Abstract
F420 is a low-potential redox cofactor that mediates the transformations of a wide range of complex organic compounds. Considered one of the rarest cofactors in biology, F420 is best known for its role in methanogenesis and has only been chemically identified in two phyla to date, the Euryarchaeota and Actinobacteria. In this work, we show that this cofactor is more widely distributed than previously reported. We detected the genes encoding all five known F420 biosynthesis enzymes (cofC, cofD, cofE, cofG and cofH) in at least 653 bacterial and 173 archaeal species, including members of the dominant soil phyla Proteobacteria, Chloroflexi and Firmicutes. Metagenome datamining validated that these genes were disproportionately abundant in aerated soils compared with other ecosystems. We confirmed through high-performance liquid chromatography analysis that aerobically grown stationary-phase cultures of three bacterial species, Paracoccus denitrificans, Oligotropha carboxidovorans and Thermomicrobium roseum, synthesized F420, with oligoglutamate sidechains of different lengths. To understand the evolution of F420 biosynthesis, we also analyzed the distribution, phylogeny and genetic organization of the cof genes. Our data suggest that although the Fo precursor to F420 originated in methanogens, F420 itself was first synthesized in an ancestral actinobacterium. F420 biosynthesis genes were then disseminated horizontally to archaea and other bacteria. Together, our findings suggest that the cofactor is more significant in aerobic bacterial metabolism and soil ecosystem composition than previously thought. The cofactor may confer several competitive advantages for aerobic soil bacteria by mediating their central metabolic processes and broadening the range of organic compounds they can synthesize, detoxify and mineralize.
Introduction
F420 serves as the cofactor in the catalysis of some of the most chemically demanding redox reactions in biology. Among them are the one-carbon reactions of methanogenesis (Thauer, 1998; Shima et al., 2000; Hagemeier et al., 2003), the biosynthesis pathways of tetracycline antibiotics (Wang et al., 2013) and the biodegradation of picrate and aflatoxins (Ebert et al., 2001; Taylor et al., 2010; Lapalikar et al., 2012). The cofactor appears to have been selected for these roles because of its unique electrochemical properties compared with the ubiquitous flavin and nicotinamide cofactors FMN (flavin mononucleotide), FAD (flavin adenine dinucleotide) and NAD(P) (nicotinamide adenine dinucleotide (phosphate)) (Walsh, 1986; Greening et al., 2016a). As a 5-deazaflavin, F420 is structurally and biosynthetically related to FMN and FAD, but exhibits distinct electrochemical properties because of several key substitutions. It has a relatively low redox potential of −340 mV under standard conditions and −380 mV under certain physiological conditions (de Poorter et al., 2005). This enables reduced F420 (F420H2) to reduce a wide range of organic compounds otherwise recalcitrant to activation (Jacobson and Walsh, 1984; Greening et al., 2016a). As an obligate two-electron carrier, the cofactor can transform alkene, alkyne, alcohol and imine groups through hydride transfer reactions (Shima et al., 2000; Hagemeier et al., 2003; Aufhammer et al., 2004; Shen et al., 2009; Wang et al., 2013).
Half a century since its discovery (Cheeseman et al., 1972), F420 is still perceived to be a rare cofactor synonymous with methanogenesis (Greening et al., 2016a). It has been confirmed to be synthesized in just two phyla to date: the Euryarchaeota (Eirich et al., 1979) and Actinobacteria (Eker et al., 1980; Daniels et al., 1985). It is thought to serve as the primary catabolic electron carrier in multiple lineages of Euryarchaeota, including representatives of the methanogenic (Eirich et al., 1979), methanotrophic (Michaelis et al., 2002; Knittel et al., 2005) and sulfate-reducing orders (Lin and White, 1986). Genomic and spectroscopic evidence suggests that the cofactor is also synthesized in the aerobic ammonia-oxidizing phylum Thaumarchaeota (Spang et al., 2012). Among bacteria, the cofactor has been chemically identified only within the Actinobacteria, where its physiological roles remain under investigation. In these organisms, F420 reduction is coupled to either glucose 6-phosphate or NADPH oxidation and hence is dependent on the pentose phosphate pathway (Greening et al., 2016a). The reduced cofactor (F420H2) is reported to enhance the metabolic flexibility of mycobacteria by facilitating the catalysis of a wide range of reductions of endogenous and exogenous organic compounds (Ahmed et al., 2015; Greening et al., 2016a). F420 also has roles in antibiotic synthesis and xenobiotic degradation in species of Streptomyces (Wang et al., 2013), Rhodococcus (Heiss et al., 2002) and Nocardioides (Ebert et al., 1999).
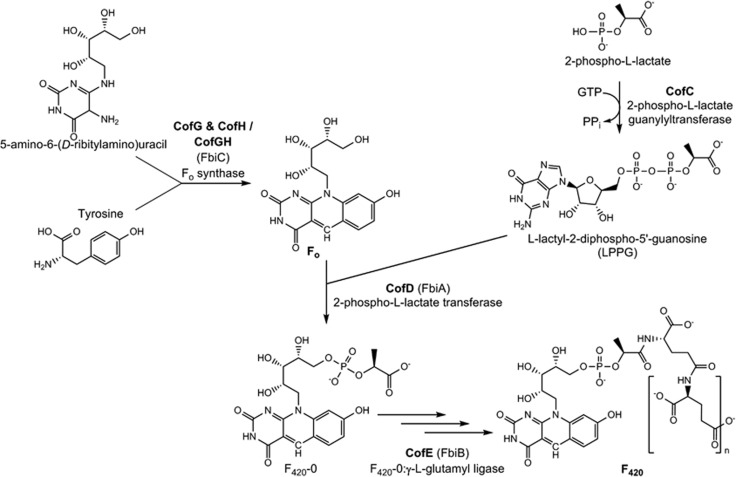
F420 is synthesized in three major steps in bacteria and archaea (Figure 1). In the first, a riboflavin precursor (5-amino-6-(D-ribitylamino)uracil) is condensed with tyrosine to form 8-hydroxy-5-deazaflavin, also known as Fo; this step is catalyzed by the radical S-adenosylmethionine enzymes CofG and CofH that are fused into a single protein in some bacteria (known as CofGH or FbiC) (Choi et al., 2002; Philmus et al., 2015). Subsequently, LPPG (L-lactyl-2-diphospho-5′-guanosine) is proposed to be synthesized from 2-phospho-L-lactate by CofC (Grochowski et al., 2008) and transferred to Fo by CofD (also known as FbiA) (Choi et al., 2001; Graupner and White, 2001; Graupner et al., 2002). The resulting LPPG sidechain is finally elongated with glutamate residues by the F420:γ-L-glutamyl ligase CofE (also known as FbiB) (Choi et al., 2001; Li et al., 2003; Nocek et al., 2007) that is fused with an FMN-dependent oxidoreductase in Actinobacteria (Bashiri et al., 2016). For reasons still not understood, the number of glutamate residues added varies between organisms, ranging from two to three in most methanogens (Gorris and van der Drift, 1994), four to five in Methanosarcina (Gorris and van der Drift, 1994) and five to seven in Mycobacterium (Bair et al., 2001). In addition to being an obligate intermediate in the F420 biosynthesis pathway, Fo is also synthesized independently by bacteria, archaea and eukaryotes for use as an antennal chromophore in DNA repair photolyases (Eker et al., 1988; Yasui et al., 1988; Kiener et al., 1989; Epple and Carell, 1998).
Figure 1.
Biosynthesis pathways for Fo and F420.
We recently proposed that F420 could be synthesized in a wider range of bacteria than currently described in the literature (Greening et al., 2016a). Our analysis of a protein superfamily, the flavin/deazaflavin oxidoreductases, identified putative F420-utilizing oxidoreductases in microorganisms other than the Euryarchaeota and Actinobacteria, including Chloroflexi, Proteobacteria and Firmicutes (Ahmed et al., 2015). In this work, we explored the distribution of the genes encoding the F420 biosynthesis enzymes CofC, CofD, CofE, CofG and CofH in public genomes and metagenomes. This revealed that the genes required to synthesize F420 are encoded in a broad range of aerobic bacteria and are widespread in soil and aquatic ecosystems. Using this information, we validated through pure culture studies that three representatives of the dominant soil phyla Proteobacteria and Chloroflexi synthesize F420. We propose that F420 is much more widely distributed in microorganisms than previously reported and present a model of the evolution of the Fo and F420 biosynthesis pathways to explain the origin and dispersal of this cofactor.
Materials and methods
Gene sequence retrieval
The amino acid sequences of all known F420 biosynthesis enzymes (CofC, CofD, CofE, CofG and CofH) represented in the NCBI (National Center for Biotechnology Information) Reference Sequence (RefSeq) database (Pruitt et al., 2007) were retrieved by Protein BLAST and PSI-BLAST (Altschul et al., 1990). The homologous proteins in Aigarchaeota, Bathyarchaeota, Geoarchaeota, Lokiarchaeota and Tectomicrobia were retrieved from the Joint Genome Institute's Integrated Microbial Genomes database (Markowitz et al., 2012). Taxonomic annotations for the sequences were obtained from the NCBI Taxonomy database and sequences duplicated at the species level were deleted. Clustering on sequence similarity networks (Atkinson et al., 2009) generated using the Enzyme Function-Initiative Enzyme Similarity Tool (Gerlt et al., 2015) were used to identify homologs of characterized proteins from nonspecific hits. In this analysis, nodes represent individual proteins and edges represent the all-versus-all BLAST E-values (Altschul et al., 1990) between them. Closely related proteins form visual clusters, allowing the identification of sequences belonging to a protein family from those belonging to related families (Atkinson et al., 2009). Final sequence sets were obtained by decreasing logE-value cutoffs until no major changes in clustering were observed with large increases in cutoff value. Final logE-value cutoffs used to identify sequences were −20 for CofC, −52 for CofE and −60 for CofD, CofG and CofH.
Evolutionary analysis
For robust phylogenetic tree construction, representative smaller sequence sets were generated by removing any sequences with >90% sequence identity using CD-HIT (Fu et al., 2012). This was first done for CofH and the same taxa as in the resulting sequence set were used for the phylogenetic analysis of the other proteins for consistency. RogueNaRok (Aberer et al., 2013) was used to remove fast-evolving sequences (all belonging to Firmicutes, Tectomicrobia, Thermoleophilia and Rubrobacteria) that otherwise caused long-branch attractions and hence unreliable evolutionary inferences (Anderson and Swofford, 2004). Sequences were aligned using MAFFT (Katoh and Standley, 2013) or MUSCLE (Edgar, 2004), with gaps and poorly aligned variable regions in the alignment removed using Gblocks (Castresana, 2000). Approximate maximum-likelihood trees were generated using Fasttree 2 (Price et al., 2010), with the JTT+CAT evolutionary model for 100 bootstrap replicates that were generated using SeqBoot in the Phylip package (Felsenstein, 2005). To analyze co-evolution of the F420 biosynthesis proteins, phylogenetic tree topologies were compared and correlated using MirrorTree (Kann et al., 2009). Genetic organization was compared using the Microbial Genomic Context Viewer (Overmars et al., 2013) and the Integrated Microbial Genomes database (Markowitz et al., 2012).
Motif analysis and homology modeling
BLAST and PSI-BLAST (Altschul et al., 1990) were used to retrieve sequences encoding probable F420-dependent oxidoreductases from the NCBI reference genomes of Oligotropha carboxidovorans, Paracoccus denitrificans and Thermomicrobium roseum. Characterized representatives of the 20 previously described F420-dependent oxidoreductase enzyme families were used as seed sequences (Greening et al., 2016a). Phyre2 (Kelley et al., 2015) was used to model protein structures based on these sequences based on solved protein structures with highest percentage amino acid sequence identity. The quality of the models at the global (full protein) and local (F420-binding site) scales were assessed using ProQ2 (Ray et al., 2012). F420-binding motifs were identified based on previous studies (Eguchi et al., 1984; Purwantini and Daniels, 1998; Aufhammer et al., 2004; Ahmed et al., 2015).
Metagenome analysis
Metagenomes were screened for the presence of F420 biosynthesis genes via a translated BLAST screen against the reference database as previously described (Greening et al., 2016b). In all, 19 publicly available metagenomes (11 ecosystem types) were randomly subsampled to an equal depth of 4 million reads with minimum read length >140 nucleotides. To remove false positives, hits within the initial screen were further sieved by removing any result with a minimum percentage identity <60 or minimum query coverage <40 amino acids.
Bacterial culturing and harvesting
O. carboxidovorans OM5 (Meyer and Schlegel, 1978) was grown on carbon monoxide oxidizer media supplemented with 36 mM sodium acetate (Meyer and Schlegel, 1978) and maintained on solid carbon monoxide oxidizer containing 1.2% (w/v) agar. P. denitrificans strain PD1222 (de Vries et al., 1989) was grown and maintained in lysogeny broth liquid media and agar. Mycobacterium smegmatis mc2155 (Snapper et al., 1990) was grown and maintained in lysogeny broth liquid media and agar supplemented with 0.05% (v/v) Tween 80. T. roseum (Jackson et al., 1973) cultures were grown in Castenholz salts solution supplemented with 5 g l−1 peptone and 2.5 g l−1 sucrose (Houghton et al., 2015). Liquid cultures (500 ml) of O. carboxidovorans, P. denitrificans and M. smegmatis were grown to early stationary-phase in 2-l Erlenmeyer flasks in a rotary incubator (200 r.p.m., 37 °C). T. roseum was grown in an equivalent manner in 1-l cultures at 130 r.p.m. and 68 °C. Cells from stationary-phase liquid cultures were harvested by centrifugation (4 °C, 10 000 g, 20 min) and the supernatant was discarded. Pellets were resuspended in 1 to 3 ml 50 mM sodium phosphate buffer (pH 7.0) and lysed by boiling. Lysates were centrifuged at 11 000 g for 10 min, and the supernatant was removed for analysis.
HPLC and liquid chromatography/mass spectrometry analysis
F420 was detected by ion-pair reversed-phase high-performance liquid chromatography (HPLC) using an Agilent (Santa Clara, CA, USA) 1200 series HPLC system equipped with an autosampler, fluorescence detector and a Poroshell 120 EC-C18 2.1 × 100 mm, 2.7 μm column. Gradients of two HPLC buffers were used, A (20 mM ammonium phosphate, 10 mM tetrabutylammonium phosphate, pH 7.0) and B (100% acetonitrile), to separate F420 species at high resolution by the length of their oligoglutamate tails. The applied gradient was 0–1 min 25% B; 1–10 min from 25% to 35% B; 10–13 min 35% B; 13–16 min from 35% to 40% B; 16–19 min from 40% to 25% B. Columns were extensively washed between sample runs, and test runs validated there was no carryover of F420. For fluorescence detection, the samples were excited at 420 nm and emission spectra were recorded between 470 and 600 nm. To verify the relative abundance of F420 species, concentrated F420 standard was purified from a recombinant F420 overexpression strain of M. smegmatis mc24517 (Bashiri et al., 2010) as previously described (Isabelle et al., 2002). The standard was lyophilized for storage, resuspended in 50 μl ultrapure water and serially diluted. The sample was analyzed on a Waters (Milford, MA, USA) LCT Premier OA-TOF (orthogonal acceleration time-of-flight) mass spectrometer. Samples were ionized by electrospray in negative ion mode at a rate of 5 μl per min with a capillary voltage of 2500 V, desolvation temperature of 150 °C and source temperature of 100 °C. Deconvolution of the mass data was facilitated by MaxEnt software (Princeton, NJ, USA).
Results
The genetic determinants of F420 biosynthesis are widely encoded by aerobic soil bacteria
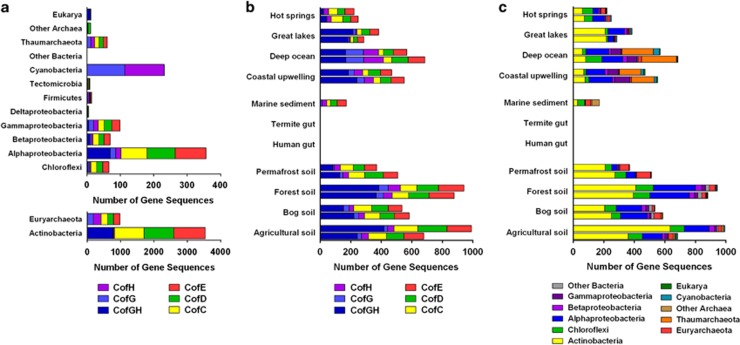
We initially retrieved the sequences of all F420 biosynthesis genes in publicly available genomes in order to understand the distribution and evolution of F420 in microorganisms (Supplementary Table S1). A total of 653 bacterial and 173 archaeal species named in the NCBI Reference Sequence database encoded all five proteins specifically required to synthesize F420 (CofC, CofD, CofE, CofG and CofH) (Supplementary Table S2).
Among archaea, the genes encoding F420 biosynthesis enzymes were unsurprisingly widespread in the Euryarchaeota, specifically in all six validated methanogenic orders, Halobacteria and Archaeoglobi. Biosynthesis genes were also detected in full suites in the Thaumarchaeota and Geoarchaeota, and in partial repertoires in the metagenome-derived incomplete genomes of Aigarchaeota, Bathyarchaeota and Lokiarchaeota representatives (Figure 2a and Supplementary Table S3). Among bacteria, F420 biosynthesis genes were present in the majority of sequenced Actinobacteria, as well as multiple species within the phyla Proteobacteria (classes: Alpha, Beta and Gamma) and Chloroflexi (classes: Thermomicrobia, Ktedonobacteria and Ardenticatenia) (Figure 2a and Supplementary Table S3). Within these phyla often predominating in soil environments, species harboring cof genes were obligate aerobes and facultative anaerobes rather than obligate anaerobes. For example, these genes were widespread in the aerobic lineages of Chloroflexi, but not anaerobic lineages such as the Dehalococcoidia. In addition, cof genes were disproportionally abundant in known soil- and marine-dwelling orders of Proteobacteria (for example, Rhizobiales and Alteromonadales) compared with host-associated orders (for example, Rickettsiales and Enterobacteriales) (Supplementary Table S1). Finally, F420 biosynthesis genes were also detected in two Firmicutes species and within the Tectomicrobia (Supplementary Table S3), a recently discovered candidate phylum with wide biosynthetic capacity (Wilson et al., 2014). Putative F420-dependent oxidoreductases from multiple families were identified in the genomes of the phyla predicted to synthesize F420 (Supplementary Table S4).
Figure 2.
Genomic and metagenomic distribution of the cof genes that encode F420 biosynthesis enzymes. The genes encoding the five known proteins specifically required for F420 synthesis are shown, namely CofC, CofD, CofE, CofG and CofH, where CofGH represents a fusion protein. (a) Distribution of cof genes by phyla in the NCBI Reference Sequence database. (b) Distribution of cof genes in 19 publicly available metagenomes by enzyme. (c) Distribution of cof genes in 19 publicly available metagenomes by phylum of the closest BLAST hit (>60% identity).
We also analyzed the distribution of the two genes (cofG and cofH) required for synthesis of Fo, both an independent chromophore and an F420 precursor (Figure 2a and Supplementary Table S3). It has been postulated that Fo is universally distributed given it is used in DNA photolyases across the three domains of life (Kiener et al., 1989; Mees et al., 2004; Petersen and Ronan, 2010). Our analysis revealed, however, that cofG and cofH genes were almost exclusively encoded by predicted F420 producers, that is, those organisms that harbor all five F420 biosynthesis genes. The only exceptions were Cyanobacteria, Chlorophyta and Streptophyta (Supplementary Table S1) that encoded the genetic determinants of Fo biosynthesis (cofG and cofH) but not F420 synthesis (cofC, cofD and cofE). These three lineages comprise oxygenic phototrophs known to encode Fo-utilizing photolyases (Mees et al., 2004; Petersen and Ronan, 2010). As previously observed (Graham et al., 2003), the cofG and cofH genes are fused into a single open reading frame (referred hereafter as cofGH) in most Actinobacteria, some Proteobacteria and phototrophic eukaryotes.
To explore the ecological role of F420, we surveyed publicly available metagenomes for the genetic determinants of F420 synthesis (Supplementary Table S5). In total, a full complement of F420 biosynthesis genes were identified in all 17 soil, water and sediment metagenomes analyzed (Supplementary Table S5). These genes were especially abundant in the aerated forest and agricultural soil samples, accounting for ~0.02% of total sequence reads (Figure 2b). Consistent with the microbial community composition of soil ecosystems (Janssen, 2006), the majority of the identified reads matched those from Actinobacteria, Alphaproteobacteria and Chloroflexi (Figure 2c). Based on metagenomes, F420 biosynthesis genes were also abundant in eight aquatic environments. In these ecosystems, genes encoding Fo synthases (cofG and cofH) were 1.4 to 1.8 times more abundant relative to those encoding F420-specific biosynthesis enzymes (cofC, cofD and cofE) (Figure 2b), suggesting that these environments harbor large communities of both Fo-utilizing phototrophs and F420-synthesizing chemotrophs (Figure 2c). F420 biosynthesis genes closely related to those encoded in Euryarchaeota were unsurprisingly common in marine sediments known to harbor high concentrations of methane-cycling archaea (Figure 2c). Consistent with their community composition, F420 biosynthesis genes were detected in very low abundance within gut ecosystems (Supplementary Table S5).
Representatives of the Proteobacteria and Chloroflexi synthesize F420
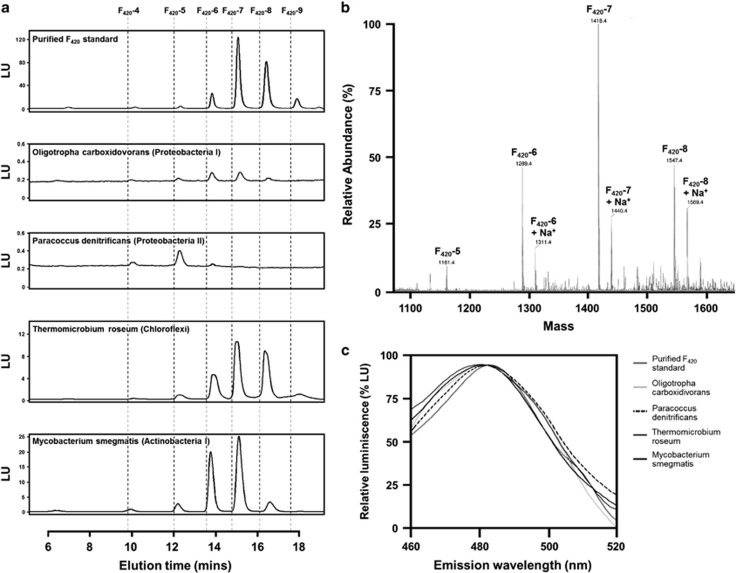
To validate these genome-based predictions, we investigated whether isolates of the phyla Proteobacteria and Chloroflexi synthesized F420. We cultured three bacteria to test for F420 production under conditions that would promote aerobic, heterotrophic growth: O. carboxidovorans, an obligate aerobe and facultative lithotroph of the alphaproteobacterial order Rhizobiales (Meyer and Schlegel, 1978); P. denitrificans, a facultative anaerobe and facultative lithotroph of the alphaproteobacterial order Rhodobacterales (de Vries et al., 1989); and T. roseum, a thermophilic obligate aerobe and obligate heterotroph of the phylum Chloroflexi (Jackson et al., 1973). F420 was detected in whole-cell lysates of all three bacteria as well as within the positive control M. smegmatis (Figure 3). In HPLC analysis, F420 species from all four organisms eluted with retention times (Figure 3a) identical to those from a purified F420 standard validated by mass spectrometry (Figure 3b). When excited at 420 nm, all samples emitted fluorescence (λmax=480 nm) (Figure 3c) with a spectrum characteristic of F420 (Eirich et al., 1978).
Figure 3.
Chemical detection of F420 in dominant soil phyla. (a) HPLC traces showing F420 from cell lysates of different species relative to a purified F420 standard. The traces show the intensity of the fluorescence emitted (λexcitation=420 nm, λemission=480 nm). The dotted lines show the times at which each species started to elute. (b) Mass spectra confirming the molecular weight and oligoglutamate side chain length of the purified F420 standard. (c) Fluorescence emission spectra of F420 from cell lysates of five different species against a purified F420 standard.
The quantity of F420 produced and distribution of oligoglutamate chain lengths differed between the strains analyzed (Table 1). Whereas T. roseum and M. smegmatis produced large quantities of F420, normalized production was ∼100-fold lower for the proteobacterial strains under comparable conditions (Figure 3a); F420 was nevertheless unambiguously and reproducibly detected in such organisms. Ion-pair HPLC was used to resolve the F420 species based on the length of their oligoglutamate side chains (Figure 3a). Mass spectrometry of the purified standard confirmed the lengths of the dominant HPLC peaks (Figure 3b). The number of glutamate residues attached differed between species, with 4 to 6 glutamates detected in P. denitrificans, 5 to 7 detected in O. carboxidivorans and 6 to 8 dominating in the T. roseum and M. smegmatis samples (Table 1).
Table 1. Length of oligoglutamate chains of F420 in the cultured organisms.
| F420 species (%) | F420-2 | F420-3 | F420-4 | F420-5 | F420-6 | F420-7 | F420-8 | F420-9 |
|---|---|---|---|---|---|---|---|---|
| F420 standard | <1 | <1 | 1 | 1 | 9 | 45 | 36 | 7 |
| Oligotropha carboxidovorans | — | 2 | 7 | 14 | 32 | 37 | 8 | — |
| Paracoccus denitrificans | — | — | 13 | 82 | 5 | — | — | — |
| Thermomicrobium roseum | — | <1 | <1 | 3 | 20 | 39 | 34 | 3 |
| Mycobacterium smegmatis | — | 1 | 2 | 5 | 35 | 50 | 7 | <1 |
The percentage amount of each F420 species compared with the total F420 present is shown.
Having established that species from the phyla Proteobacteria and Chloroflexi synthesize F420, we investigated whether any F420-dependent enzymes that have been characterized in Archaea and Actinobacteria are conserved in the genomes of P. denitrificans, O. carboxidovorans and T. roseum. We identified close homologs of Fgd (F420-reducing glucose 6-phosphate dehydrogenase) in the Chloroflexi and Fno (F420-reducing NADPH dehydrogenase) in the two Proteobacteria. Putative F420H2-dependent reductases from two superfamilies, the luciferase-like hydride transferases (Greening et al., 2016a) and flavin/deazaflavin oxidoreductases (Ahmed et al., 2015), were also encoded in the three organisms (Supplementary Table S6). Multiple sequence alignments and protein homology models confirmed the residues involved in F420 binding are almost entirely conserved in these sequences, including residues that interact with the isoalloxazine ring and oligoglutamate chain, that are not found in the flavins FAD and FMN (Supplementary Figure S1). The presence of these sequences and the high conservation of the F420-binding sequence motifs provide support that these organisms encode F420-dependent oxidoreductases.
F420 biosynthesis pathways evolved through bacterial-to-archaeal horizontal gene transfers
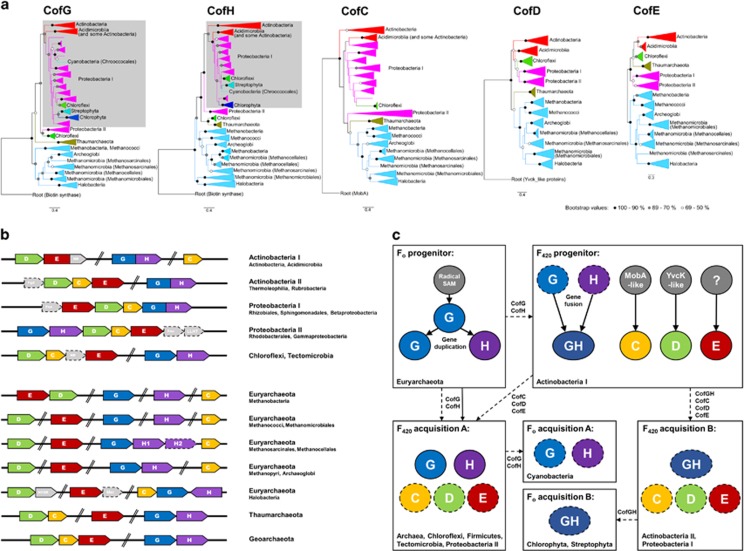
Finally, we analyzed the evolution of the F420 biosynthesis enzymes (CofC, CofD, CofE, CofG and CofH) to understand the origin and distribution of the cofactor. With respect to all five proteins, phylogenetic tree topologies (Figure 4a) and sequence similarity networks (Supplementary Figure S2) were similar. In all cases, two large clades corresponding to the euryarchaeotal and actinobacterial enzymes flanked the trees and networks, and there were smaller middle clades corresponding to homologous enzymes in Proteobacteria, Chloroflexi, Tectomicrobia, Firmicutes, Thaumarchaeota and deep-branching actinobacterial lineages such as Acidimicrobiia. A MirrorTree analysis comparing the topologies of the phylogenetic trees indicated that the five Cof enzymes co-evolved (with Pearson's correlation coefficients between 0.71 and 0.93) (Supplementary Figure S3). This is consistent with selection for all five subunits being required to produce functional F420. The Fo biosynthesis proteins CofG and CofH in Cyanobacteria have diverged further from their homologs in F420-synthesizing organisms (Supplementary Figures S2 and S4), consistent with their lack of co-evolution with the rest of the pathway.
Figure 4.
Evolution of the determinants of F420 biosynthesis. (a) Phylogenetic trees of the F420 biosynthesis proteins (CofC, CofD, CofE, CofG and CofH). Trees were rooted with a related protein family characterized by the same protein fold, except for CofE (novel protein fold), presented as an unrooted tree. Clades are labeled according to phyla, except for Actinobacteria and Euryarchaeota that are labeled by class. The Firmicutes, Tectomicrobia, Thermoleophilia and Rubrobacteria lineages have been omitted as their inclusion compromises phylogenetic inferences because of long-branch attractions. Gray-shaded regions represent the CofGH fusion proteins. CofG and CofH trees incorporating cyanobacterial sequences are presented in Supplementary Figure S4 as these sequences caused low bootstrap values at key nodes. (b) Generalized schematic of the genetic organization of the genes encoding the five enzymes specifically required for F420 biosynthesis (CofC, CofD, CofE, CofG and CofH) from five bacterial phyla (Actinobacteria, Proteobacteria, Chloroflexi, Tectomicrobia and Firmicutes) and three archaeal phyla (Euryarchaeota, Thaumarchaeota and Geoarchaeota). Fgd, F420-reducing glucose 6-phosphate dehydrogenase; Fno, F420-reducing NADPH dehydrogenase; HFDR, predicted F420H2-dependent reductase; LLHT, predicted F420H2-dependent luciferase-like oxidoreductase; Mer, methylenetetrahydromethanopterin reductase; NR, hypothetical nitroreductase. (c) Schematic representation of the proposed evolutionary origin of F420 biosynthesis genes and their acquisition by different phyla. Solid lines/circles indicate vertical acquisition, whereas dashed lines/circles indicate horizontal acquisition. The capacity for F420 biosynthesis appears to have evolved on at least two separate occasions in both the Actinobacteria (Actinobacteria I=Actinobacteria, Acidimicrobiia; Actinobacteria II=Thermoleophilia, Rubrobacteria) and Proteobacteria (Proteobacteria I=Rhizobiales, Sphingomonadales, Betaproteobacteria; Proteobacteria II=Rhodobacterales, Gammaproteobacteria).
To better understand the evolutionary origin of these proteins, phylogenetic trees were rooted with related families containing the same protein fold (Figure 4a), namely biotin synthase (a radical S-adenosylmethionine enzyme) for CofG and CofH, adenosylcobinamide guanosine transferase (MobA) for CofC and YvcK (hypothetical protein family) for CofD. CofE adopts a novel protein fold and does not share any significant sequence homology with any other known proteins (Nocek et al., 2007) and thus is presented as an unrooted tree with no evolutionary origin inferred. Our analysis indicates that CofG is likely to have originated in an ancestral archaeon. Subsequently, the functionally and structurally related CofH evolved from an ancient duplication of the cofG gene within an early methanogenic euryarchaeon. In contrast, the roots of the CofC and CofD trees indicate these enzymes originated in an early actinobacterium. These findings are consistent with the recent work suggesting cofC and cofD were among the metabolic genes acquired from bacteria during the diversification of archaea (Nelson-Sathi et al., 2015). Our observations suggest that, in contrast to the Fo synthases, enzymes specific for F420 biosynthesis evolved in bacteria, most probably an early actinobacterium, and were subsequently horizontally transferred into archaea.
The genetic organization of the cof genes is highly variable (Figure 4b). The five opening reading frames are genomically separated in two methanogenic orders (Methanococcales and Methanomicrobiales) and are partially clustered in Actinobacteria, Chloroflexi, Tectomicrobia and all other archaea. In contrast, cof genes are organized as complete operons within Proteobacteria; consistent with the presence of two distinct clusters of proteobacterial sequences in all phylogenetic trees, these operons were organized in two distinct configurations (designated Proteobacteria I and Proteobacteria II). Our analysis suggests that the cof genes initially evolved separately, but became increasingly syntenic because of selective pressures to inherit all five enzymes for F420 production. This analysis shows that the deepest-rooted branches of the CofGH fusion protein are between the classes Actinobacteria and Acidimicrobiia. This suggests the CofGH fusion occurred early in actinobacterial evolution, presumably to enhance the catalytic efficiency of the coordinated radical S-adenosylmethionine reactions required for Fo synthesis (Philmus et al., 2015). In some organisms, F420-reducing dehydrogenases (Fgd, Fno and Mer) and putative F420H2-dependent reductases also appear to be operonic with the cof genes (Figure 4b).
Discussion
Until now, the vast majority of research into F420 has focused on the roles of the cofactor in methane cycling and tuberculosis infection (Greening et al., 2016a). However, our findings demonstrate that the cofactor is widely distributed among diverse taxa and ecosystems, and is likely to play a role in a broader array of metabolic and ecological phenomena than is currently recognized. We have demonstrated, using a combination of comparative genomics and analytical chemistry approaches, that F420 is widely distributed in three of the five most dominant soil phyla (Actinobacteria, Proteobacteria and Chloroflexi) and provided genomic evidence that the cofactor is encoded in other phyla (Tectomicrobia, Firmicutes, Thaumarchaeota and Geoarchaeota). The finding that F420 biosynthesis genes are widely distributed within aerobic bacterial and archaeal taxa indicates that F420 is far from a niche methanogenic cofactor. Furthermore, the abundance of these genes in soil ecosystems suggests that F420 influences the biological and chemical composition of soils.
There are three feasible explanations for the observed distribution of F420 in biological systems: (1) F420 biosynthesis evolved pre-LUCA (last universal common ancestor); (2) F420 biosynthesis evolved in an archaeon and was acquired by bacteria; or (3) F420 biosynthesis evolved in a bacterium and was acquired by archaea. Of these, the first hypothesis is unsupported given that cof gene evolution does not parallel 16S rRNA gene phylogenies and the cofactor appears to be completely absent from proposed deep-branching bacteria (for example, Aquificae and Thermotogae) and archaea (for example, Crenarchaeota and Thermococci). The widely assumed archaea-to-bacteria hypothesis remains plausible, but is challenged by phylogenetic trees suggesting a bacterial origin for CofC and CofD. The bacteria-to-archaea hypothesis for F420 acquisition is more robustly supported by our phylogenetic trees and is also consistent with the findings of the large-scale Martin analysis on interdomain lateral gene transfers (Nelson-Sathi et al., 2015).
In Figure 4c, we present an evolutionary model of the origin and distribution of F420. Our studies strongly support that the genes required for Fo synthesis (cofG and cofH) evolved independently and probably earlier than the genes required for F420 synthesis from Fo (cofC, cofD and cofE). We hypothesize that an early methanogen synthesized Fo but not F420 using the products of ancient cofG and cofH genes. This cofactor may have initially sustained many of the redox functions now supported by F420, including hydrogenotrophic methanogenesis. Our evolutionary analysis suggests that an early actinobacterium subsequently acquired cofG and cofH (later fused into cofGH) and evolved the cofC, cofD and likely cofE genes required to produce F420, the lactyloligoglutamyl derivative of Fo. Multiple horizontal gene transfer events thereafter led to the acquisition of these genes in the archaea and other bacteria. The euryarchaeotal branches of the CofC, CofD, and CofE phylogenetic trees mirror those of 16S rRNA gene trees (Hedderich and Whitman, 2013). This is consistent with their genes being acquired from Actinobacteria early in the evolution of Euryarchaeota (Figure 4), resulting in the capacity for F420 biosynthesis being vertically inherited by the six methanogenic orders, Archaeoglobi, Halobacteria and likely anaerobic methanotrophs (ANME). Unicellular eukaryotes (Chlorophyta and Streptophyta) appear to have acquired the chromophore Fo by laterally acquiring the fused cofGH gene from Proteobacteria. The origin of the cyanobacterial cofG and cofH remains unclear because of their evolutionary distance from the rest of the sequences; scenarios involving acquisition from methanogens or Proteobacteria are both plausible.
We propose that the evolutionary driving force resulting in Fo synthesis was the need for redox cofactors with distinct electrochemical properties to flavins and nicotinamides to drive the unique reactions of hydrogenotrophic methanogenesis. By hijacking the riboflavin biosynthesis pathway with CofG and CofH, an early euryarchaeon would have produced a cofactor with the ideal electrochemistry to mediate the central reactions of methanogenesis: H2 oxidation, methenyl and methylene reduction and NADP reduction. Fo could have functioned as the primordial methanogenic redox cofactor as it has near-identical electrochemical properties to F420, including a standard redox potential of −340 mV and obligate two-electron reactivity (Walsh, 1986; Greening et al., 2016a). Furthermore, it has been demonstrated that Fo can functionally substitute for F420 in various enzymatic processes in vitro, including in the crucial methanogenesis enzymes Frh (F420-reducing hydrogenase) and Fno (F420-dependent NADP reductase) (Yamazaki and Tsai, 1980; Muth et al., 1987). The current distribution of cof genes emphasizes that deazaflavins have primarily been selected for their electrochemical and not photochemical properties. With the exception of cyanobacterial and eukaryotic phototrophs, all organisms appear to have retained cofG and cofH genes primarily to generate F420 as a redox cofactor rather than Fo as an independent chromophore. We propose that the need to produce a charged derivative of Fo, F420, served as the main selection pressure for the evolution and dispersal of CofC, CofD and CofE. The feature that distinguishes F420 from its precursor Fo is the presence of a negatively charged sidechain containing phosphate and glutamate groups (Eirich et al., 1978). Thus, whereas Fo diffuses through membranes, F420 is retained in the cytosol (Glas et al., 2009). The electrostatic properties of F420 may also facilitate higher affinity enzyme–cofactor interactions according to recent structural studies (Greening et al., 2016a). We also show here that the length of the oligoglutamate sidechain of F420 varies between phyla and progressively increases between the Euryarchaeota (Gorris and van der Drift, 1994), Proteobacteria, Actinobacteria and Chloroflexi (Table 1). We are presently investigating the reasons and mechanisms behind these variations.
F420 may confer a number of competitive advantages to these bacteria by mediating both endogenous and exogenous metabolic processes. F420 is known to serve as a redox cofactor in an increasing number of endogenous processes in mycobacteria, including central carbon metabolism (Bashiri et al., 2008), cell wall modification (Purwantini and Mukhopadhyay, 2013), antioxidant production (Ahmed et al., 2015) and possibly quinone reduction (Gurumurthy et al., 2013). Although the cofactor is synthesized in high levels under oxic conditions (Figure 2), phenotypic evidence suggests it is particularly important for survival under hypoxia (Gurumurthy et al., 2013). In this condition, mycobacteria may increasingly depend on low-potential cofactors such as F420 to maintain redox homeostasis (Berney et al., 2014; Cook et al., 2014). Many of the F420H2-dependent oxidoreductases involved in mycobacterial metabolism are also conserved in other taxa, where they may have similar roles (Supplementary Table S5). F420 is likely to be particularly important in the redox metabolism of Chloroflexi and other Actinobacteria given representatives of such phyla produce large amounts of the compound even under optimal growth conditions (Figure 3a). A central role for F420 in the metabolism of Proteobacteria seems less likely, given the levels detected of the cofactor in O. carboxidovorans (Proteobacteria I lineage) and P. denitrificans (Proteobacteria II lineage) were low (Figure 3a). It is likely that transcription of the cof genes is constitutively repressed in such organisms, but is activated in response to environmental stresses or other signals. Consistently, TetR and MarR family transcriptional repressors were identified immediately downstream of the cof operons in most Proteobacteria, including P. denitrificans and O. carboxidivorans.
The richest literature on the physiological roles of F420 in bacteria concerns the metabolism of complex organic compounds. F420H2-dependent reductases mediate the biodegradation of nitroaromatic explosives (Ebert et al., 2001), triphenyl dyes (Guerra-Lopez et al., 2007) and furanocoumarins (Taylor et al., 2010), as well as the biosynthesis of tetracycline and pyrrolobenzodiazepine antibiotics (Li et al., 2009; Wang et al., 2013). A role for F420 in equivalent processes seems particularly likely for the Chloroflexi and Tectomicrobia; both phyla contain an abundance of F420H2-dependent reductases (Supplementary Tables S3 and S6) and are reputed for their biosynthetic versatility and biodegradative capacities (Björnsson et al., 2002; Wilson et al., 2014). In turn, these F420-dependent processes are likely to affect the biological composition of soil ecosystems, for example, by shaping the antibiotics arms race. They may also influence the chemical composition of soil environments by controlling the levels of complex organic compounds produced and consumed. Better understanding of the endogenous and exogenous roles of F420 depends on defining the functions of the numerous putative F420-dependent oxidoreductases encoded in aerobic bacteria and archaea (Supplementary Table S3). In turn, further understanding the physiological and ecological significance of F420 may open opportunities in the fields of pharmaceuticals, biocatalysis and bioremediation.
Acknowledgments
We thank Dr Ghader Bashiri (University of Auckland) for providing the fbiABC overexpression plasmid, Professor William Jacobs Jr (Albert Einstein College of Medicine) for supplying Mycobacterium smegmatis mc24517 and the three anonymous reviewers for their helpful suggestions. This work was supported by Australian Research Council research Grants (DE120102673, DP130102144) awarded to CJJ; Australian National University Summer and PhD scholarships awarded respectively to BN and FHA; and a CSIRO Office of the Chief Executive Postdoctoral Fellowship awarded to CG.
Author contributions
CG, CJJ, BN, FHA, CRC, MBS, MCT, ACW, JGO, SJW and GP designed experiments. BN, FHA, CG, CRC, AB, SJW, and CJJ performed experiments. CG, CJJ, BN and FHA analyzed data. CJJ, CG, MCT, ACW, JGO and SEM supervised students and researchers. CG, CJJ, FHA, BN and JGO wrote the paper.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aberer AJ, Krompass D, Stamatakis A. (2013). Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst Biol 62: 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed FH, Carr PD, Lee BM, Afriat-Jurnou L, Mohamed AE, Hong N-S et al. (2015). Sequence-structure-function classification of a catalytically diverse oxidoredu+ctase superfamily in mycobacteria. J Mol Biol 427: 3554–3571. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson FE, Swofford DL. (2004). Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Mol Phylogenet Evol 33: 440–451. [DOI] [PubMed] [Google Scholar]
- Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. (2009). Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4: e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufhammer SW, Warkentin E, Berk H, Shima S, Thauer RK, Ermler U. (2004). Coenzyme binding in F420-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family. Structure 12: 361–370. [DOI] [PubMed] [Google Scholar]
- Bair TB, Isabelle DW, Daniels L. (2001). Structures of coenzyme F420 in Mycobacterium species. Arch Microbiol 176: 37–43. [DOI] [PubMed] [Google Scholar]
- Bashiri G, Rehan AM, Greenwood DR, Dickson JMJ, Baker EN. (2010). Metabolic engineering of cofactor F420 production in Mycobacterium smegmatis. PLoS One 5: e15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiri G, Rehan AM, Sreebhavan S, Baker HM, Baker EN, Squire CJ. (2016). Elongation of the poly-gamma-glutamate tail of F420 requires both domains of the F420:gamma-glutamyl ligase (FbiB) of Mycobacterium tuberculosis. J Biol Chem 291: 6882–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiri G, Squire CJ, Moreland NJ, Baker EN. (2008). Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem 283: 17531–17541. [DOI] [PubMed] [Google Scholar]
- Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. (2014). An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci USA 111: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnsson L, Hugenholtz P, Tyson GW, Blackall LL. (2002). Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removalc. Microbiology 148: 2309–2318. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Cheeseman P, Toms-Wood A, Wolfe RS. (1972). Isolation and properties of a fluorescent compound, Factor420, from Methanobacterium strain M.o.H. J Bacteriol 112: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-P, Bair TB, Bae Y-M, Daniels L. (2001). Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F420 biosynthesis by Mycobacterium bovis BCG. J Bacteriol 183: 7058–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-P, Kendrick N, Daniels L. (2002). Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and FO biosynthesis. J Bacteriol 184: 2420–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GM, Greening C, Hards K, Berney M. (2014) Energetics of pathogenic bacteria and opportunities for drug development. In: Poole RK (ed) Advances in Bacterial Pathogen Biology Vol. 65 Academic Press: Cambridge, MA, USA, pp 1–62. [DOI] [PubMed] [Google Scholar]
- Daniels L, Bakhiet N, Harmon K. (1985). Widespread distribution of a 5-deazaflavin cofactor in Actinomyces and related bacteria. Syst Appl Microbiol 6: 12–17. [Google Scholar]
- de Poorter LMI, Geerts WJ, Keltjens JT. (2005). Hydrogen concentrations in methane-forming cells probed by the ratios of reduced and oxidized coenzyme F420. Microbiology 151: 1697–1705. [DOI] [PubMed] [Google Scholar]
- de Vries GE, Harms N, Hoogendijk J, Stouthamer AH. (1989). Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a (nGATCn) DNA-modifying property. Arch Microbiol 152: 52–57. [Google Scholar]
- Ebert S, Fischer P, Knackmuss HJ. (2001). Converging catabolism of 2,4,6-trinitrophenol (picric acid) and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. Biodegradation 12: 367–376. [DOI] [PubMed] [Google Scholar]
- Ebert S, Rieger P-G, Knackmuss H-J. (1999). Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J Bacteriol 181: 2669–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi S, Nakata H, Nishio N, Nagai S. (1984). NADP+ reduction by a methanogen using HCOOH or H2 as electron donor. Appl Microbiol Biotechnol 20: 213–217. [Google Scholar]
- Eirich LD, Vogels GD, Wolfe RS. (1979). Distribution of coenzyme F420 and properties of its hydrolytic fragments. J Bacteriol 140: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich LD, Vogels GD, Wolfe RS. (1978). Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry 17: 4583–4593. [DOI] [PubMed] [Google Scholar]
- Eker APM, Hessels JKC, van de Velde J. (1988). Photoreactivating enzyme from the green alga Scenedesmus acutus. Evidence for the presence of two different flavin chromophores. Biochemistry 27: 1758–1765. [Google Scholar]
- Eker APM, Pol A, van der Meyden P, Vogels GD. (1980). Purification and properties of 8-hydroxy-5-deazaflavin derivatives from Streptomyces griseus. FEMS Microbiol Lett 8: 161–165. [Google Scholar]
- Epple R, Carell T. (1998). Flavin- and deazaflavin-containing model compounds mimic the energy transfer step in type-II DNA-photolyases. Angew Chemie Int Ed 37: 938–941. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (phylogeny inference package) Distributed by the author. Dep Genome Sci Univ Washington, Seattle, Version 3.
- Fu L, Niu B, Zhu Z, Wu S, Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR et al. (2015). Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim Biophys Acta 1854: 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas AF, Maul MJ, Cryle M, Barends TRM, Schneider S, Kaya E et al. (2009). The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes. Proc Natl Acad Sci USA 106: 11540–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorris LG, van der Drift C. (1994). Cofactor contents of methanogenic bacteria reviewed. Biofactors 4: 139–145. [PubMed] [Google Scholar]
- Graham DE, Xu H, White RH. (2003). Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F420 biosynthesis. Arch Microbiol 180: 455–464. [DOI] [PubMed] [Google Scholar]
- Graupner M, White RH. (2001). Biosynthesis of the phosphodiester bond in coenzyme F420 in the methanoarchaea. Biochemistry 40: 10859–10872. [DOI] [PubMed] [Google Scholar]
- Graupner M, Xu H, White RH. (2002). Characterization of the 2-phospho-L-lactate transferase enzyme involved in coenzyme F420 biosynthesis in Methanococcus jannaschii. Biochemistry 41: 3033–3037. [DOI] [PubMed] [Google Scholar]
- Greening C, Ahmed FH, Mohamed AE, Lee BM, Pandey G, Warden AC et al. (2016. a). Physiology, biochemistry, and applications of F420- and Fo-dependent redox reactions. Microbiol Mol Biol Rev 80: 451–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB et al. (2016. b). Genome and metagenome surveys of hydrogenase diversity indicate H2 is a widely-utilised energy source for microbial growth and survival. ISME J 10: 761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochowski LL, Xu H, White RH. (2008). Identification and characterization of the 2-phospho-L-lactate guanylyltransferase involved in coenzyme F420 biosynthesis. Biochemistry 47: 3033–3037. [DOI] [PubMed] [Google Scholar]
- Guerra-Lopez D, Daniels L, Rawat M. (2007). Mycobacterium smegmatis mc2 155 fbiC and MSMEG_2392 are involved in triphenylmethane dye decolorization and coenzyme F420 biosynthesis. Microbiology 153: 2724–2732. [DOI] [PubMed] [Google Scholar]
- Gurumurthy M, Rao M, Mukherjee T, Rao SPS, Boshoff HI, Dick T et al. (2013). A novel F420-dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol Microbiol 87: 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier CH, Shima S, Thauer RK, Bourenkov G, Bartunik HD, Ermler U. (2003). Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure. J Mol Biol 332: 1047–1057. [DOI] [PubMed] [Google Scholar]
- Hedderich R, Whitman W. (2013) Physiology and biochemistry of the methane-producing Archaea. In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F (eds). The Prokaryotes SE—81. Springer: Berlin, Heidelberg, pp 635–662. [Google Scholar]
- Heiss G, Hofmann KW, Trachtmann N, Walters DM, Rouvière P, Knackmuss HJ. (2002). npd gene functions of Rhodococcus (opacus) erythropolis HL PM-1 in the initial steps of 2,4,6-trinitrophenol degradation. Microbiology 148: 799–806. [DOI] [PubMed] [Google Scholar]
- Houghton KM, Morgan XC, Lagutin K, MacKenzie AD, Vyssotski M, Mitchell KA et al. (2015). Thermorudis pharmacophila WKT50.2T sp. nov., a novel isolate of class Thermomicrobia isolated from geothermal soil, and emended descriptions of Thermomicrobium roseum, Thermomicrobium carboxidum, Thermorudis peleae and Sphaerobacter thermophilus. Int J Syst Evol Microbiol 61: 2482–2490. [DOI] [PubMed] [Google Scholar]
- Isabelle D, Simpson DR, Daniels L. (2002). Large-scale production of coenzyme F420-5,6 by using Mycobacterium smegmatis. Appl Environ Microbiol 68: 5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TJ, Ramaley RF, Meinschein WG. (1973). Thermomicrobium, a new genus of extremely thermophilic bacteria. Int J Syst Evol Microbiol 23: 28–36. [Google Scholar]
- Jacobson F, Walsh C. (1984). Properties of 7,8-didemethyl-8-hydroxy-5-deazaflavins relevant to redox coenzyme function in methanogen metabolism. Biochemistry 23: 979–988. [Google Scholar]
- Janssen PH. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72: 1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann MG, Shoemaker BA, Panchenko AR, Przytycka TM. (2009). Correlated evolution of interacting proteins: looking behind the Mirrortree. J Mol Biol 385: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiener A, Husain I, Sancar A, Walsh C. (1989). Purification and properties of Methanobacterium thermoautotrophicum DNA photolyase. J Biol Chem 264: 13880–13887. [PubMed] [Google Scholar]
- Knittel K, Lösekann T, Boetius A, Kort R, Amann R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71: 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapalikar GV, Taylor MC, Warden AC, Scott C, Russell RJ, Oakeshott JG. (2012). F420H2-dependent degradation of aflatoxin and other furanocoumarins Is widespread throughout the Actinomycetales. PLoS One 7: e30114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Graupner M, Xu H, White RH. (2003). CofE catalyzes the addition of two glutamates to F420-0 in F420 coenzyme biosynthesis in Methanococcus jannaschii. Biochemistry 42: 9771–9778. [DOI] [PubMed] [Google Scholar]
- Li W, Chou S, Khullar A, Gerratana B. (2009). Cloning and characterization of the biosynthetic gene cluster for Tomaymycin, an SJG-136 monomeric analog. Appl Environ Microbiol 75: 2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XL, White RH. (1986). Occurrence of coenzyme F420 and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol 168: 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T et al. (2004). Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306: 1789–1793. [DOI] [PubMed] [Google Scholar]
- Meyer O, Schlegel HG. (1978). Reisolation of the carbon monoxide utilizing hydrogen bacterium Pseudomonas carboxydovorans (Kistner) comb. nov. Arch Microbiol 118: 35–43. [DOI] [PubMed] [Google Scholar]
- Michaelis W, Seifert R, Nauhaus K, Treude T, Thiel V, Blumenberg M et al. (2002). Microbial reefs in the black sea fueled by anaerobic oxidation of methane. Science 297: 1013–1015. [DOI] [PubMed] [Google Scholar]
- Muth E, Mörschel E, Klein A. (1987). Purification and characterization of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from the archaebacterium Methanococcus voltae. Eur J Biochem 169: 571–577. [DOI] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chavez N, Thiergart T, Janssen A et al. (2015). Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocek B, Evdokimova E, Proudfoot M, Kudritska M, Grochowski LL, White RH et al. (2007). Structure of an amide bond forming F420:γ-glutamyl ligase from Archaeoglobus fulgidus—a member of a new family of non-ribosomal peptide synthases. J Mol Biol 372: 456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmars L, Kerkhoven R, Siezen RJ, Francke C. (2013). MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genomics 14: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JL, Ronan PJ. (2010). Critical role of 7,8-didemethyl-8-hydroxy-5-deazariboflavin for photoreactivation in Chlamydomonas reinhardtii. J Biol Chem 285: 32467–32475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philmus B, Decamps L, Berteau O, Begley TP. (2015). Biosynthetic versatility and coordinated action of 5'-deoxyadenosyl radicals in deazaflavin biosynthesis. J Am Chem Soc 137: 5406–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwantini E, Daniels L. (1998). Molecular analysis of the gene encoding F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol 180: 2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwantini E, Mukhopadhyay B. (2013). Rv0132c of Mycobacterium tuberculosis encodes a coenzyme F420-dependent hydroxymycolic acid dehydrogenase. PLoS One 8: e81985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Lindahl E, Wallner B. (2012). Improved model quality assessment using ProQ2. BMC Bioinformatics 13: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang J, Zuo Y, Wang L, Sun X, Li J et al. (2009). Biodegradation of 2,4,6-trinitrophenol by Rhodococcus sp. isolated from a picric acid-contaminated soil. J Hazard Mater 163: 1199–1206. [DOI] [PubMed] [Google Scholar]
- Shima S, Warkentin E, Grabarse W, Sordel M, Wicke M, Thauer RK et al. (2000). Structure of coenzyme F420 dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea. J Mol Biol 300: 935–950. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WRJ. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4: 1911–1919. [DOI] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N et al. (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14: 3122–3145. [DOI] [PubMed] [Google Scholar]
- Taylor MC, Jackson CJ, Tattersall DB, French N, Peat TS, Newman J et al. (2010). Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol Microbiol 78: 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144: 2377–2406. [DOI] [PubMed] [Google Scholar]
- Walsh C. (1986). Naturally occurring 5-deazaflavin coenzymes: biological redox roles. Acc Chem Res 19: 216–221. [Google Scholar]
- Wang P, Bashiri G, Gao X, Sawaya MR, Tang Y. (2013). Uncovering the enzymes that catalyze the final steps in oxytetracycline biosynthesis. J Am Chem Soc 135: 7138–7141. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K et al. (2014). An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506: 58–62. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Tsai L. (1980). Purification and properties of 8-hydroxy-5-deazaflavin-dependent NADP+ reductase from Methanococcus vannielii. J Biol Chem 255: 6462–6465. [PubMed] [Google Scholar]
- Yasui A, Takao M, Oikawa A, Kiener A, Walsh CT, Eker AP. (1988). Cloning and characterization of a photolyase gene from the cyanobacterium Anacystis nidulans. Nucleic Acids Res 16: 4447–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.