Abstract
The gene encoding Migration and Invasion Inhibitory Protein (MIIP), located on 1p36.22, is a potential tumour suppressor gene in glioma. In this study, we aimed to explore the role and mechanism of action of MIIP in colorectal cancer (CRC). MIIP protein expression gradually decreased along the colorectal adenoma-carcinoma sequence and was negatively correlated with lymph node and distant metastasis in 526 colorectal tissue samples (P<0.05 for all). Analysis of The Cancer Genome Atlas (TCGA) data showed that decreased MIIP expression was significantly associated with MIIP hemizygous deletion (P=0.0005) that was detected in 27.7% (52/188) of CRC cases, and associated with lymph node and distant metastasis (P<0.05 for both). We deleted one copy of the MIIP gene in HCT-116 CRC cells using zinc finger nuclease technology and demonstrated that MIIP haploinsufficiency resulted in increased colony formation and cell migration and invasion, which was consistent with the results from siRNA-mediated MIIP knockdown in two CRC cell lines (P<0.05 for all). Moreover, MIIP haploinsufficiency promoted CRC progression in vivo (P<0.05). Genomic instability and spectral karyotyping assays manifested that MIIP haploinsufficiency induced chromosomal instability (CIN). Besides modulating the downstream proteins of APC/CCdc20, securin and cyclin B1, MIIP haploinsufficiency inhibited topoisomerase II (Topo II) activity and induced chromosomal missegregation. Therefore, we report that MIIP is a novel potential tumour suppressor gene in CRC. Moreover, we characterized the MIIP gene as a novel CIN suppressor gene, through altering the stability of mitotic checkpoint proteins and disturbing Topo II activity.
Keywords: MIIP, colorectal cancer, progression, chromosomal instability, topoisomerase
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in both men and women and is the third leading cause of cancer death in the United States [1]. Despite early screening and the introduction of new chemotherapeutic strategies, CRC survival rates have not substantially improved during the past 20 years. The 5-year survival rate is less than 10% for patients with stage IV CRC [2]. It has been commonly accepted that most CRCs undergo a neoplastic progression of the adenoma-carcinoma sequence, accompanied by an accumulation of successive genetic alterations [3, 4]. The chromosomal instability (CIN), mutator phenotype/mismatch repair, and hypermethylation phenotype (CIMP+) pathways have been demonstrated to be important molecular pathogenesis processes underlying tumorigenesis and progression of CRC [5, 6]. However, the molecular events that contribute to the development and progression of CRC are likely far more extensive than previously appreciated [7].
The gene encoding Migration and Invasion Inhibitory Protein (MIIP) is located on chromosome 1p36.22 and was found to be a potential tumour suppressor gene in glioma [8]. In our previous studies, MIIP was initially identified, in a yeast-two-hybrid screen, as a binding partner for insulin-like growth factor binding protein 2 and was shown to inhibit glioma cell invasion [9]. Our subsequent work demonstrated that MIIP can also interact with Cdc20, which is involved in the regulation of mitotic progression [8]. However, the function of MIIP in CRC remains unclear.
In this study, we provide evidence that MIIP haploinsufficiency is associated with CRC progression in TCGA and an independent cohort. The effect of MIIP haploinsufficiency/downregulation in CRC is then manifested in vitro and in vivo. Subsequent molecular studies demonstrate that MIIP haploinsufficiency leads to a weakened mitotic checkpoint and inhibits the decatenation activity of Topo II, promoting chromosomal missegregation and CIN. Therefore, we report that MIIP is a novel tumour suppressor gene and CIN suppressor gene in CRC and is a potential target for CRC treatment.
Materials and Methods
CRC patients and data collection
We collected 526 formalin-fixed, paraffin-embedded colorectal tissue samples (including 287 carcinomas, 145 adenomas, and 94 samples of histologically normal mucosa which were more than 5 cm distant from the corresponding cancer or adenoma tissues, from the Department of Pathology at Tianjin Medical University Cancer Institute and Hospital, after we received institutional review board approval from Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) and obtained consent from every patient. All of the samples were from inpatients undergoing surgical operation from January 2000 to December 2004, and none of the patients had received any chemotherapy or radiotherapy before operation. The cohort included 53 sets of matched surgical samples (carcinomas, adenomas, and non-neoplastic mucosae from the same patient). The clinical information is shown in Supplementary Table S1. Tissue microarrays (TMA) were constructed using a manual tissue microarray instrument (Beecher Instruments) equipped with a 2.0 mm punch needle, as described in our previous study [10].
Copy number data from an Affymetrix SNP 6.0 array, whole exome mutation data, and clinical information, were obtained from the open- and controlled-access tiers of the TCGA data portal, with National Cancer Institute approval. The clinical details and patient selection criteria can be found in the recent TCGA report [11]. Alignment of the sample identifiers yielded 188 CRC cases for which information was available at the time of data retrieval.
Immunohistochemical analysis
We performed an immunohistochemical analysis for MIIP on TMA sections using a rabbit anti-human MIIP antibody (1:300, HPA044948; Sigma-Aldrich, St. Louis, MO, USA), as previously described [12]. MIIP-positive samples were defined as those with brown staining in the cytoplasm. The staining intensity of MIIP was graded on a scale from 0 to 3 (0 for no staining, 1 for light yellow, 2 for yellow, and 3 for brown). The percentage of cells with immunoreactivity was scored on a scale from 0 to 3 (0 for no positive cells, 1 for estimated <25% positive, 2 for 25%-50% positive, and 3 for >50% positive). The staining intensity and percentage of immunoreactivity scores were then multiplied to obtain the total score (0, 1, 2, 3, 4, 6, or 9). MIIP expression was classified as negative (−), weakly positive (++), moderately positive (++), or strongly positive (+++) when the total score was 0, 1-3, 4-6, or 9, respectively. High MIIP expression was defined as (+++), and low expression was defined as (−), (+), or (++) (Supplementary Figure S1A).
Fluorescence in situ hybridization analysis
A dual-colour fluorescence in situ hybridization (FISH) analysis was performed on 4 μm TMA sections using a Spectrum-Green-labelled chromosome 1 centromeric probe (CEP 1) and a Spectrum-Orange-labelled locus-specific MIIP probe (Empire Genomics, Buffalo, New York, USA), as described previously [13]. Fluorescence signals were captured by a computer-controlled digital camera and processed by ProgRes CapturePro software (NatureGene, USA). Sequential digital images were captured by a stack motor for each fluorescence filter, and the resulting images were reconstructed with blue, green, and red pseudo-colours. The FISH sections were evaluated simultaneously by two observers. Typically, 30-90 cells were enumerated in each case. In the areas where cell borders were clear, the numbers of red and green signals per nucleus were evaluated. If there were two MIIP gene (red) signals in more than 80% of nuclei, the case was considered MIIP-diploid. MIIP hemizygous deletion was defined as the presence of only one red signal in at least 20% of nuclei, and MIIP homozygous deletion was defined as both red signals having been lost at least 20% of nuclei with at least one green signal.
Zinc finger nuclease-mediated MIIP gene deletion
A MIIP-knockout zinc finger nuclease kit (MIIP-ZFN) was purchased from Sigma (St. Louis, MO, USA; #CKOZFN13330-1KT). We used the Amaxa Nucleofector system to transfect HCT116 cells with MIIP-ZFN constructs (Lonza, Koln, Germany). In brief, 2 × 106 HCT116 cells per transfection were washed twice with PBS and resuspended in 100 μl of Nucleofection Solution V. MIIP-pZFN1 (2.5 μg) and MIIP-pZFN2 (2.5 μg) were mixed with the cells; the mixture was transferred to a 2 mm electroporation cuvette and nucleofected on a Nucleofector with the D032 program. The cells were then transferred to pre-warmed medium in a six-well plate and incubated for 3 days. Ninety-six single colonies were prepared from the transfected cells and seeded in a 96-well plate. Two replica plates were prepared: one was used to harvest genomic DNA and perform genotyping (ZFN primer F: TCTGGGAGGAAAGGGGTTAG; ZFN primer R: ACCAGTGGTTCAGGCTCTGT), and the other was used as a working plate. After genotyping and sequencing, the MIIP knockout clones were used to test MIIP protein expression by Western blot analysis.
Cell migration and invasion assay
Cell migration and invasion assay was performed as described previously [14].
Orthotopic CRC mouse model
Male athymic nude mice (6 weeks old) were from the specific pathogen-free animal colony of the Department of Experimental Radiation Oncology and housed in the specific pathogen-free animal facility of the Department of Veterinary Medicine at The University of Texas MD Anderson Cancer Center (Houston, Texas). All mouse experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee. HCT116 MIIP-diploid (HCT 116_MIIP +/+) and MIIP-deleted (HCT 116_MIIP +/−_1 and HCT 116_MIIP +/−_2) cells were harvested, washed in serum-free medium, and resuspended in HBSS. Each mouse (10 per group) was anesthetized with a ketamine/xylazine solution; the caecum was exteriorized using a small left abdominal flank incision, and 2 × 106 cells, suspended in 50 μl of HBSS, were injected into the caecal wall using a 30-gauge needle. To prevent leakage, a cotton swab was held gently over the injection site. The injected caecum was returned to the abdominal cavity, and the wound was closed with wound clips (CellPoint Scientific, Inc., Gaithersburg, MD). Mice were weighed weekly and monitored for primary tumour development and metastasis; they were killed when most mice demonstrated signs of morbidity. The caecum, liver, and large lymph and omentum nodes were collected for histological analysis.
Chromosomal analysis
Exponentially growing cells were exposed to colcemid (0.04 μg/ml) for 1 h at 37° C. The cells were trypsinised, washed with PBS, and resuspended in hypotonic (75 mM KCl) for 20 min at ambient temperature. Cells were fixed in a methanol and acetic acid (3:1 v/v) mixture for 15 min and washed three times in the fixative. Air-dried preparations were created, and the slides were stained with 4% Giemsa. The cells were analysed for several parameters, including chromosome aberrations (as evidenced by chromosome and chromatid-type breaks), fragments, tetraploidy, and fusion. At least 40 metaphases were analysed from each sample. Images were captured using a Nikon 80i microscope equipped with karyotyping software from Applied Spectral Imaging, Inc. (Vista, CA).
Spectral karyotyping
Spectral karyotyping was performed according to the manufacturer’s protocol using Human Paint probes (Applied Spectral Imaging, Carlsbad, CA). Images were captured using a Nikon 80i microscope equipped with spectral karyotyping software (Applied Spectral Imaging).
Co-immunoprecipitation and Western blot analysis
Co-immunoprecipitation and Western blot analysis were performed as described previously [8]. Detailed information is provided in Supplementary methods.
Topo II activity assays
DNA Topo II activity was measured by decatenating kinetoplast DNA (kDNA) with a Topo II assay kit (TopoGEN, Port Orange, FL). In brief, equal volumes of buffers A (0.5 M Tris-HCl, pH=8, 1.5 M NaCl, 100 mM MgCl2, and 5 mM dithiothreitol) and B (20 mM ATP in water) were mixed to make a fresh 5 × complete assay buffer. Each 20 μl of reaction mixture contained 4 μl of 5 × complete assay buffer, 200 ng of kDNA, and 100 ng (or 300 ng) of nuclear extract. To determine whether Topo II activity was regulated by MIIP specifically, we used a mixture of 0.25 IU of Topo IIα (TopoGEN) and different doses of purified MIIP protein (OriGene, Rockville, MD) to replace the nuclear extracts. This reaction was kept at 37° C for 30 min and stopped with 4 μl of 5×Stop buffer. The mixture was loaded in 1% agarose gel and run for 1 h at 50 V by electrophoresis. The catenated and decatenated kDNA in the gel was stained with ethidium bromide and photographed under ultraviolet illumination. Topo II activity was quantified on the basis of the amount of decatenated kDNA using Image J software from the National Institutes of Health website (http://rsb.info.nih.gov/ij/).
Statistical analysis
We used Student’s t-test, analysis of variance, chi-square test, Wilcoxon rank-sum test, and Fisher’s exact test with SPSS and R 2.10.0 software. Statistical significance was defined as P<0.05. The Benjamini-Hochberg multiple testing correction [15] was used to estimate the false discovery rate.
Results
MIIP downregulation accompanies clinical CRC tumorigenesis and progression
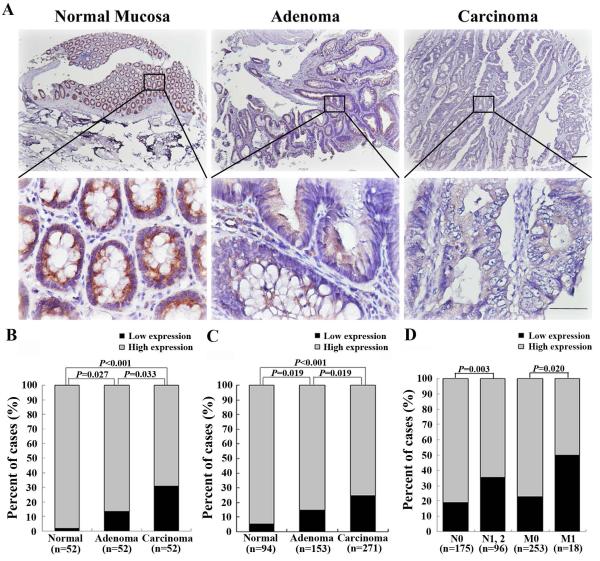
To explore the role of MIIP protein in CRC tumorigenesis and progression, we performed immunohistochemical analysis of MIIP on the TMAs derived from patient samples that included 526 cases of colorectal mucosa, adenoma, and carcinoma tissues from Tianjin Cancer Hospital. The immunohistochemical staining could be evaluated in 518 cases (271 CRCs, 153 adenomas and 94 normal mucosae) (Supplementary Figure S1B). MIIP expression was highest in non-neoplastic mucosa; the levels were lower in adenomas and lowest in carcinomas in the same patient (Figure 1A). This significantly decreased MIIP expression during the mucosa-adenoma-carcinoma sequence was confirmed not only in the 52 cases with matched samples of these three tissue types from the same patient (P<0.05, Figure 1B), but also in the whole 518 cases of colorectal tissue that could be evaluated on TMA (P<0.05, Figure 1C). Detailed clinical correlation analyses revealed that among the 271 CRC patients, a low level of MIIP expression was associated with lymph node (P=0.003) and distant metastasis (P=0.020) (Figure 1D; Supplementary Table S1).
Figure 1. MIIP protein downregulation accompanies clinical CRC development and progression.
MIIP expression was evaluated using immunohistochemical staining on TMAs. A. Respective images of MIIP expression in normal mucosa, adenoma, and adenocarcinoma from the same patient. Scale bars in the top panel, 500 μm. Scale bars in the lower panel, 100 μm. B. A statistical analysis revealed that MIIP expression was significantly decreased during the mucosa-adenoma-carcinoma sequence in the 52 sets of matched samples. C. A statistical analysis revealed that MIIP expression was highest in non-neoplastic mucosa, lower in adenoma, and lowest in carcinoma in 518 cases. D. A statistical analysis revealed that low MIIP expression was correlated with lymph node metastasis and distant metastasis in 271 CRC patients.
MIIP hemizygous deletion is associated with MIIP downregulation and CRC progression
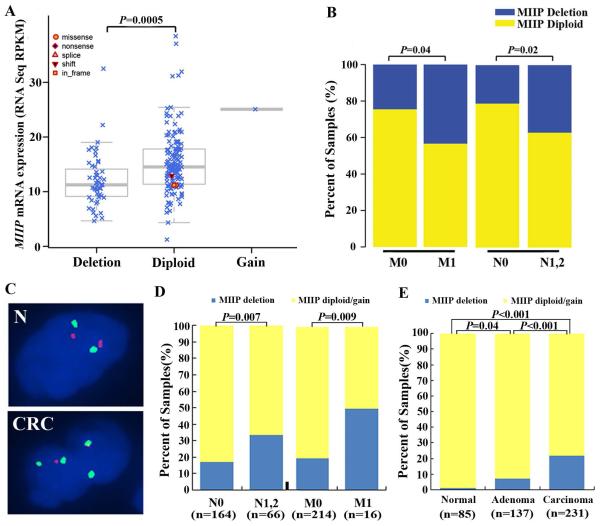
To determine the genomic status of MIIP, we performed a GISTIC analysis of copy number data derived from an Affymetrix SNP 6.0 array analysis of TCGA CRC cases. Among the 188 CRC patients with complete genomic and clinical data, MIIP mutation was detected in three diploid cases, including two missense and one frame-shift mutation (Figure 2A), while MIIP deletion was found in 52 cases (27.7%). Moreover, all of the cases with MIIP deletion had hemizygous deletions. The analysis of the relationship between MIIP deletion and expression subtypes (designated by TCGA) showed that MIIP hemizygous deletion was more frequent in CIN cases than invasive and MSI/CIMP cases (P<0.001, Supplementary Figure S2A). In addition, MIIP deletion demonstrated striking mutual exclusivity with high levels of MSI (MSI-H) (P=0.001, Supplementary Figure S2B). An analysis of the RNA-seq data in the same 188 cases revealed that MIIP transcript levels were significantly downregulated in the CRC cases with MIIP hemizygous deletion (P=0.0005, Figure 2A). MIIP hemizygous deletion was significantly associated with lymph node metastasis (P=0.04) and distant metastasis (P=0.02) (Figure 2B, Supplementary Table S2). In addition, we did not observe an association between MIIP hemizygous deletion and the deletion or mutation of other well-established genes in CRC, including TP53, APC, PIK3CA, KRAS, NRAS, TGFBR2, SMAD4, MSH6, and MSH3 (Supplementary Figure S2C), suggesting that loss of MIIP function may occur mainly through MIIP hemizygous deletion which is an important molecular event independent of the well-established genetic events in CRC.
Figure 2. MIIP hemizygous deletion is associated with MIIP downregulation and CRC progression in TCGA CRC cases and an independent cohort.
A and B. The genomic analysis of MIIP and the association between MIIP deletion and metastasis in TCGA CRC cases. Among the 188 CRC patients with complete genomic and clinical data, MIIP mutation was detected in three diploid cases, and MIIP hemizygous deletion was found in 52 cases. MIIP mRNA expression levels were significantly lower in MIIP- hemizygous cases than in MIIP-diploid CRC within the TCGA data (A). Positive associations were present between MIIP hemizygous deletion and lymph node and distant metastasis in TCGA (B). C-E. FISH analysis for 526 colorectal tissue samples in Tianjin cohort. Dual-colour FISH analysis of TMA sections using a Spectrum-Green-labelled CEP 1 and a Spectrum-Orange-labelled locus-specific MIIP probe (Empire Genomics, Buffalo, New York, USA). The FISH sections were evaluated in 30-90 cells of each case. Respective FISH images from normal mucosa and CRC (C). Two pairs of green and red signals (MIIP diploid) were detected in one nucleus of normal mucosa (N), whereas two copies of green signals and one copy of the red signal (MIIP hemizygous deletion) were detected in each nucleus of CRC cells. The statistical analysis showed that MIIP hemizygous deletion was correlated with lymph node metastasis and distant metastasis (D). Among the 453 colorectal cases available for FISH analysis on TMA, the ratio of MIIP hemizygous deletion gradually increased during the mucosa-adenoma-carcinoma sequence (E).
To validate results from the TCGA cohort, we performed FISH analysis of the MIIP gene on the TMAs (Figure 2C). FISH results could be assessed in 231 CRC, 137 adenoma and 85 normal mucosa cases. Among the 231 CRC available for FISH analysis on TMA, MIIP hemizygous deletion, MIIP diploid, and MIIP gain were detected in 50 (21.6%), 178 (77.1%), and 3 (1.3%) cases. MIIP hemizygous deletion was associated with lymph node (P=0.007) and distant metastasis (P=0.009) (Figure 2D). Among the 453 colorectal cases available for FISH analysis on TMA, MIIP hemizygous deletion, MIIP diploid, and MIIP gain were detected in 61 (13.5%), 388 (85.7%), and four (0.9%) cases. The ratio of MIIP hemizygous deletion was highest in adenocarcinoma and lowest in non-neoplastic mucosa (P<0.05 for all, Figure 2E). Moreover, among the 231 CRC patients and 453 colorectal cases available for both FISH and immunohistochemical analysis on TMA, the MIIP hemizygous deleted cases showed significantly lower MIIP expression (P<0.001 for both, Supplementary Table 3). The above results suggest that loss of MIIP function may occur mainly through MIIP hemizygous deletion and be important in CRC tumorigenesis and progression.
MIIP haploinsufficiency promotes colony formation, migration, and invasion
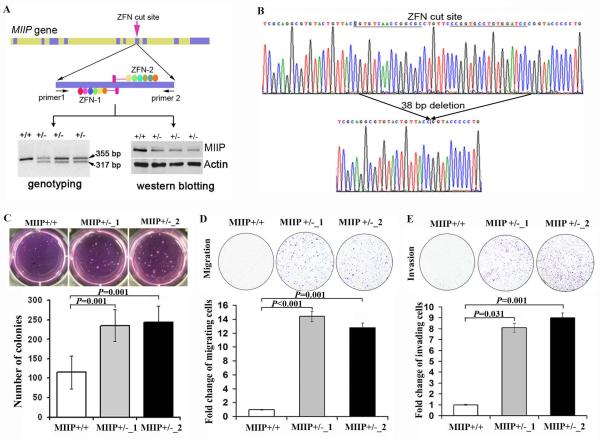
To identify the role of MIIP hemizygous deletion in CRC, we deleted MIIP in HCT116 CRC cells using ZFN technology. After genotyping 96 single clones from transfected HCT116 cells with ZFN-MIIP constructs, followed by sequencing validation, we found three with MIIP hemizygous deletion (MIIP +/−) (Figure 3A, lower left panel; Figure 3B). In all three MIIP +/− clones, MIIP expression was approximately 50% lower than in parental cells (Figure 3A, lower right panel). To obtain clones with homozygous deletion of MIIP, we further transfected the MIIP+/− clones with ZFN-MIIP constructs. We genotyped 180 clones but did not identify an MIIP clone with homozygous deletion (MIIP−/−), suggesting that MIIP is required for cell viability in HCT116 cells. Using the first two confirmed MIIP heterozygous deleted clones (MIIP +/−_1 and MIIP +/−_2), we performed a soft agar assay. Compared with HCT116 parental cells, significantly more colonies were found in both MIIP +/− clones (P=0.001 for both, Figure 3C). Both MIIP +/− HCT116 clones had augmented migration and invasion (P<0.05 for all, Figure 3D, E).
Figure 3. MIIP haploinsufficiency promotes colony formation, cell migration, and invasion.
A. ZFN-mediated MIIP deletion in HCT116 cells. Top panel: A pair of ZFNs (ZFN-1 and ZFN-2) was designed to target the MIIP gene at exon 7; each zinc finger array recognized 18 nt of the MIIP target site to ensure specificity. Primers 1 and 2, used for genotyping, are described in the Materials and Methods. Lower panel: Three clones containing MIIP deletion (MIIP +/−) were identified by genotyping (left), with identification of the 317-bp fragment. MIIP expression was lower in the three MIIP +/− clones, as determined by Western blotting analysis (right). B. MIIP haploinsufficiency was confirmed by sequencing. The data revealed a 38 bp deletion around the ZFN-targeted site. C. MIIP haploinsufficiency increased colony formation in soft agar. Respective images of HCT116 MIIP-diploid and MIIP- haploinsufficient cells after growing in soft agar for 2 weeks (top panel). The number of colonies represents the mean ± SEM for triplicate tests. D and E. MIIP haploinsufficiency increased cell migration and invasion. Respective images of HCT116 MIIP-diploid and MIIP- haploinsufficient cells migrating through an 8-μm pore size membrane (migration assay) after 20 h of incubation (D, top panel) and invading through an 8-μm pore size membrane coated with Matrigel (invasion assay) after 24 h of incubation (E, top panel). The migrating or invading cell numbers on each filter were counted, and data were plotted as the fold change by defining the number in parental cells as 1. The fold change of migrating or invading cells is the mean ± SEM for triplicate tests.
We observed a similar increase in colony formation when we used siRNA to downregulate MIIP expression in both HCT116 and DLD1 CRC cells (Supplementary Figure S3A,B). MIIP knockdown also increased the migration and invasion rates of HCT116 and DLD1 cells (Supplementary Figure S3C-3F).
MIIP haploinsufficiency promotes tumour growth and progression in an orthotopic mouse model
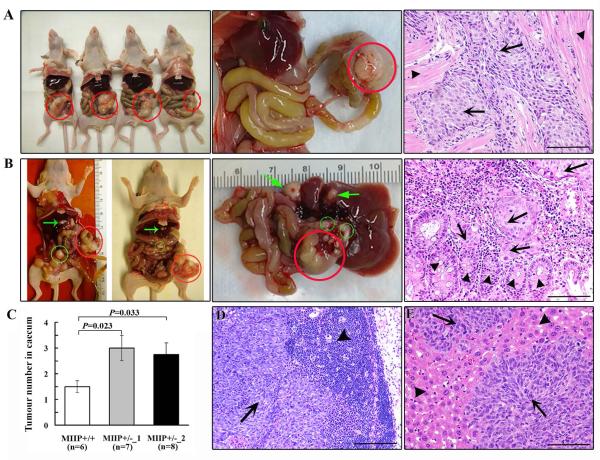
To gain deeper insight into MIIP’s role in CRC progression, we used an orthotopic mouse model. We injected HCT116 MIIP-diploid (MIIP +/+) or MIIP haploinsufficiency cells (MIIP +/−_1 or MIIP +/−_2) into the caecum of nude mice (10 mice per group, three groups). Seven that had been injected with HCT116_ MIIP +/+ cells, eight with HCT116_MIIP +/−_1 cells, and nine with HCT116_MIIP +/−_2 cells survived all surgical procedures. Tumours grew in the caecal wall at similar frequencies in the three groups: six mice (86%) in the HCT116_ MIIP +/+ group (Figure 4A), seven mice (88%) in the HCT116_MIIP +/−_1 group, and eight mice (89%) in the HCT116_MIIP +/−_2 group (Figure 4B). The MIIP +/− cells, however, produced more tumours than did the MIIP +/+ cells (Figure 4C) and resulted in more frequent lymph node and omental metastases (Figure 4B, D). Moreover, liver metastases were found only in mice injected with HCT116_MIIP +/− cells, including two mice in the HCT116_MIIP +/−_1 group and three in the HCT116_MIIP+/−_2 group (Figure 4B, E). The xenograft model provided strong evidence of the causal role of MIIP haploinsufficiency in CRC tumour growth and metastasis.
Figure 4. MIIP haploinsufficiency promotes tumour growth and progression in a CRC orthotopic mouse model.
HCT116_ MIIP+/+, HCT116_ MIIP+/−_1, and HCT116_MIIP+/−_2 cells were injected into the caecal walls of nude mice. A. Photographs of the nude mice injected with HCT116_ MIIP+/+ cells, their orthotopic tumours, and the respective histological images. Orthotopic tumours (red circle) were found, with invasion of tumour cells (arrows) in the muscularis propria of the caecal wall (arrowheads). B. In the mice injected with HCT116 MIIP- haploinsufficient cells, lymph node and omental metastases (green circles) and liver metastases (green arrows) were found in addition to orthotopic tumours (red circle). In the orthotopic tumours, HCT116_ MIIP+/−_ cells (arrows) invaded into the mucosa (arrowheads) from the caecal wall. C. MIIP haploinsufficiency induced more tumours in the cecum in the CRC orthotopic mouse model. Tumour number is the mean ± SEM. D. Histological images of lymph node metastasis in mice injected HCT116 MIIP- haploinsufficient cells. Arrows and arrowheads represent metastatic tumour cells and lymphocytes respectively in lymph nodes. E. Liver metastasis is shown in a mouse injected with HCT116 MIIP- haploinsufficient cells. Arrows represent cancer cells, and arrowheads represent liver tissue. Scale bars: 100 μm.
MIIP haploinsufficiency results in a weakened mitotic checkpoint and induces CIN in CRC
In our previous study, we demonstrated that MIIP is a key mitotic regulatory protein that modulates cyclin B1 stability by inhibiting the activity of anaphase promoting complex (APC/C), a ubiquitin ligase E3 in mitosis [8]. Besides cyclin B1, securin is one of the downstream proteins of APC/CCdc20 and critical for sister chromosome segregation and stability [16]. In this study, we determined whether MIIP targets the downstream proteins of APC/CCdc20 that are involved in mitosis in CRC cells. Overexpression of MIIP via an adenoviral expression system in CRC cells resulted in increased cyclin B1 and securin stability (Supplementary Figure S4A,B). A mitotic transition assay revealed that HCT116_MIIP+/− cells transitioned more quickly from the G2/M phase to the G1 phase than did parental cells (Supplementary Figure S4C,D).
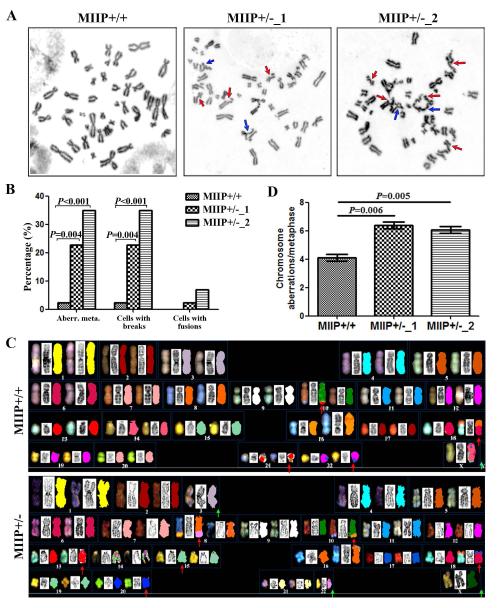
Because dysregulation of mitotic transition and sister chromosome segregation leads to CIN [17], we next determined whether MIIP haploinsufficiency induces CIN in CRC cells. HCT116 cells have MSI-H and a relatively normal chromosomal composition that represents a diploid karyotype [18]. Therefore, HCT116 cells provide an opportunity to directly test the causal relationship between MIIP haploinsufficiency and CIN. A chromosomal analysis of HCT116_parental, HCT116_MIIP +/−_1, and HCT116_MIIP +/−_2 cells identified more chromosomal aberrations and chromosomal breaks in both MIIP+/− clones than in HCT116_ MIIP+/+ cells (Figure 5A, 5B). A spectral karyotyping assay further verified that MIIP haploinsufficiency led to more chromosome aberrations (Figure 5C, 5D).
Figure 5. MIIP haploinsufficiency induces chromosome instability in CRC cells.
A and B. The genomic instability analysis for HCT116 MIIP-diploid and MIIP-haploinsufficient cells. The representative metaphase of HCT116_MIIP +/+, HCT116_MIIP +/−_1 and HCT116_MIIP +/−_2 cells (A). The red arrows indicate chromosome breaks; the blue arrows indicate chromosome fusion. Compared with MIIP-diploid cells, MIIP- haploinsufficient cells had increased chromosomal abnormalities, especially more chromosome breaks and fusions (B). At least 40 metaphases were analysed from each sample. C and D. The spectral karyotyping (SKY) analysis for HCT116 MIIP-diploid cells and MIIP-haploinsufficient cells. Example of spectral karyotypes of HCT116_MIIP +/+ and HCT116_MIIP +/− cells (C). Arrows show deletions (green) and structural aberrations (red). Compared with MIIP-diploid cells, MIIP- haploinsufficient cells had more chromosome aberrations (D). Approximately 10-15 metaphase spreads per sample were analysed in each experiment. Chromosome aberrations per metaphase is the mean ± SEM for triplicate tests.
MIIP haploinsufficiency interrupts the activity of Topo II
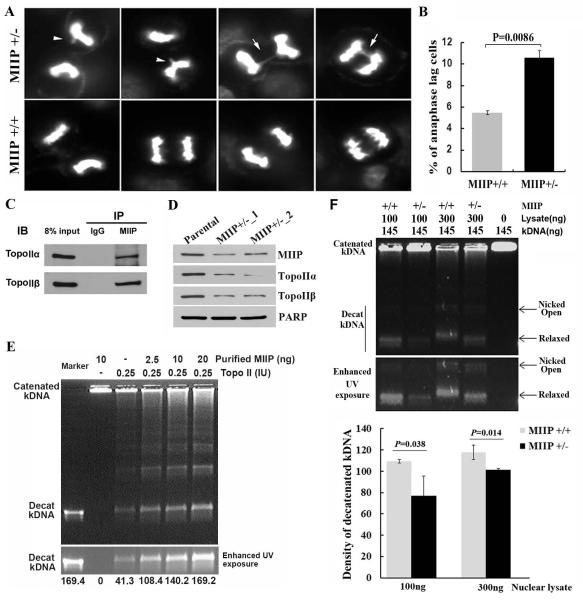
The dramatic alterations from the above chromosomal analysis and spectral karyotyping assay suggest that MIIP’s role in maintaining genome stability also involves genes that are involved in regulating chromosomal remodelling. Studies have demonstrated that Topo IIα is a critical regulator of sister chromatid unwinding and separation during mitosis [19-21]. Abnormal regulation of Topo IIα results in incorrect chromatid separation and breakage [22, 23]. At the cellular level, an anaphase bridge is a common phenotype when Topo IIα is abnormally regulated [24]. We next evaluated the anaphase cells in the HCT116_MIIP +/− and HCT116_ MIIP +/+ cell lines and observed more chromosomal bridges in the HCT116_MIIP +/− cells (Figure 6A, B). We thus sought to determine the possible regulatory relationship between MIIP haploinsufficiency and topoisomerase activity. The co-immunoprecipitation assay showed that both Topo IIα and Topo IIβ were detected in the precipitated complex with MIIP (Figure 6C). In addition, both Topo IIα and Topo IIβ were downregulated in MIIP +/− cells (Figure 6D). A Topo II activity assay was performed by decatenating kinetoplast DNA (kDNA) as a readout (the cut kDNA migrates into an agarose gel; the uncut catenated DNA remains at the top of the well). As shown in Figure 6E, when purified MIIP was added in increasing doses to the system, with a fixed amount of purified Topo II, catenated DNA was cut in a MIIP dose-dependent manner. Moreover, the density of decatenated kDNA was significantly lower in HCT116_MIIP +/− cells than in MIIP +/+ cells at two different doses of nuclear proteins (Figure 6F). Therefore, our results provide evidence that MIIP haploinsufficiency suppresses Topo II protein level and disturbs the activity of topoisomerases, which is a known cause of CIN.
Figure 6. MIIP haploinsufficiency reduces the decatenation activity of topo II and induces incorrect chromatid separation.
A. The respective images of anaphase analysis of HCT116_MIIP+/+ and HCT116_ MIIP+/− cells. B. A statistical analysis of three independent experiments showed that the percentage of abnormal anaphase cells in counted MIIP-haploinsufficient cells was significantly higher than for MIIP-diploid cells. C. Co-immunoprecipitation experiments. HCT116 nuclear extract was subjected to immunoprecipitation with an antibody to MIIP and analysed for co-immunoprecipitation of Topo IIα, and Topo IIβ. Both Topo IIα and Topo IIβ were detected in the precipitated complex. D. Western blot assay for Topo IIα and Topo IIβ of MIIP-diploid and MIIP-haploinsufficient cells. Compared with MIIP-diploid cells, both Topo IIα and Topo IIβ levels were decreased in MIIP- haploinsufficient cells. E. TopoII activity assay. Differing amounts of the purified TopoII enzyme and human recombinant MIIP protein were used, as indicated. The uncut catenated kDNA remained at the top of the well; the cut kDNA (decatenated kDNA) migrated into an agarose gel (top panel). The decatenated kDNA bands were shown in enhanced UV exposure in the middle panel. Quantification using Image J software revealed that the density of decatenated Kdna (lower panel). F. Topo II activity assay for HCT-116 MIIP-diploid and MIIP- haploinsufficient cells. The decatenated kDNA bands are shown using enhanced UV exposure in the middle panel. Quantification using Image J software revealed that the density of decatenated kDNA was significantly lower in HCT116 MIIP- haploinsufficient cells than in MIIP-diploid cells at both doses of nuclear proteins (lower panel).
Etoposide, an inhibitor of TopoII, has been used to treat CRC after failure of front-line therapy [25]. We examined the percentage of sub-G1 of HCT116_MIIP +/+ and HCT116_MIIP +/− cells after treatment with different doses of etoposide at two time points using flow cytometry. The results showed that MIIP +/− cells were more sensitive to etoposide than MIIP +/+ cells (Supplementary Figure S5).
Discussion
According to its genetic alterations, CRC can be broadly classified as having CIN or MSI [26]. CIN, which is characterized by structural or numerical chromosomal alterations, is detected in 85% of CRCs [27]. CIN can induce cancer development and progression and is associated with poor prognosis and drug resistance in CRC [28-31]. Recently, Burrell et al. identified three CIN-suppressor genes that are related to DNA replication stress on chromosome 18q in CRC [18]. MIIP is located in 1p36, which is also a region that is commonly deleted in CRC [32, 33]. In this study, we provided functional evidence showing that MIIP haploinsufficiency induced CIN in chromosomally stable HCT116 cells. In these experiments, because the MIIP gene was the only gene deleted using the ZFN technology, these results strongly suggest that MIIP haploinsufficiency is a key driving event for CIN.
CIN can be induced by defective mitotic checkpoints, abnormal chromosome attachment to the mitotic spindle, and pre-mitotic defects that affect chromosome structure [34-36]. In our previous study, we observed that MIIP modulated the stability of cyclin B1, a key mitotic checkpoint protein, by interacting with Cdc20 and inhibiting APC/CCdc20 activity [8]. Our current data demonstrated that besides cyclin B1, MIIP controlled the degradation of securin which is another downstream target of APC/CCdc20 and is critical for sister chromosome segregation and stability. MIIP haploinsufficiency would weaken the role of these mitotic check proteins, resulting in abnormal chromosomal separation. We also found that MIIP interacted with TopoII and enhanced its ability to cut double-stranded DNA to release the torsional stress accumulated during mitosis. Thus, MIIP haploinsufficiency leads to decreased TopoII activity during mitosis, ultimately causing highly stressed and intertwined chromatids and aberrant chromosomal breaks.
We detected a positive association between MIIP haploinsufficiency and metastasis in TCGA CRC cases and an independent validation cohort of CRC patients. To gain insight into whether CIN causes metastasis or metastasis causes genomic instability; we used an orthotopic mouse model and MIIP-heterozygously-deleted HCT116 cells. HCT116 itself, which contains two copies of MIIP, was not metastatic. However, when one copy of MIIP was deleted, leading to decreased MIIP expression, the cells became metastatic. Thus, our data support the notion that specific genetic alterations can promote cancer metastasis [26]. However, because we have also demonstrated that MIIP inhibits cellular migration and invasion, MIIP haploinsufficiency may promote cancer metastasis independent of CIN. Genomic instability and metastasis may be two parallel processes that are intertwined.
In summary, in this study, the combined genomic analyses, in vitro molecular studies, and an in vivo mouse model provided strong evidence that MIIP is a potential tumour suppressor gene in CRC tumorigenesis and progression. Moreover, loss of MIIP function may occur mainly through MIIP haploinsufficiency. MIIP haploinsufficiency led to deleterious unbalance in stabilities of key mitotic checkpoint proteins and reduced activity of Topo II, thus causing aberrant mitosis and chromosome missegregation and finally induced CIN. Given the common loss of 1p36 in multiple cancer types and the importance of CIN as a hallmark of cancer [37-43], MIIP haploinsufficiency is likely a common driver event in cancer development and progression. Understanding the mechanisms regulating MIIP may improve the clinical management of all CIN cancers. Our data in this study suggests that MIIP loss may expose a vulnerability of CRC cells to Etoposide. Further investigations of the modulation of MIIP in the Topo network will help us identify CRC patients who are responsive to Topo inhibitors thus improving the therapeutic efficacy.
Supplementary Material
Acknowledgements
We thank Brittany Parker for her critical review of and comments on this manuscript and Ms. Ann Sutton in the Department of Scientific Publications for editing this manuscript. This work was partially supported by the National Nature Science Foundation of China (81472263), the National Key Clinical Specialist Construction Programs of China (No. 2013-544), a grant from the Tianjin Municipal Science and Technology Commission (14JCYBJC27500), the National Institutes of Health grant U24CA143835, MD Anderson’s National Cancer Institute Cancer Center Support Grant (CA16672), a grant from the MD Anderson Sister Institute Network Fund, a grant from the James McDonald Foundation, a grant from the Asian Fund for Cancer Research, and a grant from the National Foundation for Cancer Research. Y. Sun was supported by The A. Lavoy Moore Endowment Fund T. Chen was supported by a fellowship of the China Education Council and the National Nature Science Foundation of China (81502523). D. Yang was an Odyssey Fellow at MD Anderson and is supported by The Harold C. and Mary L. Daily Endowment Fund.
Footnotes
The authors declare that no conflict of interest exists.
Author Contributions: Yan Sun and Ping Ji: acquisition of data; analysis and interpretation of data; drafting of the manuscript; and statistical analysis. Tao Chen, Xinhui Zhou, Limei Hu, Dianren Xia, Yanxue Liu and Asha S. Multani: acquisition of data; and analysis and interpretation of data. Da Yang and Yuexin Liu: statistical analysis. Ilya Shmulevich, Raju Kucherlapati, Scott Kopetz, Anil K. Sood, Stanley R. Hamilton and Baocun Sun: critical revision of the manuscript for important intellectual content; administrative, technical, or material support; study supervision. Yan Sun and Wei Zhang: study concept and design; analysis and interpretation of data; obtained funding; administrative, technical, or material support; study supervision.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Chew MH, Teo JY, Kabir T, et al. Stage IV colorectal cancers: an analysis of factors predicting outcome and survival in 728 cases. J Gastrointest Surg. 2012;16:603–612. doi: 10.1007/s11605-011-1725-1. [DOI] [PubMed] [Google Scholar]
- 3.Ilyas M, Straub J, Tomlinson IP, et al. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35:1986–2002. doi: 10.1016/s0959-8049(99)00298-1. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 5.Simons CC, Hughes LA, Smits KM, et al. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol. 2013;24:2048–2056. doi: 10.1093/annonc/mdt076. [DOI] [PubMed] [Google Scholar]
- 6.Yamagishi H, Kuroda H, Imai Y, et al. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer. 2016;35:4. doi: 10.1186/s40880-015-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo E, Ramamoorthy R, Oukrif D, et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J Pathol. 2013;229:441–448. doi: 10.1002/path.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji P, Smith SM, Wang Y, et al. Inhibition of gliomagenesis and attenuation of mitotic transition by MIIP. Oncogene. 2010;29:3501–3508. doi: 10.1038/onc.2010.114. [DOI] [PubMed] [Google Scholar]
- 9.Song SW, Fuller GN, Khan A, et al. IIp45, an insulin-like growth factor binding protein 2 (IGFBP-2) binding protein, antagonizes IGFBP-2 stimulation of glioma cell invasion. Proc Natl Acad Sci U S A. 2003;100:13970–13975. doi: 10.1073/pnas.2332186100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Yokoi K, Li H, et al. NGAL expression is elevated in both colorectal adenoma-carcinoma sequence and cancer progression and enhances tumorigenesis in xenograft mouse models. Clin Cancer Res. 2011;17:4331–4340. doi: 10.1158/1078-0432.CCR-11-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Sun Y, Ji P, et al. MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol. 2014;233:308–318. doi: 10.1002/path.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun B, Sun Y, Wang J, et al. The diagnostic value of SYT-SSX detected by reverse transcriptase-polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) for synovial sarcoma: a review and prospective study of 255 cases. Cancer Sci. 2008;99:1355–1361. doi: 10.1111/j.1349-7006.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Hu L, Zheng H, et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015;235:25–36. doi: 10.1002/path.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Royal Statistical Society Series B. 1995;57:289. [Google Scholar]
- 16.Waizenegger IC, Hauf S, Meinke A, et al. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 17.Nojima H. Cell cycle checkpoints, chromosome stability and the progression of cancer. Hum Cell. 1997;10:221–230. [PubMed] [Google Scholar]
- 18.Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MJ, Martin BA, Gootz TD, et al. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J Biol Chem. 1991;266:14585–14592. [PubMed] [Google Scholar]
- 20.Chen GL, Yang L, Rowe TC, et al. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 21.Hashash N, Johnson AL, Cha RS. Topoisomerase II- and condensin-dependent breakage of MEC1ATR-sensitive fragile sites occurs independently of spindle tension, anaphase, or cytokinesis. PLoS Genet. 2012;8:e1002978. doi: 10.1371/journal.pgen.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warsi TH, Navarro MS, Bachant J. DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol Biol Cell. 2008;19:4421–4433. doi: 10.1091/mbc.E08-05-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LH, Schwarzbraun T, Speicher MR, et al. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawlaty MM, Malureanu L, Jeganathan KB, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeremic B, Acimovic L, Mijatovic L. Carboplatin and etoposide in advanced colorectal carcinoma. A phase II study. Cancer. 1993;71:2706–2708. doi: 10.1002/1097-0142(19930501)71:9<2706::aid-cncr2820710903>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 27.Hung KE, Chung DC. New insights into the molecular pathogenesis of colorectal cancer. Drug Discov Today Dis Mech. 2006;3:439. doi: 10.1016/j.ddmec.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoler DL, Chen N, Basik M, et al. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci U S A. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih IM, Zhou W, Goodman SN, et al. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–822. [PubMed] [Google Scholar]
- 30.Nowak MA, Komarova NL, Sengupta A, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci U S A. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe T, Kobunai T, Yamamoto Y, et al. Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J Clin Oncol. 2012;30:2256–2264. doi: 10.1200/JCO.2011.38.6490. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Yanoshita R, Konishi M, et al. Suppression of tumourigenicity in human colon carcinoma cells by introduction of normal chromosome 1p36 region. Oncogene. 1993;8:2253–2258. [PubMed] [Google Scholar]
- 33.Bomme L, Bardi G, Pandis N, et al. Chromosome abnormalities in colorectal adenomas: two cytogenetic subgroups characterized by deletion of 1p and numerical aberrations. Hum Pathol. 1996;27:1192–1197. doi: 10.1016/s0046-8177(96)90314-7. [DOI] [PubMed] [Google Scholar]
- 34.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2011;19:433–444. doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen A, van der Burg M, Szuhai K, et al. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 36.Crasta K, Ganem NJ, Dagher R, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caren H, Fransson S, Ejeskar K, et al. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–1424. doi: 10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichimura K, Vogazianou AP, Liu L, et al. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene. 2008;27:2097–2108. doi: 10.1038/sj.onc.1210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loukopoulos P, Shibata T, Katoh H, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98:392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez AA, Lambers AR, Lancaster JM, et al. Allele loss on chromosome 1p36 in epithelial ovarian cancers. Gynecol Oncol. 2001;82:94–98. doi: 10.1006/gyno.2001.6175. [DOI] [PubMed] [Google Scholar]
- 41.Tsukamoto K, Ito N, Yoshimoto M, et al. Allelic loss on chromosome 1p is associated with progression and lymph node metastasis of primary breast carcinoma. Cancer. 1998;82:317–322. [PubMed] [Google Scholar]
- 42.Girgis AH, Iakovlev VV, Beheshti B, et al. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer Res. 2012;72:5273–5284. doi: 10.1158/0008-5472.CAN-12-0656. [DOI] [PubMed] [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.