Abstract
Juvenile dermatomyositis (JDM) is a chronic inflammatory myopathy and vasculopathy driven by genetic and environmental influences. Here, we investigated the genetic underpinnings of an analogous, spontaneous disease of dogs also termed dermatomyositis (DMS). As in JDM, we observed a significant association with a haplotype of the major histocompatibility complex (MHC) (DLA-DRB1*002:01/-DQA1*009:01/-DQB1*001:01), particularly in homozygosity (P-val = 0.0001). However, the high incidence of the haplotype among healthy dogs indicated that additional genetic risk factors are likely involved in disease progression. We conducted genome-wide association studies in two modern breeds having common ancestry and detected strong associations with novel loci on canine chromosomes 10 (P-val = 2.3X10-12) and 31 (P-val = 3.95X10-8). Through whole genome resequencing, we identified primary candidate polymorphisms in conserved regions of PAN2 (encoding p.Arg492Cys) and MAP3K7CL (c.383_392ACTCCACAAA>GACT) on chromosomes 10 and 31, respectively. Analyses of these polymorphisms and the MHC haplotypes revealed that nine of 27 genotypic combinations confer high or moderate probability of disease and explain 93% of cases studied. The pattern of disease risk across PAN2 and MAP3K7CL genotypes provided clear evidence for a significant epistatic foundation for this disease, a risk further impacted by MHC haplotypes. We also observed a genotype-phenotype correlation wherein an earlier age of onset is correlated with an increased number of risk alleles at PAN2 and MAP3K7CL. High frequencies of multiple genetic risk factors are unique to affected breeds and likely arose coincident with artificial selection for desirable phenotypes. Described herein is the first three-locus association with a complex canine disease and two novel loci that provide targets for exploration in JDM and related immunological dysfunction.
Author summary
Juvenile dermatomyositis (JDM) is an autoimmune disease of the skin and muscle influenced by both genetic and environmental components. Although genes independent of the MHC are thought to contribute to disease pathogenesis, their identification has been complicated by a paucity of biological samples, disease heterogeneity, and genetically diverse subjects. A naturally occurring inflammatory disease of domestic dogs, also termed dermatomyositis (DMS), is analogous to JDM and is the only animal model available for genetic study. We first determined that, as in JDM, a particular MHC haplotype confers susceptibility to DMS. Capitalizing on the genetic isolation of dog breeds and extremely low MHC diversity within affected breeds, we used multibreed genome-wide association studies to identify two novel loci. Through genome resequencing and additional genotyping, we uncovered highly associated polymorphisms in conserved positions of PAN2 and MAP3K7CL. Analysis of three-locus genotypes revealed uniquely high frequencies among affected breeds and nine allelic combinations that confer moderate or high risk for DMS. The pattern of disease probability illustrates the presence of gene-gene interaction, as well as an inverse correlation between age of onset and number of risk alleles. This study highlights the utility of canine models for mapping susceptibility loci in complex diseases and detecting patterns of genetic interactions obscured in diverse human populations.
Introduction
Juvenile dermatomyositis (JDM) is an autoimmune vasculopathy that causes a characteristic skin rash (heliotrope rash across the eyelids and Gottron’s papules on the bony prominences) and proximal muscle weakness [1]. It is the most frequently diagnosed childhood inflammatory myopathy, comprising 80% of all cases [1] and affecting 3.2 in every million children between the ages of 2 and 17 within the USA [2]. Prognosis is positively correlated with early diagnosis and swift treatment with corticosteroids and/or immunosuppressants [1,3]. While treatment of JDM is much improved overall, many children still suffer from chronic flare-ups [1].
Though the etiology is unknown, JDM is thought to be triggered by exposure to a virus or other environmental agent. Manlhiot et al. [4] reported that 71% of JDM patients had a clinical history consistent with infection prior to symptoms. Investigations into the class II major histocompatibility complex (MHC), TNF, and IL1 identified several susceptibility and protective alleles, but their collective contribution to pathogenesis is poorly understood [5–8]. Recent genome-wide association studies (GWASs) to identify additional susceptibility loci in JDM confirmed a strong association with the MHC but failed to detect novel major risk factors, likely because of a paucity of biological samples and genetically heterogeneous populations [9,10].
In domestic dogs, an inflammatory vasculopathy of the skin and muscle, also termed dermatomyositis (DMS), is clinically, histologically, and immunologically similar to JDM and provides the only animal model available to study genetic risk factors [11–16]. The earliest clinical signs of DMS are crusting and scaling on the face, ears, tail tip, and/or across the bony prominences of the limbs and feet [17–19] (S1 Fig). Alopecia and more extensive skin lesions may develop over time, resulting in dermal scarring associated with erythema and mottled pigmentation [17,19]. Lesions persist for weeks to months, and may or may not chronically recur [17]. Muscle wasting manifests as atrophy of the head musculature; difficulty eating, drinking, and swallowing; and an atypical, high-stepping gait [17,19].
Similar to JDM, DMS is an immune-mediated disease [13,18,20] that typically develops following an environmental trigger, such as vaccination or viral infection, and is exacerbated by subsequent stressors like exposure to UV light [13,17,21,22]. Anecdotal reports indicate that rabies vaccination, parvovirus infection, owner surrender, or mistreatment commonly precede disease onset. Consistent with an environmental trigger, age at onset is variable with many cases occurring between seven weeks and six months of age, but others not developing until well into adulthood [17–19,23].
DMS is diagnosed almost exclusively in the genetically [24] and phenotypically similar collie and Shetland sheepdog breeds, suggesting the presence of a strong heritable component(s) arising from ancestors common to both breeds. A 1980s study of disease transmission in the collie eliminated simple Mendelian modes of inheritance [14]. In two test matings, an affected male collie sired litters from an affected collie and a healthy Labrador retriever. All six collie puppies were affected with variable degrees of severity, while three of the four mixed breed puppies developed milder forms of DMS. Retrospective pedigree analyses of the collie sire and dam showed a complete absence of affected ancestors [14].
The availability of a naturally-occurring canine model provides a new opportunity for the identification of genetic risk factors of JDM. The conserved genomic backgrounds of genetically isolated dog breeds have enabled detection of risk loci in complex diseases that are often obscured by heterogeneity within human cohorts [25–30]. Here, we evaluated class II MHC haplotypes, performed multibreed GWASs, and generated whole genome and transcriptome sequencing data to dissect the genetic basis of DMS. We uncovered common polymorphisms of the MHC and two novel loci that are strongly associated with DMS, as well as patterns of allelic inheritance that explain 93% of cases studied. A genetic test is now available to determine the likelihood of a dog developing DMS and to facilitate breeding decisions that avoid progeny having high-risk genotypes.
Results and discussion
Association of a major histocompatibility complex haplotype
Given the involvement of MHC genes in JDM, we first determined alleles of the highly polymorphic canine MHC class II dog leukocyte antigen (DLA) genes: DLA-DRB1, -DQA1, and -DQB1. Two locus (DLA-DRB1 and -DQB1) and three locus (DLA-DRB1, -DQA1, and -DQB1) haplotypes were first generated for 50 collies and 117 Shetland sheepdogs, respectively. Because all observed haplotypes contained a unique DLA-DRB1 allele, the 355 remaining dogs were genotyped for this locus only and the haplotype was inferred (Table 1).
Table 1. Frequency of DLA-DRB1/-DQA1/-DQB1 haplotypes in collies and Shetland sheepdogs.
| COLLIE | |||||
| DLA-DRB1*/-DQA1*/-DQB1* | Cases (2n = 80) | Controls (2n = 370) | OR | 95% CI | P-value |
| 002:01/009:01/001:01 | 74 | 353 | 0.59 | 0.23–1.56 | 0.40 |
| 006:01/050:11/007:01 | 0 | 3 | - | - | - |
| 015:01/009:01/001:01 | 6 | 14 | 2.06 | 0.77–5.54 | 0.23 |
| SHETLAND SHEEPDOG | |||||
| DLA-DRB1*/-DQA1*/-DQB1* | Cases (2n = 184) | Controls (2n = 410) | OR | 95% CI | P-value |
| 002:01/009:01/001:01 † | 158 | 301 | 2.20 | 1.38–3.52 | 0.0010 |
| 015:01/009:01/001:01 | 1 | 6 | 0.37 | 0.04–3.08 | 0.4454 |
| 023:01/003:01/005:01 | 24 | 102 | 0.45 | 0.28–0.73 | 0.0011 |
| Other haplotypes | 1 | 1 | - | - | - |
The number of times each haplotype was observed is reported for cases and controls with odds ratios (OR), 95% confidence intervals, and Fisher’s exact two-tailed P-values. Significant statistics are bolded.
†71 out of 92 cases and 109 out of 205 controls were homozygous for the 002:01/009:01/001:01 haplotype (OR = 2.98, 95% CI = 1.70–5.21, P-val = 0.0001).
We observed remarkably low DLA diversity among collies, with only three haplotypes present in 225 collies worldwide. This lack of heterogeneity precluded detection of associations with DMS, as 91% of collies were homozygous for the haplotype DLA-DRB1*002:01/-DQA1*009:01/-DQB1*001:01. In 297 Shetland sheepdogs, we identified two predominant haplotypes, 002:01/009:01/001:01 and 023:01/003:01/005:01. The former was over-represented among cases (P-val = 0.0010, OR = 2.20), primarily because of increased homozygosity (P-val = 0.0001, OR = 2.98). We therefore conclude that 002:01/009:01/001:01 is a risk factor for DMS and that homozygosity confers increased susceptibility. Under the assumption that the causal alleles derive from an ancestor common to both breeds, we extrapolate the observed DLA risk to collies. The high frequency of the DLA risk haplotype in both populations indicates that additional loci must influence disease progression.
Genome-wide association studies reveal signals on chromosomes 10 & 31
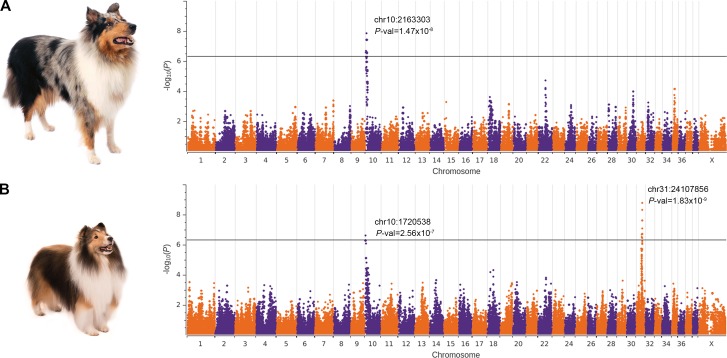
We conducted an independent GWAS for each breed, using a total of 97 cases (31 collies, 66 Shetland sheepdogs), 68 controls (23 collies, 45 Shetland sheepdogs), and 98,520 SNPs after filtering. In collies, a single signal (P-val = 1.47X10-8) composed of 17 SNPs at the centromeric end of chromosome 10 exceeded Bonferroni significance (Fig 1A). In Shetland sheepdogs, this association was replicated (P-val = 2.56X10-7), and a second signal (P-val = 1.83X10-9) composed of 11 SNPs surpassing Bonferroni significance was detected on chromosome 31 (Fig 1B). Both associations persisted in a combined breed analysis (chr10: P-val = 2.3X10-12, chr31: P-val = 3.95X10-8) (S1 Table, S2 Fig); although the breeds possessed a common haplotype on chromosome 31, the Shetland sheepdogs appeared to drive this association. No associated SNPs were detected near the MHC loci on chromosome 12, likely a result of high homogeneity in our cohort and poor SNP coverage on the array [25].
Fig 1. Manhattan plots of DMS GWASs in two breeds.
(A) Collies: 31 cases and 23 controls. (B) Shetland sheepdogs: 66 cases and 45 controls. The –log10P-values (y-axis) for 98,520 Illumina SNPs are plotted against chromosome position (x-axis). The threshold for Bonferroni significance is shown as a black horizontal line. The P-value and position (canFam3) of the lead SNPs are reported.
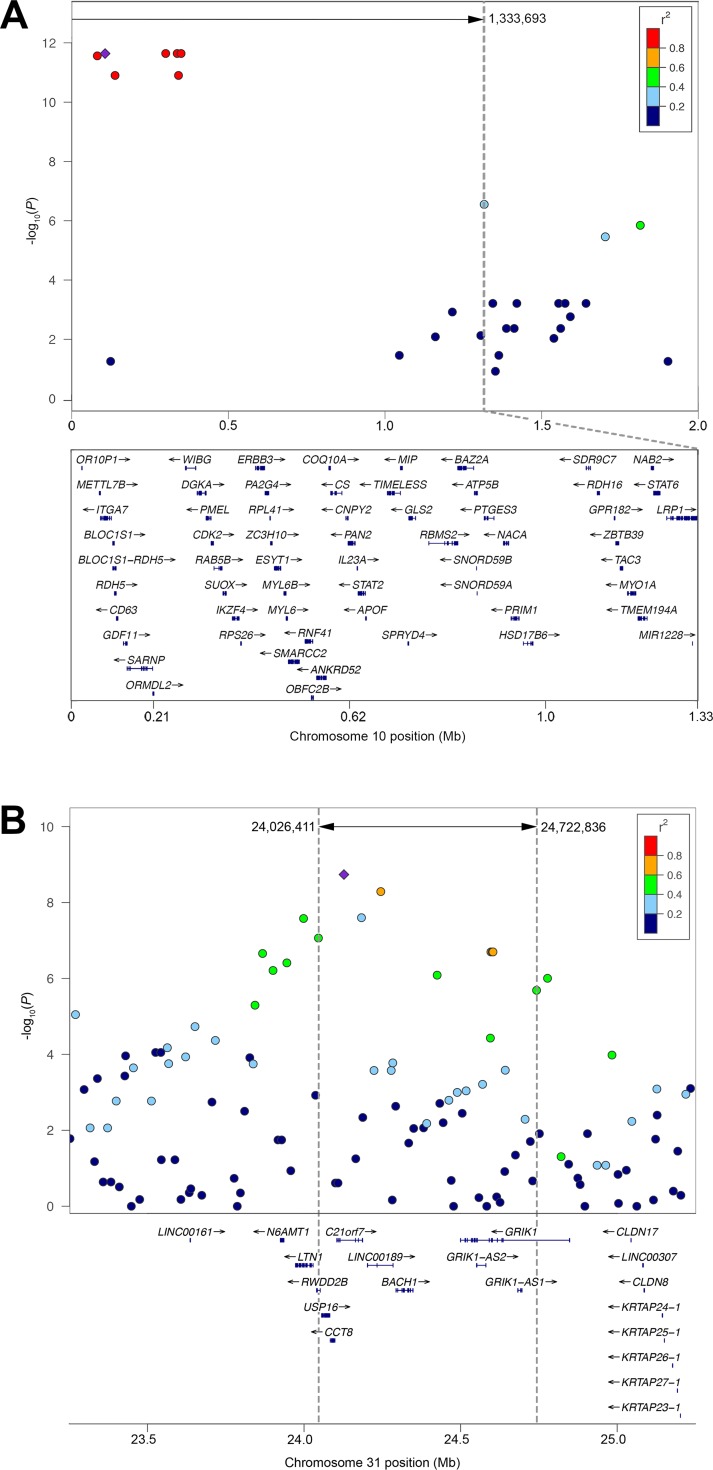
On chromosome 10, 97% of all affected dogs were homozygous or heterozygous for the risk alleles of the lead SNPs. On chromosome 31, 88% of affected Shetland sheepdogs, but only 39% of affected collies, shared the risk alleles of the lead SNPs. As neither locus appeared to be independently necessary for disease development, we surveyed the extent of regional linkage disequilibrium (LD) to demarcate candidate intervals of ~1.33 Mb on chromosome 10 (Fig 2A) and ~696 kb on chromosome 31 (Fig 2B), harboring ~65 and 6 genes, respectively. The large size of the chromosome 10 region is attributed to lower recombination rates near the centromere and a dearth of informative SNPs.
Fig 2. Regional plots depicting LD at two associated loci.
Association results (-log10P) for (A) combined breed GWAS on chromosome 10 and (B) Shetland sheepdog GWAS on chromosome 31 are color-coded based on pairwise LD (r2) with the lead SNPs (purple diamond). Dotted lines demarcate the candidate interval. Genes within the region (based on hg18) are shown below the plot with chromosome positions reported in canFam3. On chromosome 10, the gene track is zoomed in to represent the candidate interval only; 4 genes were omitted from the figure (DNAJC14, LOC440104, MMP19, and SLC39A5).
Identification of candidate variants in PAN2 and MAP3K7CL
Whole genome resequencing was performed for four affected dogs (three collies and one Shetland sheepdog) and two unaffected collies, resulting in 17X to 41X coverage. Variants were filtered for those lying within our delineated intervals (chr10:1–1,333,693; chr31:24,026,411–24,722,836) and following the inheritance pattern of the most significantly associated SNPs in the affected dogs (S2 and S3 Tables). Five intergenic and two intronic variants were unique to these breeds (i.e., not present in the Boxer reference genome, dbSNP, or 27 whole genome sequences from 16 other breeds); however, most were in repetitive regions and none were in conserved positions. Thus, the pathogenic variants were likely to be common polymorphisms, so we next prioritized variants within predicted exons and splice sites of genes expressed in skin for further study.
To confirm exon/intron boundaries predicted by Ensembl 79 and establish expression of variants in affected tissue, we generated RNAseq data. We obtained a minimum of 89 million reads per tissue from active skin lesions of two affected dogs (one collie and one Shetland sheepdog) and skin from one unaffected Australian shepherd dog. All genes expressed in skin within the candidate regions were also expressed in affected tissues. Seventeen exonic variants were expressed; seven of these were nonsynonymous and evaluated using in silico programs [31–33] (Table 2).
Table 2. Expressed exonic changes on chromosomes 10 and 31.
| Position | Gene | A1/A2 | AA change | Poly-Phen2 | PANTHER | SIFT | P-value |
|---|---|---|---|---|---|---|---|
| CHROMOSOME 10 | Cases n = 93 Controls n = 63 | ||||||
| 95042 | ITGA7 | T/C | Val/Val | - | - | - | - |
| 121835 | RDH5 | G/A | Ala/Ala | - | - | - | - |
| 222809 | MMP19 | T/G | Gln/Pro | 0 | 91 | 0.43 | - |
| 331135 | SUOX | A/G | His/Arg | 0 | 85 | N/A | - |
| 565958 | ANKRD52 | G/C | Ser/Cys | 0.92 | 361 | 0.02 | 1.93X10-17 |
| 627760 | PAN2 | G/A | Arg/Cys | 1 | 1628 | 0 | 1.93X10-17 |
| 1127082 | RDH16 | C/T | Ala/Ala | - | - | - | - |
| 1228277 | NAB2 | G/C | Gly/Arg | 0 | 85 | N/A | - |
| 1239562 | STAT6 | G/A | Thr/Met | 0.98 | 361 | 0.05 | 1.52X10-4 |
| 1286150 | LRP1 | C/T | Asn/Asn | - | - | - | - |
| 1333693 | LRP1 | G/A | Gln/Gln | - | - | - | - |
| CHROMOSOME 31 | Cases n = 63 Controls n = 45 | ||||||
| 24068039 | CCT8 | T/C | Ala/Ala | - | - | - | - |
| 24132273 | MAP3K7CL | Indel/- | 5′ UTR | N/A | N/A | N/A | 2.09X10-11 |
| 24132343 | MAP3K7CL | A/C | 5′ UTR | N/A | N/A | N/A | 1 |
| 24292521 | BACH1 | A/G | Asn/Asp | 0.007 | 324 | 0.21 | - |
| 24294659 | BACH1 | A/G | Glu/Glu | - | - | - | - |
| 24295208 | BACH1 | A/G | Pro/Pro | - | - | - | - |
Alleles 1 (A1 = minor allele) and 2 (A2 = reference allele) are reported. Amino acid changes are based on the dog reference genome. P-values are reported for matched populations. Variants with PolyPhen2 scores ranging from 0.85–1, PANTHER preservation time (in millions of years) >450my, and SIFT scores ranging from 0–0.05 are considered deleterious/probably damaging. Variants more strongly associated with DMS than the most associated SNPs from the array are bolded.
We genotyped a subset of our mapping population for three nonsynonymous SNPs on chromosome 10 (ANKRD52 g.565958G>C, PAN2 g.627760G>A, and STAT6 g.1239562G>A) that were predicted to be deleterious or probably damaging by more than one in silico program. The ANKRD52 and PAN2 variants were more strongly associated with DMS than the lead SNP. These variants were in perfect linkage disequilibrium with each other; however, PAN2 g.627760G>A was assigned damaging scores with higher confidence by in silico programs (Table 2). We therefore focused further studies on PAN2 g.627760G>A, encoding p.Arg492Cys (XP_531635.3), although ANKRD52 cannot be excluded. On chromosome 31, we genotyped Shetland sheepdogs for a SNP (g.24132343A>C) and an indel (c.383_392ACTCCACAAA>GACT, XM_846337.4), both located in a 5′ non-coding exon of MAP3K7CL. Only the indel was associated with DMS (Table 2, S3 Fig). In an expanded, combined population (132 affected and 390 unaffected collies and Shetland sheepdogs), both the PAN2 (P-val = 2.08X10-35) and MAP3K7CL (P-val = 1.45X10-33) variants were highly associated with DMS (S4 Table).
PAN2 (or USP52) encodes the catalytic subunit of the poly(A) nuclease deadenylation complex (PAN2-PAN3) and is one of two exonucleases involved in mRNA degradation in eukaryotes [34,35]. Deadenylation plays a role in translational regulation of inflammatory response [36]. Independent of this function, PAN2 also stabilizes HIF1A transcripts via their 3′-UTR, which contain AU-rich elements (AREs), and may be involved in regulating other transcripts having AREs [37]. HIF1A, a key regulator of inflammation [38], and other ARE-containing transcripts, such as IL-6 [39], are upregulated in JDM [40, 41]. PAN2 is widely expressed and highly evolutionarily conserved [35]; human (NP_001120932.1) and dog (XP_013972628.1) amino acid sequences share 98% identity.
MAP3K7CL (also known as TAK1L or C21orf7) is a poorly studied kinase gene that is transcriptionally active in immunological tissues and expressed primarily in peripheral blood leukocytes [42,43]. Human (NP_001273546.1) and dog (XP_013965340.1) MAP3K7CL protein sequences share >90% identity. The transcription factors RUNX3 and EP300 bind the 5′ non-coding exon of human MAP3K7CL (UCSC Genome Browser ENCODE Transcription Factor ChIP-seq track). In this exon, the c.383_392ACTCCACAAA>GACT indel causes the loss of six conserved base pairs, omitting a RUNX3 binding motif (P-val = 2.07X10-3 from TOMTOM [44]) (S3 Fig). RUNX3 has known roles in inflammatory response (e.g., thymopoiesis [45,46] and the TGF-β signaling cascade [47]), and it has been directly implicated in a number of immune-related diseases [48–50]. Furthermore, SNPs disrupting RUNX binding motifs in target genes confer susceptibility to autoimmune rheumatic diseases, including psoriasis [51] and systemic lupus erythematosus [52].
Analysis of three-locus genotypes reveal gene-gene interactions
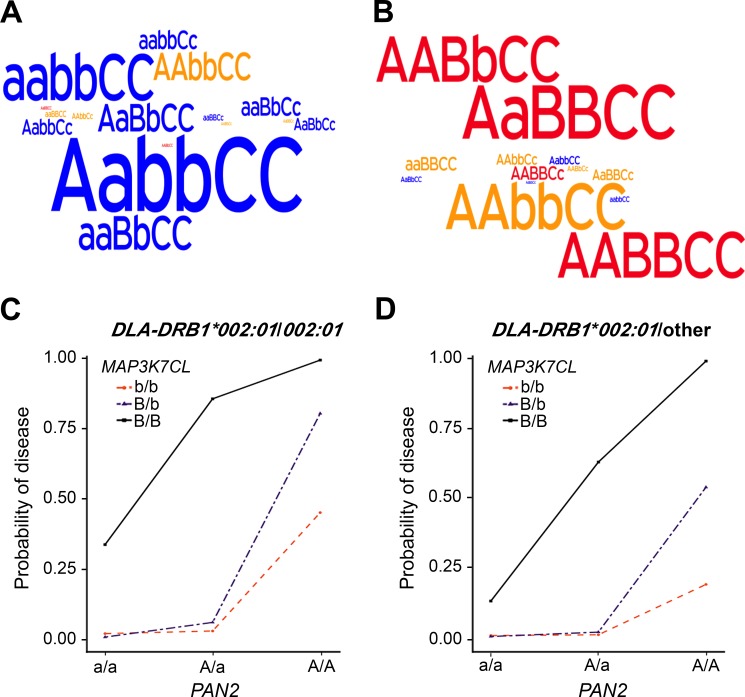
We next considered three-locus genotypes in our expanded, combined population (132 affected and 390 unaffected dogs) where A = the PAN2 variant encoding p.Arg492Cys, B = MAP3K7CL c.383_392ACTCCACAAA>GACT, and C = DLA-DRB1*002:01, lower-case letters denote wild type alleles (c represents any alternate allele of DLA-DRB1). Only 4% of dogs possessed a three-locus genotype with cc, barring further analysis of those nine genotypes. We considered nine of the remaining genotypes to be low-risk, as less than 6% of dogs with these allelic combinations had DMS (Table 3). Among healthy dogs (Fig 3A), the most frequently observed genotypes were AabbCC (24%) and aabbCC (15%). Based on penetrance, we classified five genotypes as conferring moderate risk (33–50%) and four as high risk (90–100%) for DMS. All cases possessed at least two risk alleles and all but one were homozygous for at least one risk allele. The most common genotypes of affected dogs (Fig 3B) were AaBBCC (20% of cases), followed by AAbbCC, AABbCC, and AABBCC (17% of cases each).
Table 3. Distribution of three-locus genotypes in 132 cases and 390 controls with penetrance, 95% confidence intervals, and P-values.
| Genotype | Collies | Shetland sheepdogs | Combined | |||||
|---|---|---|---|---|---|---|---|---|
| Cases (n = 40) | Controls (n = 185) | Cases (n = 92) | Controls (n = 205) | Penetrance (%) | Risk* | 95% Confidence Interval | P-value** | |
| aabbcc | 0 | 1 | 0 | 1 | - | - | 0–0.842 | 1 |
| aabbCc | 0 | 5 | 0 | 18 | - | low | 0–0.148 | 0.0023 |
| aabbCC | 1 | 43 | 1 | 17 | - | low | 0.004–0.112 | 0 |
| Aabbcc | 0 | 0 | 0 | 3 | - | - | 0–0.708 | 0.577 |
| AabbCc | 0 | 3 | 1 | 15 | - | low | 0.001–0.260 | 0.060 |
| AabbCC | 2 | 71 | 2 | 23 | - | low | 0.011–0.101 | 0 |
| aaBbcc | 0 | 0 | 0 | 5 | - | low | 0–0.522 | 0.399 |
| aaBbCc | 0 | 1 | 0 | 21 | - | low | 0–0.154 | 0.002 |
| aaBbCC | 0 | 10 | 0 | 32 | - | low | 0.084 | 0 |
| AaBbcc | 0 | 0 | 0 | 1 | - | - | 0–0.975 | 1 |
| AaBbCc | 0 | 0 | 0 | 14 | - | low | 0–0.232 | 0.028 |
| AaBbCC | 0 | 13 | 2 | 23 | - | low | 0.006–0.177 | 0.002 |
| AAbbcc | 1 | 0 | 0 | 0 | - | - | 0.025–1 | 0.253 |
| AAbbCc | 4 | 6 | 1 | 1 | 42 | moderate | 0.152–0.723 | 0.194 |
| AAbbCC | 15 | 31 | 7 | 3 | 39 | moderate | 0.265–0.532 | 0.021 |
| aaBBcc | 0 | 0 | 0 | 1 | - | - | 0–0.975 | 1 |
| aaBBCc | 0 | 0 | 0 | 6 | - | low | 0.0459 | 0.348 |
| aaBBCC | 0 | 0 | 6 | 7 | 46 | moderate | 0.192–0.749 | 0.107 |
| AaBBcc | 0 | 0 | 2 | 1 | - | - | 0.094–0.992 | 0.160 |
| AaBBCc | 0 | 0 | 5 | 5 | 50 | moderate | 0.187–0.813 | 0.136 |
| AaBBCC | 3 | 0 | 23 | 3 | 90 | high | 0.762–0.978 | 0 |
| AABbcc | 0 | 0 | 3 | 0 | - | - | 0.292–1 | 0.016 |
| AABbCc | 0 | 0 | 2 | 4 | 33 | moderate | 0.043–0.777 | 0.647 |
| AABbCC | 11 | 1 | 11 | 1 | 92 | high | 0.730–0.990 | 0 |
| AABBcc | 0 | 0 | 0 | 0 | - | - | - | - |
| AABBCc | 0 | 0 | 7 | 0 | 100 | high | 0.590–1 | 0.0001 |
| AABBCC | 3 | 0 | 19 | 0 | 100 | high | 0.846–1 | 0 |
Significant values in bold.
*Risk interpretations were only made for three-locus genotypes observed at least five times.
**P-values are calculated for each genotype compared to the population as a whole (0.253, 132 affected dogs out of 522 total dogs).
Fig 3. Three-locus genotype disease probabilities and frequencies (132 cases, 390 controls).
Word clouds of three-locus genotypes illustrate frequency differences between (A) controls and (B) cases with high-risk genotypes in red, moderate-risk in yellow, and low-risk in blue. Probability of disease (y-axis) for all combinations of PAN2 (x-axis) and MAP3K7CL genotypes are plotted in dogs (C) homozygous and (D) heterozygous for DLA-DRB1*002:01.
Interestingly, only affected dogs possessed AABBCc or AABBCC (n = 29), indicating that DMS is fully penetrant in dogs having these combinations. Epistasis plots illustrated that genotypes with at least one a or b allele confer a lower probability of disease when a c allele is present, compared to their CC counterparts (Fig 3, compare 3C and 3D). The plots also depicted a greater probability of disease than expected under a strictly additive model, providing evidence for additive-by-additive epistasis between the chromosome 10 and 31 loci [53,54]. We noted at least one ARE in MAP3K7CL, presenting a mechanism for interaction with PAN2. No difference in gene interactions was observed between the sexes (S4 Fig).
Information regarding age at onset or diagnosis was available for 91 dogs. We compared dogs having two, three, or four risk alleles across PAN2 and MAP3K7CL and observed an inverse correlation between age of onset and number of risk alleles (S5 Fig). Dogs having four risk alleles developed DMS at a significantly younger median age (5 months) than did dogs with only two risk alleles (18.5 months). The complete penetrance of AABB genotypes, combined with an early age of onset, suggest that these dogs may be hypersensitive to commonplace environmental stimuli (e.g., routine puppy vaccinations).
Collies and Shetland sheepdogs have uniquely high frequencies of associated alleles
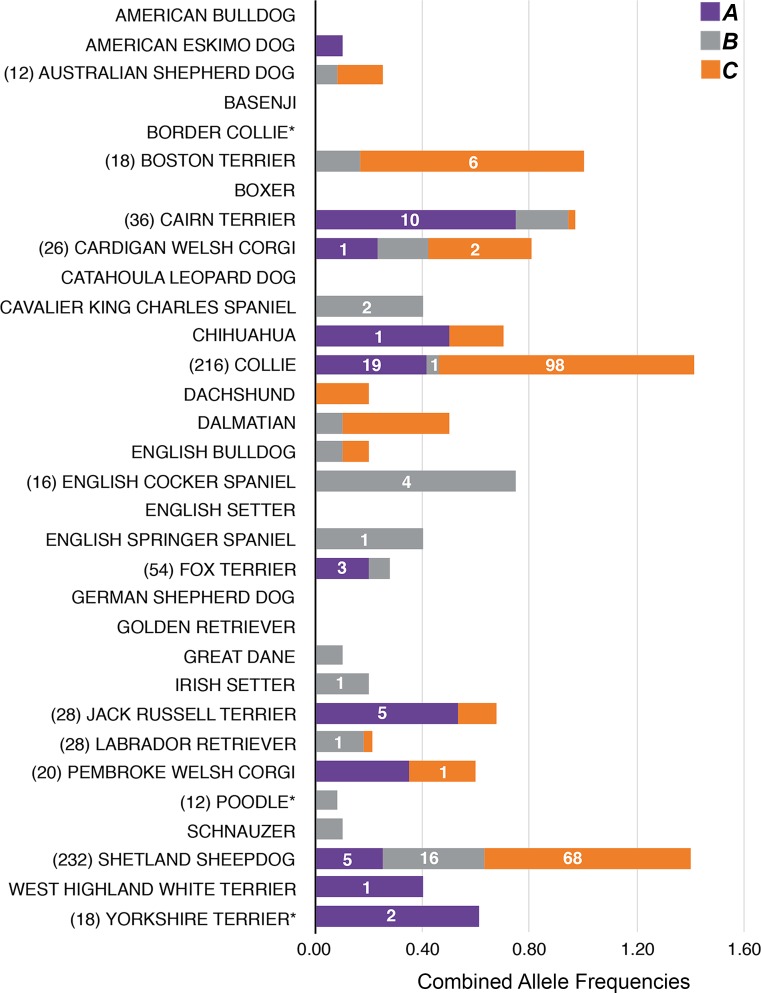
All three identified variants associated with DMS are polymorphisms present in several breeds, raising the question: why are other breeds rarely, if ever, affected by DMS? We genotyped five or more unrelated individuals from each of 30 diverse breeds for all three loci (Fig 4). The only other breeds to possess all three risk alleles were Cardigan Welsh corgis and Cairn terriers. Three Jack Russell terriers had moderate-risk genotypes (AAbbCc), as did one Cardigan Welsh corgi (AABbCc); both breeds are occasionally diagnosed with dermatomyositis-like disease [55,56]. None of the 229 individuals possessed a high-risk genotype (S5 Table). Interestingly, Labrador retrievers had both B and C, which could have enabled moderate or high risk genotypes (AaBBCc or AaBBCC) in puppies from the outcross mating described by Haupt et al. [14].
Fig 4. Frequencies of DMS-associated polymorphisms across breeds.
The number of alleles used to calculate the frequencies is shown to the left of the breed name, if > 10. Risk allele frequencies (x-axis) for each gene are represented by a single, segmented colored bar: the PAN2 variant encoding p.Arg492Cys is represented in purple, MAP3K7CL c.383_392ACTCCACAAA>GACT in gray, and DLA-DRB1*002:01 in orange. The number of homozygous dogs observed is reported on the corresponding color bar. Asterisks denote breeds lacking DLA-DRB1*002:01 in this study, but which were previously reported to possess the allele [57].
Combined frequencies of risk alleles in other breeds were dramatically lower than those observed among collie and Shetland sheepdog populations, and homozygosity for a risk allele (a characteristic of all moderate- to high-risk genotypes) was rare. Additionally, breeds having a high frequency of one risk allele had few or no risk alleles at the other loci. For example, Cairn terriers had a high frequency of A (75%) but low frequencies of B (19%) and C (3%), and no high- or moderate-risk genotypes were observed among 18 individuals. These findings suggest that independently the polymorphisms are neither deleterious nor selected against.
It is likely that recent artificial selection for phenotypes shared by collies and Shetland sheepdogs led to increased frequencies of A. We propose that persistence of A in these two breeds is attributed to linkage disequilibrium (D′ = 0.998) between wildtype PAN2 (a) and another chromosome 10 allele, Merle of PMEL. In heterozygosity, Merle causes a popular pigmentation pattern (see collie in Fig 1A), but homozygosity for the allele results in severe hypopigmentation often with auditory and ocular defects [58]. Wildtype PMEL occurred on chromosomes with either A or a, whereas Merle was only found in conjunction with a. Accordingly, the Merle phenotype was underrepresented in affected dogs (P-value = 0.0018), 64% of which were homozygous for A. Consistent selection for heterozygosity (but not homozygosity) for Merle would simultaneously encourage maintenance of both PAN2 alleles. To our knowledge, there are no loci on chromosome 31 under positive selection for maintenance of a characteristic phenotype of collies and/or Shetland sheepdogs.
Across five collie genomes, we observed a 1.2 Mb selective sweep on chromosome 12 (S6 Fig) encompassing the MHC class II loci and leading to near fixation of the 002:01/009:01/001:01 haplotype. We suggest that essentially all purebred collies have increased susceptibility for DMS, conferred by homozygosity for allele C. The Shetland sheepdog population has retained a less common second haplotype that permits heterozygosity at the DLA loci, associated with a lower risk for developing DMS. Ironically, this reduced risk may have masked the presence of otherwise high-risk genotypes (i.e., AABb and AaBB) and hindered selection against A and B alleles.
We observed striking allele frequency differences between the two affected breeds at PAN2 and MAP3K7CL: collies had a higher frequency of A, 42% (25% in Shetland sheepdogs), whereas Shetland sheepdogs had a higher frequency of B, 38% (5% in collies) (Fig 4). Consequently, the frequency of observed allelic combinations varied between the breeds. The most common genotypes in healthy dogs were aaBbCC in Shetland sheepdogs, 16% (5% in collies), and AabbCC in collies, 38% (11% in Shetland sheepdogs). Among affected dogs, AaBBCC predominated in Shetland sheepdogs, 25% (8% in collies), whereas AAbbCC was the most frequent genotype in diseased collies, 38% (8% in Shetland sheepdogs). The latter finding is interesting given that AAbbCC is only a moderate-risk genotype. Among collies having this genotype, 67% were unaffected by age 8, whereas only 30% of Shetland sheepdogs with this genotype were disease-free. This discrepancy in disease probabilities between breeds was unique to this genotype. Further studies will be necessary to determine if other loci confer additional risk for or protection from DMS.
The contribution of alleles from multiple loci explains the spontaneous appearance of the disease in lines with no prior history [14] and has hindered elimination of DMS in the absence of a genetic test. For example, a mating between two healthy dogs having low risk genotypes (e.g., AaBbCC x AaBbCC) can produce puppies with low, moderate, or high risk for DMS. This study has led to the first three-locus genetic test for a complex disease of dogs, which will allow breeders to carefully reduce the frequency of A and B among collie and Shetland sheepdog populations while preserving genetic diversity and desirable breed characteristics.
In a canine model of JDM, we have identified a complex pattern of causation involving three independent loci, two of which offer new targets for exploration in JDM. Furthermore, these data provide support for the involvement of genetic risk factors independent of the MHC in human inflammatory myopathies. While further experiments are necessary to determine the exact contribution of the chromosome 10 and 31 loci, our findings suggest that DMS may result from an inability to properly regulate inflammatory response. This work highlights the utility of the canine model for unraveling genetic susceptibility conferred by common polymorphisms and/or gene-gene interactions in complex diseases.
Methods
Ethics statement
All samples were obtained with informed consent according to protocols approved by the Clemson University Institutional Review Board (IBC2008-17) and IACUC (2012–039).
Study population
Three populations were assembled: DMS-affected dogs (92 Shetland sheepdogs, 40 collies), control dogs for GWAS (45 Shetland sheepdogs, 23 collies), and unaffected dogs (160 Shetland sheepdogs, 162 collies). Affected dogs were diagnosed either through histopathology (75 Shetland sheepdogs, 30 collies) or by a veterinarian based on clinical signs and elimination of demodectic mange, a differential diagnosis. Pedigrees were collected when available; some samples were obtained from affected dogs surrendered to rescue organizations without paperwork. Twenty percent of affected dogs were collected internationally, and in each case a regionally-matched control sample was obtained.
Control dogs for GWAS were eight years of age or older at time of collection, had no known family history of DMS, had no personal history of skin disease, and were unrelated to other study participants within two (most often three) generations. Pedigrees were obtained from all control dogs. The population of unaffected dogs had no clinical signs of DMS and were eight years of age or older at time of collection. This subset was collected without regard to family history of DMS, presence of other skin disorders, or relationship to other study participants. Archival samples from 229 dogs of 30 other breeds were not closely related to each other to our knowledge.
Whole blood or buccal cells were obtained from each dog, and genomic DNA was isolated according to the Puregene DNA Isolation protocol (Gentra). Skin punch biopsies from active lesions or healthy tissue were also obtained from one Shetland sheepdog, one collie, and one Australian shepherd dog and preserved in RNAlater (Ambion).
DLA class II genotyping
The hypervariable regions of DLA-DRB1, -DQA1 and -DQB1 were sequenced and genotyped according to protocols previously described [59–61]. DLA-DQA1 was largely uninformative in collies and was inferred. Association of a haplotype or allele state with DMS was assessed through Fisher’s exact tests, using VassarStats (Web Site for Statistical Computation, Vassar College, Poughkeepsie, NY).
Genome-wide association and LD analyses
Genotyping was performed for 166 dogs (85 males and 81 females) using the Illumina CanineHD BeadChip, containing 173,662 SNPs. One sample having call rates <95% was excluded. SNPs having call rates <95%, minor allele frequencies <5%, and/or significant deviation from Hardy-Weinberg equilibrium (P-value <0.0001) in the control dogs were excluded. No evidence of genomic inflation was observed in the combined Shetland sheepdog and collie analysis (λ = 1.04). Fisher’s exact P-values were calculated under a dominant model.
LD pairwise analysis was performed to calculate r2 values, which were plotted using LocusZoom [62]. We calculated r2 values between SNPs on chromosome 10 using all controls. For chromosome 31, r2 values were calculated using only control Shetland sheepdogs because no association was detected in collies alone. Assuming that the pathogenic variant would be in high LD with the lead SNP, candidate regions were characterized by SNPs with pairwise r2 ≥ 0.6 and defined by the first flanking associated SNPs in lower LD. Filtering and statistical analyses were conducted with SNP & Variation Suite v8 (SVS, Golden Helix). All chromosome positions throughout the text are reported in CanFam3.1.
Whole genome resequencing
Three affected and two control collies were selected for whole genome resequencing along with one affected Shetland sheepdog. Genomic DNA fragments of approximately 500bp were gel size selected for each sample and sequenced on two lanes of an Illumina HiSeq 2000, generating 2x100 (collies) and 2x125 bp (Shetland sheepdog) paired-end reads. 531 to 997 million total reads were generated for the five collies. Over one billion total reads were generated for the Shetland sheepdog. Paired reads were aligned to the indexed reference (CanFam3.1) with Bowtie2 [63] under sensitive parameters. The alignments were sorted and indexed with SAMtools [64,65] to be visualized in the Interactive Genomics Viewer [66].
RNA isolation and sequencing
Total RNA was extracted from 30-40mg of skin punch biopsy tissue from active lesions (collie and Shetland sheepdog) or healthy skin (Australian shepherd) using the ToTALLY RNA kit (Ambion), according to the manufacturer’s protocols. RNA samples were treated with DNase to remove DNA contamination, using the DNA-free kit (Ambion). Samples were quantitated on a NanoDrop 1000 spectrophotometer (Fisher Scientific).
Three RNAseq libraries were constructed per dog using normalized total RNA and the manufacturer's protocol for one of the following: TruSeq RNA library prep kit v2.0 (Illumina) or TruSeq stranded total RNA library prep kit (Illumina). An Agilent Bioanalyzer 2100 was used for size validation. Each sample was sequenced at 2x125 bp paired-end on an Illimina HiSeq 2500 to a depth of at least 22 million reads.
FastQC from the Babraham Institute was used to assess read quality before and after preprocessing by Trimmomatic [67], which removed low quality bases and remaining sequence adapters. Trimmed reads for each sample were aligned to CanFam3.1 using gsnap [68], and SAMtools [64,65] was used to generate sorted and indexed bam files.
Variant filtering and genotyping
SAMtools and BCFtools [64,65] were used to generate variant call files for each sample, which were analyzed in SVS. On chromosome 10 (1–1,333,693 bp) variants that were heterozygous in affected collies 1 & 2 and homozygous in affected collie 3 and the affected Shetland sheepdog (i.e., reference/reference or alternate/alternate), consistent with the allele states at the lead SNPs, were selected. On chromosome 31 (24,026,411–24,722,836 bp), all homozygous variants in the Shetland sheepdog were selected. Ensembl 79 was used to identify variants lying within predicted exons and 10 bp flanking sequences to capture splice sites. RNAseq data were manually inspected in IGV to determine whether predicted exonic variants were expressed in the affected dogs. Alternate variants were investigated using dbSNP and 27 genomes of 16 other breeds (either sequenced as part of ongoing studies or shared by other research groups) to determine whether any were unique to collies and Shetland sheepdogs. SIFT [31], PolyPhen [32], and PANTHER [33] were used to predict the impact of the amino acid substitutions. Genotyping of variants in additional dogs was accomplished by restriction digest assays or Sanger sequencing (S6 Table).
Age of onset
Because age is not distributed as a normal random variable, we made use of the Weibull distribution, a density commonly used for time-to-event data [69]. Our goal was to estimate the mean and median age of onset, along with their confidence intervals, to facilitate comparisons across the three genotypic groups.
Taking advantage of a Bayesian strategy, we assume yij ~ Weibull (r, ), where yij is the observed age at diagnosis for the j-th (j = 1, 2, …, ni) dog in the i-th (i = 1,2, …,3) genotypic class and bi is the effect of the i-th genotypic class. With this representation of the scale (r) and shape () parameters of the Weibull density, the median of the i-th genotypic class is log(2)1/r and the mean of the i-th genotypic class is Γ(1 + 1/r).
In addition, we assumed a prior density for the unknown genetic parameters (bi) to have a null mean and common variance σ (i.e., (bi ~ N(0, σ)). We consider the weakly informative prior for σ ~ Cauchy(0,25) [70]. For the scale parameter, r, we assume the weakly informative prior of r ~ exponential(0.001). The Monte Carlo Markov Chain (MCMC) sampling process was run in 4 chains through the public-domain software Stan [71], with each chain based on 50,000 total samples, and the first 20,000 were removed as part of the warm-up process, then thinned to every 20th sample, resulting in a MCMC sample of 6,000 values [71]. Convergence to the posterior density was evaluated by the Gelman-Rubin test statistic, where values less than 1.05 indicate that the MCMC sampling process was adequate to the data evaluated [72].
Genetic interaction analyses
All affected (n = 132) and healthy (n = 390) dogs were used to investigate possible interactions between the PAN2 variant encoding p.Arg492Cys and MAP3K7CL c.383_392ACTCCACAAA>GACT in dogs homozygous for 002:01/009:01/001:01 vs. dogs heterozygous for the haplotype. Three-locus genotypes observed in fewer than five dogs were excluded. For a given disease state (cases vs. controls) and a given pair of genotypic classes i (i = A/A, A/a, or a/a) and j (j = B/B, B/b, or b/b), ncases ij ~ Binomial(ncases ij + ncontrols ij, pij) where ncases ij is the number of observed cases in the combination of the genotypic class of locus i and that of locus j, ncontrols ij is the number of unaffected dogs in the combination of genotypic class of locus i and that of locus j. Finally, pij is the probability of disease for the combined genotypic classes of i and j.
Estimation of the unknown elements of our model must ensure estimates of pij within the interval [0, 1], recognizing that several genotypic classes have zero or few cases. We utilized a hierarchical Bayesian framework facilitated through the public-domain software Stan [71] to address this problem [73]. Stan can be accessed through the public-domain language R [74]. Log-odds was used to estimate pij, i.e., log(pij /(1- pij)) = intercept + addAi + domAi+ addBj + domBj + add x addij + add x domij + dom x addij + dom x domij, where intercept represents a term common to all genotypic classes, addAi, addBj domAi and domBj represent the additive and dominance terms respectively for loci A and B, and add x addij + add x domij + dom x addij + dom x domij represent the four possible epistatic interaction terms for all possible additive and dominance combinations [53].
We assumed a prior density for the intercept and unknown genetic parameters, with null mean and common variance σ (i.e., N(0, σ)). Subsequently, we consider the weakly informative prior for σ ~ Cauchy(0,25) [70]. We used the same MCMC parameters here as described above.
Chromosome 12 selective sweeps
We identified all SNPs on chromosome 12 present in five collie genomes (3 cases and 2 controls) and used a creeping window size ≤1 Mb to identify runs of homozygosity [75]. Windows containing fewer than 50 SNPs were excluded and gaps >10kb between SNPs were ignored. The heterozygosity (Hp) statistic was calculated for all windows and Z-transformed, making the average Hp value equal to zero and the standard deviation equal to 1. The–ZHp distribution was plotted in R to show putatively selected regions greater than 3.4 standard deviations from the mean.
PMEL
D' between PMEL and PAN2 was calculated as a measure of LD using all dogs for which coat pattern phenotypes were available (112 cases, 385 controls). A total of 98 dogs (10 cases and 88 controls) were described as merle-patterned, a semi-dominant trait caused by the Merle (M) allele of PMEL, and assumed to possess the Mm genotype. Computation of D', along with a test of significance from zero, was facilitated through the package genetics [76], a public domain program that is part of the R language [74].
Accession numbers
SNP data are available in dbSNP under BioProject number PRJNA338128. All whole genome and transcriptome data generated for this study were deposited in SRA genomes under accesssion number SRP081080. Accession numbers for eight of the 27 other breeds used in variant filtering are SRX1360633, SRX1360635, SRX1360637, SRX1360639, SRX1022256, SRX1022262, SRX1022286, and SRP081080.
Supporting information
Canine dermatomyositis is a vasculopathy that initially manifests as cutaneous lesions across the bony prominences of the face, tail tip, limbs, and feet, shown here. Some dogs develop alopecia and more extensive lesions over time, resulting in dermal scarring associated with erythema and mottled pigmentation.
(PDF)
The –log10P-values (y-axis) for 98,520 Illumina SNPs are plotted against chromosome position (x-axis). The threshold for Bonferroni significance is shown as a black horizontal line. The P-value and position (canFam3) of the lead SNPs are reported. The Q-Q plot is boxed in purple and plots observed vs. expected Fisher’s exact –log10P-values. The inflation factor (λ) is shown on the Q-Q plot.
(PDF)
UCSC 100 Vertebrates track for human chr21:29,130,846–29,130,860 showing the G insertion and seven base pair deletion created by MAP3K7CL indel (Dog c.383_392ACTCCACAAA>GACT). Bases in gray differ from the dog reference sequence. The canine sequence is highlighted in yellow. The RUNX3 binding motif is underlined.
(PDF)
Probability of disease (y-axis) for all combinations of PAN2 (x-axis) and MAP3K7CL genotypes are plotted in dogs (73 affected and 145 unaffected males; 59 affected and 245 unaffected females) (top) homozygous and (bottom) heterozygous for DLA- DRB1*002:01.
(PDF)
Median age of onset is plotted for genotypes consisting of 2 (AAbb, aaBB, AaBb), 3 (AABb, AaBB), and 4 (AABB) risk alleles at PAN2 and MAP3K7CL. CC and Cc genotypes were combined for analyses. Number of individuals is shown to right.
(PDF)
ZH(p) values for all creeping windows containing 50 or more SNPs are plotted against chromosome position. Creeping windows are ≤1 Mb.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors wish to thank Dr. Christine Rees and Sherry Lindsey for contributing samples, as well as the many dog owners and veterinarians. We thank Drs. Gary Johnson and Kathryn Meurs for providing genomes from unaffected dogs. We thank Drs. Mike Vaughan, Michael Sehorn, and Stephen Kresovich for critical reading of the manuscript.
Data availability
SNP data are available in dbSNP under BioProject number PRJNA338128. Whole genome and transcriptome data were deposited in SRA genomes under accesssion number SRP081080.
Funding Statement
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R15AR062868 (www.niams.nih.gov), Collie Health Foundation (www.colliehealth.org), American Shetland Sheepdog Association (www.assa.org), and AKC Canine Health Foundation (www.akcchf.org). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rider LG, Nistala K. The juvenile idiopathic inflammatory myopathies: pathogenesis, clinical and autoantibody phenotypes, and outcomes. J Intern Med. 2016;280:24–38. 10.1111/joim.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendez EP, Lipton R, Ramsey-Goldman R, Roettcher P, Bowyer S, Dyer A, et al. US incidence of juvenile dermatomyositis, 1995–1998: results from the National Institute of Arthritis and Musculoskeletal and Skin Diseases Registry. Arthritis Rheum. 2003;49:300–305. 10.1002/art.11122 [DOI] [PubMed] [Google Scholar]
- 3.Bowyer SL, Blane CE, Sullivan DB, Cassidy JT. Childhood dermatomyositis: factors predicting functional outcome and development of dystrophic calcification. J Pediatr. 1983;103:882–888. [DOI] [PubMed] [Google Scholar]
- 4.Manlhiot C, Liang L, Tran D, Bitnun A, Tyrrell PN, Feldman BM. Assessment of an infectious disease history preceding juvenile dermatomyositis symptom onset. Rheumatology 2008;47:526–529. 10.1093/rheumatology/ken038 [DOI] [PubMed] [Google Scholar]
- 5.Reed AM, Stirling JD. Association of the HLA-DQA1*0501 allele in multiple racial groups with juvenile dermatomyositis. Hum Immunol. 1995;44:131–135. [DOI] [PubMed] [Google Scholar]
- 6.Pachman LM, Liotta-Davis MR, Hong DK, Kinsella TR, Mendez EP, Kinder JM, et al. TNFα-308A allele in juvenile dermatomyositis: Association with increased production of tumor necrosis factor α, disease duration, and pathologic calcifications. Arthritis Rheum. 2000;43:2368–2377. [DOI] [PubMed] [Google Scholar]
- 7.Mamyrova G, O'Hanlon TP, Monroe JB, Carrick DM, Malley JD, Adams S, et al. Immunogenetic risk and protective factors for juvenile dermatomyositis in Caucasians. Arthritis Rheum. 2006;54:3979–3987. 10.1002/art.22216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamyrova G, O'Hanlon TP, Sillers L, Malley K, James-Newton L, Parks CG et al. Cytokine gene polymorphisms as risk and severity factors for juvenile dermatomyositis. Arthritis Rheum. 2008;58:3941–3950. 10.1002/art.24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller FW, Cooper RG, Vencovský J, Rider LG, Danko K, Wedderburn LR, et al. Genome-wide association study of dermatomyositis reveals genetic overlap with other autoimmune disorders. Arthritis Rheum. 2013;65:3239–3247. 10.1002/art.38137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller FW, Chen W, O'Hanlon TP, Cooper RG, Vencovsky J, Rider LG, et al. Genome-wide association study identifies HLA 8.1 ancestral haplotype alleles as major genetic risk factors for myositis phenotypes. Genes Immun. 2015;16:470–480. 10.1038/gene.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargis AM, Haupt KH, Hegreberg GA, Prieur DJ, Moore MP. Familial canine dermatomyositis. Initial characterization of the cutaneous and muscular lesions. Am J Pathol. 1984;116:234–244. [PMC free article] [PubMed] [Google Scholar]
- 12.Hargis AM, Haupt KH, Prieur DJ, Moore MP. Animal model of human disease. Dermatomyositis. Familial canine dermatomyositis. Am J Pathol. 1985;120:323–325. [PMC free article] [PubMed] [Google Scholar]
- 13.Haupt KH, Prieur DJ, Hargis AM, Cowell RL, McDonald TL, Werner LL, et al. Familial canine dermatomyositis: clinicopathologic, immunologic, and serologic studies. Am J Vet Res. 1985;46:1870–1875. [PubMed] [Google Scholar]
- 14.Haupt KH, Prieur DJ, Moore MP, Hargis AM, Hegreberg GA, Gavin PR, et al. Familial canine dermatomyositis: clinical, electrodiagnostic, and genetic studies. Am J Vet Res. 1985;46:1861–1869. [PubMed] [Google Scholar]
- 15.Nagaraju K, Plotz PH. Animal models of myositis. Rheum Dis Clin N Am. 2002;28:917–933. [DOI] [PubMed] [Google Scholar]
- 16.Avci P, Sadasivam M, Gupta A, De Melo WC, Huang YY, Yin R, et al. Animal models of skin disease for drug discovery. Expert Opin Drug Discov. 2013;8:331–355. 10.1517/17460441.2013.761202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkle GA, Chrisman CL, Gross TL, Fadok V, Werner LL. Dermatomyositis in collie dogs. Comp Contin Educ Pract Vet. 1985;7:185–192. [Google Scholar]
- 18.Hargis A, Mundell A. Familial canine dermatomyositis. Compend Contin Educ Vet. 1992;4:855–864. [Google Scholar]
- 19.Scott DW, Miller WH, Griffin CE. Congenital and hereditary defects In: Muller and Kirk’s Small Animal Dermatology, 6th ed. Philadelphia: Saunders, 2000. pp. 940–946. [Google Scholar]
- 20.Wahl JM, Clark LA, Skalli O, Ambrus A, Rees CA, Mansell JL, et al. Analysis of gene transcript profiling and immunobiology in Shetland sheepdogs with dermatomyositis. Vet Dermatol. 2008;19:52–58. 10.1111/j.1365-3164.2008.00655.x [DOI] [PubMed] [Google Scholar]
- 21.Hargis AM, Prieur DJ, Haupt KH, McDonald TL, Moore MP. Prospective study of familial canine dermatomyositis: correlation of the severity of dermatomyositis and circulating immune complex levels. Am J Pathol. 1986;123:465–479. [PMC free article] [PubMed] [Google Scholar]
- 22.Rider LG, Wu L, Mamyrova G, Targoff IN, Miller FW. Environmental factors preceding illness onset differ in phenotypes of the juvenile idiopathic inflammatory myopathies. Rheumatology 2010;49:2381–2390. 10.1093/rheumatology/keq277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dourmishev LA, Dourmishev AL. Dermatomyositis: Advances in Recognition, Understanding and Management. New York: Springer; 2009. [Google Scholar]
- 24.vonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010;464:898–902. 10.1038/nature08837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilbe M, Jokinen P, Truvé K, Seppala EH, Karlsson EK, Biagi T et al. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet. 2010;42:250–254. 10.1038/ng.525 [DOI] [PubMed] [Google Scholar]
- 26.Karlsson EK, Sigurdsson S, Ivansson E, Thomas R, Elvers I, Wright J et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013;14:R132 10.1186/gb-2013-14-12-r132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karyadi DM, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, Parker HG et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 2013;9:e1003409 10.1371/journal.pgen.1003409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang R, Noh HJ, Wang D, Sigurdsson S, Swofford R, Perloski M, et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 2014;15:R25 10.1186/gb-2014-15-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivansson EL, Megquier K, Kozyrev SV, Murén E, Körberg IB, Swofford R, et al. Variants within the SP110 nuclear body protein modify risk of canine degenerative myelopathy. Proc Natl Acad Sci. 2016;113:E3091–100. 10.1073/pnas.1600084113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melin M, Rivera P, Arendt M, Elvers I, Murén E, Gustafson U, et al. Genome-wide analysis identifies germ-line risk factors associated with canine mammary tumours. PLoS Genet. 2016;12:e1006029 10.1371/journal.pgen.1006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 32.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2232. 10.1093/bioinformatics/btw222 [DOI] [PubMed] [Google Scholar]
- 34.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001;104:377–386. [DOI] [PubMed] [Google Scholar]
- 35.Wolf J, Passmore LA. mRNA deadenylation by Pan2-Pan3. Biochem Soc Trans. 2014;42:184–187. 10.1042/BST20130211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weill L, Belloc E, Bava FA, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. 10.1038/nsmb.2311 [DOI] [PubMed] [Google Scholar]
- 37.Bett JS, Ibrahim AF, Garg AK, Kelly V, Pedrioli P, Rocha S, et al. The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. Biochem J. 2013;451:185–194. 10.1042/BJ20130026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity 2014;41:518–528. 10.1016/j.immuni.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preuße C, Allenbach Y, Hoffmann O, Goebel HH, Pehl D, Radke J, et al. Differential roles of hypoxia and innate immunity in juvenile and adult dermatomyositis. Acta Neuropathol Commun. 2016;4:45 10.1186/s40478-016-0308-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilgic H, Ytterberg SR, Amin S, McNallan KT, Wilson JC, Koeuth T, et al. Interleukin-6 and type I interferon-regulated genes and chemokines mark disease activity in dermatomyositis. Arthritis Rheum. 2009;60:3436–3446. 10.1002/art.24936 [DOI] [PubMed] [Google Scholar]
- 42.Li J, Ji C, Yang Q, Chen J, Gu S, Ying K, et al. Cloning and characterization of a novel human TGF-beta activated kinase-like gene. Biochem Genet. 2004;42:129–137. [DOI] [PubMed] [Google Scholar]
- 43.Kilpinen S, Ojala K, Kallioniemi O. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS One 2010;5:e15068 10.1371/journal.pone.0015068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA. 2003;100:7731–7736. 10.1073/pnas.1232420100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Djuretic IM1, Cruz-Guilloty F, Rao A. Regulation of gene expression in peripheral T cells by Runx transcription factors. Adv Immunol. 2009;104:1–23. 10.1016/S0065-2776(08)04001-7 [DOI] [PubMed] [Google Scholar]
- 47.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. 10.1038/sj.emboj.7600085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E, et al. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci USA. 2004;101:16016–16021. 10.1073/pnas.0407180101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo C, Yao F, Wu K, Yang L, Zhang X, Ding J. Chromatin immunoprecipitation and association study revealed a possible role of Runt-related transcription factor 3 in the ulcerative colitis of Chinese population. Clin Immunol. 2010;135:483–489. 10.1016/j.clim.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 50.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. 10.1038/ng.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helms C, Cao L, Krueger JG, Wijsman EM, Chamian F, Gordon D, et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet. 2003;35:349–356. 10.1038/ng1268 [DOI] [PubMed] [Google Scholar]
- 52.Prokunina L, Castillejo-López C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–669. 10.1038/ng1020 [DOI] [PubMed] [Google Scholar]
- 53.Hallgrímsdóttir IB, Yuster DS. A complete classification of epistatic two-locus models. BMC Genet. 2008;9:17 10.1186/1471-2156-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackay TR. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 2014;15:22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker WM, Foster RA. Cutaneous vasculitis in five Jack Russell Terriers. Vet Dermatol. 1996;7:109–115. [DOI] [PubMed] [Google Scholar]
- 56.Ihrke PJ. Ischemic skin disease in the dog. 31st World Small Animal Veterinary Congress (2006).
- 57.Kennedy LJ, Barnes A, Short A, Brown JJ, Lester S, Seddon J, et al. Canine DLA diversity: 1. New alleles and haplotypes. Tissue Antigens 2007;69:272–288. 10.1111/j.1399-0039.2006.00779.x [DOI] [PubMed] [Google Scholar]
- 58.Clark LA, Wahl JM, Rees CA, Murphy KE. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA 2006;103:1376–1381. 10.1073/pnas.0506940103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans JM, Cox ML, Huska J, Li F, Gaitero L, Guo LT, et al. Exome sequencing reveals a nebulin nonsense mutation in a dog model of nemaline myopathy. Mamm Genome 2016;27:495–502. 10.1007/s00335-016-9644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy LJ, Davison LJ, Barnes A, Short AD, Fretwell N, Jones CA, et al. Identification of susceptibility and protective major histocompatibility complex haplotypes in canine diabetes mellitus. Tissue Antigens 2006;68:467–476. 10.1111/j.1399-0039.2006.00716.x [DOI] [PubMed] [Google Scholar]
- 61.Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SGE. IPD—the immuno polymorphism database. Nucleic Acids Res. 2013;41:1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–7. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009;25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein JP, Moeschberger ML. Survival analysis: techniques for censored and truncated data. New York: Springer; 2005. [Google Scholar]
- 70.Gelman A. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 2006;1:515–534. [Google Scholar]
- 71.Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B, Betancourt M, et al. Stan: A probabilistic programming language. J Stat Softw. 2016;Forthcoming [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gelman A, Rubin D. Inference from iterative simulation using multiple sequences. Statist Sci. 1992;7:457–472. [Google Scholar]
- 73.Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. Bayesian Data Analysis, 3rd ed. Broken Sound Parkway: CRC Press Taylor & Francis Group; 2014. [Google Scholar]
- 74.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2015. [Google Scholar]
- 75.Qanbari S, Strom TM, Haberer G, Weigend S, Gheyas AA, Turner F, et al. A high resolution genome-wide scan for significant selective sweeps: an application to pooled sequence data in laying chickens. PLoS One 2012;7:e49525 10.1371/journal.pone.0049525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warnes G, Gorjanc G, Leisch F, Man M. Genetics: Population Genetics. R package version 1.3.8.1 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Canine dermatomyositis is a vasculopathy that initially manifests as cutaneous lesions across the bony prominences of the face, tail tip, limbs, and feet, shown here. Some dogs develop alopecia and more extensive lesions over time, resulting in dermal scarring associated with erythema and mottled pigmentation.
(PDF)
The –log10P-values (y-axis) for 98,520 Illumina SNPs are plotted against chromosome position (x-axis). The threshold for Bonferroni significance is shown as a black horizontal line. The P-value and position (canFam3) of the lead SNPs are reported. The Q-Q plot is boxed in purple and plots observed vs. expected Fisher’s exact –log10P-values. The inflation factor (λ) is shown on the Q-Q plot.
(PDF)
UCSC 100 Vertebrates track for human chr21:29,130,846–29,130,860 showing the G insertion and seven base pair deletion created by MAP3K7CL indel (Dog c.383_392ACTCCACAAA>GACT). Bases in gray differ from the dog reference sequence. The canine sequence is highlighted in yellow. The RUNX3 binding motif is underlined.
(PDF)
Probability of disease (y-axis) for all combinations of PAN2 (x-axis) and MAP3K7CL genotypes are plotted in dogs (73 affected and 145 unaffected males; 59 affected and 245 unaffected females) (top) homozygous and (bottom) heterozygous for DLA- DRB1*002:01.
(PDF)
Median age of onset is plotted for genotypes consisting of 2 (AAbb, aaBB, AaBb), 3 (AABb, AaBB), and 4 (AABB) risk alleles at PAN2 and MAP3K7CL. CC and Cc genotypes were combined for analyses. Number of individuals is shown to right.
(PDF)
ZH(p) values for all creeping windows containing 50 or more SNPs are plotted against chromosome position. Creeping windows are ≤1 Mb.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
SNP data are available in dbSNP under BioProject number PRJNA338128. Whole genome and transcriptome data were deposited in SRA genomes under accesssion number SRP081080.