reveal how single miRNAs (i.e. miR-33) can target multiple components of the reverse cholesterol transport (RCT) pathway and how key genes involved in HDL metabolism are under the control of many miRNAs (i.e ABCA1). They also demonstrate the potential of miRNA-based therapeutics to raise HDL-C, which is inversely associated with the development of atherosclerosis and its downstream complications [1–3].
Figure 2. miRNAs that regulate HDL metabolism.
The figure represents the initiator, miRNA target genes and the overall effect of miRNAs that control HDL metabolism.
miRNA regulation of ABCA1
ABCA1 and ABCG1 are sterol-sensitive genes that are transcriptionally controlled by the liver X receptors (LXRs) [30]. These nuclear hormone receptors form heterodimers with the retinoic-X-receptors (RXRs), leading to the transcriptional activation of both ABC transporters. Under cellular cholesterol excess, ABCA1 and ABCG1 facilitate the efflux of cholesterol from macrophages by lipidating nascent and mature HDL to generate larger HDL particles destined for clearance by the liver. Hepatic ABCA1 also plays a critical role in the biogenesis of HDL particles [31–33]. Given its importance in regulating HDL-C, many groups have studied the regulation of this transporter. In particular, several groups have shown that ABCA1 expression is highly regulated at the post-transcriptional level by multiple miRNAs, including miR-33, miR-144, miR-758, miR-26, miR-106b, miR-27, miR-145, miR-33*, and miR-10b [13–17, 34–40] (Figure 2). These miRNAs have been shown to repress ABCA1 expression and function in a variety of different cell types, including macrophages, neurons, pancreatic beta-cells, enterocytes, and hepatocytes. As the relevant importance of the aforementioned miRNAs in regulating ABCA1 expression is likely to be dictated by physiological stimuli and the tissue-specific expression of each miRNA, further studies in vivo are warranted to understand the miRNA-dependent regulation of ABCA1. Nonetheless, work over the past years has highlighted the importance of two miRNAs, miR-33 and miR-144, in regulating ABCA1 expression and HDL levels in mice and non-human primates [13–17, 41, 42].
miR-33 was the first miRNA described to regulate hepatic ABCA1 expression and plasma HDL-C in vivo. miR-33a/b are lipid-responsive miRNAs embedded within the introns of the SREBP2 and SREBP1 genes, respectively, and are co-transcribed along with their host genes under conditions that increase SREBP activation. miR-33 represses the expression of genes involved in cholesterol export and HDL biogenesis, including ABCA1 and ABCG1 [14, 15, 17]. Since the first reports describing the role of the miR-33 family in modulating ABCA1 expression, multiple groups have gone on to assess the efficacy of anti-miR-33 therapy to increase circulating HDL-C levels [14, 15, 17]. Inhibition of miR-33 was shown to induce the expression of hepatic ABCA1, where it increased plasma HDL, but also in macrophages, where it enhanced RCT and the regression of atherosclerosis. Genetic deletion of miR33 also resulted in sustained increases in HDL, as well as decreased atherosclerosis progression in ApoE−/− mice [43]. Intriguingly, anti-miR-33 therapy has also been shown to increase ABCA1 expression and plasma HDL-C levels in two separate studies using non-human primates [41, 42]. Despite these promising data, a recent study has demonstrated that miR-33 null mice develop obesity and dyslipidemia when fed a high fat diet [44]
In addition to miR-33, two different groups recently showed that miR-144 modulates ABCA1 expression in hepatocytes and macrophages, as well as cellular cholesterol efflux [13, 16]. While the two studies report that miR-144 expression is transcriptionally induced by different nuclear receptors (LXR or FXR), both studies show that changes in hepatic miR-144 influence hepatic ABCA1 expression and circulating HDL in vivo. Taken together, these studies highlight the physiologic importance of miR-144 in regulating plasma levels of HDL.
Aside from directly binding to the 3’UTR of ABCA1 and decreasing its expression, miRNAs have also been shown to modulate ABCA1 levels by post-transcriptionally regulating LXR, a transcriptional activator of ABCA1 expression. miR-1, miR-206, and miR-155 have been shown to directly target LXR in vitro [45, 46]. In particular, miR-1 and miR-206 suppress lipogenesis by inhibiting LXRα and its target genes, SREBP1, acetyl-CoA (ACC), and fatty acid synthase (FAS). Interestingly, miR-26 is inhibited in cells treated with LXR agonists, suggesting that miR-26 may cooperate with LXR to transcriptionally activate ABCA1 [47]. RXRα is also regulated by miRNAs, including miR-128-2 and miR-24 [34]. Both miRNAs have been reported to inhibit RXRα expression, thus inhibiting LXR-induced ABCA1 expression. Overall these data highlight the complex transcriptional and post-transcriptional network that governs the expression of ABCA1.
miRNAs targeting SRB1
SRB1 mediates the final step of RCT by delivering HDL-C to the liver for bile acid synthesis and cholesterol excretion. Its important contribution to the removal of plasma cholesterol is evidenced by loss-of-function studies in mice, which demonstrate that absence of SRB1 impairs RCT and results in massive atherosclerosis. While the transcriptional activation of SRB1 is well-characterized, recent in silico analyses have shed light on the importance of SRB1 regulation by miRNAs. In this regard, miR-455, miR-125a, miR-185, miR-96, and miR-223 have been shown to directly bind to the 3’UTR of SRB1 and suppress its expression [48, 49]. Accordingly, overexpression of these miRNAs reduces SRB1 protein levels and HDL-C uptake in vitro, while antagonism of miR-455, miR-125a, miR-185, miR-96, and miR-233 have the opposite effect. Interestingly, the levels of miR-96 and miR-185 inversely correlate with SRB1 in the livers of high-fat-diet-fed ApoE−/− mice [49]. Furthermore, miR-185 has also been reported to negatively regulate the expression of SREBP2, suggesting a role for this miRNA in also controlling LDL-C through the regulation of the SREBP2-dependent gene, LDLR [12]. Indeed, future in vivo studies are needed to clarify the role of this miRNA and others in regulating SRB1 expression, HDL uptake, and RCT.
CONCLUSIONS
High levels of plasma LDL-C and reduced levels of HDL-C are associated with increased coronary artery disease (CAD) risk. As a result, statins have been widely used for lowering circulating LDL-C. Despite this, statins are not efficient in a significant number of individuals and secondary effects, including Rhabdomyolysis have been associated with their consumption. Most of the drugs used to increase plasma HDL-C levels, such as CETP inhibitors, have failed reduce CAD risk. The recent findings that manipulating miRNA expression might reduce circulating LDL-C or increase plasma HDL-C levels open new avenues for the treatment of dyslipidemias. Indeed, work over the past few years has demonstrated that the inhibition of miR-33 results in a significant increase of plasma HDL-C in mouse and non-human primates and protection against the progression of atherosclerosis in atheroprone-mouse models. Moreover, a recent report from Hussain’s group demonstrated that prolonged delivery of miR-30c mimics markedly reduces plasma LDL-C levels and atherogenesis in mice. These studies suggest that manipulating miRNA expression in vivo may be a useful therapeutic strategy for treating dyslipidemia and related cardiovascular disorders.
KEY POINTS.
miRNAs regulate lipoprotein metabolism.
Dyslipidemia is a major cause of atherosclerotic vascular disease.
Anti-miR-33 therapy enhances reverse cholesterol transport.
A number of novel miRNAs, including miR-96/182/183, miR-30c and miR-185 regulate LDL metabolism.
Inhibition of miRNAs is a promising strategy for treating cardiometabolic disorders.
Acknowledgments
CF-H laboratory is supported by the National Institutes of Health (R01HL107953 and R01HL106063) and The Leducq Foundation.
Footnotes
CONFLICTC OF INTEREST
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. MicroRNAs in metabolic disease. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:178–185. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends in endocrinology and metabolism: TEM. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 9.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10. Soh J, Iqbal J, Queiroz J, et al. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nature medicine. 2013;19:892–900. doi: 10.1038/nm.3200. This study reports that miR-30c, an intronic miRNA located within the NFYC gene regulates VLDL secretion, therebt controlling circulating LDL-C. Of note, miR-30 in vivo delivery reduces plasma LDL levels and inhibits the progression of atherosclerosis in mice.
- 11. Jeon TI, Esquejo RM, Roqueta-Rivera M, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell metabolism. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. This study uncovers a novel mechanism by which SREBP increases the expression of miRNAs that regulates its own activity.
- 12. Yang M, Liu W, Pellicane C, et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. Journal of lipid research. 2014;55:226–238. doi: 10.1194/jlr.M041335. This study reports that miR-185, a miRNA regulated by SREBP1 regulates cholesterol biosynthesis via SREBP2 inhibition.
- 13. de Aguiar Vallim T, Tarling E, Kim T, et al. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300648. This study demonstrates that FXR transcription factor induce the expression of miR-144 to control HDL biogenesis
- 14.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramirez CM, Rotllan N, Vlassov AV, et al. Control of Cholesterol Metabolism and Plasma HDL Levels by miRNA-144. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300626. This study demonstrates that LXR transcription factor induce the expression of miR-144, thereby regulating cellular cholesterol efflux and plasma HDL-C levels
- 17.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 20.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 21.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of clinical investigation. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown MS, Goldstein JL. Familial hypercholesterolemia: defective binding of lipoproteins to cultured fibroblasts associated with impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:788–792. doi: 10.1073/pnas.71.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 24.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang T, Espenshade PJ, Wright ME, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 26.Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. The Journal of biological chemistry. 2009;284:5885–5895. doi: 10.1074/jbc.M807906200. [DOI] [PubMed] [Google Scholar]
- 27.Abifadel M, Varret M, Rabes JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nature genetics. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tontonoz P. Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors. Cold Spring Harbor symposia on quantitative biology. 2011;76:129–137. doi: 10.1101/sqb.2011.76.010702. [DOI] [PubMed] [Google Scholar]
- 31.Bodzioch M, Orso E, Klucken J, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nature genetics. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 32.Brooks-Wilson A, Marcil M, Clee SM, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nature genetics. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 33.Rust S, Rosier M, Funke H, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nature genetics. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 34.Adlakha YK, Khanna S, Singh R, et al. Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRalpha expression and cholesterol homeostasis. Cell death & disease. 2013;4:e780. doi: 10.1038/cddis.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goedeke L, Vales-Lara FM, Fenstermaker M, et al. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Molecular and cellular biology. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. This study demonstrates that the passenger strand of miR-33 cooperate with the guide for regulating lipid homeostasis.
- 36.Kang MH, Zhang LH, Wijesekara N, et al. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:2724–2732. doi: 10.1161/ATVBAHA.113.302004. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Yoon H, Ramirez CM, et al. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez CM, Davalos A, Goedeke L, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirasaki T, Honda M, Shimakami T, et al. MicroRNA-27a Regulates Lipid Metabolism and Inhibits Hepatitis C Virus Replication in Human Hepatoma Cells. J Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Xia M, Yan X, et al. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circulation research. 2012;111:967–981. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 41.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rottiers V, Obad S, Petri A, et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Science translational medicine. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. This study demostrate for the first time that LNA-antisense oligonucleotides designed to target only the seed sequence is effective to inhibit the effect of a family of miRNAs. This work also reports that inhibiting both miR-33a and miR-33b increases markedly plasma HDL-C levels.
- 43.Horie T, Ono K, Horiguchi M, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horie T, Nishino T, Baba O, et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nature communications. 2013;4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller AM, Gilchrist DS, Nijjar J, et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PloS one. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong D, Huang G, Zhang Y, et al. MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cellular signalling. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Sun D, Zhang J, Xie J, et al. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS letters. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 48.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Molecular and cellular biology. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L, Jia XJ, Jiang HJ, et al. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Molecular and cellular biology. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. This is the first study that reports miRNAs that targets SRB1, thus suppresin HDL-C uptake.