Age-associated molecular changes are deleterious and causally linked with aging and may affect life span through diet.
Keywords: aging, lifespan, damage, molecular changes, mice, Yeast, flies
Abstract
Transition through life span is accompanied by numerous molecular changes, such as dysregulated gene expression, altered metabolite levels, and accumulated molecular damage. These changes are thought to be causal factors in aging; however, because they are numerous and are also influenced by genotype, environment, and other factors in addition to age, it is difficult to characterize the cumulative effect of these molecular changes on longevity. We reasoned that age-associated changes, such as molecular damage and tissue composition, may influence life span when used in the diet of organisms that are closely related to those that serve as a dietary source. To test this possibility, we used species-specific culture media and diets that incorporated molecular extracts of young and old organisms and compared the influence of these diets on the life span of yeast, fruitflies, and mice. In each case, the “old” diet or medium shortened the life span for one or both sexes. These findings suggest that age-associated molecular changes, such as cumulative damage and altered dietary composition, are deleterious and causally linked with aging and may affect life span through diet.
INTRODUCTION
Aging is an inevitable biological process characterized by a progressive decrease in physiological function (1–4). Transition through the aging process is associated with numerous molecular changes, such as altered gene expression and metabolite levels, somatic mutations and epimutations, and accumulated molecular damage. The age-dependent accumulation of damage has long been considered a general cause of aging. Many previous studies focused on particular forms of damage, such as oxidative damage, mutations, errors in transcription or translation, and damage to metabolites, as well as on cumulative damage, but it has been difficult to prove its causal role in aging (5–8). Nevertheless, many researchers agree that molecular damage, in the form of either particular damage types or cumulative damage, contributes to the aging process.
However, models of aging that argue against this idea have also been proposed (1, 9). While acknowledging that damage does accumulate as a function of age, but these models predict that this damage has no direct impact on the aging process. Instead, it is proposed that aging may be caused by hyperfunction or continued development because of excessive biological activities, leading to organ pathologies, before the damage can exert its deleterious effects. Hyperfunction and continued development are, in part, reflected in altered gene expression and metabolite levels. These models further expose the fact that the impact of either damage or other molecular changes on the aging process is difficult to characterize and quantify. Because the forms of damage are so numerous, and because they vary depending on factors such as genotype, environment, and diet, how could one address the causal role of damage in aging?
Studies in the past two decades have shown that a great variety of factors and pathways can modulate aging, adjusting life span through genetic, pharmacological, or nutritional manipulation. In particular, previous studies identified two canonical nutrient-sensing pathways: mammalian or mechanistic target of rapamycin (mTOR) and insulin/insulin-like growth factor signaling (10, 11). During aging, continuous stimulation of these pathways leads to a higher risk of aging-related diseases and a decreased life span via reduced autophagy, increased protein aggregation and proteotoxicity, inflammation, reduced expression of antioxidant proteins, mitochondrial dysfunction, and other mechanisms (12–15). Modulation of these pathways is thought to affect the rate of aging and postpone the advent of aging-related diseases across a wide range of taxa from yeast to mice (12). One of the most intriguing aspects of these nutrient-sensing pathways is that they are regulated by nutrients (that is, carbohydrates, proteins, and lipids). This nutritional relationship suggests the possibility that these pathways directly or indirectly regulate the patterns of age-related gene expression and molecular damage (both intrinsic damage and damage associated with diet and environment).
Here, we used species-specific culture media or diets based on the lysates or tissues of young and old organisms, which differed in age-related molecular changes. We examined the long-term effects of these media and diets on the life span of three model organisms: yeast (Saccharomyces cerevisiae), fruitflies (Drosophila melanogaster), and mice (Mus musculus). We observed a decreased life span in yeast, fruitflies, and female mice under the dietary regimens based on old organisms. We discuss the implications of these findings.
RESULTS
Testing the effects of age-related changes on life span of model organisms
To examine how age-related molecular changes, such as cumulative damage and altered molecular composition of cells and tissues, influence the life span of organisms, we reasoned that these changes might affect life span when the age-related compounds were introduced through diet. We also considered that the effect may be more direct if the introduced dietary items mimic closely the specific age-related molecular forms that accumulate in particular organisms throughout their life spans. Although only a fraction of these dietary forms may be expected to be taken up by living organisms, the long-term effect of this treatment may be significant enough to affect life span. To test this possibility, we prepared media or diets based on the biochemistry of organisms of different ages, applied them to the same or closely related living species, and examined their life span. Three model organisms were used: yeast cells, which were maintained on the media containing the lysates of young or old yeast; fruitflies, which were subjected to diets whose nonsugar components (for example, proteins and micronutrients) were replaced with the extracts of young or old fruitflies; and mice, which were fed diets containing the skeletal muscle of young or old deer (Fig. 1). Hereafter, “young” media or diets refer to the media and diets based on young organisms, and “old” media and diets refer to those based on old organisms.
Fig. 1. Testing the effects of age-related changes on life span in three model organisms.
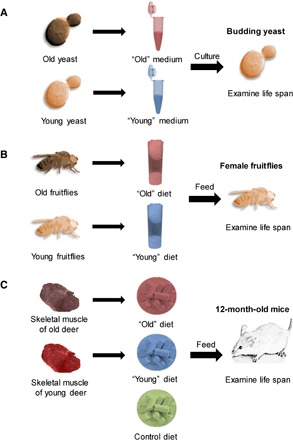
Experimental design of this study included (A) yeast (S. cerevisiae), (B) fruitflies (D. melanogaster), and (C) mice (M. musculus).
Modulation of yeast life span by molecular components derived from young versus old yeast
The budding yeast S. cerevisiae is a classical model organism of aging. Many genetic factors and dietary and pharmacological interventions that affect its life span also modulate the life span of higher eukaryotes (16). Yeast lysate is a common component of cell culture media used in S. cerevisiae experiments. To prepare the media containing the lysates of young and old yeast, we subjected yeast cells to chronological aging (Fig. 2A and fig. S1A), on a large scale (1.5 liters of cell culture for each time point), to generate sufficient biomass for subsequent aging assays. During growth, yeast cells initially metabolize glucose to ethanol and, after glucose depletion, switch to ethanol utilization. Upon depletion of this carbon source, cells stop dividing and show a time-dependent decrease in viability (17). We collected young cells at the beginning of the phase when cells stopped dividing (3 days in stationary phase) and old cells at the point when the cells started losing viability (8 days in stationary phase) (Fig. 2A). We found that cell lysates prepared from these young and old cells could supply all vital components for growth of live yeast cells, with the exception of glucose, which was added as a carbon source to the media. We determined optimal amounts of cell lysates by varying their amounts (for example, lower lysate amounts resulted in shortened life span, likely due to insufficient nutrients) (fig. S2A), also ensuring that the life span of cells grown on these lysates was similar to the one obtained on regular yeast media.
Fig. 2. Age of yeast cells, which are used as components of growth media, modulates the replicative life span of S. cerevisiae.
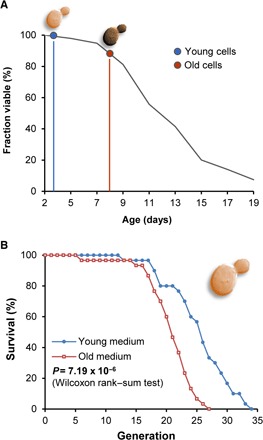
(A) Sampling of cells for media preparation. Wild-type (WT) yeast cells were subjected to chronological aging. Cells were grown in YPD medium, and after the culture reached the stationary phase, cells were collected on days 3 (blue, young) and 8 (red, old). Viability of yeast cells as a function of chronological age was assessed and is also shown in the figure. (B) Replicative life span of yeast cells grown on culture media containing the lysates of young (blue; n = 30) and old (red; n = 30) cells. See table S1 for life-span analysis of individual dietary groups.
Analysis of the replicative life span (Fig. 2B, fig. S1B, and table S1) revealed that cells grown on the young media had an increased maximum and mean life span compared to those grown on the media based on the same amounts of the old lysate (P = 7.19 × 10−6, Wilcoxon rank sum test). This analysis was also confirmed by comparing the Gompertz fit parameters of mortality curves for yeast on young and old media [P = 9.89 × 10− 15 for the difference in Gompertz α and P = 0.0018 for the difference in value of initial mortality log(M0); fig. S3, A and B]. The goodness of Gompertz fit for the mortality curve of yeast on young and old media was R2 = 0.6337 and R2 = 0.9497, respectively. We consistently observed this life-span effect using different batches of chronologically old and young cells. We also found that the largest amounts of old lysate led to a decrease in life span (fig. S2A), consistent with the idea that the age-related molecular changes affect the aging process in yeast cells.
These data suggested that the old lysate harbors factors that accelerate the aging process in yeast or is deficient in factors that decelerate the process. To obtain further insights, we separated the lysates into low–molecular weight (LMW) and high–molecular weight (HMW) fractions (fig. S2, B and C) and tested the influence of each fraction on the life span. When the LMW and HMW fractions were recombined separately for young and old, the life span was shorter in the old + old medium than in the young + young medium (fig. S2D), suggesting that the separation procedure did not alter the aging-modifying properties. Old LMW and young HMW showed a decreased life span compared to young LMW in combination with old HMW (fig. S2E). Thus, the LMW fraction had a stronger influence on the life span under our experimental conditions.
Modulation of fruitfly life span by diets containing components of young and old flies
In the fruitfly study, we prepared old diets by collecting a large number of freshly deceased flies (n = 5000, average life span = 45 days), which were allowed to age and die naturally in a fly incubator under standard laboratory conditions. We also sacrificed 3- to 5-day-old flies (n = 5000) for the young diet group. Both groups of flies were treated identically, and we observed no overall changes in total amino acid levels (fig. S4). The lysates were prepared by freezing the flies in liquid nitrogen and grounding them to dust, which was briefly agitated in water. The preparations were then centrifuged, and the supernatants were scored for protein levels. By measuring the protein concentration, we matched the general nutrient requirement of dietary yeast, which is frequently used for live fly culture and life-span studies. Dietary preparations for laboratory fly culture also include sucrose as a carbohydrate source, in addition to the autolyzed yeast. In these diets, the individual amounts of sugar and yeast are each represented as 1X. Thus, to optimize the diets prepared from fly lysates (which replaced dietary yeast), we examined the life span of flies with various amounts of fly lysates mixed with fixed 1X or 0.5X amount of carbohydrates, in comparison with the commonly used 1X yeast/sucrose media (Fig. 3A). The dietary composition of sugar plus fly lysate (young) at protein concentrations matching 1X levels of yeast yielded the same life span as did the 1X sugar/yeast diet and was chosen for subsequent studies. Using this standardization, we next prepared fly lysate–containing diets using old and young flies. Both were adjusted for protein concentration, which matched protein levels in the 1X yeast fly diets (Fig. 3A and table S2). Subjecting young, mated female D. melanogaster to these diets revealed that the life span was significantly lower in the old diet group compared with the young diet group (mean life-span difference: −12.8%; P = 0.024, log-rank test; Fig. 3B and table S2). Thus, similar to the study in yeast, we found that the life span of fruitflies was decreased by the old diet. However, the Gompertz fit did not show a statistically significant difference between Gompertz parameters of mortality curves for fruitflies on old and young diets (P = 0.508; fig. S3, C and D). The goodness of Gompertz fit for mortality curve of flies on young and old diets was R2 = 0.4825 and R2 = 0.6033, respectively.
Fig. 3. Age of fruitflies, whose lysates are used as a dietary source of protein, modulates the life span of female D. melanogaster.
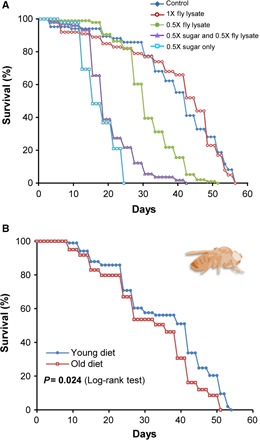
(A) Survival curves of female fruitflies reared on diets containing various carbohydrate and protein sources: Control (1X sugar and 1X yeast; n = 85), 1X fly lysate (1X sugar and 1X fly lysate; n = 100), 0.5X fly lysate (1X sugar and 0.5X fly lysate; n = 96), 0.5X sugar and 0.5X fly lysate (n = 105), and 0.5X sugar only (n = 95). The diet based on 1X fly lysate supports the normal life span of D. melanogaster. (B) Kaplan-Meier survival curves of female fruitflies subjected to diets containing lysates of young (n = 121) or old flies (n = 96). The log-rank test was used for statistical analysis. See table S2 for life-span analysis of individual dietary groups.
Modulation of life span of mice by diets containing young and old skeletal muscle
In the case of mice, we adopted a dietary intervention strategy that used skeletal muscle derived from animals of different age. For the long-term survival experiment, large quantities of diets were required, which would be difficult if mouse tissue were used as the diet source. Instead, the skeletal muscle of farmed red deer (Cervus elaphus; maximum life span, 31.5 years) was chosen because of the large amount of this tissue and the availability of animals differing in age. Skeletal muscle of freshly sacrificed young adult (3-year-old) and old (25-year-old) male red deer were obtained and immediately frozen. To test the nutritional composition of the tissues, proximate analysis was performed following freeze-drying and pulverization. The composition of protein and fat was found to be different in young (90.0% protein and 2.1% fat) and old (57.8% protein and 38.0% fat) muscles (table S3), in agreement with previous studies, which found that the muscle fat increases during aging in mammals (18). To accommodate this difference, we adjusted AIN-76A diet to control for both fat and protein levels (table S4). Diet composition may affect the rate of aging (19), and the balance of macronutrients may influence longevity and late-life health in ad libitum mice (20). Therefore, all experimental diets used in our study were isoenergetic (4.5 kcal/g) and contained the same proportion of protein (20.3%) and fat (18.1%).
We divided 12-month-old mice into three dietary groups: control group (n = 27), young diet group (n = 30), and old diet group (n = 30). These mice were maintained on each diet ad libitum until the end of their lives. We found no difference in the weight of mice among the groups (fig. S5A), as well as among females and males (fig. S5, B and C). The survival curves of mice fed young and old diets are shown in Fig. 4. Compared to the young diet group, the old group exhibited a trend toward decreased life span (mean life-span difference, −6.1%), which did not reach statistical significance (P = 0.2, log-rank test; Fig. 4A). However, when the survival curves were separated by sex, the old diet group displayed a significantly shorter life span compared to the young diet group in females (P = 0.475, log-rank test); the mean life span of the old diet females decreased by 13.3% compared with that of the young diet females (Fig. 4B and table S5). There was no difference in the survival between the young and old diet groups of males (P = 0.87, log-rank test; Fig. 4C). These results were also confirmed by the Gompertz fit analysis of mortality curves produced in the corresponding experiments (fig. S3, E to H). Namely, the Gompertz rate α was different for female mice on young and old diets (P = 0.0184), whereas for male mice on young and old diets we did not find a statistically significant difference (P = 0.238). When the group that was fed a control diet was compared with the young or old diet group, there was no significant difference in life span (by log-rank test) among either females or males (fig. S5, D to I). Together, the data suggest that the diet containing the old skeletal muscle may shorten the mouse life span, although sexual dimorphism was apparent under the conditions used.
Fig. 4. Age of the skeletal muscle used as a component of diet may modulate the life span of mice.
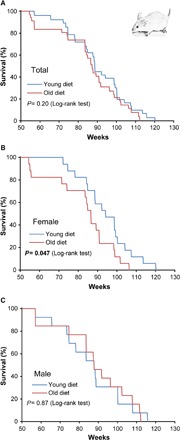
Kaplan-Meier survival curves of the indicated dietary groups containing young and old skeletal muscles. (A) All mice. (B) Females only. (C) Males only. Data were from 30 mice in the young diet group (17 females and 13 males) and 30 mice in the old group (17 females and 13 males). The log-rank test was used for statistical analysis. See table S5 for life-span analysis of individual dietary groups.
To examine the probable causes of death and progression of pathological lesions, we performed a full histopathological analysis of mice at their time of death. The control diet group generally had a lower tumor incidence compared with the old diet group (33.3% and 66.7%, respectively; P = 0.014; fig. S6A). In addition, this group had a less diverse spectrum of tumors than either the young or old diet group (fig. S6D). When the incidence of tumors was separated by sex, the old diet group displayed a higher incidence of tumors compared to the control diet group in males (27.3% and 83.3%, respectively; P = 0.006; fig. S6C). Adenoma was the major contributing neoplastic lesion (fig. S6F). In females, there was no significant difference in tumor incidence among the groups, although the tumor spectrum was somewhat different (fig. S6, B and E). Thus, neoplastic disease did not seem to be a major contributor to the difference in life span in females. The major lesion of nonneoplastic disease in all groups was systemic or localized amyloidosis (table S7), a known cause of morbidity and mortality in ICR mice (21), because these mice are susceptible to senile ApoA2 (22). There was no significant difference in the amyloidosis lesions between the dietary groups.
To determine the long-term effect of diets on gut microbiota, we sequenced 16S ribosomal RNA (rRNA) of fecal samples from female mice at 270 days after starting each diet. The coverage [8152 ± 2125 (mean ± SD) reads per sample] allowed us to analyze α diversity of samples. More than 700 species were detected when analyzed by 4000 reads per sample (fig. S7A). At the 4000-read level (fig. S7B), the fecal microbiota of the old diet group showed lower α diversity (724 species per 4000 reads) than that of the young diet group (P < 0.05, 852 species per 4000 reads), and α diversity of the control diet group was closer to that of the young diet group (801 species per 4000 reads, P = 0.30). The overall pattern of fecal microbiota was also compared among the three diet groups. According to β diversity, the control diet group was close to the young diet group, whereas the old diet group was more distant (fig. S7C). The relative abundance of some bacterial groups differed significantly (table S8). For example, the genus Ruminococcus (fig. S7D) was less abundant in the old diet group than in the control (P < 0.05) and young (P = 0.14) groups.
DISCUSSION
It is commonly thought that aging is caused by molecular damage that leads to an age-associated decrease in the organism’s fitness, age-related pathologies, and, ultimately, death. However, the myriad of damage forms that accumulate in organisms with age make it difficult to confirm a causal relationship experimentally. Although it is known that particular damage forms (for example, DNA damage and oxidative damage) in laboratory mutant organisms are associated with reduced life span, these damage forms rarely reach the high levels used in experimental investigations under normal, physiological conditions. Life span may also be modulated by a combinatorial effect of many damage forms known to increase with age (23). It has also been argued that molecular damage may not be relevant to aging at all, because hyperfunction due to excessive gene activities may lead to pathology and kill organisms first, that is, before the damage can exert its deleterious effects (9). These excessive activities are associated with altered or dysregulated gene expression. Although it may also be argued that hyperfunction is not mutually exclusive with molecular damage, a causal role in aging of either molecular damage or excessive gene functions has not yet been established.
To address this issue from a different perspective, we designed experiments to examine the effect of age-related molecular changes through a dietary approach. We hypothesized that these molecular changes (for example, accumulated molecular damage) may be taken up by organisms from diet. If so, the long-term effect of the altered age-associated molecular state may be evaluated by analyzing the life spans of organisms fed diets (or grown on media) that contain (or not) these molecular changes. We tested this idea by using three model organisms of aging: yeast, fruitflies, and mice. Yeast cells were grown on the media containing the lysates prepared from chronologically young or old yeast, fruitflies were fed diets based on the homogenates of young or old flies, and mice were fed diets that included the skeletal muscle of young or old deer. We adjusted the conditions of these experiments so that the life spans on these diets were similar to those observed on control diets or conditions, to ensure that the diets were not deficient in critical components.
For each model organism tested, we observed a longer life span on the young diet (or medium) compared to the old diet (medium). In yeast, the replicative life span was analyzed, and the difference was statistically significant between the young and old media groups. In fruitflies, we only analyzed females (because we were limited by the number of old flies that we could collect that could sustain viability of live flies throughout the entire life span), and the difference in life span was also statistically significant between the young and old diet groups. In the case of mice, the regular AIN-76A purified diet was modified to include skeletal muscle of young (3-year-old) or old (25-year-old) deer. Because of time and diet limitations, we used 12-month-old mice (rather than weaning mice) as experimental subjects. Under these conditions, we observed no difference in life span among males, but a statistically significant decrease in life span in female mice, when fed the old diet. None of the conditions tested throughout this study showed an opposite effect (of increased life span when fed old diet) in any of the model organisms.
As previously reported (24), the gut microbiota changes with age. For example, human centenarians (>100 years old) have lower α diversity than either elderly (70 years old) or young people. Such age-associated changes in α diversity may be related to the higher diversity of the young diet group (female mice, fig. S7B). In addition, Ruminococcus (Ruminococcus lactaris and Ruminococcus obeum) decrease in abundance with human age (24), and centenarians have lower levels of these species than either elderly or young people. A similar reduction in Ruminococcus was observed in our study in the old diet group of female mice (fig. S7D). These data suggest that diet-induced changes in the gut microbiota may be influenced by, and may also have a role in, aging.
It is of interest that the effects observed in our study were relatively minor (that is, the difference in life span was on the order of 10%). We interpret this finding as an indication that diet, and more generally environment, is only one of the contributors to the aging process. Other major contributors, such as intrinsic factors, genotype, and stochastics, presumably account for the remaining control of life span, but they were not examined in the study. It is also possible that the relatively small effect observed was due to the possibility that internal molecular changes have a stronger impact than changes introduced through diet.
Our study has several caveats. First, the contributions of various forms of molecular change to life-span control could not be defined. For example, both cumulative damage and nutritional composition likely differed between young and old diets. This was already apparent from the analysis of fat in young and old skeletal muscle tissue in the mouse experiment. To account for this difference, we prepared isocaloric diets that had the same protein and lipid content. Nevertheless, the nature of age-related changes is such that it is impossible to account for all nutritional differences between young and old muscles, because these tissues presumably also differ in the components other than proteins, lipids, and carbohydrates. Likewise, fruitfly aging is associated with profound quantitative changes in transcriptome (25), metabolome (23), protein levels (26), and proteasome function (27). Studies in Drosophila indicate that individual nutrients and, in particular, protein/amino acids are important regulators of life span (28). Similarly, marked changes at all levels occur during aging in yeast. Thus, full control of macronutrients and micronutrients is not possible when the effects of age-related molecular changes are examined.
A second caveat is that, in the mouse study, the life span of males was not affected by the diet, and the mortality of mice did not match the incidence of pathology. One possible explanation is that the contribution of age-associated changes in our experimental setup was insufficient to observe the effect of the old diet in males, due either to the diet composition or to the fact that 12-month-old, as opposed to young, mice were used. In addition, gender specificity in the effects of treatments that extend life span was previously observed in both mice and fruitflies. In mice, the effects of deletion of insulin substrate receptor I (29) and the mTOR inhibitor (30) were found to be more robust in females than in males. Similarly, reduced activity of the insulin-signaling pathway (31) and the mTOR pathway by deletion of S6K (32) extended the life span only in females. Sexual dimorphism is repeatedly observed in studies of rapamycin-mediated life-span extension (33–35).
A third caveat is our inability to pinpoint the contribution of various individual components of the diet to the life span–shortening effect. In yeast, where we were not limited by the availability of young and old media, we were able to test the effects of LMW and HMW fractions and found that the LMW fraction exerted a stronger effect. This was expected because this fraction is expected to contain the most significant amounts of soluble, nutritionally accessible damaged molecules. We did not pursue further fractionation of yeast media, because the effect on life span is cumulative and is unlikely to be defined by a single component. It is also likely that the contribution of various dietary components to life-span shortening is different in different species. We could not test the contributions of various dietary components in flies and mice because of the limited diet amounts available for these experiments.
Despite these caveats, we were encouraged by the consistent effect we observed when the same experimental design was applied to the three model organisms of aging. The fact that, in each species, the old diet (or growth medium) was associated with reduced life span argues that the age-related changes are deleterious and directly affect the aging process. The idea that age-associated damage and/or nutrient composition contribute to aging is not new, but it had not previously been thoroughly examined. For example, previous studies found that abnormal amino acids in the diet do not reduce life span (36). However, to our knowledge, none of the previous studies analyzed the impact of molecular changes as a whole or examined the effect of these changes across several model organisms.
In summary, our report supports the idea of a causal link between age-related molecular changes and the aging process, suggesting that these changes are deleterious. Moreover, these changes (for example, accumulated damage and altered nutritional composition) may affect living organisms when age-damaged molecules are introduced through diet. Finally, this study also strengthens the case for the impact of cumulative changes, as opposed to its individual forms, as a key factor in aging.
MATERIALS AND METHODS
Yeast experiments
WT (BY4741) yeast cells were grown overnight in 10 ml of yeast peptone dextrose (YPD) liquid medium. Five hundred microliters of overnight culture was transferred to 1.5 liters of YPD medium. Growth was monitored by measuring the OD600 (optical density at 600 nm) over the course of 2 days. After 2 days, the same culture was subjected to chronological aging. Viability was measured every 2 days by growth on YPD plates at 30°C, and the colonies were counted after 3 days and plotted as percent survival. Cells were collected at certain time points by centrifugation of 1.5-liter culture of young or old cells, washed with water, and frozen at −80°C. The pH of cell culture medium changed somewhat (6.4 versus 7.2) during chronological aging, although it is known that this does not affect the replicative life span (37). To prepare yeast cell extracts, frozen yeast cells were thawed, and 10 g of glass beads and 5 ml of water were added. Using a bead beater, cells were disrupted for 15 min on ice. The supernatant was cleared by centrifugation at 15,000 rpm for 30 min at 4°C. Cell lysates were then passed through syringe filters (0.25 μm) and normalized according to protein concentration (determined using the bicinchoninic acid method). For plate preparation, 2% agar was mixed with water (v/v), autoclaved, and cooled to ~50°C. Next, cell lysates (50 mg total) and 2 ml of sugar (2 mg/ml) were added to the final volume of 20 ml of agar mix and poured onto plates. Cells were grown on YPD plates overnight at 30°C, then transferred to the lysate plates, and incubated overnight before the experiment. For replicative aging assays, 30 virgin cells were chosen, and new buds (daughters) from these virgin cells were removed and discarded as they formed every 2 hours. This process continued until cells ceased dividing. Life span was determined as the number of daughter cells that each mother cell generated.
Dietary experiment with fruitflies
D. melanogaster (Canton-S strain; females) was used in preparing the diet and life-span studies. Sugar and yeast in the Drosophila laboratory culture medium provide carbohydrate and protein sources, respectively, which are essential macronutrients. In this medium, the individual amounts of sugar and yeast are represented at 1X concentration. To evaluate the effects of fly lysate on life span and to compare it with the traditionally used yeast diet, we performed control experiments that used several media sources adjusted for identical protein concentration, which we sourced from yeast and flies. To prepare diets consisting of flies as a component of diet, we froze them in liquid nitrogen and ground them to dust, which was later agitated and allowed to sit in water for 15 min at room temperature. The preparation was centrifuged, and the supernatant was scored for protein concentration using the DC Protein Assay Kit (Bio-Rad). Titrations were used, and the effects of different levels of diet components on life span were determined (Fig. 3A). Liquid diets containing fruitflies were stored as frozen aliquots at −80°C. Food changes were performed every 3 days using 200 μl of freshly thawed medium that was applied atop of agarose bedding and allowed to dry for several hours. Life-span experiments were performed according to standard methods: Cultures were maintained at 12-hour light/12-hour dark cycle at 25°C with 50% relative humidity. Dead flies were counted every 3 days during food changes. To evaluate the effects of young and old diets on life span, we tested the effects of lysates prepared from old and young flies according to the above procedures. Old flies were obtained during large-scale aging experiments, using naturally dying flies collected between 30 and 60 days from the beginning of the life-span study (n = 5000, average life span = 45 days). Specimens were collected during food changes and immediately placed at −80°C for storage. Young flies (3 to 5 days old; n = 5000) were killed instantly and frozen after 2 to 3 days. This provided a control for the old flies, which could start to decompose after dying. Diets were then prepared as described above in parallel for young and old diets.
Dietary experiment with mice
Experimental procedures performed on mice were approved and carried out according to the guidelines established by the Institutional Animal Care and Use Committee (IACUC) of Ewha Womans University (IACUC No. 2012-01-038). Eighty-seven 11-month-old Hsd:ICR (CD-1) mice (37 males and 50 females) were purchased from Harlan. The mice were housed in a facility with 12-hour light cycle and maintained on a standard purified mouse diet (CA.170481, Harlan) for 1 month before the start of the experiment. The animals were then randomly assigned to three different diet groups: control (11 males and 16 females), young diet (13 males and 17 females), and old diet (13 males and 17 females) groups. Food and water were available ad libitum throughout the acclimation and experimental periods. Experimental diets included skeletal muscles of young adult (3-year-old) and old (25-year-old) male red deer (C. elaphus). Fresh tissues were obtained from slaughtered deer and immediately frozen. To verify the nutritional composition of young and old muscles, we performed proximate analysis following freeze-drying and pulverizing the frozen tissues (table S3). Three isoenergetic experimental diets with the protein source from the muscles of young and aged deer were then produced (table S4). The control diet contained casein and beef tallow, commonly used in rodent diets, as protein and fat sources. All diets were based on AIN-76A purified diet (CA.170481, Harlan) and contained the same proportion of protein (20.3%) and fat (18.1%). Diets were stored at 4°C and distributed to cages once per week. Body weights were measured on a weekly basis for 15 weeks from the starting time point of the dietary experiment. Survival curves were plotted using the Kaplan-Meier method, which includes all available animals at each time point.
Necropsy and pathology
Full histopathological analyses of all mice were performed at their time of death. Mice were considered to be at the end of life and euthanized by carbon dioxide inhalation when they were moribund and demonstrated one or more clinical signs suggesting imminent death: nonresponsive to being touched, labored breathing, and failure to eat or drink. Mice found dead in cages were also submitted for necropsy. Liver, kidney, spleen, thymus, brain, colon, heart, and lung plus any visible abnormalities were preserved in 10% neutral phosphate-buffered formalin. Fixed tissues were embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin. Tissues were examined microscopically and assessed as to the probable cause of death. The determination was based on the pathology in the tissue sections, degree of tissue involvement, severity of changes, and whether the effects were expected to contribute or lead to the death of the animal.
DNA extraction and 16S rRNA sequencing
Bacterial genomic DNA was extracted from the fecal samples (female mice; day 270 after starting the diet) using the NucleoSpin Soil kit (Macherey-Nagel). To amplify the V4 region of the 16S rRNA gene, we used bar-coded primer sets containing 5′-GGACTACHVGGGTWTCTAAT-3′ or 5′-GGACTACHVGGGTWTCTAAT-3′. Bacterial genomic DNA (5 ng) was used as a template for polymerase chain reaction (PCR) with reaction buffer [25 mM Mg2+, 200 μM deoxynucleotide triphosphate each, 0.75 U of Ex Taq (TaKaRa), and bar-coded primers (5 pmol each)]. We carried out PCR under the following conditions: 94°C for 3 min, 35 cycles of amplification (94°C for 45 s, 55°C for 1 min, and 72°C for 90 s), and 72°C for 10 min. The same amount of each amplified DNA was pooled for construction of sequencing libraries. Sequencing libraries were prepared by using the NEBNext Ultra DNA Library Prep Kit (New England BioLabs, catalog no. E7370S) and sequenced for paired-end 250–base pair reads on the Illumina MiSeq.
Microbiota analysis
MiSeq reads were quality-filtered and demultiplexed according to bar codes. The processed paired reads were assembled, and the assembled reads were further used for operational taxonomic unit (OTU) picking in Quantitative Insights Into Microbial Ecology (QIIME 1.6.0) pipelines (38). Greengenes database (gg_ptus-13_8-release version, 97% nucleotide identity) (39) was used for OTU picking. On the basis of the assigned OTU, α/β-diversity analyses and taxonomy summarization were performed using the QIIME pipelines. For a rarefaction curve of observed species, we used 4000 reads per sample. Principal components analysis was performed, with selected bacterial groups showing significantly different relative abundance at the genus level between any two groups.
Statistical analyses
Unless otherwise stated, two-tailed Student’s t test assuming unequal variances was used. For life-span analyses, statistical analyses were performed using the JMP (version 10) software (SAS Institute Inc.). Log-rank tests were used for mouse and fruitfly survival analyses, with stratification by sex for comparisons that pooled data across sex (mice). Wilcoxon rank sum test was used for the analyses of the yeast replicative life span. Differences between the two groups were evaluated by t test, and nominal P values were reported without adjustment for multiple comparisons. Gompertz fit of mortality curves was performed to the two-parametric exponential Gompertz law M(t) = M0eαt, where M0 is the initial mortality rate and α is the Gompertz exponent, as the amount of data produced in all three experiments was insufficient to construct a robust fit to the three-parametric Gompertz-Makeham law M(t) = λ + M0eαt. To perform the fit, a natural logarithm of mortality was taken, and the result was subjected to the robust LAR (least absolute residuals) fit to a linear two-parametric dependence log(M) = log(M0) + αt to minimize the effect of outliers. Fisher’s exact test, programmed in R, was used to compare proportions of mice between groups. In all cases, P values of ≤0.05 were considered significant.
Acknowledgments
Funding: This work was supported by the NIH; the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Republic of Korea (no. 914005-04); and 2014 Research Grant (C1011640-01-01) from Kangwon National University, Republic of Korea. Author contributions: S.-G.L. carried out the main studies with mice, A.K. with yeast, and A.S.A. with flies. D.I.P., E.J.S., D.-M.G., G.-D.J., J.Y.H., E.B.K., and D.-Y.K. conducted the experiments or analyzed the data. V.N.G. designed and supervised the project. S.-G.L. and V.N.G. wrote the paper with contributions from other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1601833/DC1
fig. S1. Growth curve and independent replicates of life-span analysis in yeast.
fig. S2. Replicative life span in yeast experiment.
fig. S3. Gompertz fits of survival curves in different experiments.
fig. S4. Age-associated changes in amino acid levels in fruitflies.
fig. S5. Change of body weights and life span of the control group compared with the young and old diet groups in mice.
fig. S6. Percentage of mice with tumor and tumor spectrum in the different diet groups.
fig. S7. Fecal microbiota of female mice at 270 days after starting the diet.
table S1. Replicative life-span analysis of yeast cells grown on young and old diets.
table S2. Life-span analysis from survival curves in the young and old diet groups in fruitflies.
table S3. Proximate composition (%, w/w) of muscles from young and old red deer.
table S4. Ingredients, energy, and macronutrient content of experimental diets in the mice study.
table S5. Life-span analysis from survival curves in the young and old dietary groups in mice.
table S6. Life-span analysis from survival curves of the control group compared with the young and old diet groups.
table S7. Pathology at time of death in all experimental diet groups.
table S8. Relative abundance of selected bacterial groups found in fecal samples of female mice.
REFERENCES AND NOTES
- 1.Gems D., Partridge L., Genetics of longevity in model organisms: Debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood T. B. L., Understanding the odd science of aging. Cell 120, 437–447 (2005). [DOI] [PubMed] [Google Scholar]
- 3.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijg J., Campisi J., Puzzles, promises and a cure for ageing. Nature 454, 1065–1071 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rattan S. I. S., Increased molecular damage and heterogeneity as the basis of aging. Biol. Chem. 389, 267–272 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Golubev A. G., Random necessity, transcription initiation, induction of differentiation and need for randomness. Biokhimiia 61, 1303–1319 (1996). [PubMed] [Google Scholar]
- 7.Gladyshev V. N., Aging: Progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell 15, 594–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladyshev V. N., The origin of aging: Imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet. 29, 506–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blagosklonny M. V., Aging and immortality: Quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle 5, 2087–2102 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Wullschleger S., Loewith R., Hall M. N., TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Bartke A., Sun L. Y., Longo V., Somatotropic signaling: Trade-offs between growth, reproductive development, and longevity. Physiol. Rev. 93, 571–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenyon C. J., The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Fontana L., Partridge L., Longo V. D., Extending healthy life span—From yeast to humans. Science 328, 321–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson S. C., Rabinovitch P. S., Kaeberlein M., mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuervo A. M., Autophagy and aging: Keeping that old broom working. Trends Genet. 24, 604–612 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denoth Lippuner A., Julou T., Barral Y., Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 38, 300–325 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Kaeberlein M., Lessons on longevity from budding yeast. Nature 464, 513–519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolden-Hanson T., Marck B. T., Smith L., Matsumoto A. M., Cross-sectional and longitudinal analysis of age-associated changes in body composition of male Brown Norway rats: Association of serum leptin levels with peripheral adiposity. J. Gerontol. 54, B99–B107 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Grandison R. C., Piper M. D. W., Partridge L., Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solon-Biet S. M., McMahon A. C., Ballard J. W. O., Ruohonen K., Wu L. E., Cogger V. C., Warren A., Huang X., Pichaud N., Melvin R. G., Gokarn R., Khalil M., Turner N., Cooney G. J., Sinclair D. A., Raubenheimer D., Le Couteur D. G., Simpson S. J., The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 19, 418–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelhardt J. A., Gries C. L., Long G. G., Incidence of spontaneous neoplastic and nonneoplastic lesions in Charles River CD-1® mice varies with breeding origin. Toxicol. Pathol. 21, 538–541 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Gruys E., Tooten P. C. J., Kuijpers M. H. M., Lung, ileum and heart are predilection sites for AApoAII amyloid deposition in CD-1 Swiss mice used for toxicity studies. Pulmonary amyloid indicates AApoAII. Lab. Anim. 30, 28–34 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Avanesov A. S., Ma S., Pierce K. A., Yim S. H., Lee B. C., Clish C. B., Gladyshev V. N., Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. eLife 3, e02077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkïla J., Monti D., Satokari R., Franceschi C., Brigidi P., De Vos W., Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLOS ONE 5, e10667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou S., Meadows S., Sharp L., Jan L. Y., Jan Y. N., Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 13726–13731 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming J. E., Quattrocki E., Latter G., Miquel J., Marcuson R., Zuckerkandl E., Bensch K. G., Age-dependent changes in proteins of Drosophila melanogaster. Science 231, 1157–1159 (1986). [DOI] [PubMed] [Google Scholar]
- 27.Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., Miura M., Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 29, 1095–1106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatar M., Post S., Yu K., Nutrient control of Drosophila longevity. Endocrinol. Metab. 25, 509–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selman C., Lingard S., Choudhury A. I., Batterham R. L., Claret M., Clements M., Ramadani F., Okkenhaug K., Schuster E., Blanc E., Piper M. D., Al-Qassab H., Speakman J. R., Carmignac D., Robinson I. C. A., Thornton J. M., Gems D., Partridge L., Withers D. J., Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 22, 807–818 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., Flurkey K., Nadon N. L., Wilkinson J. E., Frenkel K., Carter C. S., Pahor M., Javors M. A., Fernandez E., Miller R. A., Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P. C., Cervera P., Le Bouc Y., IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Selman C., Tullet J. M. A., Wieser D., Irvine E., Lingard S. J., Choudhury A. I., Claret M., Al-Qassab H., Carmignac D., Ramadani F., Woods A., Robinson I. C. A., Schuster E., Batterham R. L., Kozma S. C., Thomas G., Carling D., Okkenhaug K., Thornton J. M., Partridge L., Gems D., Withers D. J., Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller R. A., Harrison D. E., Astle C. M., Baur J. A., Boyd A. R., de Cabo R., Fernandez E., Flurkey K., Javors M. A., Nelson J. F., Orihuela C. J., Pletcher S., Sharp Z. D., Sinclair D., Starnes J. W., Wilkinson J. E., Nadon N. L., Strong R., Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J. Gerontol. 66, 191–201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamming D. W., Ye L., Katajisto P., Goncalves M. D., Saitoh M., Stevens D. M., Davis J. G., Salmon A. B., Richardson A., Ahima R. S., Guertin D. A., Sabatini D. M., Baur J. A., Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Bokov A., Gelfond J., Soto V., Ikeno Y., Hubbard G., Diaz V., Sloane L., Maslin K., Treaster S., Réndon S., van Remmen H., Ward W., Javors M., Richardson A., Austad S. N., Fischer K., Rapamycin extends life and health in C57BL/6 mice. J. Gerontol. 69, 119–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. L. Strehler, Time, Cells, and Aging (Demetriades Brothers, 1999). [Google Scholar]
- 37.Wasko B. M., Carr D. T., Tung H., Doan H., Schurman N., Neault J. R., Feng J., Lee J., Zipkin B., Mouser J., Oudanonh E., Nguyen T., Stetina T., Stetina A., Shemorry A., Lemma M., Kaeberleinl M., Buffering the pH of the culture medium does not extend yeast replicative lifespan. F1000Research 2, 2–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L., Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1601833/DC1
fig. S1. Growth curve and independent replicates of life-span analysis in yeast.
fig. S2. Replicative life span in yeast experiment.
fig. S3. Gompertz fits of survival curves in different experiments.
fig. S4. Age-associated changes in amino acid levels in fruitflies.
fig. S5. Change of body weights and life span of the control group compared with the young and old diet groups in mice.
fig. S6. Percentage of mice with tumor and tumor spectrum in the different diet groups.
fig. S7. Fecal microbiota of female mice at 270 days after starting the diet.
table S1. Replicative life-span analysis of yeast cells grown on young and old diets.
table S2. Life-span analysis from survival curves in the young and old diet groups in fruitflies.
table S3. Proximate composition (%, w/w) of muscles from young and old red deer.
table S4. Ingredients, energy, and macronutrient content of experimental diets in the mice study.
table S5. Life-span analysis from survival curves in the young and old dietary groups in mice.
table S6. Life-span analysis from survival curves of the control group compared with the young and old diet groups.
table S7. Pathology at time of death in all experimental diet groups.
table S8. Relative abundance of selected bacterial groups found in fecal samples of female mice.


