Abstract
Site-directed scarless mutagenesis is an essential tool of modern pathogenesis research. We describe an optimized two-step protocol for genome editing in Salmonella enterica serovar Typhimurium to enable multiple sequential mutagenesis steps in a single strain. The system is based on the λ Red recombinase-catalyzed integration of a selectable antibiotics resistance marker followed by replacement of this cassette. Markerless mutants are selected by expressing the meganuclease I-SceI which induces double-strand breaks in bacteria still harboring the resistance locus. Our new dual-functional plasmid pWRG730 allows for heat-inducible expression of the λ Red recombinase and tet-inducible production of I-SceI. Methyl-accepting chemotaxis proteins (MCP) are transmembrane chemoreceptors for a vast set of environmental signals including amino acids, sugars, ions and oxygen. Based on the sensory input of MCPs, chemotaxis is a key component for Salmonella virulence. To determine the contribution of individual MCPs we sequentially deleted seven MCP genes. The individual mutations were validated by PCR and genetic integrity of the final seven MCP mutant WRG279 was confirmed by whole genome sequencing. The successive MCP mutants were functionally tested in a HeLa cell infection model which revealed increased invasion rates for non-chemotactic mutants and strains lacking the MCP CheM (Tar). The phenotype of WRG279 was reversed with plasmid-based expression of CheM. The complemented WRG279 mutant showed also partially restored chemotaxis in swarming assays on semi-solid agar. Our optimized scarless deletion protocol enables efficient and precise manipulation of the Salmonella genome. As demonstrated with whole genome sequencing, multiple subsequent mutagenesis steps can be realized without the introduction of unwanted mutations. The sequential deletion of seven MCP genes revealed a significant role of CheM for the interaction of S. Typhimurium with host cells which might give new insights into mechanisms of Salmonella host cell sensing.
Introduction
The ability for precise manipulation of bacterial genomes is of utmost importance in modern microbiological research. Although there is a long history of manipulating bacterial genomes, the application of phage-derived recombinases constitute a breakthrough in bacterial genetics [1, 2]. These “recombineering” strategies jointly exploit the ability of phage Rac RecE/T or of phage λ Red recombinases to use DNA fragments of less than 40 bp as substrates for homologous recombination [3]. Direct integration of these homologous sequences within short oligonucleotides is a great advantage which makes cloning of helper plasmids obsolete.
Over the years a vast number of different strategies and ever more refined protocols have been developed with a clear trend towards “scarless” genome manipulations [4–9]. These techniques are usually based on two steps starting with the integration of a selectable marker (e.g. an antibiotic resistance gene) followed by seamless replacement of that particular marker. For the second step efficient methods to select for loss of the marker are required. Here, accumulation of toxic metabolites based on tetAR [6, 10], sucrose sensitivity utilizing the sacB gene [7, 9], rpsL-mediated streptomycin sensitivity in resistant hosts [11, 12], the CcdA/CcdB toxin-antitoxin system [13] or I-SceI induced double-strand breaks (DSB) [4, 5, 14–17] were successfully used. The meganuclease I-SceI of S. cerevisiae has an unusually long recognition site of 18 bp which is statistically not present in bacterial genomes [18]. Mechanistically, the I-SceI site is co-integrated into the genome with the antibiotic resistance cassette during the first recombination step. Expression of the I-SceI enzyme after the second step selects for successful recombinants. Thus the precise and independent regulated expression of λ Red recombinase and I-SceI is a prerequisite for maximum efficiency and reliability of this method. Whereas the above mentioned methods rely on double-stranded DNA (dsDNA) as substrate for recombination, the chromosomal integration of short single-stranded DNA (ssDNA) oligonucleotides has also been demonstrated [3]. Recombineering of ssDNA requires only the function of λ Beta/RecT ssDNA binding proteins and functions without selectable or counter-selectable markers [19, 20]. However, methyl-directed mismatch repair reduces efficiency of the method and thereby increasing the effort of screening for correct clones [21].
Recombineering techniques are not only limited by the efficiency of the applied systems but also in general by the amenability of microorganisms to the Red/Rec recombinases. Alternative approaches are explored not only to avoid this limitation but to make genome editing even more efficient. The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and its associated protein, Cas9) system promises many advantages and is tremendously successful for editing eukaryotic genomes. Unfortunately, its application in bacteria is still limited. Amongst other reasons this is mainly due to the lack of a non-homologous end joining mechanism for DNA repair in most bacteria (reviewed in [22]). Nevertheless, successful CRISPR/Cas9-mediated genome editing including the introduction of deletions, insertions, and point mutations, was demonstrated when combined with λ Red recombinase functions [23]. The system was especially promoted for introducing multiple genome modifications since it does not rely on the cyclic integration and excision of a selectable marker [24]. However, the method requires careful design of the specific sequences for the gene-targeting protospacer adjacent motif (PAM) in order to prevent potential off-target activity. In addition, the PAM sequence together with the single guide RNA has to be supplied on a plasmid which needs to be cloned and, ideally, sequence-verified for each target gene to achieve high efficiency in bacteria [8, 23, 24]. Until further optimization of CRISPR/Cas9 system for bacteria, refined scarless recombineering protocols are an efficient and cost-effective tool for genome editing. Based on our previously developed method [4] we optimized the system to enhance efficiency and enable fast and reliable sequential modifications of the Salmonella Typhimurium (STM) genome.
We wanted to demonstrate the functionality of the optimized protocol in STM deleting seven genes encoding for methyl-accepting chemotaxis proteins (MCPs). MCPs are sensor molecules which respond to a variety of environmental cues including amino acids, sugars, ions and oxygen. Receptor signaling is initiated by reversible ligand binding at the periplasmic domains of dimeric MCPs [25]. Upon activation the receptor-bound kinase CheA phosphorylates the response regulator CheY. Through binding to the flagellar motor complex CheY influences the direction of flagellar rotation. As a result environmental signals perceived by MCPs control chemotactic swimming (reviewed in [26]). Salmonella expresses homologs to the E. coli MCPs Tsr, Tar, Trg and Aer. Tsr and Tar detect the amino acids serine and aspartate, respectively [27, 28]. Although the Salmonella Tar homolog CheM shows only 79% sequence identity with Tar of E. coli it was also demonstrated to bind and respond to aspartate [29, 30]. Trg is responsible for the sensing of the sugars glucose, galactose and ribose [31]. Alterations of the redox potential can be detected by Aer [32]. In contrast to the above mentioned MCPs the chemotaxis sensors Tap, Tip, McpA, McpB and McpC are only found in Salmonella. Tap was shown to sense citrate and phenol [33] whereas McpB/C mediate a repellent response towards L-cystine [34]. Currently no function for McpA [35], lacking a transmembrane domain, and Tip which is devoid of the periplasmic sensor domain is known.
We used a thoroughly defined set of MCP mutants to investigate the impact of individual MCPs on the ability of Salmonella to invade HeLa cells. Our infection experiments revealed an increased invasion rate for mutants lacking CheM indicating a detrimental effect of CheM-mediated chemotaxis in this infection model.
Results and discussion
Design and construction of system components
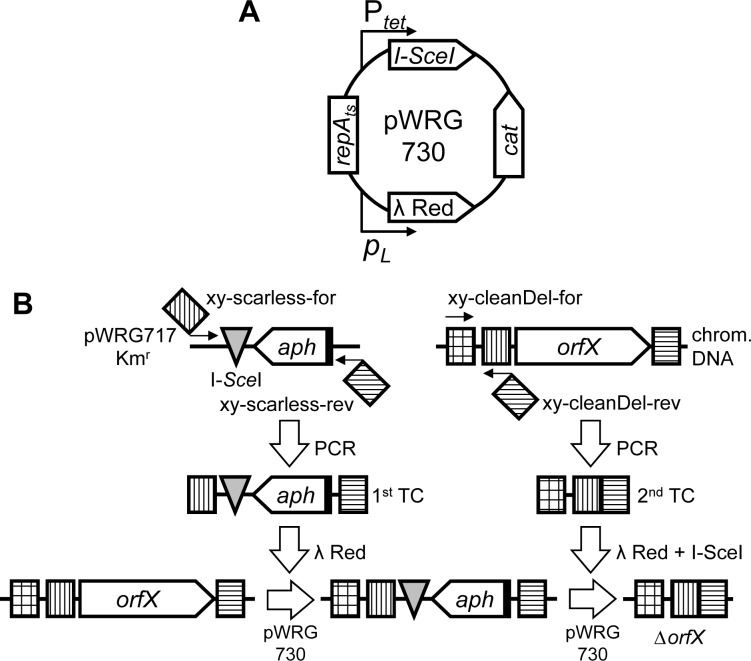
In our previously published system [4] the arabinose-inducible expression of the λ Red recombinase was combined with the meganuclease I-SceI under control of the tetA promoter in plasmid pWRG99 [36]. Although the system allowed for efficient generation of scarless deletions or single nucleotide exchanges, it required curing and re-transformation of pWRG99 between each recombination step [4]. We speculated that leaky expression of the λ Red proteins from the PBAD promoter in the absence of glucose [37] could have selected for inactive recombinases. Therefore we reasoned that a differently regulated λ Red expression plasmid might circumvent this problem. The pSC101-based pSIM5 harbors the phage λ pL operon comprising the genes exo, bet and gam under the control of the temperature-sensitive repressor CI857 that enables heat-inducible expression of λ Red recombinase functions [38]. Moreover, the pSIM5 plasmid allows for simple curing by its temperature-sensitive repAts origin of replication. From the functional perspective pSIM5 exhibited 10- to 60-fold higher recombination efficiency compared to the arabinose-inducible λ Red expression plasmid pKD119, which is similar to pKD46 except for the tetracycline resistance gene [38]. The combined λ Red/I-SceI expression plasmid pWRG730 was constructed by integrating the tetracycline-inducible I-SceI expression cassette of pWRG99 into the highly efficient pSIM5 vector (Fig 1A).
Fig 1. Overview of the method.
(A) Schematic representation of the functional units of plasmid pWRG730. The operon containing λ Red recombinase functions is under control of the heat-inducible phage-derived promoter pL. Expression of the I-SceI meganuclease is controlled by a tetracycline-inducible promoter (Ptet). A chloramphenicol resistance cassette (cat) is used for selection purposes. Due to its temperature-sensitive origin of replication (repAts) the plasmid can be easily cured at elevated growth temperatures. (B) Representation of the two-step scarless deletion methodology. A kanamycin resistance cassette (aph) is amplified together with an I-SceI cleavage site (grey triangle) from pWRG717 with two 60-mer primers each containing site-specific homology extensions at their 5’-ends (striped squares). Chromosomal integration of this first targeting construct (TC) is achieved by λ Red recombinase expression from pWRG730. The 2nd TC is also generated by PCR using chromosomal DNA as template and contains a direct fusion of up- and downstream homology regions. After genomic integration of the 2nd TC using λ Red recombinase, successful recombinants are selected by I-SceI expression from pWRG730. A detailed description of the method can be found in the main text.
Together with pWRG730 new FRT-free template plasmids were constructed based on pBluescript II SK+. Flp recombinase target (FRT) sites as present on the old template plasmid pWRG100 negatively interfered with DSB-based counterselection [4]. To circumvent this problem, the kanamycin resistance cassette of pKD4 [36] was integrated in two orientations relative to the I-SceI cleavage site thereby producing pWRG717 (Fig 1B) and pWRG832 (not shown). To provide an alternative selection marker, a spectinomycin resistance cassette originating from pDL1098 [39] was cloned in both orientations which resulted in pWRG829 and pWRG865 (data not shown). Although all four template vectors are suitable for the introduction of deletions, heterologous DNA or nucleotide exchanges, we routinely used the template vectors with the resistance gene in reversed orientation compared to the I-SceI cleavage site (pWRG717, pWRG829) (Fig 1B and not shown) to minimize polar effects in the first recombination step. The vectors with the resistance cassette in same orientation to the I-SceI cleavage site enable the simple integration of the antibiotic resistance gene in an artificial operon structure to serve as a reporter gene as described for Vibrio cholerae [39].
Sequential deletion of MCP genes
To evaluate the functionality of the system we decided to sequentially delete all genes encoding for known MCPs in S. Typhimurium strain NCTC 12023. The MCP genes were deleted in the following order: aer, tcp, tsr, trg, cheM (tar), mcpC (STM14_3893) and mcpB (STM14_3817) [35]. Each scarless deletion involved two successive recombination steps: (i) integration of a kanamycin resistance cassette amplified from pWRG717 and (ii) replacement of that resistance cassette by a PCR fragment containing the fused flanking regions of the gene to be deleted (Fig 1B) [4]. Similar strategies have been described before but with Red recombinase and I-SceI functionality provided on different plasmids [5] or requiring multiple rounds of culturing in selective medium for maximum efficiency [17]. In contrast to our first approach [4] the new plasmid pWRG730 encoding the recombinase and I-SceI meganuclease could be maintained within the bacteria during the whole sequential gene deletion process through selection with chloramphenicol (Cm) and keeping the cells at 30°C.
Primers ‘xy-scarless-for’ and ‘xy-scarless-rev’ were used for amplification of an I-SceI cleavage site together with a kanamycin resistance cassette from template vector pWRG717 to produce the 1st targeting construct (TC) for recombination. Whereas the 20 bases at the 3’ ends of the primers were designed to bind to the template vectors, the 40 bases at each 5’ end are homologous to regions up- and downstream of ‘orfX’ and thus determine the genome integration site (Fig 1B). Electrocompetent Salmonella with heat-induced λ Red recombinase were transformed with the 1st TC and transformants were selected on kanamycin (Km) and Cm-containing LB agar plates. Proper integration of the I-SceI/kanamycin cassette and replacement of each MCP gene was verified in both directions by PCR. The TC for the second recombination step was also generated by PCR. The construct represents a fusion of upstream and downstream homologous regions of the target deletion site. The homologous upstream sequence was amplified from wild-type (WT) chromosomal DNA as template using a short upstream-binding ‘xy-cleanDel-for’ primer and a 60-mer ‘xy-cleanDel-rev’ primer. The downstream homology region comprises the 40 bases of the 5’ end of ‘xy-cleanDel-rev’ (Fig 1B). This PCR-based approach to obtain a 2nd TC is much more flexible and cost-effective for introducing deletions or for site-directed mutagenesis compared to phosphorylated oligonucleotides [4]. PCR using 60-mer primers with 40 bases homology extensions would be also the means of choice for insertion of heterologous DNA sequences. Alternatively, synthetic DNA with compatible terminal homology regions can be used in the 2nd recombination step hence providing maximal flexibility. This step was selected on anhydrotetracycline (AHT) containing LB agar plates which induced I-SceI expression from pWRG730 allowing only the growth of successful recombinants devoid of the I-SceI site (Fig 1B).
Verification of mutants
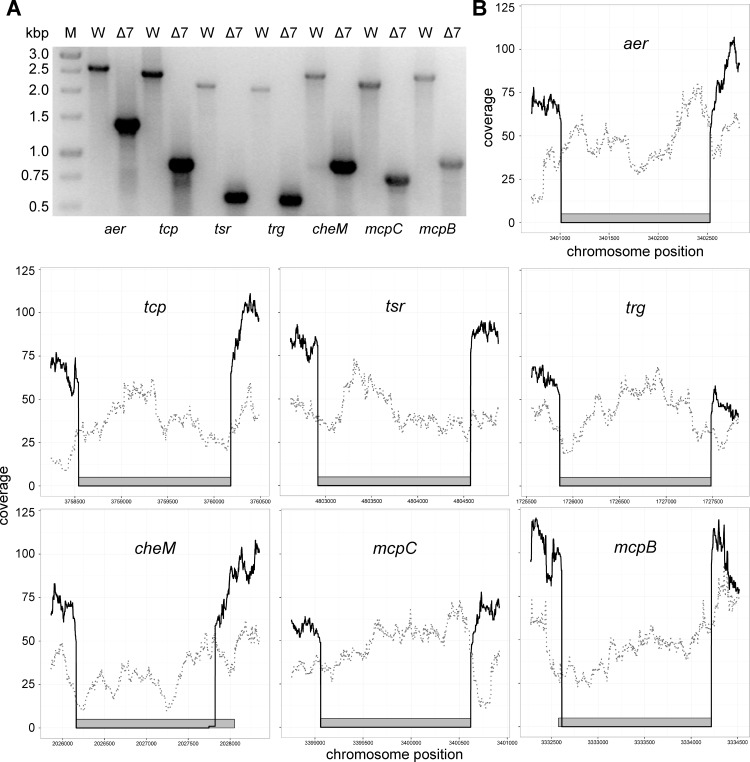
Successful scarless deletion of each MCP gene was verified after the 2nd recombination step by PCR using primers which bind up- and downstream of the site of deletion, respectively. Using these primer combinations, mutant alleles should result in shorter fragments compared to the WT situation. The theoretical fragment lengths for mutant and WT of each MCP gene are listed in Table 1. Starting from the single deletion strain WRG246 Δaer all scarless mutants were checked whether their MCP alleles corresponded to the expected genotype. Agarose gels that summarize the individual allele types for each of the MCP genes of the seven sequential deletion strains are depicted in S1 Fig. No PCR fragments were observed for the ‘aer’ locus in the Δ6 (WRG277) and Δ7 (WRG279) strains. Since the mcpC gene is located adjacent to aer, deletion of mcpC removed the reverse primer binding site in these mutants. All other observed fragments were of the expected size as listed in Table 1. Fig 2A shows a direct comparison of the PCR products originating from WT genomic DNA or from WRG279 lacking all seven MCP genes (Δ7) which confirmed the absence of the MCP genes in the Δ7 strain. Finally, Sanger sequencing of the PCR fragments verified the expected nucleotide sequence of all MCP deletion sites (data not shown).
Table 1. Expected fragment sizes of verification PCRs.
| Locus | Forward primer | Reverse primer | Fragment WT [bp] | Fragment after deletion [bp] |
|---|---|---|---|---|
| aer | Aer-Delcheck-for2 | McpC-Delcheck-rev | 2605 | 1370 |
| tcp | Tcp-Delcheck-for | Tcp-Delcheck-rev | 2150 | 509 |
| tsr | Tsr-Delcheck-for | Tsr-Delcheck-rev | 2203 | 544 |
| trg | Trg-Delcheck-for | Trg-Delcheck-rev | 2153 | 530 |
| cheM (tar) | CheM-Delcheck-for | CheM-Delcheck-rev | 2555 | 896 |
| mcpC | McpC-Delcheck-for | McpC-Delcheck-rev | 2330 | 768 |
| mcpB | McpB-Delcheck-for2 | McpB-Delcheck-rev | 2414 | 813 |
Fig 2. Genomic characterization of the WRG279 (Δ7 MCP) mutant.
(A) Agarose gel showing PCR fragments generated using the primers as listed in Table 1 with either wild type (W) or WRG279 (Δ7) chromosomal DNA as a template. Shorter PCR products of the expected sizes confirmed gene deletion for the seven loci in WRG279. M = DNA marker, band sizes are shown in kbp (B) Next generation sequencing coverage data of strain WRG279 (black line) and NCTC 12023 WT (dotted gray line) is shown for the targeted MCP genes (gray rectangles) including 300 nucleotides before and after each coding sequence. For strain WRG279 no sequencing reads were obtained for the deleted genes and thus a lack of coverage was observed at the respective nucleotide positions of the wild type sequence.
Expression of highly efficient phage recombinases such as λ Red can cause unwanted recombination events within the chromosome [40]. This is especially evident if “scars” e.g. from recombinases such as Flp or Cre accumulate in the genome during multi-step mutagenesis protocols. It has been shown that a set of mutated recombination sites provides one possible but laborious solution to circumvent this problem [41]. There is no need for such amendments using a scarless protocol as presented in this study. Given the mutagenic activity of extended high-level expression of λ Red recombinase [40], it is very important to strictly limit λ Red expression when multiple successive mutation steps are carried out in the same bacterial background. With the pSIM5-based Red expression plasmid pWRG730 10–15 minutes of heat induction is sufficient for efficient recombination [42].
Despite all these precautions we could not completely exclude accumulation of mutations in the strain WRG279. To address this, Illumina-based whole genome sequencing of our NCTC 12023 WT laboratory strain and of the Δ7 MCP mutant WRG279 was carried out. The obtained sequencing reads were mapped to the published genome sequence of STM ATCC 14028S which is isogenic with strain NCTC 12023. In contrast to NCTC 12023 WT, no sequencing reads of WRG279 were mapped to the deleted MCP genes (Fig 2B). Furthermore, the coverage data of WRG279 confirmed the precise deletion sites as determined by the homology regions of the 2nd TC (not shown). The ATCC 14028S genome data was also used as a reference to identify single nucleotide polymorphisms (SNPs). From the total 23 SNPs identified comparing ATCC 14028S with NCTC 12023 and WRG279 (data not shown) only two were unique for WRG279 (Table 2). One SNP resulted in a silent substitution within STM14_2710 and was detectable in all MCP mutants (Table 2). The other SNP was located within the putative ribosome binding site (RBS) of the yceB gene (STM14_1333) and appeared only in the last three mutants generated (Table 2). YceB is a predicted outer membrane lipoprotein and found to be downregulated in a resistant Salmonella strain isolated after a single challenge with nalidixic acid. Thus, this isolate exhibited additional resistance to tetracycline and chloramphenicol [43]. In contrast the WRG279 mutant showed minimal inhibitory concentrations (MICs) for both antibiotics similar to the parental NCTC 12023 WT strain in broth dilution assays (S2 Fig). However, modulation of YceB activity beyond antibiotic resistance due to altered protein levels cannot be excluded. These results argue against the yceB RBS mutation as an adaptation to the prolonged exposure to chloramphenicol, AHT, or both. Furthermore, both identified mutations are at chromosomal loci distantly located from the sites of recombination which makes their emergence unlikely to be directly linked to the recombination procedures applied. These low impact mutations might rather reflect natural genomic plasticity from prolonged laboratory handling as shown recently for chronic Salmonella infections [44, 45].
Table 2. Distribution of single nucleotide polymorphisms unique for WRG279.
| Strain | Position* 1,208,402 RBS of yceB (STM14_1333) | Position* 2,345,688 synonymous mutation within STM14_2710 |
|---|---|---|
| NCTC 12023 WT | A | G |
| WRG246 | A | A |
| WRG255 | A | A |
| WRG260 | A | A |
| WRG264 | A | A |
| WRG269 | G | A |
| WRG277 | G | A |
| WRG279 | G | A |
* reference: ATCC 14028S genome.
In summary our data highlight that the optimized system based on the dual-functional pWRG730 in combination with the pWRG717-derived kanamycin resistance cassette allows for fast and reliable manipulation of the Salmonella genome. This highly efficient tool is likely applicable in other bacteria amenable for the Red recombinase system, for example E. coli [36], Shigella spp. [46], Yersinia enterocolitica [47], Y. pestis [9], Pseudomonas aeruginosa [7] or Pantoea ananatis [48].
Functional characterization of the MCP deletion strains
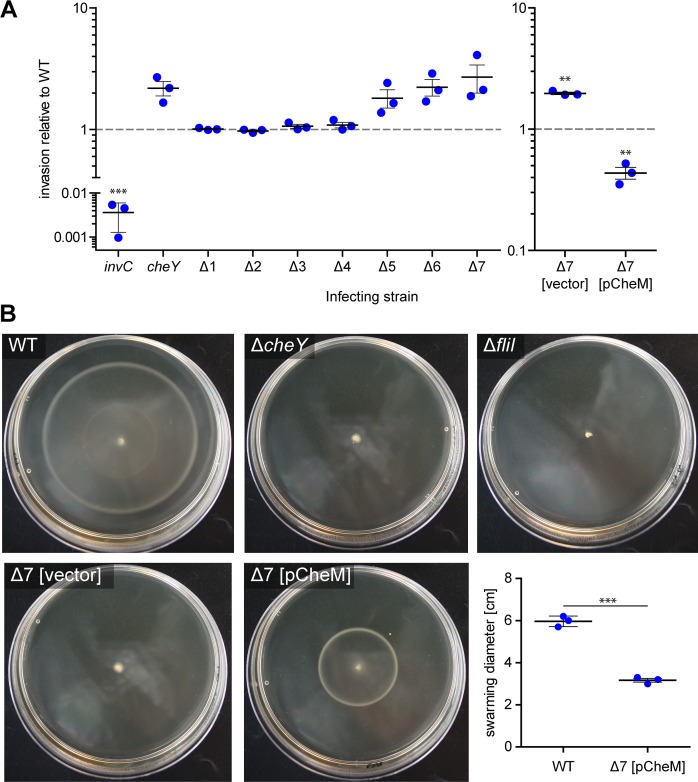
Having confirmed the genetic integrity of the mutants we went on to functionally characterize the strains. It has been demonstrated previously that bacterial motility is an important factor for efficient invasion of host cells [49, 50]. Actively swimming bacteria encounter shear forces which bring them within close proximity of the host cell surface. This “near surface swimming” promotes Salmonella-cell interactions and cooperative invasion of membrane ruffles [51]. By utilizing a cheY mutant which uncouples motility from chemotaxis [26] it was demonstrated that directed swimming is not required for near surface swimming [51]. However, in vivo results using streptomycin-pretreated mice underlined the importance of chemotaxis as a major virulence function besides motility [52]. We set out to test whether the lack of multiple MCPs influences Salmonella invasion in a HeLa-based infection model. Quantification of intracellular bacteria was done after one hour and was normalized to STM WT (set to 1). Very low amounts of intracellular bacteria were detected for an invC deletion mutant harboring a non-functional SPI-1 encoded type three secretion system (T3SS-1) (Fig 3A, left panel). Our data revealed an approximately 2-fold increased invasion rate for the cheY mutant suggesting an inhibitory effect of chemotaxis on HeLa invasion (Fig 3A, left panel). This result is in line with previous observations where “smooth” swimming mutants such as cheY or cheA exhibited increased invasion capabilities in HEp-2 cells [49]. In contrast a “tumbling only” cheB mutant was shown to have lower tissue culture invasion rates [49] presumably due to decreased near surface swimming. Next we used the set of successive MCP deletion mutants to elucidate the impact of specific chemotactic signaling on HeLa cell invasion. Surprisingly, we found a clear phenotypical separation of two groups of mutants. The first group comprising mutants Δ1 (WRG246 Δaer) to Δ4 (WRG264 Δaer, Δtcp, Δtsr, Δtrg) exhibited invasion rates very similar to WT whereas the remaining mutants, which lack five to seven MCP genes, showed an elevated invasion comparable to a cheY mutant (Fig 3A, left panel).
Fig 3. Functional characterization of the WRG279 mutant.
(A) HeLa cells were infected with different STM strains and relative invasion rates compared to STM WT were calculated after one hour of infection. An invC mutant lacking a functional T3SS-1 was used as a negative control for invasion and a motile but non-chemotactic cheY mutant was included to evaluate the impact of directed motility. The Δ1 to Δ7 strains represent sequential MCP deletions as follows: Δ1 = WRG246 Δaer; Δ2 = WRG255 Δaer, Δtcp; Δ3 = WRG260 Δaer, Δtcp, Δtsr; Δ4 = WRG264 Δaer, Δtcp, Δtsr, Δtrg; Δ5 = WRG269 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM; Δ6 = WRG277 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC; Δ7 = WRG279 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC, ΔmcpB. The right panel shows the invasion rates of the Δ7 strain complemented with pCheM (pWRG847) or transformed with the empty vector pWSK29 (vector). Statistical significance was calculated using a one sample t test against the hypothetical value 1.0 and was defined as ** for p < 0.01 and *** for p < 0.001. (B) Swarming phenotypes of different Salmonella strains as indicated on LB soft agar plates. Depicted is one representative out of three similar experiments. The diagram in the lower right panel shows the diameter of the swarming rings of S. Typhimurium WT and the Δ7 MCP mutant complemented with a CheM expression plasmid or a vector control as described in (B). Data of three independent biological replicates including means and standard deviations are shown. Statistical significance was calculated using a two-tailed paired Student’s t test and was defined as *** for p < 0.001.
Deletion of the cheM gene is the common feature of mutants Δ5 (WRG269) to Δ7 (WRG279). The observed phenotypes suggested that only CheM (Tar) is responsible for chemotactic signaling in the HeLa infection model supported by previous results obtained with HEp-2 cells [49]. To investigate this effect in more detail we complemented the Δ7 MCP strain WRG279 with the low-copy number plasmid pCheM (pWRG847) harboring the cheM gene under control of its natural promoter. While WRG279 transformed with the empty vector pWSK29 exhibited an elevated invasion similar to the cheY mutant, invasion of WRG279 [pCheM] was significantly decreased compared to WT (Fig 3A, right panel). We thus hypothesize that the six to eight plasmid copies per cell [53] led to an increased expression level of cheM which in turn fostered CheM-dependent chemotactic signaling with higher tumbling rates. CheM can directly sense aspartate [29]. Aspartate is not a component of the DMEM medium but small amounts of different amino acids are present in the FCS used during infection. The cell- and receptor density-dependent signaling preference of Tar-Tsr receptor complexes for either aspartate or serine could explain the apparent lack of chemotactic response from Tsr in our assays [54]. In the HeLa infection model aspartate or other so far uncharacterized CheM ligands might be liberated from the host cells thereby triggering a chemotactic response of Salmonella.
In a further set of experiments we wanted to characterize the MCP mutant strains in swarming assays on soft agar plates. As expected we observed a decrease in swarming diameter with more MCP genes deleted which reflects their increasing incapacity to perceive chemotactic signals (S3 Fig). Interestingly, the mutant WRG269 expressing only the MCPs McpB and McpC did show residual chemotactic movement whereas the successive WRG277, with mcpC deleted, was incapable of directed motility (S3 Fig). Like for WRG277, no chemotactic swarming could be observed for mutants lacking cheY or for the non-motile fliI mutant (Fig 3B). The Δ7 mutant WRG279 harboring the empty vector pWSK29 was phenotypically indistinguishable from the cheY mutant. Because both mutants are still motile they exhibited a slightly “blurred” inoculation site, which is most likely the result of “tumbly” swimming (Fig 3B). Introduction of the CheM complementation plasmid pCheM in WRG279 partially restored its chemotactic capacity (Fig 3B, lower right panel). Intriguingly, a single swarming ring could be detected for the CheM-complemented Δ7 MCP mutant which diameter roughly corresponded to the inner (CheM/Tar) ring observed for STM WT (Fig 3B). For STM WT two concentric rings were observed in some experiments which expanded over time to the agar plate periphery (Fig 3B). It has been demonstrated for E. coli that the outer ring corresponds to cells sensing serine through Tsr and the inner ring is composed of bacteria which sense aspartate by Tar [27, 28].
Our results and those of others [49] suggest that functional chemotaxis alone is disadvantageous for Salmonella invasion of HeLa and HEp-2 cells in vitro. However, the in vivo environment might be far more complex with chemotaxis being an important virulence factor [52, 55]. The optimized mutagenesis protocol described in the present study enabled us to efficiently generate a set of mutants lacking up to seven MCP genes. Further functional characterization of this set of MCP mutants in vitro identified CheM/Tar as the only MCP responding to chemotactic signals in a HeLa-based infection model. Future in vivo testing of these and other mutants, successfully generated by the developed procedure, might help to decipher the environmental signals Salmonella responds to during natural infection.
Materials and methods
Bacterial strains and plasmids
All strains used are listed in Table 3. Bacteria were routinely grown in LB media supplemented with 50 μg/mL carbenicillin (Cb) (Carl Roth, Mannheim, Germany), 25 μg/mL kanamycin (Km) (Carl Roth), 10 μg/mL chloramphenicol (Cm) (Carl Roth), 50 μg/mL spectinomycin (Sp) (Carl Roth) or 100 ng/mL anhydrotetracycline (AHT) (# 37919 Sigma-Aldrich, Schnelldorf, Germany) if required. Table 4 gives an overview of all the plasmids used in this study.
Table 3. Strains used in this study.
| Strain | Relevant characteristic(s) | Source or Reference |
|---|---|---|
| MvP818 | NCTC 12023 ΔinvC FRT | [56] |
| MvP1212 | NCTC 12023 ΔcheY FRT | [57] |
| MvP1213 | NCTC 12023 ΔfliI FRT | [58] |
| NCTC 12023 | Wild type, Nals, isogenic to ATCC 14028 | NCTC, Colindale, UK |
| WRG244 | NCTC 12023 Δaer::I-SceI aph, Kmr | This study |
| WRG246 | NCTC 12023 Δaer | This study |
| WRG247 | NCTC 12023 Δaer Δtcp::I-SceI aph, Kmr | This study |
| WRG255 | NCTC 12023 Δaer Δtcp | This study |
| WRG259 | NCTC 12023 Δaer Δtcp Δtsr::I-SceI aph, Kmr | This study |
| WRG260 | NCTC 12023 Δaer Δtcp Δtsr | This study |
| WRG263 | NCTC 12023 Δaer Δtcp Δtsr Δtrg::I-SceI aph, Kmr | This study |
| WRG264 | NCTC 12023 Δaer Δtcp Δtsr Δtrg | This study |
| WRG266 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM::I-SceI aph, Kmr | This study |
| WRG269 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM | This study |
| WRG276 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM ΔmcpC::I-SceI aph, Kmr | This study |
| WRG277 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM ΔmcpC | This study |
| WRG278 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM ΔmcpC ΔmcpB::I-SceI aph, Kmr | This study |
| WRG279 | NCTC 12023 Δaer Δtcp Δtsr Δtrg ΔcheM ΔmcpC ΔmcpB | This study |
Table 4. Plasmids used in this study.
| Plasmid | Relevant characteristic(s) | Source or Reference |
|---|---|---|
| pDL1098 | Temperature-sensitive mTn10 delivery vector, Cmr, Spr | [39] |
| pKD4 | aph resistance cassette flanked by FRT sites, λ Pir dependent replication, Kmr, Apr | [36] |
| pSIM5 | temperature-sensitive replication (30°C) and Red recombinase expression (42°C), Cmr | [38] |
| pWRG99 | pKD46 [36] derivative, temperature-sensitive replication (30°C), arabinose-inducible expression of Red recombinase, Tet-inducible expression of I-SceI, Apr | [4] |
| pWRG717 | pBluescript II SK+ derivative, aph resistance cassette and I-SceI cleavage site, Kmr, Apr | This study |
| pWRG730 | pSIM5 [38] derivative, temperature-sensitive replication (30°C) and Red recombinase expression (42°C), Tet-inducible expression of I-SceI, Cmr | This study |
| pWRG829 | pBluescript II SK+ derivative, aad9 resistance cassette and I-SceI cleavage site, Spr, Apr | This study |
| pWRG832 | pWRG717 derivative, aph resistance cassette reversed, Kmr, Apr | This study |
| pWRG841 | PcheM::cheM in pCRII-TOPO, Kmr, Apr | This study |
| pWRG847 | pCheM; PcheM::cheM in pWSK29, Apr | This study |
| pWRG865 | pWRG829 derivative, aad9 resistance cassette reversed, Spr, Apr | This study |
| pWSK29 | Low-copy-number vector, Apr | [53] |
PCR and cloning
All primers used for cloning are listed in S1 Table. For construction of the I-SceI aph template plasmid pWRG717, an I-SceI cleavage site was fused to the kanamycin resistance cassette of pKD4 by PCR using primers XhoI-aph-for2 and Aph-I-SceI-KnpI-rev2. The PCR fragment was cloned via XhoI/KpnI in pBluescript II SK+ (Agilent Technologies, Waldbronn, Germany). For construction of all other plasmids including the spectinomycin template plasmid pWRG829, assembly cloning of PCR fragments was used [59]. Primers pWSK29-Gbs-for and pKD4-Gbs-rev were used with pWRG717 as template to obtain a PCR fragment containing the vector and the I-SceI cleavage site. A spectinomycin resistance cassette was amplified with primers pSK-aad9-Gbs-for and pKD-aad9-Gbs-rev from plasmid pDL1098 [39] and subsequently combined with the first PCR product to obtain pWRG829. In template plasmids pWRG832 and pWRG865 the antibiotic resistance cassettes are in reversed orientation compared to pWRG717 and pWRG829, respectively. Here, pBluescript II SK+ was amplified with primers pWSK29-Gbs-for and -rev. The resistance cassettes were amplified from pWRG717 and pWRG829 with primer pairs Aph-I-SceI-pSK-Gbs-for/Aph-pSK-Gbs-rev2 and Aad9-I-SceI-pSK-Gbs-for/Aad9-pSK-Gbs-rev, respectively. A two-fragment assembly of each of the resistance cassettes with the vector PCR fragment led to the final plasmids. The heat-inducible Red recombinase expression plasmid pSIM5 [38] was linearized by PCR using primers pSIM-Gbs-for and pSIM-Gbs-rev. The tetracycline-inducible I-SceI expression cassette was amplified with primers pSIM-TetR-Gbs-for2 and pSIM-I-SceI-Gbs-rev using pWRG99 [4] as template. The two PCR fragments were combined by assembly cloning resulting in plasmid pWRG730. The region containing the cheM promoter and coding sequence was amplified from chromosomal DNA using primers CheM-Delcheck-for and CheM-pWSK-Gbs-rev. The PCR fragment was blunt-cloned in pCR II-TOPO (Thermo Fisher Scientific, Karlsruhe, Germany) resulting in plasmid pWRG841 which was subsequently digested with KpnI and XbaI. The cheM-containing fragment was gel-purified and cloned in the similarly-digested pWSK29 to obtain pWRG847 (pCheM).
Generation of mutants
All primers used to amplify the kanamycin resistance cassette from pWRG717 and to obtain a TC from Salmonella genomic DNA are listed in S1 Table. The desalted primers were purchased from Integrated DNA Technologies (Munich, Germany). For amplification of the first targeting construct forward primers consisting of the 3’ sequence 5’-AGGGTTTTCCCAGTCACGAC-3’, which binds to all pBluescript II SK+ -based template vectors, and a 5’ 40 bases sequence homologous to the genomic target site were used. The 60-mer reverse primers were similarly designed with the following 3’-located sequence binding to the template vectors: 5’-TGCTTCCGGCTCGTATGTTG-3’. Overnight (O/N) cultures of Salmonella harboring pWRG730 were grown at 30°C in LB supplemented with 10 μg/mL chloramphenicol. O/N cultures were re-inoculated 1:100 in fresh medium and grown aerated to an OD600 of 0.3 to 0.5. Red recombinase expression was induced for 12.5 minutes in a shaking water bath at 42°C [42]. After that bacteria were immediately put on ice and electro-competent cells were prepared essentially as described before [4]. Cells were transformed with 100–500 ng purified 1st TC using a Micropulser device (Bio-Rad, Munich, Germany) at ‘EC2’ setting. Successful recombinants were selected on LB plates containing chloramphenicol (plasmid pWRG730) and kanamycin (aph cassette from pWRG717 template) and kept at 30°C to preserve pWRG730. Colony-PCRs with suitable primers were routinely used to check for correct insertion of the resistance cassette within the genome in both directions. For subsequent removal of the resistance cassette competent cells were prepared from confirmed mutants still harbouring pWRG730 as described above. After transformation a 10-fold dilution series of the cells up to 10−4 was prepared in LB and plated on LB agar containing chloramphenicol and AHT. Plates were kept O/N at 30°C and large colonies were picked and purified again on LB agar plates containing chloramphenicol and AHT. Successful deletion of a MCP gene was confirmed with PCR using primers binding to the flanking regions of the gene (see Table 1) and subsequent Sanger sequencing of the PCR products (data not shown). After going through the desired number of deletion cycles, plasmid pWRG730 was cured from the bacteria by O/N incubation at 42°C.
Whole genome sequencing and mapping
Genomic DNA of strain NCTC 12023 wild type (WT) and the isogenic 7x MCP mutant WRG279 was prepared from O/N cultures in LB medium using a GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, Schnelldorf, Germany) according to manufacturer’s instructions. One ng of genomic DNA of each strain was fragmented using the Nextera sample preparation kit (Illumina, San Diego, CA, USA) and sequenced on a MiSeq (Illumina) machine running in paired end mode with 300 bp read length. All raw sequence reads of BioProject PRJNA355390 (http://www.ncbi.nlm.nih.gov/bioproject/) are available through SRA (http://www.ncbi.nlm.nih.gov/sra) accessions SRR5062192 (WT) and SRR5062193 (WRG279).
Consensus sequences for the two genomes were determined utilizing a custom in-house analysis pipeline as described earlier [60]. Briefly, MiSeq reads were mapped to the published genome sequence of S. Typhimurium strain ATCC 14028S (accession: CP001363) by a combination of BWA-SW version 0.7.13-r1126 [61] and SAMtools 0.1.19 [62]. VarScan 2.3 [63] was utilized for consensus calling. After mapping sequencing coverage was extracted using SAMtools 1.3.1 and visualized with ‘ggplot2’ [64] from ‘R’ 3.3.0 [65]. SNPs were extracted from the NCTC 12023 WT and WRG279 genomes obtained by reference-based mapping using a custom in-house Python script. Regions containing SNPs unique for WRG279 or sites of MCP deletion were PCR-amplified and subjected to Sanger sequencing (GATC Biotech, Cologne, Germany) using suitable primers listed in S1 Table. Sequence data is available through BioProject PRJNA355390.
Minimal inhibitory concentration assay
O/N cultures of test strains were diluted 1:100 in fresh LB and grown at 37°C to an OD600 between 0.5 and 0.7. After adjusting the cultures to an OD600 of 0.0002 (approximately 2 × 105 bacteria/mL) in fresh 2-fold concentrated LB, 100 μl were added to each well of a 96-well plate containing 100 μl of increasing concentrations of chloramphenicol or tetracycline in distilled water. The plates were incubated at 37°C in a humid chamber for 16 h and absorbance was measured at 600 nm (Tecan Infinite M1000). The MICs were calculated using ‘R’ as described before [66].
Cell culture and infection
HeLa cells (LGC Standards, Wesel, Germany) were grown in DMEM (Biowest, Germany) supplemented with 10% FCS, sodium pyruvate and 2 mM GlutaMax (Thermo Fisher Scientific, Karlsruhe, Germany) under humidified atmosphere with 5% CO2. Gentamicin protection assays were essentially carried out as described previously [67]. Briefly, 5 × 104 HeLa per well were seeded in 24-well plates (Cell-star, Greiner bio-one, Frickenhausen, Germany) 24 h prior infection. Bacterial O/N cultures grown in LB supplemented with appropriate antibiotics were reinoculated 1:31 in fresh medium and grown aerobically for another 3.5 h. An inoculum corresponding to a multiplicity of infection (MOI) of 10 was prepared in DMEM and used to infect the HeLa cells for 25 min. After the cells were washed thrice with PBS, 500 μl of DMEM containing 100 μg/mL gentamicin was applied to each well to kill remaining extracellular bacteria. After one hour of incubation the cell layers were washed again with PBS and then lysed for 10 min with PBS containing 1% Elugent (Merck Millipore, Darmstadt, Germany) and 0,0625% Antifoam B (Sigma-Aldrich, Schnelldorf, Germany) to liberate the intracellular bacteria. Serial dilutions of the inoculum and the lysates were plated on Mueller Hinton (MH) plates to determine the colony-forming units. Based on the inoculum the percentage of invasive bacteria was calculated and subsequently normalized to WT.
Swarming assay
Swarming of different Salmonella strains was assessed on LB semi-solid agar plates (LB with 5 g/L NaCl, 0.5% agar). A small amount (0.2 μl) of bacterial O/N cultures was applied onto the center of LB soft agar plate and incubated for six hours at 37°C. The diameters of the swarm colonies were measured and the plates were photographed.
Supporting information
Primer combinations as listed in Table 1 were used. One exception was the ‘aer’ locus where primer ‘Aer-Delcheck-rev2’ instead of ‘McpC-Delcheck-rev’ was used producing a 1087 bp fragment. Due to deletion of the reverse primer binding site during replacement of mcpC and mcpB, no product was observed (*) for WRG277 (6) and WRG279 (7). 1 = WRG246 Δaer; 2 = WRG255 Δaer, Δtcp; 3 = WRG260 Δaer, Δtcp, Δtsr; 4 = WRG264 Δaer, Δtcp, Δtsr, Δtrg; 5 = WRG269 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM; 6 = WRG277 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC; 7 = WRG279 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC, ΔmcpB, M = DNA marker, band sizes indicated in kbp.
(TIF)
MICs for chloramphenicol (left) and tetracycline (right) were determined for either S. Typhimurium NCTC 12023 wild type (WT) or the isogenic mutant WRG279 lacking seven MCP genes in broth dilution assays. Data of three independent biological replicates done in duplicates with means and standard deviations are shown. n.s. = not significant as calculated using a two-tailed unpaired Student’s t test.
(EPS)
(A) Swarming phenotypes on LB soft agar plates of different Salmonella MCP mutants as indicated. Depicted is one representative out of three similar experiments. (B) Diameters of the swarming rings with means and standard deviations of the different Salmonella strains from (A) for three independent biological replicates are shown. Statistical significance compared to WT was calculated using a two-tailed paired Student’s t test and was defined as * for p < 0.05 and *** for p < 0.001.
(TIF)
(XLSX)
Acknowledgments
We thank Monique Duwe for technical support during genome sequencing of strain WRG279.
Data Availability
All sequencing data files are available through BioProject (http://www.ncbi.nlm.nih.gov/bioproject/) PRJNA355390 and SRA (http://www.ncbi.nlm.nih.gov/sra), e.g. NGS sequencing reads can be accessed through SRR5062192 and SRR5062193.
Funding Statement
This project was funded in part by the 'Deutsche Forschungsgemeinschaft' (www.dfg.de) grant GE 2533/2-2 and an intramural research grant of the Robert Koch Insitute (www.rki.de) to RGG.
References
- 1.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2014;106:1 16 1–39. [DOI] [PubMed] [Google Scholar]
- 2.Pines G, Freed EF, Winkler JD, Gill RT. Bacterial Recombineering: Genome Engineering via Phage-Based Homologous Recombination. ACS Synth Biol. 2015;4(11):1176–85. 10.1021/acssynbio.5b00009 [DOI] [PubMed] [Google Scholar]
- 3.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–99. 10.1016/S0076-6879(06)21015-2 [DOI] [PubMed] [Google Scholar]
- 4.Blank K, Hensel M, Gerlach RG. Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One. 2011;6(1):e15763 Epub 2011/01/26. PubMed Central PMCID: PMC3021506. 10.1371/journal.pone.0015763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MM, Layton SL, Jiang T, Cole K, Hargis BM, Berghman LR, et al. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 2007;7:59 Epub 2007/09/19. PubMed Central PMCID: PMC2096622. 10.1186/1472-6750-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlach RG, Jäckel D, Hölzer SU, Hensel M. Rapid oligonucleotide-based recombineering of the chromosome of Salmonella enterica. Appl Environ Microbiol. 2009;75(6):1575–80. Epub 2009/01/20. PubMed Central PMCID: PMC2655478. 10.1128/AEM.02509-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang R, Liu J. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol. 2010;10(1):209. Epub 2010/08/05. PubMed Central PMCID: PMC2924854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reisch CR, Prather KL. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci Rep. 2015;5:15096 PubMed Central PMCID: PMC4604488. 10.1038/srep15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun W, Wang S, Curtiss R, 3rd. Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl Environ Microbiol. 2008;74(13):4241–5. Epub 2008/05/20. PubMed Central PMCID: PMC2446500. 10.1128/AEM.00940-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlinsey JE. λ-Red genetic engineering in Salmonella enterica serovar Typhimurium In: Hughes KT, Maloy SR, editors. Advanced Bacterial Genetics: Use of Transposons and Phage for Genomic Engineering. Methods Enzymol. 421. 2007/03/14 ed. New York: Academic Press; 2007. p. 199–209. [DOI] [PubMed] [Google Scholar]
- 11.Rivero-Müller A, Lajić S, Huhtaniemi I. Assisted large fragment insertion by Red/ET-recombination (ALFIRE)—an alternative and enhanced method for large fragment recombineering. Nucleic Acids Res. 2007;35(10):e78 Epub 2007/05/23. PubMed Central PMCID: PMC1904275. 10.1093/nar/gkm250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4(1):1 Epub 2003/01/18. PubMed Central PMCID: PMC149363. 10.1186/1471-2199-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Bian X, Xia L, Ding X, Müller R, Zhang Y, et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res. 2014;42(5):e37 PubMed Central PMCID: PMC3950717. 10.1093/nar/gkt1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DJ, Bingle LE, Heurlier K, Pallen MJ, Penn CW, Busby SJ, et al. Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol. 2009;9:252 Epub 2009/12/17. PubMed Central PMCID: PMC2796669. 10.1186/1471-2180-9-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27(22):4409–15. PubMed Central PMCID: PMC148724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40(2):191–7. Epub 2006/03/11. [DOI] [PubMed] [Google Scholar]
- 17.Yu BJ, Kang KH, Lee JH, Sung BH, Kim MS, Kim SC. Rapid and efficient construction of markerless deletions in the Escherichia coli genome. Nucleic Acids Res. 2008;36(14):e84 Epub 2008/06/24. PubMed Central PMCID: PMC2504295. 10.1093/nar/gkn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteilhet C, Perrin A, Thierry A, Colleaux L, Dujon B. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990;18(6):1407–13. Epub 1990/03/25. PubMed Central PMCID: PMC330504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98(12):6742–6. Epub 2001/06/07. PubMed Central PMCID: PMC34423. 10.1073/pnas.121164898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–8. PubMed Central PMCID: PMC4590770. 10.1038/nature08187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantino N, Court DL. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100(26):15748–53. PubMed Central PMCID: PMC307639. 10.1073/pnas.2434959100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo ML, Leenay RT, Beisel CL. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng. 2016;113(5):930–43. PubMed Central PMCID: PMC4816669. 10.1002/bit.25851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyne ME, Moo-Young M, Chung DA, Chou CP. Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Appl Environ Microbiol. 2015;81(15):5103–14. PubMed Central PMCID: PMC4495200. 10.1128/AEM.01248-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronda C, Pedersen LE, Sommer MO, Nielsen AT. CRMAGE: CRISPR Optimized MAGE Recombineering. Sci Rep. 2016;6:19452 PubMed Central PMCID: PMC4726160. 10.1038/srep19452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33(1):9–19. PubMed Central PMCID: PMC2890293. 10.1016/j.tibs.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5(12):1024–37. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- 27.Adler J. Chemotaxis in bacteria. Science. 1966;153(3737):708–16. [DOI] [PubMed] [Google Scholar]
- 28.Edwards JC, Johnson MS, Taylor BL. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol Microbiol. 2006;62(3):823–37. PubMed Central PMCID: PMC1858650. 10.1111/j.1365-2958.2006.05411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blat Y, Eisenbach M. Tar-dependent and -independent pattern formation by Salmonella typhimurium. J Bacteriol. 1995;177(7):1683–91. PubMed Central PMCID: PMC176793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodziej AF, Tan T, Koshland DE Jr. Producing positive, negative, and no cooperativity by mutations at a single residue located at the subunit interface in the aspartate receptor of Salmonella typhimurium. Biochemistry. 1996;35(47):14782–92. 10.1021/bi961481v [DOI] [PubMed] [Google Scholar]
- 31.Kondoh H, Ball CB, Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979;76(1):260–4. PubMed Central PMCID: PMC382918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibikov SI, Miller AC, Gosink KK, Parkinson JS. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J Bacteriol. 2004;186(12):3730–7. PubMed Central PMCID: PMC419962. 10.1128/JB.186.12.3730-3737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K, Imae Y. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci U S A. 1993;90(1):217–21. PubMed Central PMCID: PMC45631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazova MD, Butler MT, Shimizu TS, Harshey RM. Salmonella chemoreceptors McpB and McpC mediate a repellent response to L-cystine: a potential mechanism to avoid oxidative conditions. Mol Microbiol. 2012;84(4):697–711. PubMed Central PMCID: PMC4285363. 10.1111/j.1365-2958.2012.08051.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, et al. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188(6):2233–43. PubMed Central PMCID: PMC1428135. 10.1128/JB.188.6.2233-2243.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. PubMed Central PMCID: PMC18686. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–30. PubMed Central PMCID: PMC177145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–15. 10.1016/j.gene.2006.04.018 [DOI] [PubMed] [Google Scholar]
- 39.McDonough E, Lazinski DW, Camilli A. Identification of in vivo regulators of the Vibrio cholerae xds gene using a high-throughput genetic selection. Mol Microbiol. 2014;92(2):302–15. PubMed Central PMCID: PMC4005888. 10.1111/mmi.12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11 PubMed Central PMCID: PMC317293. 10.1186/1471-2199-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter Z, Delneri D. New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast. 2010;27(9):765–75. 10.1002/yea.1774 [DOI] [PubMed] [Google Scholar]
- 42.Lee SC, Wang W, Liu P. Construction of gene-targeting vectors by recombineering. Methods Mol Biol. 2009;530:15–27. PubMed Central PMCID: PMC3706932. 10.1007/978-1-59745-471-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowd SE, Killinger-Mann K, Brashears M, Fralick J. Evaluation of gene expression in a single antibiotic exposure-derived isolate of Salmonella enterica Typhimurium 14028 possessing resistance to multiple antibiotics. Foodborne Pathog Dis. 2008;5(2):205–21. 10.1089/fpd.2007.0062 [DOI] [PubMed] [Google Scholar]
- 44.Octavia S, Wang Q, Tanaka MM, Sintchenko V, Lan R. Genomic Variability of Serial Human Isolates of Salmonella enterica Serovar Typhimurium Associated with Prolonged Carriage. J Clin Microbiol. 2015;53(11):3507–14. PubMed Central PMCID: PMC4609693. 10.1128/JCM.01733-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marzel A, Desai PT, Goren A, Schorr YI, Nissan I, Porwollik S, et al. Persistent Infections by Nontyphoidal Salmonella in Humans: Epidemiology and Genetics. Clin Infect Dis. 2016;62(7):879–86. PubMed Central PMCID: PMC4787607. 10.1093/cid/civ1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranallo RT, Barnoy S, Thakkar S, Urick T, Venkatesan MM. Developing live Shigella vaccines using λ Red recombineering. FEMS Immunol Med Microbiol. 2006;47(3):462–9. 10.1111/j.1574-695X.2006.00118.x [DOI] [PubMed] [Google Scholar]
- 47.Trülzsch K, Sporleder T, Igwe EI, Rüssmann H, Heesemann J. Contribution of the major secreted Yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model. Infect Immun. 2004;72(9):5227–34. Epub 2004/08/24. PubMed Central PMCID: PMC517446. 10.1128/IAI.72.9.5227-5234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katashkina JI, Hara Y, Golubeva LI, Andreeva IG, Kuvaeva TM, Mashko SV. Use of the λ Red-recombineering method for genetic engineering of Pantoea ananatis. BMC Mol Biol. 2009;10:34 PubMed Central PMCID: PMC2682490. 10.1186/1471-2199-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones BD, Lee CA, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60(6):2475–80. PubMed Central PMCID: PMC257184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khoramian-Falsafi T, Harayama S, Kutsukake K, Pechere JC. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb Pathog. 1990;9(1):47–53. [DOI] [PubMed] [Google Scholar]
- 51.Misselwitz B, Barrett N, Kreibich S, Vonaesch P, Andritschke D, Rout S, et al. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog. 2012;8(7):e1002810 PubMed Central PMCID: PMC3406100. 10.1371/journal.ppat.1002810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stecher B, Hapfelmeier S, Müller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72(7):4138–50. PubMed Central PMCID: PMC427403. 10.1128/IAI.72.7.4138-4150.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–9. [PubMed] [Google Scholar]
- 54.Kalinin Y, Neumann S, Sourjik V, Wu M. Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J Bacteriol. 2010;192(7):1796–800. PubMed Central PMCID: PMC2838042. 10.1128/JB.01507-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivera-Chávez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9(4):e1003267 PubMed Central PMCID: PMC3630101. 10.1371/journal.ppat.1003267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerlach RG, Cláudio N, Rohde M, Jäckel D, Wagner C, Hensel M. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol. 2008;10(11):2364–76. 10.1111/j.1462-5822.2008.01218.x [DOI] [PubMed] [Google Scholar]
- 57.Wille T, Barlag B, Jakovljevic V, Hensel M, Sourjik V, Gerlach RG. A gateway-based system for fast evaluation of protein-protein interactions in bacteria. PLoS One. 2015;10(4):e0123646 PubMed Central PMCID: PMC4391838. 10.1371/journal.pone.0123646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bender JK, Wille T, Blank K, Lange A, Gerlach RG. LPS structure and PhoQ activity are important for Salmonella Typhimurium virulence in the Galleria mellonella infection model. PLoS One. 2013;8(8):e73287 PubMed Central PMCID: PMC3738532. 10.1371/journal.pone.0073287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 60.Steglich M, Nitsche A, von Müller L, Herrmann M, Kohl TA, Niemann S, et al. Tracing the Spread of Clostridium difficile Ribotype 027 in Germany Based on Bacterial Genome Sequences. PLoS One. 2015;10(10):e0139811 PubMed Central PMCID: PMC4596877. 10.1371/journal.pone.0139811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. PubMed Central PMCID: PMC2828108. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. PubMed Central PMCID: PMC2723002. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22(3):568–76. PubMed Central PMCID: PMC3290792. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2016. [Google Scholar]
- 65.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016.
- 66.Erhardt M, Mertens ME, Fabiani FD, Hughes KT. ATPase-independent type-III protein secretion in Salmonella enterica. PLoS Genet. 2014;10(11):e1004800 PubMed Central PMCID: PMC4230889. 10.1371/journal.pgen.1004800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt WD, et al. Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol. 2007;9(7):1834–50. 10.1111/j.1462-5822.2007.00919.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer combinations as listed in Table 1 were used. One exception was the ‘aer’ locus where primer ‘Aer-Delcheck-rev2’ instead of ‘McpC-Delcheck-rev’ was used producing a 1087 bp fragment. Due to deletion of the reverse primer binding site during replacement of mcpC and mcpB, no product was observed (*) for WRG277 (6) and WRG279 (7). 1 = WRG246 Δaer; 2 = WRG255 Δaer, Δtcp; 3 = WRG260 Δaer, Δtcp, Δtsr; 4 = WRG264 Δaer, Δtcp, Δtsr, Δtrg; 5 = WRG269 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM; 6 = WRG277 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC; 7 = WRG279 Δaer, Δtcp, Δtsr, Δtrg, ΔcheM, ΔmcpC, ΔmcpB, M = DNA marker, band sizes indicated in kbp.
(TIF)
MICs for chloramphenicol (left) and tetracycline (right) were determined for either S. Typhimurium NCTC 12023 wild type (WT) or the isogenic mutant WRG279 lacking seven MCP genes in broth dilution assays. Data of three independent biological replicates done in duplicates with means and standard deviations are shown. n.s. = not significant as calculated using a two-tailed unpaired Student’s t test.
(EPS)
(A) Swarming phenotypes on LB soft agar plates of different Salmonella MCP mutants as indicated. Depicted is one representative out of three similar experiments. (B) Diameters of the swarming rings with means and standard deviations of the different Salmonella strains from (A) for three independent biological replicates are shown. Statistical significance compared to WT was calculated using a two-tailed paired Student’s t test and was defined as * for p < 0.05 and *** for p < 0.001.
(TIF)
(XLSX)
Data Availability Statement
All sequencing data files are available through BioProject (http://www.ncbi.nlm.nih.gov/bioproject/) PRJNA355390 and SRA (http://www.ncbi.nlm.nih.gov/sra), e.g. NGS sequencing reads can be accessed through SRR5062192 and SRR5062193.