Abstract
Aims
Bacillus halodurans C-125 is a Gram-positive bacterium that was the first alkaliphilic species to have its genome completely sequenced. Despite its many years as a model for alkaliphily and source of industrially important enzymes, genetic manipulation of B. halodurans C-125 remains difficult, and therefore we sought to develop a robust method to allow routine transformation of this organism.
Methods and Results
A Plasmid Artificial Modification system (PAM system, Yasui et al. 2008) for B. halodurans C-125 was created that increases transformation efficiency by 10 to 1000 fold. Also, recovering transformed protoplasts on succinate nutrient agar yields faster, more robust colony recovery than on the traditional recovery media. Combining these two techniques often allows recovery of transformants in as little as 48 hours.
Conclusions
Use of the B. halodurans C-125 PAM system and succinate nutrient agar greatly improve the efficiency and speed of protoplast transformation of B. halodurans C-125.
Significance and Impact of the Study
These techniques allow routine genetic manipulation of B. halodurans C-125, a model alkaliphilic bacterium with important industrial properties.
Keywords: plasmid artificial modification (PAM), protoplast transformation, methylase
Text
While genetic manipulation of Escherichia coli, Bacillus subtilis, and other model bacteria is routine, the study of other, non-model bacteria is often hindered by the lack of reliable and rapid genetic transformation methods. One such bacterium is Bacillus halodurans C-125 (ATCC# BAA-125), a model alkaliphile and source of a several enzymes of industrial importance (Takami and Horikoshi, 2000). B. halodurans C-125 was the first alkaliphilic bacterium to have its genome sequenced, and is among the best studied of alakliphilic bacilli (Takami et al. 2000). Despite this history, there are only two reports of successful transformations of strain C-125 in the literature (Kudo et al. 1990; Hamamoto et al. 1994), with a similarly small number for related strains (e.g. Martinez et al. 2002; Crampton et al. 2007). The scarcity of published methods is likely due to the difficulty of transforming B. halodurans.
Most published methods use protoplast transformation (Kudo et al. 1990), but we found that transformation often occurs several orders of magnitude less efficiently than reported, especially for large plasmids (data not shown). Additionally, colonies often need five or more days to appear, during which time minor imperfections in the agar can lead to crystal formation and precipitation of media components, both of which impede protoplast recovery. To improve the speed and efficiency of transformation, we have made two key changes to the standard protocol.
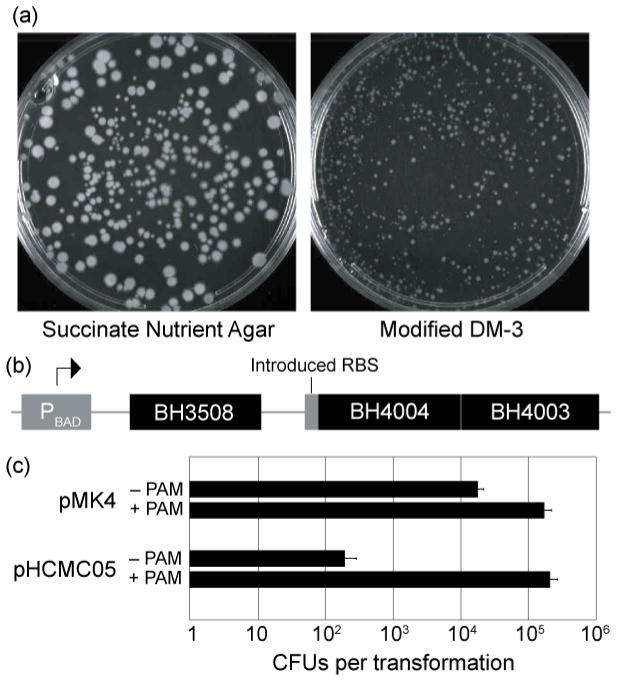
First, we replaced the modified DM-3 regeneration media with nutrient agar [per liter, 5 g peptone (Difco), 3 g beef extract (Lab Scientific), 15 g agar (Difco)] supplemented with 0.5 mol l−1 sodium succinate (pH 7.8 at 23°C). Although this resulted in a slight drop in protoplast recovery, the colonies grew larger and faster than when cultured using the original medium at the same pH (Fig. 1a). Additionally, succinate-nutrient agar (SNA) did not show any precipitate or crystal formation on the agar surface, even after extended incubation at 30°C.
Figure 1. Methods for improving transformation of Bacillus halodurans C-125.
(a) Protoplasts recovered on succinate nutrient agar (SNA, left) yield larger, more robust colonies than those on modified DM-3 medium (right). Colonies also appear two to three days sooner on SNA compared to modified DM-3 (data not shown). (b) Schematic of the Plasmid Artificial Modification (PAM) system for B. halodurans C-125. The three genes shown were inserted into the multiple cloning site of pBAD33 in-frame with each other and with an introduced ribosome binding site to ensure translation of the downstream genes. BH4003 and BH4004 were inserted as a unit, and their orientation reflects the native direction of transcription. (c) PAM treatment results in a 10-fold increase of colony-forming units (CFUs) for pMK4 (a ~5600 bp shuttle vector) and a 1000-fold increase for pHCMC05 (a ~8300 bp inducible expression vector). Error bars show the standard error of three independent transformations with 500 ng of plasmid. Both pMK4 and pHCMC05 were acquired from the Bacillus Genetic Stock Center.
Second, we created a Plasmid Artificial Modification (PAM) system specific for B. halodurans C-125, according to published methods (Yasui et al. 2008). In brief, we used the REBASE database (NEB, http://rebase.neb.com) to identify three putative methylases in the B. halodurans C-125 genome: BH3508, BH4003, and BH4004. We then cloned these into the multiple cloning site of plasmid pBAD33 (Fig. 1b), resulting in plasmid pPAMC125. Plasmids to be transformed into B. halodurans were first transformed into E. coli Top10 cells (Invitrogen) harboring pPAMC125, then harvested via miniprep with a QIAPrep spin miniprep kit (Qiagen). Plasmids treated this way are pre-methylated, protecting them from the cognate restriction enzymes once inside the target cell. This protection results in a 10- to 1000-fold increase in transformation efficiency relative to untreated controls (Fig. 1c).
Protoplasts were transformed according to the following protocol, modified from a method published previously (Kudo et al. 1990). Luria-Bertani broth (LB; USB Corporation) was adjusted to pH ~10.5 by the addition of 1% (w/v, final concentration) Na2CO3 after autoclaving. SMM and ASMMP buffers are as described previously (Kudo et al. 1990).
B. halodurans C-125 cells were grown to mid-exponential phase (OD 0.4 to 0.8) in 20 ml LB (pH ~10.5) at 37°C and harvested by centrifuging for 10 minutes at 12,000 g in a Beckman Avanti J-30I centrifuge with a JA-30.50 rotor. Cells were resuspended in 2 ml ASMMP and added to 10 mg of lysozyme (Sigma-Aldrich) dissolved in 100 μl 1x SMM. Cells were then gently agitated for one hour at 37°C. The resulting protoplasts were harvested by centrifugation at 2600 g for 10 minutes in a Tomy MRX-150 microcentrifuge and washed once with 1 ml ASMMP. Protoplasts were then resuspended in ASMMP to an OD equivalent of ~10 (as calculated from the culture OD at harvest). 150 μl of protoplast suspension was added to 5 μl plasmid stock mixed with 5 μl of 2x SMM. The cell-plasmid suspension was then added to 450 μl of 30% (w/v) PEG 8000 (Sigma-Aldrich) in 1x SMM and mixed gently by inverting the tube. After two minutes, 1.5 ml ASMMP was added and the cells were pelleted at 2600 g for 10 minutes as described above.
Cells were resuspended in 300 μl ASMMP and gently agitated for 90 minutes at 30°C to allow expression of the resistance marker. For the experiments shown in Fig. 1c, serial dilutions in ASMMP were plated on succinate nutrient agar with 3 μg ml−1 chloramphenicol and incubated at 30°C in partially sealed plastic bags to prevent desiccation. The first colonies appeared after 48–72 hours, although we allowed recovery to continue for six days to capture all transformants. Incubation at 37°C could sometimes recover colonies in as little as 36 hours, but resulted in reduced colony counts and occasional crystal formation on the surface of the agar (data not shown).
We also tested these methods on the related B. halodurans strains A-59 and DSM 497 (ATCC # 21591 and 27557, respectively). While both similarly benefited from recovery on SNA, PAM treatment of plasmids had no effect (data not shown). These strains probably contain different restriction-methylation systems, such that the methylases from strain C-125 provide no benefit. We did observe that even without PAM treatment, strains A-59 and DSM 497 transformed at near the frequency of C-125 with PAM treatment, such that there is no need for a PAM system for most laboratory applications.
In summary, we have modified the protoplast transformation method for B. halodurans C-125 to increase the speed and robustness of protoplast recovery. We also created a Plasmid Artificial Modification system that boosts transformation efficiency by up to three orders of magnitude. Plasmid pPAMC125 is available from the Bacillus Genetic Stock Center (http://www.bgsc.org), and should be useful to any researchers seeking to genetically transform strain C-125. The use of succinate nutrient agar may prove a simpler and attractive alternative to traditional protoplast recovery media regardless of species.
References
- Crampton M, Berger E, Reid S, Louw M. The development of a flagellin surface display expression system in a moderate thermophile, Bacillus halodurans Alk36. Appl Microbiol Biotechnol. 2007;75:599–607. doi: 10.1007/s00253-007-0869-0. [DOI] [PubMed] [Google Scholar]
- Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- Kudo T, Hino M, Kitada M, Horikoshi K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J Bacteriol. 1990;172:7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez MA, Delgado OD, Breccia JD, Baigorí MD, Siñeriz F. Revision of the taxonomic position of the xylanolytic Bacillus sp. MIR32 reidentified as Bacillus halodurans and plasmid-mediated transformation of B. halodurans. Extremophiles. 2002;6:391–395. doi: 10.1007/s00792-002-0269-4. [DOI] [PubMed] [Google Scholar]
- Takami H, Horikoshi K. Analysis of the genome of an alkaliphilic Bacillus strain from an industrial point of view. Extremophiles. 2000;4:99–108. doi: 10.1007/s007920050143. [DOI] [PubMed] [Google Scholar]
- Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K, Kano Y, Tanaka K, Watanabe K, Shimizu-Kadota M, Yoshikawa H, Suzuki T. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 2009;37:e3. doi: 10.1093/nar/gkn884. [DOI] [PMC free article] [PubMed] [Google Scholar]