Abstract
Precise separation of spermatogonial stem cells (SSCs) from progenitor spermatogonia that lack stem cell activity and are committed to differentiation remains a challenge. To distinguish between these spermatogonial subtypes, we identified genes that exhibited bimodal mRNA levels at the single-cell level among undifferentiated spermatogonia from Postnatal Day 6 mouse testes, including Tspan8, Epha2, and Pvr, each of which encode cell surface proteins useful for cell selection. Transplantation studies provided definitive evidence that a TSPAN8-high subpopulation is enriched for SSCs. RNA-seq analyses identified genes differentially expressed between TSPAN8-high and -low subpopulations that clustered into multiple biological pathways potentially involved in SSC renewal or differentiation, respectively. Methyl-seq analysis identified hypomethylated domains in the promoters of these genes in both subpopulations that colocalized with peaks of histone modifications defined by ChIP-seq analysis. Taken together, these results demonstrate functional heterogeneity among mouse undifferentiated spermatogonia and point to key biological characteristics that distinguish SSCs from progenitor spermatogonia.
Keywords: epigenetics, single-cell, spermatogonial stem cells, surface marker, transcriptome, transplantation
INTRODUCTION
Conventional gene expression analyses yield aggregate results from thousands or millions of cells, and therefore obscure specific subtypes within a heterogeneous cell population. This poses a particular challenge for studies of rare stem cells, such as the spermatogonial stem cells (SSCs) that support the seminiferous epithelium in the mammalian testis. SSCs are undifferentiated male germ cells that facilitate ongoing sperm production and male fertility by producing progeny cells that will either self-renew to maintain the stem cell pool or initiate differentiation yielding committed progenitors that will ultimately produce spermatozoa [1–5]. Mechanisms underlying distinctions between these cell fates remain poorly understood, in part because SSCs are extremely rare (∼3000 per adult mouse testis [6]) and no endogenous marker has been described that can effectively facilitate their prospective purification (reviewed in [7]). However, experimental confirmation of SSC activity can be accomplished retrospectively using a functional transplantation assay that measures the ability of transplanted cells to seed spermatogenesis in a recipient testis [8, 9].
Recently, Chan et al. [10] reported a transgenic mouse line in which Id4 gene regulatory sequences restrict EGFP expression to a small fraction of undifferentiated spermatogonia enriched for SSCs. However, even this ID4-EGFP+ subpopulation of spermatogonia appears to be heterogeneous for SSCs and progenitors committed to differentiation. We reasoned that further resolution of differential gene expression among cells within the ID4-EGFP+ spermatogonial subpopulation could be used to further enhance enrichment of SSCs. Our recent single-cell analysis of gene expression among spermatogonia in the Postnatal Day 6 (P6) mouse testis revealed three clusters of spermatogonia displaying distinct gene expression signatures [11]. Here, we exploited these gene expression differences to recover two discrete subpopulations of ID4-EGFP+ spermatogonia based on high or low expression of TSPAN8, a sortable cell surface marker. Transplantation analysis showed that these subpopulations differ in SSC content (enriched in ID4-EGFP+/TSPAN8High cells), and subsequent studies of gene expression, histone modification, and DNA methylation patterns provided unprecedented insight into molecular characteristics of the SSC-enriched subpopulation. Our results indicate that functionally distinct subtypes of undifferentiated spermatogonia are present in the P6 mouse testis, and that gene expression differences between these spermatogonial subtypes reflect developmentally relevant differences in cell fate characteristic of SSCs and committed progenitor spermatogonia.
MATERIALS AND METHODS
Animals and Testis Cell Isolations
All experiments utilizing animals were approved by the Institutional Animal Care and Use Committees of the University of Texas at San Antonio (Assurance A3592-01), East Carolina University (Assurance A3469-01), or Washington State University (Assurance A3485-01), and were performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Testes from P6 F1 offspring (at least two animals per experiment) from a cross between Id4-eGfp (LT-11B6 [10]) and either C57BL/6J or B6;129S-Gt (ROSA)26Sor/J [12]; both from The Jackson Laboratory) were used to generate suspensions of cells following enzymatic digestion, as described previously [11, 13–15].
Flow Cytometry and Fluorescence-Activated Cell Sorting
Testis cell suspensions were used for flow cytometry and fluorescence-activated cell sorting (FACS), essentially as described previously [11]. Briefly, cells were suspended (5–20 × 106 cells/ml) in ice-cold Dulbecco PBS (DPBS) containing 10% FBS (DPBS + S), labeled with antibodies (Supplemental Table S1; Supplemental Data are available online at www.biolreprod.org), and subjected to flow cytometry using an LSRII cytometer (BD) or FACS using either a FACS Aria (BD) or SY3200 (Sony). Positive antibody labeling was determined by comparison to staining with isotype control antibodies (Supplemental Table S1). Positive ID4-EGFP epifluorescence was determined by comparison to testis cells from P6 Id4-eGfp− littermates. For discrimination of dead cells, we used either propidium iodide (Biolegend) or LIVE/DEAD Fixable Violet or Near-IR Dead Cell Stain Kits (ThermoFisher Scientific).
Cell Cycle Analyses
For cell cycle analysis, cells were suspended in DPBS + S and treated with 50 μM verapamil (Sigma-Aldrich) for 5 min at 37°C. Subsequently, cells were labeled with 5–10 μM Vybrant DyeCycle Violet Stain (ThermoFisher Scientific) for an additional 30 min at 37°C. Cells were then cooled on ice for 5 min and labeled with antibodies, as noted above, except that all washes and antibody incubations were performed with ice-cold DPBS + S containing 50 μM verapamil and 5 μM Vybrant DyeCycle Violet Stain. Evaluation of cell staining was performed utilizing an LSRII cytometer and cell cycle state was determined from these data using FlowJo v.10.0.7 with the Cell Cycle Univariate analysis [16]. Results were from four independent labeling experiments.
Immunostaining of Id4-eGFP testes
P6 Id4-eGFP+ testes were stained for TSPAN8 and EPHA2 proteins, as described previously [10]. Primary and secondary antibodies are noted in Supplemental Table S1, and sections were counterstained with either phalloidin-635 (Thermofisher Scientific) or 4′,6-diamidino-2-phenylindole (Vector Laboratories). Primary antibodies were omitted as a negative control.
SSC Transplantation
Cells from Id4-eGfp+/Rosa-LacZ F1 hybrid pups were sorted and transplanted into the seminiferous tubules of busulfan-treated recipient mice as described previously [15]. Briefly, sorted cell suspensions were diluted in medium to 1 × 106 cells/ml and ∼10 μl was microinjected into the seminiferous tubules of each adult 129×C57 F1 hybrid busulfan-treated (60 mg/kg) recipient mouse testis. One testis of each recipient received TSPAN8High cells, and the contralateral testis received TSPAN8Low cells. Presence of donor-derived colonies of spermatogenesis was detected ∼2–3 mo posttransplantation by staining with X-Gal, and spermatogenic colonies were counted. Results shown are from 30 recipient testes and four replicate cell-sorting and transplant experiments.
RNA-seq
Sorted cells were pelleted, counted (Supplemental Table S2), and subjected to direct cDNA synthesis using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech Laboratories), per the manufacturer recommendations, with nine cycles of amplification (Supplemental Table S3). Using 250 pg input cDNA, we prepared Nextera XT dual-index libraries with modifications from manufacturer recommendations: a) tagmentation was performed with 2.5 μl Tagment DNA buffer, 1.25 μl Amplification Tagment Mix, and 1.25 μl cDNA for 10 min at 55°C, ramp to 10°C, and immediate addition of 1.25 μl NT buffer; and b) PCR amplification with index primers was performed with the entire 6.25 μl of Tagmentation reaction mix plus 3.75 μl Nextera PCR Mix with recommended cycling conditions and 60-sec extension. Libraries were qualified for fragment size and distribution on a 2100 Bioanalyzer (522 ± 6 bp; Table S3), pooled at equal molarity, and subjected to rapid-mode Illumina HiSeq2500 sequencing (paired-end 100 bp) at the University of Texas Southwestern Medical Center Genomics and Microarray Core. Resulting FASTQ files from each sample were merged, trimmed, and quality was confirmed with FASTQC. Trimmed FASTQs were aligned to the mouse genome (mm9) with TopHat v2.0.12 and Bowtie v2.2.3.0, and transcript abundance was determined with Cufflinks [17]. Raw and processed data were submitted to NIH Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) databases under accession number GSE83311. Transcript abundance values (FPKM) for each gene in each sample were normalized and used for differential gene expression analysis, as described previously [18], producing normalized expression counts. We considered genes with normalized expression counts of >2 to be expressed above the detection threshold. Statically significant differences in transcript abundance between samples were determined using an associative t-test [18]. For gene ontology (GO) analyses, lists of differentially expressed genes were analyzed by Ingenuity Pathway Analysis (build 377306M, content version 27216297, 3/16/2016; Qiagen). Hierarchical clustering heatmaps were generated using ComplexHeatmap (v1.10.2) and R (v3.3.0) with genes clustered on Ward linkage based on the Euclidean distance [19] and samples clustered based on the Spearman rank correlation coefficient [20]. For some analyses, the gene dendograms were split into six clusters according to k means.
Quantitative RT-PCR
Sorted cells were pelleted and suspended in lysis buffer for the RNAqueous-Micro Total RNA Isolation kit (ThermoFisher Scientific) and RNA was extracted according to manufacturer recommendations. Genomic DNA was removed with the Turbo DNA-free kit (ThermoFisher Scientific). Complementary DNA was synthesized from DNase-treated RNA, as described previously [21], using SuperScript III reverse transcriptase (ThermoFisher Scientific) and oligo-dT18 priming. All Taqman assays and primer sets were validated for 90%–100% efficiency (Supplemental Table S4). For quantitative PCR, reactions were carried out in triplicate for each sample and primer set using Power SYBR green PCR master mix (ThermoFisher Scientific) on a 7300 Real-Time PCR System (Applied Biosystems). For two transcripts, Aldh1a3 and Dmrt2, TaqMan assays (Mm00474049_m1 and Mm00659912_m1) were used with TaqMan Universal PCR master mix. The relative mRNA abundance for each gene of interest was calculated using the ΔΔCt method, where Actb cDNA amplification was used for normalization to determine the fold-change value (2−ΔΔCt), and significant differences between samples were identified using t-tests.
Reduced-Representation Bisulfite Sequencing
Snap-frozen cell pellets from five independent sorts of P6 ID4-EGFP+/TSPAN8High and TSPAN8Low subpopulations were pooled and used for genomic DNA isolation and reduced-representation bisulfite sequencing (RRBS) using the Methyl-MidiSeq service (Zymo Research). Libraries were prepared from 300 ng of genomic DNA digested with the BfaI, MseI, and MspI restriction enzymes, and the fragments produced were ligated to preannealed adapters containing 5-methylcytosine instead of cytosine. Adapter-ligated fragments were filled in and 3′-terminal-A extended, and purified. Bisulfite treatment of the fragments was done using the EZ DNA Methylation–Lightning kit (Zymo Research). PCR was performed and the size and concentration of the fragments were confirmed on an Agilent 2200 TapeStation, then sequenced on an Illumina Hiseq 2500 (paired-end 50 bp). Sequence reads were analyzed using a Zymo Research proprietary analysis pipeline. The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T. Fisher exact test or t-test was performed for each CpG site that has at least five reads covered. In addition, promoter, gene body, and CpG island annotations were added for each CpG. Raw and processed data were submitted to NIH GEO and SRA databases under accession number GSE83422.
Chromatin Immunoprecipitation-seq
Snap-frozen cell pellets from eight independent sorts of the ID4-EGFP+/TSPAN8High and TSPAN8Low subpopulations were pooled and subjected to low-input chromatin immunoprecipitation (ChIP)-seq service (Active Motif). Cells were fixed with 1% formaldehyde for 15 min, quenched with 0.125 M glycine, and cell lysates were sonicated to shear the chromatin to an average length of 300–500 bp. Genomic DNA (input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for decrosslinking, followed by ethanol precipitation. ChIP reactions contained chromatin from either 9000 cell equivalents (H3K4me3) or 34 000 cell equivalents (H3K27me3 and H3K27Ac), assuming 6.6 pg chromatin/cell. Chromatin was precleared with protein-A-agarose beads (ThermoFisher Scientific) and then used for ChIP using 4 μg of antibodies against H3K4me3, H3K27ac, or H3K27me3 (Supplemental Table S1). Immune complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. Input or ChIP'd genomic DNA were subsequently used to prepare Illumina sequencing libraries by the standard consecutive enzymatic steps of end polishing, dA addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on a NextSeq 500 (75 bp, single end; Illumina). Reads were aligned to the mouse genome (mm9) using BWA (default settings), retaining only uniquely mapped reads (mapping quality ≥25) for further analysis. Alignments were extended in silico at their 3′ ends to a length of 200 bp, and assigned to 32-nt bins along the genome. Enriched genomic regions for each histone mark were identified using SICER (FDR 1E-10, gap size = 600 bp [22]). For comparative tracks, genomic positions were converted to the mm10 assembly/annotation by liftOver. Raw and processed data were submitted to NIH GEO and SRA databases under accession number GSE83422.
RESULTS
Genes with Bimodal Expression Patterns Define Subpopulations of Undifferentiated Spermatogonia
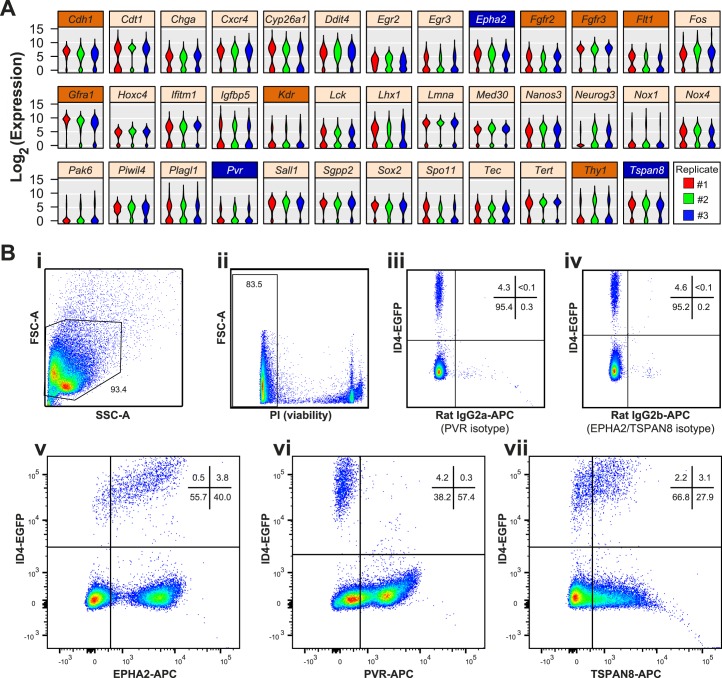
Previously reported single-cell quantitative RT-PCR (qRT-PCR) data from 229 individual P6 ID4-EGFP+ undifferentiated spermatogonia [11] were examined for genes exhibiting bimodal expression patterns indicative of subgroups within the EGFP+ spermatogonial population. For any given gene, this bimodal pattern was characterized by a population of cells with detectable mRNA levels and another population without detectable mRNA. While many of the 189 genes we previously examined exhibited uniform mRNA levels (see examples highlighted in [11]), 38 genes exhibited bimodal mRNA abundance patterns (Fig. 1A), 10 of which encoded cell surface proteins, and for three of which (EPHA2, PVR, and TSPAN8), robust antibodies could be used with flow cytometry and FACS to further fractionate the EGFP+ spermatogonial population (Fig. 1B, Supplemental Fig. S1). Importantly, none of these markers was restricted to ID4-EGFP+ spermatogonia in the testis (Fig. 1B, vi–vii, bottom-right quadrants, Supplemental Fig. S2). Immunofluorescence staining for TSPAN8 and EPHA2 in sections of P6 Id4-eGfp testes confirmed heterogeneous expression of these proteins among EGFP+ spermatogonia in vivo (Supplemental Fig. S2). Thus, these sortable markers can be used to subdivide ID4-EGFP+ spermatogonia into multiple subpopulations.
FIG. 1.
Bimodal mRNA abundance predicts subpopulations of P6 ID4-EGFP+ undifferentiated spermatogonia. A) Violin plots depict bimodal patterns of mRNA abundance among individual P6 ID4-EGFP spermatogonia based on single-cell qRT-PCR analyses (data are from [11]; the original publication details examples of genes not exhibiting bimodal abundance [e.g., Ddx4, Dazl, Itga6, Zbtb16]). Each violin is a histogram that shows log2-transformed Ct values for each replicate cell preparation (three violins per gene), where width (x-axis) at each expression level (y-axis) is indicative of the relative proportion of cells with that degree of mRNA abundance. Shown are results from 38 genes (from a total of 189 examined) that exhibited this bimodal pattern of mRNA abundance. Of the 10 bimodal genes encoding cell surface proteins, 7 (orange label bars) lacked suitable antibodies for flow cytometry or had already been investigated (e.g., GFRA1, THY1), but robust antibodies were available for the proteins encoded by Epha2, Pvr, and Tspan8 (blue label bars). B) Flow cytometry was used to characterize antibody staining for EPHA2, PVR, and TSPAN8 among testis cells from P6 Id4-eGfp mice. Density dot plots show sequential cell pregating: cells were selected based on light scatter characteristics (FSC-A × SSC-A; i) and viability (propidium iodide negative; ii). iii and iv) 4%–5% of viable single cells exhibited positive EGFP+ epifluorescence (compared with Id4-eGfp-negative littermates, not shown). EGFP+ spermatogonia exhibited either minimal background staining with isotype control antibodies (iii and iv) or abundant positive staining with antibodies against EPHA2 (v), PVR (vi), or TSPAN8 (vii). Dots denote individual cells and blue-to-red coloring scale is indicative of increasing cell number/density. The proportion of cells in gates and quadrants are noted.
Cell Cycle State Does Not Account for Bimodal Expression and Heterogeneity among Undifferentiated Spermatogonia
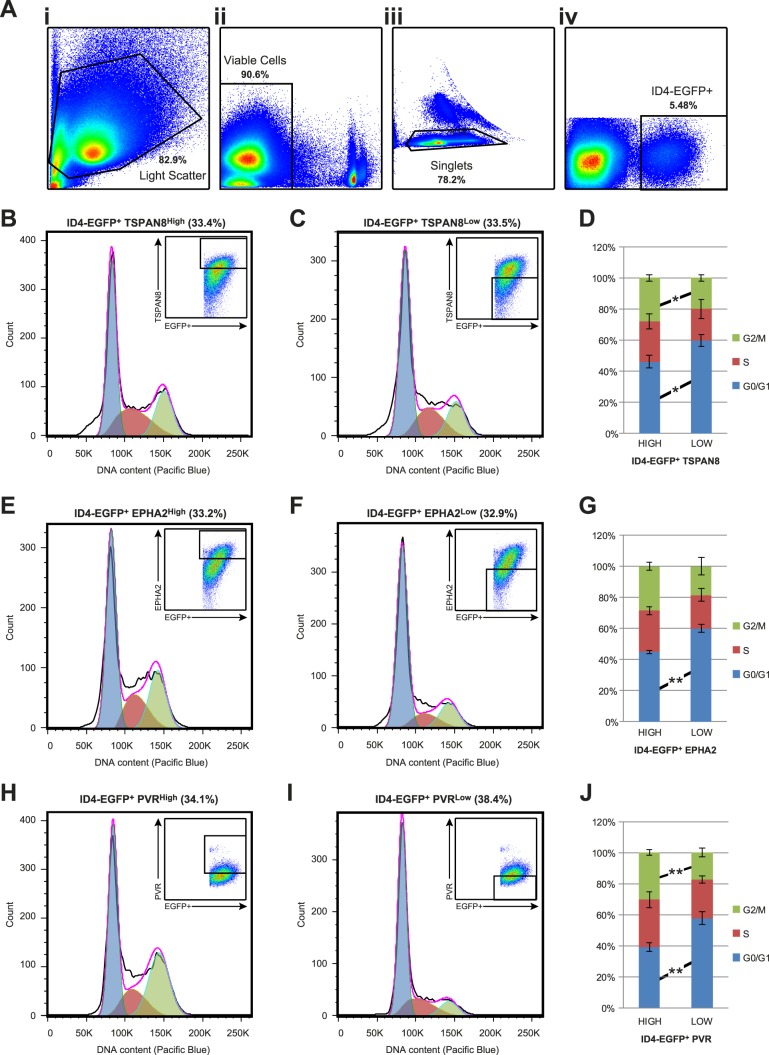
One possible explanation for gene expression heterogeneity among cells in a population is variance as a function of the cell cycle [23–25]. To address this possibility, we performed DNA content cell cycle analyses of live P6 ID4-EGFP+ spermatogonia (Fig. 2A) exhibiting higher or lower expression of TSPAN8 (TSPAN8High vs. TSPAN8Low; Fig. 2, B–D), EPHA2 (EPHA2High vs. EPHA2Low; Fig. 2, E–G), or PVR (PVRHigh vs. PVRLow; Fig. 2, H–J). All subpopulations showed a greater proportion of cells in G0/G1 than any other phase of the cell cycle (Fig. 2, B–J). Small, but statistically significant, differences in the proportion of cells in G1/G0 and G2/M phases were observed between members of each subpopulation pair (Fig. 2, D, G, and J). Specifically, the subpopulation exhibiting higher marker expression also exhibited proportionally fewer cells in G0/G1 phase of the cell cycle (Fig. 2, D, G, and J). However, despite exhibiting opposing marker staining/expression levels, the greatest difference in cell cycle state was 1.67-fold (proportion of PVRhigh/low cells in G2/M). Thus, the relatively subtle differences in cell cycle state do not account for the significant, bimodal gene expression differences between each spermatogonial subpopulation pair (e.g., presence vs. absence of mRNA for a given gene). This result supports our contention that gene expression heterogeneity is not simply the product of stochastic differences in cell cycle phase [26], but rather is indicative of bona fide subpopulations within the ID4-EGFP+ population.
FIG. 2.
Cell cycle state of ID4-EGFP+ spermatogonial subpopulations. A) Cell cycle state among P6 ID4-EGFP+ subpopulations was characterized using flow cytometry and density dot plots demonstrate sequential cell pregating based on: light scatter characteristics (FSC-A × SSC-A; i), viability (propidium iodide negative; ii), single cells (iii), and ID4-EGFP+ (iv). Cells stained with antibodies against TSPAN8 (B–D), EHPA2 (E–G), and PVR (H–J) were used for DNA content analysis with the Vybrant DyeCycle Violet Stain. Histograms indicating cell number (y-axis) and DNA content (x-axis) are shown for EGFP+ spermatogonia with the top one-third (based on cell number) most intensely stained cells (B, E, and H) and marker-positive cells with the bottom one-third (based on cell number) weakest positive staining intensity (C, F, and I). Insets in each panel show dot plots with EGFP intensity, antibody staining intensity, and selection gates for histograms. The percentage of ID4-EGFP+ cells shown in each histogram is noted above the histogram. Transparent blue, red, and green curves in each histogram show the Gaussian functions corresponding to 2N (G1/G0), 4 > N > 2 (S), and (4N) G2/M fractions of each population, which serve as estimates of the proportion of gated cells in each cell cycle phase. D, G, and J) Mean ± SEM from results of four replicate staining experiments for each marker are shown in the stacked bar graphs. In each graph, significant differences in cell cycle state between subpopulations were determined by Student t-tests and are noted between bars (*P < 0.05; **P < 0.01).
TSPAN8 Expression Correlates with Stem Cell Activity among P6 ID4-EGFP+ Spermatogonia
Differences in gene expression often correlate with differences in cellular function. The most significant function of SSCs is the capacity to initiate, maintain, and/or regenerate spermatogenesis. To determine if subpopulations of P6 ID4-EGFP+ spermatogonia differ in SSC content, we performed transplantation studies using TSPAN8High and TSPAN8Low subpopulations (Fig. 3A) to assess the capacity of each to seed spermatogenesis following transplant into testes of busulfan-treated recipients, as described previously [15]. We selected the TSPAN8 marker for sorting, because it yielded the most equitable subpopulations (Fig. 1) that showed the least difference in cell cycle state between subpopulations that might otherwise confound interpretation of transplant data (Fig. 2; only 1.3-fold difference in G1/G0; [26]).
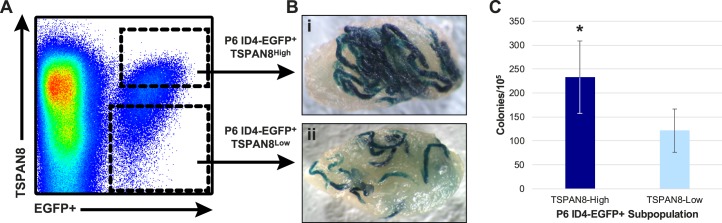
FIG. 3.
Regenerative (SSC) activity of TSPAN8High and TSPAN8Low subpopulations of P6 ID4-EGFP+ spermatogonia. To assess SSC activity among subpopulations of ID4-EGFP+ spermatogonia at P6, cells from Id4-eGfp x RosaLacZ testes were labeled with TSPAN8 antibodies, and TSPAN8High and TSPAN8Low were sorted (A) and used for SSC transplantation by efferent duct injection. Representative X-gal-stained recipient testes (2–3 mo posttransplant) are shown for TSPAN8High (Bi) and TSPAN8Low (Bii) subpopulations of P6 ID4-EGFP+ spermatogonia. C) Mean ± SEM colony counts (per 105 cells injected) from 30 recipient testes and 4 replicate cell-sorting experiments are shown. Statistically significant differences in colonization rate between TSPAN8High and TSPAN8Low subpopulations of P6 ID4-EGFP+ spermatogonia were determined using a Student t-test (*P < 0.05).
Testicular cells were recovered from P6 F1 hybrid pups hemizygous for the Id4-eGfp and Rosa-LacZ transgenes to facilitate purification of spermatogonia and tracking donor-derived spermatogenesis following transplantation, respectively. FACS was used to recover subpopulations made up of the highest (TSPAN8High) and lowest (TSPAN8Low) thirds (based on cell number) across the range of TSPAN8 marker expression from a population of undifferentiated spermatogonia marked by ID4-EGFP (Fig. 3A). Subpopulations of ID4-EGFP+/TSPAN8High and ID4-EGFP+/TSPAN8Low spermatogonia were sorted and transplanted into contralateral testes of busulfan-treated 129×C57 F1 hybrid recipient mice to evaluate the regenerative capacity of each. At 2 mo after transplantation, X-gal staining of the recipient testes was used to quantify colonies of donor-derived spermatogenesis, as described previously [15]. The ID4-EGFP+/ TSPAN8High subpopulation demonstrated a statistically significant, nearly 2-fold greater colonization capacity than the ID4-EGFP+/ TSPAN8Low subpopulation (Fig. 3, B and C).
The P6 ID4-EGFP+/TSPAN8High and TSPAN8Low Spermatogonial Subpopulations Are Distinguished by Differential Gene Expression Patterns
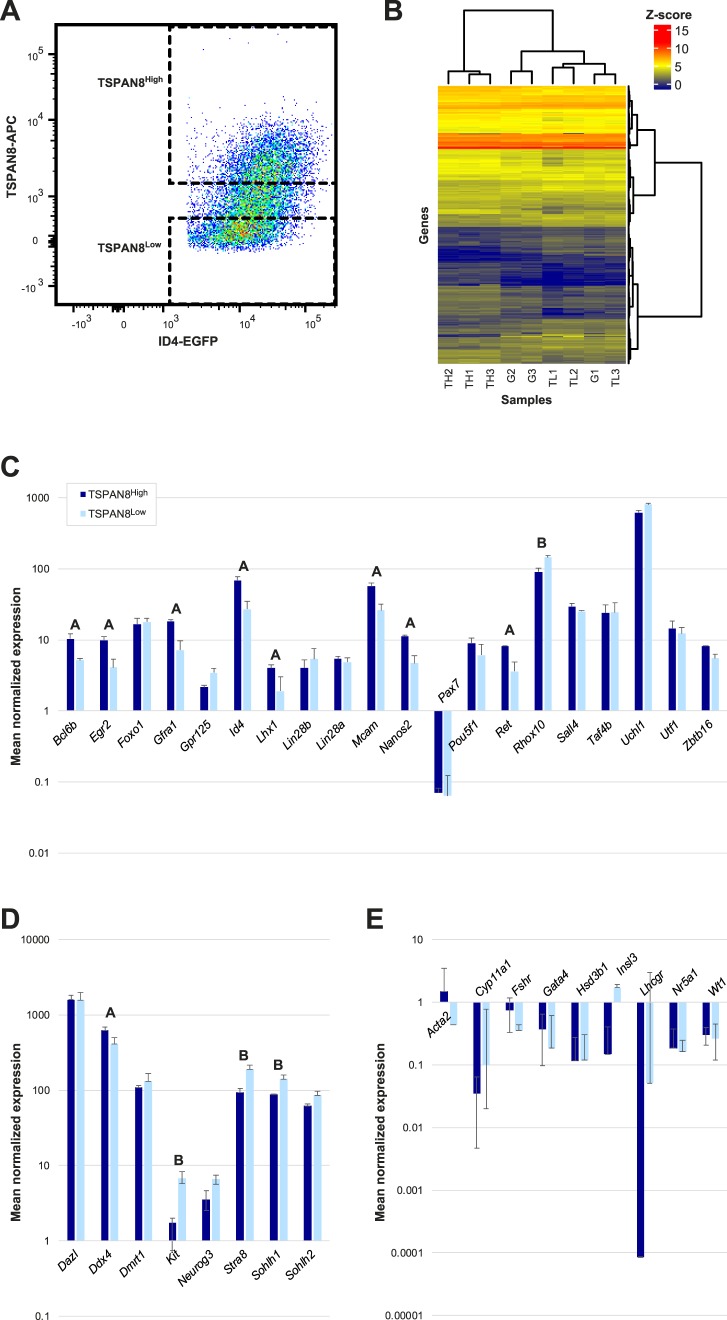
To identify gene pathways or networks associated with SSC function, we performed RNA-seq on sorted ID4-EGFP+/TSPAN8High and ID4-EGFP+/TSPAN8Low spermatogonial subpopulations (Fig. 4A, Supplemental Tables S2 and S5). Expression of an average of 9494, 9588, and 9519 genes was detected above threshold (counts ≥2) in unfractionated ID4-EGFP+ spermatogonia, ID4-EGFP+/TSPAN8High, or TSPAN8Low subpopulations, respectively (Supplemental Table S5). As expected, pairwise comparisons of average gene expression counts revealed considerable similarity between the sorted cell populations (Supplemental Fig. S3). However, the three replicate preparations of ID4-EGFP+/ TSPAN8High spermatogonia clustered together and apart from the three replicate preparations of the TSPAN8Low spermatogonia (Fig. 4B). With the exception of Pax7, expression of every previously reported spermatogonial marker that we examined was detected in both the TSPAN8High and TSPAN8Low subpopulations (Fig. 4, C and D, Supplemental Tables S6 and S7). None of the nine somatic cell-specific genes that we examined were expressed at detectable levels above threshold in either spermatogonial subpopulation (Fig. 4E, Supplemental Tables S6 and S7), confirming the purity of these sorted subpopulations.
FIG. 4.
Transcriptome characteristics of P6 ID4-EGFP+ spermatogonia and TSPAN8High and TSPAN8Low subpopulations. RNA-seq was performed to further characterize P6 ID4-EGFP+ spermatogonia and subpopulations using cells isolated from Id4-eGfp transgenic testes. A) FACS was used to isolate all EGFP+ or subpopulations based on antibody staining for TSPAN8. Sort gates for subpopulations are shown in the density dot plot. Total EGFP+ selection was as shown in Figure 2A, iv. Three replicate preparations of Id4-eGfp testis cells were used for these experiments. B) Unsupervised hierarchical clustering was performed to examine the similarity between samples using normalized gene expression data results. The heatmap shows global Z-score (see legend scale) for the top 500 genes with an average expression >2 and the lowest P-adjusted value are shown for comparisons between the three replicate samples of total ID4-EGFP+ (G1, G2, G3) and three replicates each of ID4-EGFP+/TSPAN8High (TH1, TH2, TH3) and ID4-EGFP+/TSPAN8Low (TL1, TL2, TL3) subpopulations. The gene dendrogram (vertical) indicates Ward linkage (Euclidean distance) and the sample dendrogram (horizontal) represents the Spearman correlation coefficient. Mean ± SD mRNA levels (normalized expression counts) are shown for each ID4-EGFP+ subpopulation for undifferentiated spermatogonial marker genes (C), pan germ cell and differentiating spermatogonial marker genes (D), and markers of testicular somatic cell types (E). Note that the y-axis is presented in Log10 scale. Statistically significant differences (P ≤ 0.0001) between the TSPAN8High and TSPAN8Low subpopulations are noted above the adjacent pairs of bars: A (greater mRNA levels in TSPAN8High); B (greater mRNA levels in TSPAN8Low). Expression counts of <2 were considered undetectable.
To determine if the differences in gene expression that we detected between ID4-EGFP+/ TSPAN8High and TSPAN8Low subpopulations correlated with their significant differences in stem cell activity, we first compared mRNA levels of known markers of undifferentiated and differentiating spermatogonia in each subpopulation. This included genes previously reported to be functionally linked with SSC activity. Thus, among the 20 markers of undifferentiated spermatogonia examined, eight were significantly up-regulated in the TSPAN8High subpopulation relative to the TSPAN8Low subpopulation (Fig. 4C). These included Bcl6b and Id4, which are required for SSC self-renewal [27–29], Nanos2, which is required to inhibit SSC differentiation [30–32], Gfra1 and Ret, which form the signaling-competent GDNF receptor required for SSC self-renewal [33–35], and Mcam, which encodes a protein that enriches for SSCs [36]. Reciprocally, transcripts encoding three markers of spermatogonial differentiation were significantly elevated in the TSPAN8Low subpopulation relative to the TSPAN8High subpopulation (Fig. 4D), including Kit, Sohlh1, and Stra8 [37–43]. Expression of Rhox10, which was recently reported to be essential for normal SSC specification [44], mirrored markers of spermatogonial differentiation (Fig. 4C). These gene expression differences align with the difference that we observed in stem cell capacity within each subpopulation, as indicated by the functional transplantation assay (Fig. 3).
Differentially-Expressed Pathways Distinguish P6 ID4-EGFP+ Subpopulations
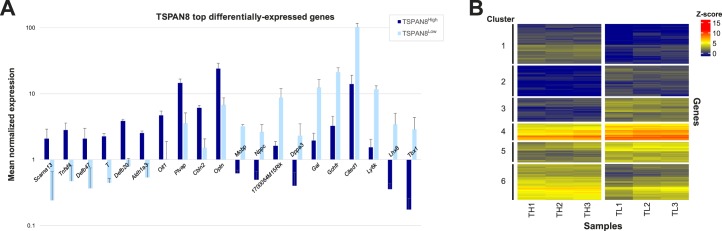
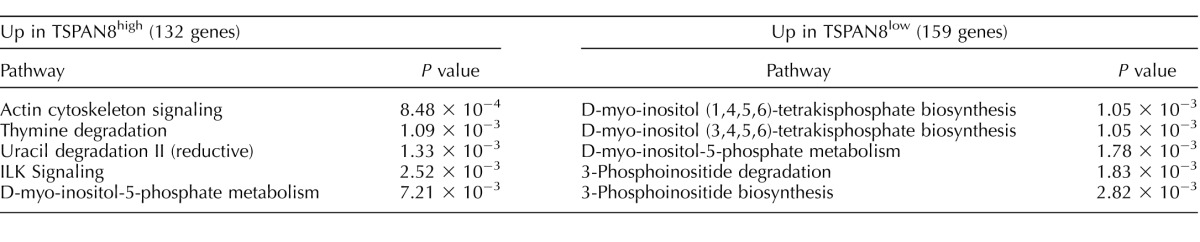
In total, we detected differential expression (≥2-fold expression difference) of 289 genes between the two subpopulations, including 132 genes expressed at higher levels in the ID4-EGFP+/TSPAN8High subpopulation and 157 genes expressed at higher levels in the ID4-EGFP+/TSPAN8Low subpopulation (Fig. 5, A and B, Supplemental Table S7). A subset of these differentially expressed genes was validated by independent qRT-PCR studies (Supplemental Fig. S4). We used k-means clustering to group these differentially expressed genes by similarity in absolute abundance, as well as direction and amplitude of differences between the subpopulations (Fig. 5B, Supplemental Table S7). We then used GO analyses to interrogate biologically significant differences indicated by the groups of genes differentially expressed in the ID4-EGFP+/TSPAN8High and TSPAN8Low subpopulations. In particular, we sought to identify mechanistic clues that might help explain the observed difference in regenerative activity (donor-derived spermatogenic colony formation from transplanted SSCs) between these subpopulations. Among the 132 genes that were significantly more abundant in the TSPAN8High subpopulation, actin cytoskeleton signaling was the top (lowest P value) canonical pathway (Actn2, Fgf9, Fgf12, Ppi4k2a, Nckap1, and Matk; Table 1, Supplemental Table S8). Elevated expression of Dpysl2 and Upb1 was also observed in TSPAN8High cells. These genes encode enzymes that catalyze the last two steps of the pyrimidine degradation pathway, which is responsible for thymidine degradation and reductive uracil degradation (Table 1, Supplemental S8). This aligns with previous results showing rare spermatogonia immunostained with antibodies for DPYSL2 in adult rhesus monkey testes [45]. Genes associated with the integrin-linked kinase (ILK) signaling pathway, involved in cell adhesion signaling, proliferation, and motility, were also upregulated in the TSPAN8High dataset (Table 1, Supplemental Table S8). Lastly, elevated levels of Cdc25b, Ppfibp2, Pip4k2a, and Sirpa, which are all part of the D-myo-inositol-5-phosphate metabolic pathway, were upregulated in the TSPAN8High subpopulation (Table 1, Supplemental Table S8). Loss of heterozygosity for a particular allelic variant of the human SIRPA gene was recently associated with nonobstructive azoospermia in a large cohort of patients [46], which may point to involvement of this gene in maintenance of SSCs.
FIG. 5.
Differential gene expression between the TSPAN8High and TSPAN8Low subpopulations of P6 ID4-EGFP+ spermatogonia. Significant differences were observed in levels of genes with an average expression value of >2 between the ID4-EGFP+/TSPAN8High and ID4-EGFP+/TSPAN8Low spermatogonia populations. Genes with statistically significant differences in levels between populations (P < 0.0001) and at least 2-fold difference in levels between ID4-EGFP+ spermatogonial populations were considered to be differentially expressed. A) The top differentially expressed genes (fold-change) are shown for comparisons between TSPAN8High and TSPAN8Low. Graphs portray the mean ± SD mRNA levels (normalized expression counts) of the noted genes for each ID4-EGFP+ subpopulation. All bar pairs are significantly different (P < 0.0001) between the TSPAN8High and TSPAN8Low subpopulations. B) Hierarchical clustering using only the differentially expressed genes was used to confirm separation between individual sample replicates and group differentially expressed genes. Heatmaps of global Z-score for the differentially expressed genes are shown for comparisons between the three replicates each of the ID4-EGFP+/TSPAN8High (TH1, TH2, TH3) and ID4-EGFP+/TSPAN8Low (TL1, TL2, TL3) subpopulations. GO analyses of these differentially expressed genes are found in Table 1 and Supplemental Table S8. Horizontal clustering (samples) is based on Euclidean distance (dendrogram not shown) and vertical clustering (genes) is based on Spearman correlation coefficient. K means clustering (six clusters) was then used to group genes with like expression patterns. Cluster labels (1–6) denote gene groups that can be found in Supplemental Table S7. GO analyses of clusters are found in Supplemental Table S10. Note: a parallel analysis with 1.5-fold differences in mRNA abundance is shown in Supplemental Figure S5 and Supplemental Table S9.
TABLE 1.
GO analysis of genes differentially expressed 2-fold or greater.
In the TSPAN8Low subpopulation, several genes involved in D-myo-inositide- and 3-phosphoinositide metabolism were more abundantly expressed (Ppp1r16b, Ppp1r14a, Ppm1k, Ppp1r14b, Dusp2; Table 1, Supplemental Table S8). These genes encode a series of protein phosphatases and phosphatase regulators that are involved in varied cellular processes, including signal transduction and mitochondria permeability transition pore function [47–50]. Our finding that genes encoding different regulators of phospho-inositide metabolism were upregulated in each subpopulation points to the potential for functional reciprocity in this pathway between SSC-enriched and -depleted cell populations. GO analyses of the genes found in each TSPAN8 gene cluster (Fig. 5B, Supplemental Table S7) demonstrated segregation of some of these biological functions to particular gene clusters (Supplemental Table S10). For instance, ILK signaling was the top pathway among genes in cluster 1, while genes encoding different enzymes involved in the pyrimidine degradation pathway were upregulated in both clusters 1 and 6 (Fig. 5B, Supplemental Table S10). Cluster 6 was headlined by Gfra1 and Ret (Fig. 5B, Supplemental Table S10), which are both required for SSC self-renewal and were significantly elevated among ID4-EGFP+/ TSPAN8High cells [34, 35]. This cluster also contained genes known to regulate glycolysis and lactolysis, suggesting that these metabolic pathways are prominent among SSC-enriched cells (Supplemental Table S10). Genes involved in the phospho-inositide metabolism pathways elevated among cells in the ID4-EGFP+/ TSPAN8Low subpopulation were divided between clusters 3 and 4 (Fig. 5B, Supplemental Table S10). This expression pattern mirrored genes involved in retinoic acid biosynthesis and signaling (Crabp1, Rbp1, Tnfsf10), which promotes spermatogonial differentiation [5, 40, 41, 51, 52], suggesting a role for D-myo-inositide- and 3-phosphoinositide metabolism in spermatogonial differentiation.
Epigenetic Programming Targets Promoters of Genes Differentially Expressed in ID4-GFP+/TSPAN8High and ID4-GFP+/TSPAN8Low Cells
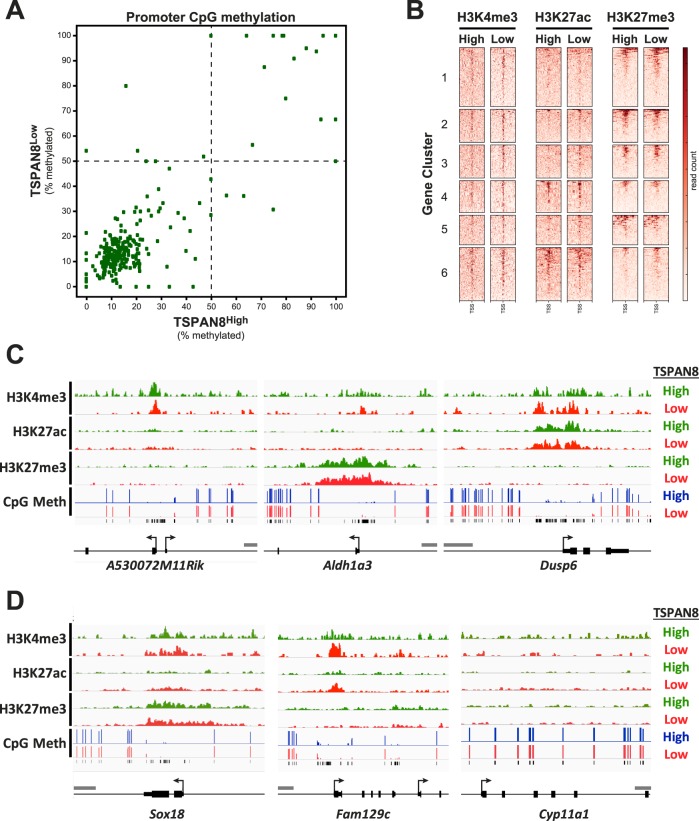
To investigate potential molecular mechanisms that might contribute to the regulation of differential gene expression in these two spermatogonial subtypes, we investigated genome-wide DNA methylation and histone modification patterns in populations of ID4-GFP+/ TSPAN8High and TSPAN8Low cells. Sorted subpopulations were prepared by FACS, as described above, and analyzed for genome-wide DNA methylation patterns by Methyl-MidiSeq (Zymo Research). This RRBS-based approach used multiple restriction enzymes to provide coverage of approximately 17 million CpG dinucleotides in the mouse genome (Supplemental Table S11), including those in gene promoters, gene bodies, and CpG islands and intergenic regions (Supplemental Fig. S6). Overall, we observed very few differences in either genome-wide (Supplemental Fig. S7) or promoter region-specific DNA methylation patterns between the two subpopulations (Fig. 6A). Notably, a preponderance of genes differentially expressed between the two spermatogonial subpopulations (237/289) were hypomethylated in the promoter region in both subpopulations, regardless of the specific subpopulation in which each gene was upregulated (Fig. 6A, bottom left quadrant; Supplemental Table S12).
FIG. 6.
Epigenetic programming of genes differentially expressed between TSPAN8High and TSPAN8Low subpopulations. Genes that were found to have significantly different mRNA levels between the TSPAN8 subpopulations of P6 ID4-EGFP+ spermatogonia were subjected to analysis of DNA methylation and posttranslational histone modifications using genome-wide sequencing approaches. A) Reduced representation bisulfite methyl-Seq was used to profile ∼17 million CpGs in genomic DNA from either pooled TSPAN8High (horizontal axis) or pooled TSPAN8Low (vertical axis) spermatogonia. Shown is a scatter plot of promoter CpG methylation over a 1-kbp window spanning the transcriptional start sites (TSSs) of differentially expressed genes (±0.5 kb from the TSS) in both cell populations. The percent CpG methylation (0% = unmethylated; 100% = fully methylated) was determined by averaging the methylation state for any detected CpGs falling within the promoter window. A total of 12 genes for which CpGs were not detected in at least one sample were excluded from this plot. Dashed lines indicate 50% methylation in each sample. Each dot represents one differentially expressed gene promoter. Of the 277 differentially expressed genes for which CpG methylation data were available for both populations, many were hypomethylated (238 genes, bottom left quadrant), and some were hypermethylated (32 genes, upper right quadrant) in both spermatogonial subtypes. Very few of the differentially expressed genes (seven genes) showed corresponding differential levels of DNA methylation in their promoter regions. B) Heatmaps showing stacked ChIP-seq reads centered on TSSs of differentially expressed genes (±10 kbp) are shown for H3K4me3, H3K27ac, and H3K27me3 using chromatin isolated from TSPAN8High (High) and TSPAN8Low (Low) subpopulations. Genes are grouped by clusters based on differential expression, as shown in Figure 5. Heatmap scale is shown at the right. C and D) Histogram tracks showing ChIP-seq (H3K4me3, H3K27ac, and H3K27me3; ChIP'd DNA abundance) and methyl-seq (CpG methylation, % 5meC) results for exemplary genes. Results from TSPAN8High (green or blue) and TSPAN8Low (red) subpopulations are shown and sample identity is indicated to the right. Detected CpGs are noted by black tick marks below the methyl-seq data. Gene annotations are noted below the tracks (TSS = bent arrow; UTR = short bar; coding exon = tall bar). Gray scale bar = 1 kbp. C) Three different patterns of histone modifications are shown: i) peaks of H3K4me3 only (e.g., A530072M11Rik); ii) peaks of H3K27me3 only (e.g., Aldh1a3); and iii) simultaneous peaks of H3K4me3 and H3K27ac (e.g., Dusp6). Each occurred in numerous differentially expressed genes in both spermatogonial subtypes, regardless of the subtype in which the gene was up- or downregulated. In each case, peaks of histone modifications colocalized with hypomethylated promoter domains. D) Sox18 exemplified genes with simultaneous peaks of H3K4me3 and H3K27me3 characteristic of bivalent genes; Fam129c was a rare example of a gene that displayed differentially intense peaks of two active histone modifications—H3K4me3 and H3K27ac—that correlated with the differential expression levels of this gene in the two spermatogonial subtypes, and Cyp11a1 exemplified genes that are repressed in both spermatogonial subtypes and show DNA hypermethylation and an absence of any peaks of histone modifications in the promoter region.
ChIP-seq (Active Motif) detected genome-wide patterns of three histone modifications, H3K4me3, H3K27ac, and H3K27me3, and revealed thousands of peaks for these marks in both TSPAN8High and TSPAN8Low cells (Fig. 6B, Supplemental Table S13). We detected ChIP-Seq peaks indicative of one or more of these three histone modifications in the promoter regions of a majority of the 289 genes differentially expressed in the TSPAN8High and TSPAN8Low spermatogonial subtypes (Fig. 6B). In the promoter regions of a majority of the differentially expressed genes, we detected similar histone modification patterns in both spermatogonial subtypes (Fig. 6, C and D). For instance, some gene promoters were marked uniquely by peaks of H3K4me3 (active gene mark; e.g., A530072M11Rik; Fig. 6C), while other gene promoters were marked only by peaks of H3K27me3 (repressive gene mark; e.g., Aldh1a3; Fig. 6C), and still other gene promoters were marked by both H3K4me3 and H3K27ac (both active marks; e.g., Dusp6; Fig. 6C). We also observed genes with simultaneous peaks of H3K4me3 and H3K27me3 characteristic of bivalent genes (e.g., Sox18; Fig. 6D). In a small number of the differentially expressed genes, we observed subtle differences with respect to the preponderance of one or more of these histone modifications (e.g., at the Fam129c gene [Fig. 6D], which showed higher peaks for H3K4me3 and H3K27ac in the TSPAN8Low cells, in agreement with higher mRNA levels in TSPAN8Low cells). Lastly, genes that were repressed in both spermatogonial subpopulations typically exhibited an absence of any peaks of histone modifications in the promoter regions (e.g., Cyp11a1; Fig. 6D). There was also significant concordance between regions of DNA hypomethylation and histone modification peaks over gene promoters in a large majority of the differentially expressed genes (Fig. 6, C and D).
DISCUSSION
Spermatogenesis is maintained throughout adulthood by SSCs that must balance self-renewal and differentiation to sustain the pool of stem cells and simultaneously meet the biological demand for sperm production required for normal male fertility in mammals. However, the mechanisms leading to formation of the SSC pool and the subsequent response of these cells to signals that induce self-renewal or differentiation are poorly understood, largely because of technical challenges that have prevented precise, selective investigations of SSCs. In part, this is due to the extreme rarity of SSCs (∼0.1% of adult mouse spermatogonia are SSCs [6, 53]), but this also reflects the fact that it has been impossible to precisely and prospectively identify and selectively recover purified subpopulations of SSCs and progenitors, respectively, from among the heterogeneous pool of undifferentiated spermatogonia in the testes of any mammalian species (reviewed in [7]). Based on experimental procedures that have been optimized for use with the mouse (e.g., transgenesis, gene knockouts, genome editing), there has been recent progress toward methods to cleanly separate SSCs and progenitor spermatogonia in the mouse testis [11, 29, 54]. Ultimately, however, methods to purify spermatogonial subtypes from domestic animals or humans will be needed to facilitate translation of this technology to agricultural or clinical settings, respectively. To this end, endogenously expressed markers offer the most promising option, because, to the extent that expression patterns of these markers are conserved, translation of sorting technology from the mouse to other mammalian species should be straightforward.
In the present study, we mined our recently published data describing gene expression in single, undifferentiated spermatogonia from the P6 mouse testis [11] to identify novel endogenous markers that can facilitate a more precise and complete subdivision of SSC and progenitor subpopulations, and that have the potential to be used to select subpopulations of SSCs and progenitors from testes of other mammalian species, including humans. We focused on a marker encoded by the Tspan8 gene, which exhibited bimodal expression among P6 ID4-EGFP+ spermatogonia. Differential labeling with an antibody to this cell surface protein defined subsets of P6 ID4-EGFP+ spermatogonia, and these subpopulations displayed differing phenotypic characteristics, the most critical of which was differential enrichment for regenerative capacity indicative of functional SSCs that can seed spermatogenesis following transplantation to a recipient testis. A functional assay of this sort provides the only reliable approach to confirm the accuracy of any putative marker that might be used to identify or selectively recover SSCs. Here, we have shown that different levels of the endogenous cell surface protein, TSPAN8, can be used in conjunction with ID4-EGFP+ labeling to selectively recover subpopulations of mouse undifferentiated spermatogonia that are relatively enriched for (TSPAN8High) or depleted of (TSPAN8Low) functional SSCs.
In the P6 testis, prospermatogonia that were present at earlier developmental stages (as late as P4) have entirely converted to either undifferentiated or differentiating spermatogonia that will contribute to the first wave of spermatogenesis [55–57]. We have previously reported that Kit mRNA is detectable in most P6 ID4-EGFP+ spermatogonia, yet the presence of Kit mRNA was not predictive of the presence of KIT protein [11]. In the present study, TSPAN8low subpopulation of P6 ID4-EGFP+ spermatogonia investigated here exhibited enhanced Kit levels, consistent with a relatively more differentiated phenotype than the TSPAN8high subpopulation. However, it is impossible to discern whether this enhanced Kit mRNA among ID4-EGFP+/TSPAN8low spermatogonia is indicative of prospermatogonial descendants that are contributing to the first wave or spermatogonia initiating differentiation from a stem cell state. Additional lineage-tracing studies would be necessary to provide more definitive evidence of the developmental origin and trajectory of these cells.
It should be noted that TSPAN8 expression was detected in both germ and somatic cell types in mouse pup testes. Thus, because TSPAN8 is not a germ cell-specific marker, we used coselection with ID4 as a spermatogonia-specific marker plus the TSPAN8 marker (high or low) to recover undifferentiated spermatogonia subdivided into SSC-enriched and SSC-depleted subpopulations. In addition, while the TSPAN8Low subpopulation was depleted of regenerative activity relative to the TSPAN8High subpopulation, SSCs were not absent. This may indicate that some SSCs were selected in the TSPAN8Low, due to proteolytic cleavage of the TSPAN8 antigen. Alternatively, this may indicate that we captured immediate progeny of SSCs that were just entering the transition between the stem cell and progenitor states, which would be associated with a transition from the TSPAN8High to the TSPAN8Low phenotype. Presumably, a subpopulation of ID4-EGFP−/TSPAN8− germ cells would be most completely depleted of SSCs, but we have yet to test this prediction.
Heterogeneity among cells within a population, similar to our observations here and previously [11], could potentially arise from flux in phenotypic states within a population that allows for metastable variants to emerge and retain the same function. Conceptually, if the two subpopulations of ID4-EGFP+ spermatogonia (TSPAN8high and TSPAN8low) spermatogonia represent transient and interchangeable phenotypic states in vivo, then the same should be true following transplantation into the recipient testis environment. However, our data revealed significant differences in colonization activity between each subpopulation, arguing that these represent stable states that do not oscillate phenotypically. Additional studies will be necessary to determine the extent and direction of any functional transition between the TSPAN8high and TSPAN8low states.
We clearly observed significant enrichment of SSCs in the ID4-EGFP+/ TSPAN8High subpopulation. The formation of 233 colonies/105 cells transplanted translates to a stem cell concentration of 1:21 cells (presuming a colonization efficiency of 5%). This concentration is more than 5-fold higher than the purity of SSCs previously obtained by GFRA1 selection (45 colonies/105 cells transplanted = 1:111 SSC purity [58]) and nearly twice the SSC purity obtained by THY-1 selection (124 colonies/105 cells transplanted = 1:40 SSC purity [59]). While multiparameter sorting of donor spermatogonia from cryptorchid adult mouse testes has yielded a greater enrichment of SSCs (343 colonies/105 cells transplanted = 1:15 SSC purity [60]), it is likely that donor spermatogonia from normal donor testes will be needed for most applications. Significant enrichment of normal adult mouse SSCs (1:6 SSC purity [36]) has also been achieved by selecting for cells bearing the cell-surface phenotype CD9+/EPCAMlow/MCAM+/ KIT−, but it is not known if this approach is effective in prepubertal testes or in other mammalian species. Thus, the protocol we used to enrich for SSCs on the basis of ID4-EGFP+/ TSPAN8High labeling is the most effective method of SSC enrichment from normal prepubertal testes in vivo reported to date, and is potentially translatable to primates.
One possible contributor to differential regenerative activity by SSCs upon transplantation is cell cycle state. Previously, cultured spermatogonia in G1/G0 were shown to have enhanced colonization activity than those in S/G2-M due to enhanced transit across the blood-testis barrier in transplant studies [26]. While it is not known if freshly isolated spermatogonia exhibit these same characteristics, we observed 1.9-fold more regenerative activity among TSPAN8high spermatogonia, despite 1.3-fold fewer cells in G0/G1 (compared with the TSPAN8low subpopulation). Thus, it is possible that differential cell cycle state may have partially muted our detection of differences in SSC content between TSPAN8 subpopulations.
The distinct subpopulations of undifferentiated spermatogonia selected using the TSPAN8-based approach we report here were further defined by our gene expression analysis, which allowed us to detect significant differential gene expression (289 genes) between these subpopulations. In particular, we observed upregulation of genes involved in SSC self-renewal (Bcl6b and Id4 [27–29]) and inhibition of SSC differentiation (Nanos2 [30, 31, 61]) in the TSPAN8High spermatogonial subpopulation, and upregulation of genes associated with spermatogonial differentiation (Kit, Sohlh1 [37–39]) in the TSPAN8Low subpopulation. These results align with our observation that the TSPAN8High subpopulation is enriched for SSC activity relative to the TSPAN8Low subpopulation, because SSC regenerative capacity is found in self-renewing spermatogonia, but lost from differentiating spermatogonia (reviewed in [4]). Thus, the differences in gene expression that we have delineated between the TSPAN8High and TSPAN8Low subpopulations provide unique signatures that can potentially distinguish self-renewing (SSC) from differentiating (committed progenitor) spermatogonia, and this distinction can facilitate future studies to more thoroughly define the molecular mechanisms underlying these alternative cell fates. A potential contributor to regulation of spermatogonial fate is TSPAN8 itself, given that cell selection on the basis of high TSPAN8 expression distinguished an SSC-enriched population. TSPAN8 is known to interact with other cell surface proteins that are expressed by undifferentiated spermatogonia, including α6-integrin [62], E-cadherin [63], and EpCAM [64]. It is tempting to speculate that TSPAN8 may somehow interface with signals mediated by these proteins to affect spermatogonial fate or function, similar to its role in other systems [65, 66].
GO analyses of differentially expressed genes between these two subpopulations identified several specific pathways up-regulated in the ID4-EGFP+/TSPAN8High subpopulation including ILK signaling. ILK signaling has been implicated in promotion of regenerative capacity and inhibition of differentiation in mesenchymal stem cells, breast cancer stem cells, and human pluripotent stem cells [67–70]. Our finding, that this pathway is upregulated in the TSPAN8High subpopulation, suggests that a similar mechanism may contribute to the self-renewal capacity of SSCs. The identification of more comprehensive pathways responsible for the alternative fates of self-renewal or differentiation of SSCs or their immediate progeny will require a combination of additional transcriptome analyses accompanied by genetic studies in which expression of one or more of these genes is perturbed, followed by assessment of SSC function by transplantation or steady-state lineage tracing.
Differential gene expression can be regulated at the level of transcription by multiple epigenetic mechanisms, including DNA methylation and histone modifications, as well as binding of transcription factors. Our analysis of DNA methylation associated with genes that are differentially expressed between ID4-EGFP+/TSPAN8High and TSPAN8Low cells showed a strong trend toward DNA hypomethylation in promoter regions of these genes in both subpopulations, regardless of the particular subpopulation in which each gene was up- or downregulated. Many of these genes also showed concordant peaks of one or more of the three histone modifications examined (H3K4me3, H3K27ac, and H3K27me3) in both spermatogonial subpopulations, regardless of which subtype showed higher or lower expression of the corresponding gene. Although H3K4me3 and H3K27ac are typically associated with actively expressed genes, whereas H3K27me3 is associated with repressed genes [71], we did not observe a strict correlation between the presence or absence of any of these modifications and up- or down-regulation of the corresponding gene in either subtype for most of the genes differentially expressed in the TSPAN8High and TSPAN8Low subpopulations. Exceptions include two examples shown in Figure 6, C and D, in which peaks representing H3K427me3 and H3K427ac were higher in the TSPAN8Low cells at both the Dusp6 and Fam129c genes. We did detect the simultaneous presence of peaks of H3K4me3 (normally an active mark) and H3K27me3 (normally a repressive mark) in association with some genes in both subpopulations. Genes with this bivalent pattern of active and repressive marks are marked similarly to the set of “poised genes” that have been previously described in the mammalian germ line [71–73], but differ from those genes by virtue of being expressed in these developing germ cells.
Our data are consistent with the notion that ID4-EGFP+/TSPAN8High and ID4-EGFP+/TSPAN8Low spermatogonia are germline subtypes that share a common developmental origin and remain biologically very similar to each other in the mouse pup testis. Thus, it appears that genes that are differentially expressed in these two spermatogonial subtypes are similarly potentiated for transcriptional activity by the absence of DNA methylation and presence of various histone modifications in promoter regions. This suggests that these epigenetic mechanisms play a permissive role in predisposing transcription of these genes in both subtypes, but do not directly regulate the differential expression of these genes between subtypes. However, patterns of hypomethylated DNA were clearly correlated with, and may predispose the presence of regulatory histone modifications in promoter regions of, many of these same 289 differentially expressed genes in the two spermatogonial subtypes. It is likely that additional mechanisms, including differential binding of transcription factors and/or posttranscriptional processes, contribute to a combinatorial molecular mechanism that regulates differential expression of these genes in these subtypes.
Given that TSPAN8 is an endogenously produced cell surface protein, our results raise the possibility that TSPAN8 selection might form the basis of a useful strategy for sorting highly enriched populations of SSCs and/or committed progenitor spermatogonia in mammalian species for which transgenic reporters (such as Id4-eGfp) are not available. For instance, the endogenous TSPAN8 marker might afford a novel opportunity to determine if differential expression of this cell surface protein also marks distinct subpopulations of undifferentiated spermatogonia in the primate testis. Thus, based on our finding that the TSPAN8 marker can be used to distinguish spermatogonial subtypes in the immature mouse testis, we can now initiate more thorough analyses of the analogous spermatogonial subtypes in the developing testis in nonhuman primates and humans. Definitive assignment of a functional relevance to selection of TSPAN8+ or negative cells in nonrodent species would require transplantation of the selected cells to measure their functional capacities. Autologous transplant of spermatogonia has been achieved in nonhuman primates [74], and could be used to assess potential enrichment of regenerative SSCs in TSPAN8-selected subpopulations. Alternatively, conserved transcriptomes shown to be differentially associated with SSC or progenitor spermatogonia in the mouse should provide insight into the development and functional capacity of SSCs in other species, including humans.
Overall, the results of our study define a novel strategy based on the use of antibody staining for TSPAN8 to fractionate populations of ID4-EGFP+ spermatogonia that can be used to select the most highly enriched population of SSCs from the mouse pup testis reported to date. This strategy also facilitated identification of novel gene expression differences distinguishing SSCs from other undifferentiated spermatogonia, which point to biological pathways that distinguish spermatogonia with regenerative capacity (SSCs) from those that have initiated, or are about to initiate, differentiation (progenitors). Finally, our studies may open the door to interrogate the functional significance of spermatogonial heterogeneity broadly among nonrodent mammalian species, including primates, which may facilitate selection of SSCs in the clinic for treatment of male infertility [75].
ACKNOWLEDGMENT
The authors are grateful for bioinformatics support from Armando Rodiguez and Zhiwei Wang in the University of Texas at San Antonio (UTSA) Computational System Biology Core, the UTSA Genomics Core Facility, and access to the University of Texas Health Science Center at San Antonio Flow Cytometry Shared Resource Facility directed by Karla Gorena and Dr. Benjamin J. Daniel. We also thank Drs. Brigida A. Rusconi and Mark Eppinger from the UTSA Department of Biology for thoughtful discussions about (and assistance with) bioinformatics analyses.
Footnotes
Supported by National Institutes of Health grants GM092334 and HD078679 to J.R.M., HD061665 to J.M.O., HD062687 to B.P.H., HD072552 to C.B.G., CA054174, MD007591, and RR025767, National Science Foundation grant DBI-1337513 to B.P.H., the Max and Minnie Tomerlin Voelcker Fund, the Helen Freeborn Kerr Charitable Foundation, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, and the University of Texas at San Antonio. A portion of these data were presented at the American Society of Andrology's 40th Annual Meeting, April 18-21, 2015, The Little America Hotel, Salt Lake City, Utah. Raw and processed data were submitted to National Institutes of Health Gene Expression Omnibus and Sequence Read Archive databases under accession numbers GSE83311 and GSE83422.
REFERENCES
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QE, Oatley JM. Spermatogonial stem cell functions in physiological and pathological conditions. Curr Top Dev Biol. 2014;107:235–267. doi: 10.1016/B978-0-12-416022-4.00009-3. [DOI] [PubMed] [Google Scholar]
- Busada JT, Geyer CB. The role of retinoic acid (RA) in spermatogonial differentiation. Biol Reprod. 2016;94:10. doi: 10.1095/biolreprod.115.135145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano MC. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol Reprod. 2003;69:701–707. doi: 10.1095/biolreprod.103.016352. [DOI] [PubMed] [Google Scholar]
- Valli H, Phillips BT, Orwig KE, Gassei K, Nagano MC. Spermatogonial stem cells and spermatogenesis. In: Plant TM, Zeleznik AJ, editors. Knobil and Neill's Physiology of Reproduction, vol. 1, 4th ed. New York: Academic Press; 2015. pp. 595–635. In. (eds.) [Google Scholar]
- Avarbock MR, Brinster CJ, Brinster RL. Reconstitution of spermatogenesis from frozen spermatogonial stem cells. Nat Med. 1996;2:693–696. doi: 10.1038/nm0696-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, McCarrey JR. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod. 2015;92:54. doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- Nagano M, Ryu BY, Brinster CJ, Avarbock MR, Brinster RL. Maintenance of mouse male germ line stem cells in vitro. Biol Reprod. 2003;68:2207–2214. doi: 10.1095/biolreprod.102.014050. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol. 2006;419:259–282. doi: 10.1016/S0076-6879(06)19011-4. [DOI] [PubMed] [Google Scholar]
- Watson JV, Chambers SH, Smith PJ. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 1987;8:1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I, Lefkovits I. Internal standard-based analysis of microarray data. Part 1: analysis of differential gene expressions. Nucleic Acids Res. 2009;37:6323–6339. doi: 10.1093/nar/gkp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh F, Legendre P. Ward's hierarchical agglomerative clustering method: which algorithms implement Ward's criterion? J Classif. 2014;31.3:274–295. [Google Scholar]
- Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. New York: Cambridge University Press; 2008. [Google Scholar]
- Hermann BP, Hornbaker KI, Maran RR, Heckert LL. Distal regulatory elements are required for Fshr expression, in vivo. Mol Cell Endocrinol. 2007;260–262:49–58. doi: 10.1016/j.mce.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WC, Wu PH, Phillip JM, Khatau SB, Choi JM, Dallas MR, Konstantopoulos K, Sun SX, Lee JS, Hodzic D, Wirtz D. Functional interplay between the cell cycle and cell phenotypes. Integr Biol (Camb) 2013;5:523–534. doi: 10.1039/c2ib20246h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A, Dennis L, Danaher P, Finak G, Krouse M, Wang A, Webster P, Beechem J, Gottardo R. Modeling bi-modality improves characterization of cell cycle on gene expression in single cells. PLoS Comput Biol. 2014;10:e1003696. doi: 10.1371/journal.pcbi.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kanatsu-Shinohara M, Shinohara T. Cell-cycle-dependent colonization of mouse spermatogonial stem cells after transplantation into seminiferous tubules. J Reprod Dev. 2014;60:37–46. doi: 10.1262/jrd.2013-083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85:347–356. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Xu Q, Zhao D, Degui CC. Id4 marks spermatogonial stem cells in the mouse testis. Sci Rep. 2015;5:17594. doi: 10.1038/srep17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Shirakawa T, Ohbo K, Sada A, Wu Q, Hasegawa K, Saba R, Saga Y. RNA Binding protein Nanos2 organizes post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells. Dev Cell. 2015;34:96–107. doi: 10.1016/j.devcel.2015.05.014. [DOI] [PubMed] [Google Scholar]
- Sada A, Hasegawa K, Pin PH, Saga Y. NANOS2 acts downstream of glial cell line-derived neurotrophic factor signaling to suppress differentiation of spermatogonial stem cells. Stem Cells. 2012;30:280–291. doi: 10.1002/stem.790. [DOI] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science. 2009;325:1394–1398. doi: 10.1126/science.1172645. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- He Z, Jiang J, Hofmann MC, Dym M. Gfra1 silencing in mouse spermatogonial stem cells results in their differentiation via the inactivation of RET tyrosine kinase. Biol Reprod. 2007;77:723–733. doi: 10.1095/biolreprod.107.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitt J, Singh D, Zhang C, Chen LC, Folmer J, Shokat KM, Wright WW. The in vivo response of stem and other undifferentiated spermatogonia to the reversible inhibition of glial cell line-derived neurotrophic factor signaling in the adult. Stem Cells. 2012;30:732–740. doi: 10.1002/stem.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Morimoto H, Shinohara T. Enrichment of mouse spermatogonial stem cells by melanoma cell adhesion molecule expression. Biol Reprod. 2012;87:139. doi: 10.1095/biolreprod.112.103861. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T, Nishikawa S. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–699. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361:301–312. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Arnold S, Kent T, Mitchell D, Isoherranen N, Griswold MD. Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol Reprod. 2015;92:37. doi: 10.1095/biolreprod.114.126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod. 2010;83:783–790. doi: 10.1095/biolreprod.110.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod. 2008;79:35–42. doi: 10.1095/biolreprod.107.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod. 2008;78:537–545. doi: 10.1095/biolreprod.107.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, Bettegowda A, Lake BB, Zhao AH, Skarbrevik D, Babajanian E, Sukhwani M, Shum EY, Phan MH, Plank TD, Richardson ME, Ramaiah M, et al. The homeobox transcription factor RHOX10 drives mouse spermatogonial stem cell establishment. Cell Rep. 2016;17:149–164. doi: 10.1016/j.celrep.2016.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xia Y, Wang G, Zhou T, Guo Y, Zhang C, An X, Sun Y, Guo X, Zhou Z, Sha J. In-depth proteomic analysis of whole testis tissue from the adult rhesus macaque. Proteomics. 2014;14:1393–1402. doi: 10.1002/pmic.201300149. [DOI] [PubMed] [Google Scholar]
- Lu C, Xu M, Wang R, Qin Y, Wang Y, Wu W, Song L, Wang S, Shen H, Sha J, Miao D, Hu Z, et al. Pathogenic variants screening in five non-obstructive azoospermia-associated genes. Mol Hum Reprod. 2014;20:178–183. doi: 10.1093/molehr/gat071. [DOI] [PubMed] [Google Scholar]
- Eto M, Ohmori T, Suzuki M, Furuya K, Morita F. A novel protein phosphatase-1 inhibitory protein potentiated by protein kinase C: isolation from porcine aorta media and characterization. J Biochem. 1995;118:1104–1107. doi: 10.1093/oxfordjournals.jbchem.a124993. [DOI] [PubMed] [Google Scholar]
- Lu G, Ren S, Korge P, Choi J, Dong Y, Weiss J, Koehler C, Chen JN, Wang Y. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev. 2007;21:784–796. doi: 10.1101/gad.1499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czikora I, Kim KM, Kasa A, Becsi B, Verin AD, Gergely P, Erdodi F, Csortos C. Characterization of the effect of TIMAP phosphorylation on its interaction with protein phosphatase 1. Biochimie. 2011;93:1139–1145. doi: 10.1016/j.biochi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csortos C, Czikora I, Bogatcheva NV, Adyshev DM, Poirier C, Olah G, Verin AD. TIMAP is a positive regulator of pulmonary endothelial barrier function. Am J Physiol Lung Cell Mol Physiol. 2008;295:L440–L450. doi: 10.1152/ajplung.00325.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol. 2015;397:140–149. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci U S A. 2015;112:E2347–E2356. doi: 10.1073/pnas.1505683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch RAJ, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- Aloisio GM, Nakada Y, Saatcioglu HD, Pena CG, Baker MD, Tarnawa ED, Mukherjee J, Manjunath H, Bugde A, Sengupta AL, Amatruda JF, Cuevas I, et al. PAX7 expression defines germline stem cells in the adult testis. J Clin Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl. 1981;4:475–493. doi: 10.1111/j.1365-2605.1981.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction. 2011;142:145–155. doi: 10.1530/REP-10-0431. [DOI] [PubMed] [Google Scholar]
- Ebata KT, Zhang X, Nagano MC. Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Mol Reprod Dev. 2005;72:171–181. doi: 10.1002/mrd.20324. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod. 2004;71:722–731. doi: 10.1095/biolreprod.104.029207. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009;336:222–231. doi: 10.1016/j.ydbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Herlevsen M, Schmidt DS, Miyazaki K, Zoller M. The association of the tetraspanin D6.1A with the alpha6beta4 integrin supports cell motility and liver metastasis formation. J Cell Sci. 2003;116:4373–4390. doi: 10.1242/jcs.00760. [DOI] [PubMed] [Google Scholar]
- Greco C, Bralet MP, Ailane N, Dubart-Kupperschmitt A, Rubinstein E, Le NF, Boucheix C. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res. 2010;70:7674–7683. doi: 10.1158/0008-5472.CAN-09-4482. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zoller M. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- Yue S, Mu W, Erb U, Zoller M. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget. 2015;6:2366–2384. doi: 10.18632/oncotarget.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, North HA, Sahni V, Jeong SJ, Mcguire TL, Berns EJ, Stupp SI., Kessler JA. beta1-Integrin and integrin linked kinase regulate astrocytic differentiation of neural stem cells. PLoS One. 2014;9:e104335. doi: 10.1371/journal.pone.0104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton PJ, Klim JR, Hernandez BA, Koonce CH, Kamp TJ, Kiessling LL. Signals from the surface modulate differentiation of human pluripotent stem cells through glycosaminoglycans and integrins. Proc Natl Acad Sci U S A. 2014;111:18126–18131. doi: 10.1073/pnas.1409525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu EC, Kulp SK, Huang HL, Tu HJ, Salunke SB, Sullivan NJ, Sun D, Wicha MS, Shapiro CL, Chen CS. Function of integrin-linked kinase in modulating the stemness of IL-6-abundant breast cancer cells by regulating gamma-secretase-mediated Notch1 activation in caveolae. Neoplasia. 2015;17:497–508. doi: 10.1016/j.neo.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JR, Creskey MC, Muradia G, Bell GI, Sherman SE, Gao J, Stewart DJ, Cyr TD, Hess DA, Rosu-Myles M. Brief report: elastin microfibril interface 1 and integrin-linked protein kinase are novel markers of islet regenerative function in human multipotent mesenchymal stromal cells. Stem Cells. 2016;34:2249–2255. doi: 10.1002/stem.2385. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci U S A. 2013;110:16061–16066. doi: 10.1073/pnas.1315204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, McCarrey JR, Page DC. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet. 2016 doi: 10.1038/ng.3591. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G, Peterson K, Masterson K, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Orwig KE. Translating spermatogonial stem cell transplantation to the clinic. In: Orwig KE, Hermann BP, editors. Male Germline Stem Cells: Developmental and Regenerative Potential, 1st ed. New York: Humana Press; 2011. pp. 227–253. In: (eds.) [Google Scholar]