Abstract
Members of the order Methanomicrobiales are abundant, and sometimes dominant, hydrogenotrophic (H2-CO2 utilizing) methanoarchaea in a broad range of anoxic habitats. Despite their key roles in greenhouse gas emissions and waste conversion to methane, little is known about the physiological and genomic bases for their widespread distribution and abundance. In this study, we compared the genomes of nine diverse Methanomicrobiales strains, examined their pangenomes, reconstructed gene flow and identified genes putatively mediating their success across different habitats. Most strains slowly increased gene content whereas one, Methanocorpusculum labreanum, evidenced genome downsizing. Peat-dwelling Methanomicrobiales showed adaptations centered on improved transport of scarce inorganic nutrients and likely use H+ rather than Na+ transmembrane chemiosmotic gradients during energy conservation. In contrast, other Methanomicrobiales show the potential to concurrently use Na+ and H+ chemiosmotic gradients. Analyses also revealed that the Methanomicrobiales lack a canonical electron bifurcation system (MvhABGD) known to produce low potential electrons in other orders of hydrogenotrophic methanogens. Additional putative differences in anabolic metabolism suggest that the dynamics of interspecies electron transfer from Methanomicrobiales syntrophic partners can also differ considerably. Altogether, these findings suggest profound differences in electron trafficking in the Methanomicrobiales compared with other hydrogenotrophs, and warrant further functional evaluations.
Introduction
Methanogens consist of at least nine (Mihajlovski et al., 2008; Sakai et al., 2008; Evans et al., 2015; Nobu et al., 2016) recognized orders and are divided into two major categories based on whether or not they contain cytochromes. Methanogens of the Methanosarcinales order contain cytochromes and use substrates such as acetate, methanol, methylamines and H2/CO2. Methanosarcinales have been extensively studied for their biochemistry and genetics (Ferry, 2011). All other orders of methanogens lack cytochromes and typically reduce CO2 using H2, formate and secondary alcohols (Thauer et al., 2008), although some are capable of reducing methylated substrates with H2 (Fricke et al., 2006; Brugère et al., 2014). Much of what is known about cytochrome-deficient methanogens comes from biochemical and genetic experiments performed on members of the Methanobacteriales and Methanococcales orders (Ferry, 2011), compared with which the cytochrome-deficient Methanomicrobiales are highly understudied.
Comparative genomic analysis across a microbial group (that is, pangenome analysis) has been proven valuable to identify functions that over time have led to niche specialization at broad (Luo et al., 2013) or fine (Lee and Marx, 2012; Youngblut et al., 2015) phylogenetic levels. Genome expansion can lead to metabolic versatility, or the ability to take advantage of high nutrient availability, or other adaptations not directly related to metabolism (Ochman et al., 2000; Dobrindt et al., 2004), while genome downsizing in free-living microbes can cause specialization by natural selection eliminating unnecessary genes during colonization or residency in an environment (Lee and Marx, 2012). Comparative genomics studies within different orders of methanogens (Methanosarcinales, Methanocellales and Methanomassiliicoccales) have shown major differences in genome content between members, and evidenced metabolic adaptation to low substrate (H2) concentrations, evidenced novel energy conservation pathways and evidenced genomic adaptations to different environments (Maeder et al., 2006; Borrel et al., 2014; Lyu and Lu, 2015; Youngblut et al., 2015).
Methanogens of the order Methanomicrobiales are widespread across terrestrial, freshwater, marine and bioreactor ecosystems (Garcia et al., 2006). Phylotypes associated with this order have been found highly abundant or dominant across very distinct conditions in anoxic soils of wetlands, lakes or river sediments, peatlands, waste-treating bioreactors, among others (Bräuer et al., 2006; Auguet et al., 2009; Bialek et al., 2013). Although these methanogens have a narrow set of metabolic substrates (Garcia et al., 2006), their broad distribution indicates its members have differentially adapted to contrasting conditions ranging from low to high: pH, nutrients, salinity, oxidative stress, temperature and others. How do diverse members of the order Methanomicrobiales succeed in such a wide range of conditions? Answers to this question are needed to better understand their activity and predict potential changes in their role in CH4 atmospheric emissions from natural or engineered environments or for technological purposes (Purwantini et al., 2014).
Although the study of Methanomicrobiales genomics has received little attention, one early comparative analysis of three genomes available then revealed that the Methanomicrobiales represent a distinct class (class II) with their own functional and phylogenetic differentiations among methanogens (Anderson et al., 2009a). Functional adaptations are likely across the diverse Methanomicrobiales and such can be explored among multiple genome sequences of strains from different habitats that have recently become available (detailed in Table 1). In this work, we developed the comparative analyses of nine Methanomicrobiales genomes to identify the order's putative genomic adaptations to contrasting environments.
Table 1. Methanomicrobiales strains analyzed in this study.
| Strain | Habitat | pH | tgen(h) | Mbp | % GC | Accession number(s) | Ref. |
|---|---|---|---|---|---|---|---|
| Methanoregulaceae family | |||||||
| Methanoregula boonei 6A8 | Acidic bog | 5.1 | 44 | 2.54 | 54.5 | CP000780 | Bräuer et al. (2011) |
| Methanoregula formicica SMSP | Anaerobic sludge | 7.4 | 24 | 2.82 | 56.2 | CP003167 | Yashiro et al. (2011) |
| Methanosphaerula palustris E1-9c | Rich fen | 5.7 | 19 | 2.92 | 58.9 | CP001338 | Cadillo-Quiroz et al. (2009) |
| Methanolinea tarda NOBI-1 | Sewage sludge | 7.0 | 144 | 2.05 | 56.3 | AGIY01000000 | Imachi et al. (2008) |
| Methanomicrobiaceae family | |||||||
| Methanoculleus marisnigri JR1 | Marine sediment | 6.4 | NA | 2.48 | 62.1 | CP000562 | Romesser et al. (1979) |
| Methanolacinia petrolearia SEBR 4847 | Off shore oilfield | 7.0 | 10 | 2.84 | 47.4 | CP002117 | Ollivier et al. (1997) |
| Methanoplanus limicola M3 | Mud of saline swamp | NA | NA | 3.20 | 42.0 | AHKP01000000 | Wildgruber et al. (1982) |
| Methanospirillaceae family | |||||||
| Methanospirillum hungatei JF-1 | Sewage sludge | ≈7.0 | 17 | 3.54 | 45.0 | CP000254 | Ferry et al. (1974) |
| Methanocorpusculaceae family | |||||||
| Methanocorpusculum labreanum Z | Tar pit | 7.0 | 12.5 | 1.80 | 50.0 | CP000559 | Zhao et al. (1989) |
Abbreviations: tgen(h), generation time (hours); Mbp, genome size in Mbp; NA, not available. pH optima are for growth in pure culture.
Materials and methods
Genomes, protein clustering and phylogenetic inference
All annotated genome sequences (protein and nucleotide) were obtained from NCBI under accession numbers detailed in Table 1. Protein sequences were compared reciprocally using blastp (Altschul et al., 1990) with an e-value cutoff of 1e−10 and filtered for alignments that spanned at least 70% the length of the longer of the two proteins. Protein sequences were grouped into homologous clusters based on negative log transformations of their e-values with a ceiling of 200 using MCL version 11-294 (van Dongen, 2000). Based on proml (Felsenstein, 1989) phylogenetic inferences, core protein clusters containing paralogs were further refined manually by splitting robust branching groupings where appropriate.
Phylogenetic inference
Phylogenetic inference was performed using three data sets: (i) small subunit (SSU) rRNA, (ii) large subunit rRNA and (iii) core predicted protein clusters. To reduce phylogenetic noise in the core protein clusters data set, we omitted all clusters that at any point of our analyses had more than one copy per genome and filtered out clusters whose corresponding nucleotide codon alignment showed evidence of substitution saturation (Xia and Lemey, 2009). The remaining 437 protein clusters (Supplementary Data 1) were aligned and concatenated for analyses.
Sequence and codon alignments were performed with Clustalw (Larkin et al., 2007) and Pal2nal v14 (Suyama et al., 2006), respectively. Gapped positions were removed with T-coffee (Notredame et al., 2000). The nucleotide index of substitution saturation test was done with Dambe 5.3 (Xia et al., 2003, 2013). Parameters for phylogenetic inference from nucleotide or protein alignments were estimated using jModelTest (Darriba et al., 2012) and ProtTest3 (Darriba et al., 2011), respectively. Phylogenetic inferences based on proml, protpars, dnaml and dnapars were done using the Phylip package (Felsenstein, 1989). Mega5 (Tamura et al., 2011) was used to perform neighbor-joining phylogenetic inference. The Bayesian Markov chain Monte Carlo method was implemented with MrBayes 3.2 (Ronquist et al., 2012). MrBayes was run using two independent runs with a burn-in value of 25% until the standard deviations of split frequencies remained below 0.01.
Assessment for positive selection
The topology presented in Figure 1a was used in the assessment of all core orthologous protein clusters for positive selection. Codon alignments were assessed for positive selection on every branch with codeml from the PAML 4.6 package (Yang, 2007), using two maximum likelihood reconstructions of protein evolution: one with neutral evolution model (null hypothesis) and the other using a model specifying a proportion of sites subject to positive selection (alternative selection hypothesis). Comparisons of likelihood ratio test between the two models were the criteria for detecting positive selection in loci as detailed elsewhere (Xu et al., 2011). The Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995) was applied to control the false positive detection rate to 1%.
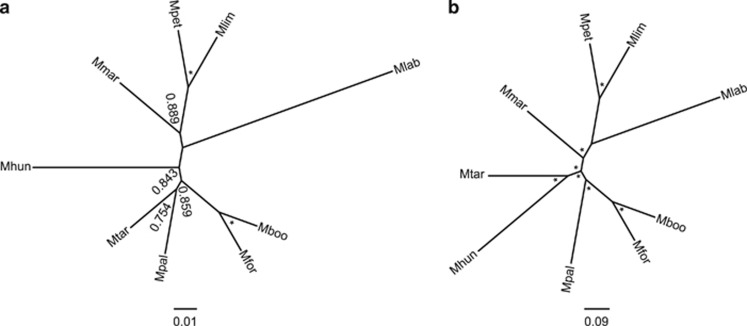
Figure 1.
Phylogeny of the Methanomicrobiales. (a) Neighbor-joining (NJ) reconstruction of the small subunit rRNA gene. (b) Bayesian reconstruction of core genome amino-acid sequences. Bootstrap support for NJ reconstruction is shown next to nodes. Asterisks adjacent to nodes indicated either 100% bootstrap support or a calculated posterior probability of 1. Mboo, M. boonei; Mfor, M. formicica; Mpal, M. palustris; Mtar, M. tarda; Mmar, M. marisnigri; Mpet, M. petrolearia; Mlim, M. limicola; Mhun, M. hungatei; Mlab, M. labreanum.
Reconstruction of gene flow dynamics
Reconstruction of gene flow dynamics was performed using COUNT (Csurös, 2010). Three categories of gamma distributed parameters were used for optimizing the rate parameters and gain, loss and duplication rates were allowed to vary between lineages.
Identification of horizontal gene transfer candidates
Putative horizontal gene transfer (HGT) candidates were screened for using three contrasting methods based on (i) detecting phylogenetically atypical proteins, (ii) detecting biases in dinucleotide frequencies and (iii) detecting biases in codon usage. Phylogenetically atypical protein sequences were detected using DarkHorse v1.4 (Podell and Gaasterland, 2007), using varying parameters to detect HGT acquisitions of different ages. When using DarkHorse, three filter thresholds (2%, 5% and 10%) were used and the definition of self was varied between four hierarchical levels of classification: (i) strain/species, (ii) genus, (iii) family and (iv) order. A normalized lineage probability index cutoff of 0.38 was applied, which was adequately stringent to examine HGT only from outside the domain Archaea. When a protein had a normalized lineage probability index <0.38 in at least two of the three filter thresholds, it was flagged as an HGT candidate. The age of HGT acquisition was estimated using the most recent definition of self at which the protein was flagged as an HGT candidate. A complementary analysis exploring the possible HGT history of a membrane-bound heterodisulfide reductase (HdrED) in Methanosphaerula palustris used a normalized lineage probability index cutoff of <0.85. IslandPath-DIMOB was used to detect HGT related with the co-occurrence of abnormal dinucleotide frequencies and mobility genes (Hsiao et al., 2003). SIGI-HMM as implemented in the Colombo package (Waack et al., 2006) was used to detect HGT based on codon usage bias. For analysis of the Methanolinea tarda NOBI-1 draft genome assembly, only contigs greater than 100 kb in length were analyzed by IslandPath-DIMOB.
Inference of gene function
Functional annotations were parsed from the genbank format genome sequences. Functions assigned by KEGG orthology were used as annotations for the genomes in KEGG (Ogata et al., 1999). Clusters of orthologous groups (COG) functional terms were parsed from the arCOG database (Wolf et al., 2012). For genomes not in the arCOG database, the category assigned was the best hit of a blastp query against the arCOG database using an e-value cutoff of 1e–10 and with at least 70% sequence overlap. Where necessary, the specificity of monovalent cation transporters was assessed using blast comparisons against experimentally verified transporters from bacteria.
Results
Pangenome and phylogenomics of the Methanomicrobiales
A total of 6749 predicted protein clusters were identified as part of the Methanomicrobiales' pangenome. Between 1012 and 1642 protein, clusters were shared by each pairwise combination of strains (Supplementary Table 1). In all, 832 were common to all strains, representing 30–51% of their coding sequences, and are hereafter termed the Methanomicrobiales' protein coding core genome. In all, 699 of the core clusters were present as single orthologs in all strains; 2429 clusters were found in more than one but not in all strains, while from 244 to 532 clusters per strain were found to be unique to each of the nine strains (Supplementary Data 1). Rarefaction curves showed the core genome is well defined with the current data set, while the variable portions of the pangenome will increase with the addition of more strains (Supplementary Figure 1).
The phylogeny within the order was examined since evolutionary history is inextricably linked with genomic level adaptive processes. The strains had between a minimum of 85% and a maximum of 96% SSU RNA identity on a pairwise basis. Phylogenies constructed with several data sets (small or large subunit rRNA genes, and concatenated amino-acid sequences of 437 core orthologous clusters; Supplementary Data 1) and methods (detailed in Materials and methods) showed overall similar tree topologies with some incongruences (Figure 1; Supplementary Figure 2). The grouping of the Methanoregulaceae strains together is supported but the exact branching order of M. palustris and M. tarda is unclear. The placement of Methanoculleus in the Methanomicrobiaceae family is uncertain, as Methanoculleus marisnigri did not always branch with Methanoplanus limicola and Methanolacinia petrolearia.
Gene flow dynamics in the Methanomicrobiales pangenome
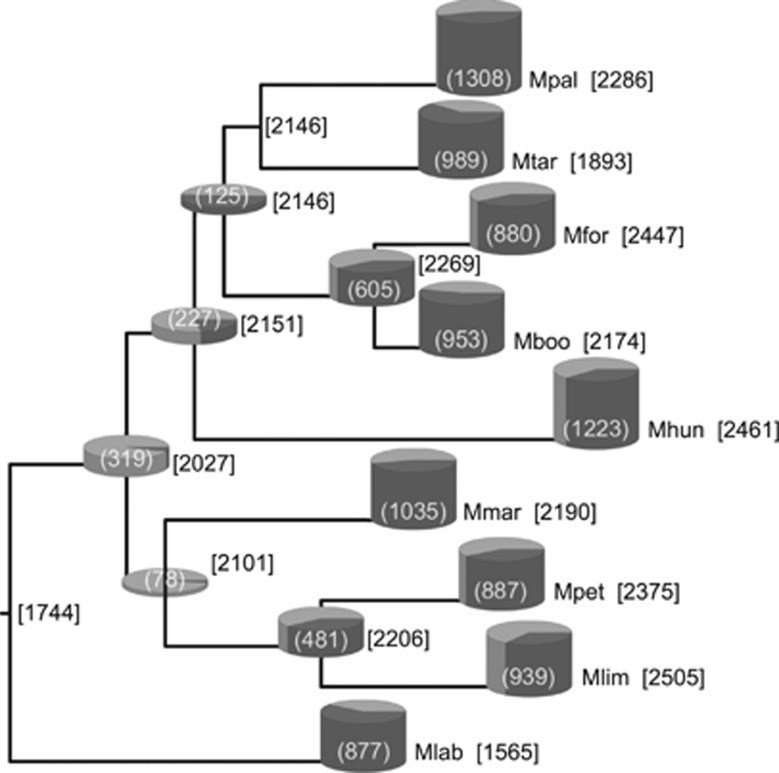
The rates of gene acquisitions and losses between phylogenetic nodes (in Figure 1a) are presented in Figure 2. The Methanomicrobiales progenitor was inferred to possess 1744 protein clusters. Since the origin of the order, Methanocorpusculum labreanum has undergone a net loss of protein clusters while the other strains evidenced net gains, although some alternated between periods of net gain and net loss (Figure 2). Since the estimation of gene loss and gain rates relies on a robust phylogenetic inference, this analysis was repeated using all unique tree topologies inferred while assessing the order's phylogenetic structure (Supplementary Figure 2). The same main findings were consistently observed across iterations, although exact gene gain and loss rates varied to a minor extent depending on the tree topology used (for instance, 1725–1779 protein clusters were estimated for the progenitor ancestor).
Figure 2.
Reconstruction of the ancestral genome content of the Methanomicrobiales. Numbers in square brackets indicate the protein cluster content of the adjacent node or extant species. The number presented in parentheses represents the combined gene gains and losses inferred to have occurred since the previous node. Gains (black portion) and losses (white portions) are proportionally scaled in cylinders. The absence of a cylinder indicates that <0.5 gains plus losses were inferred to have occurred. Abbreviations defined in Figure 1.
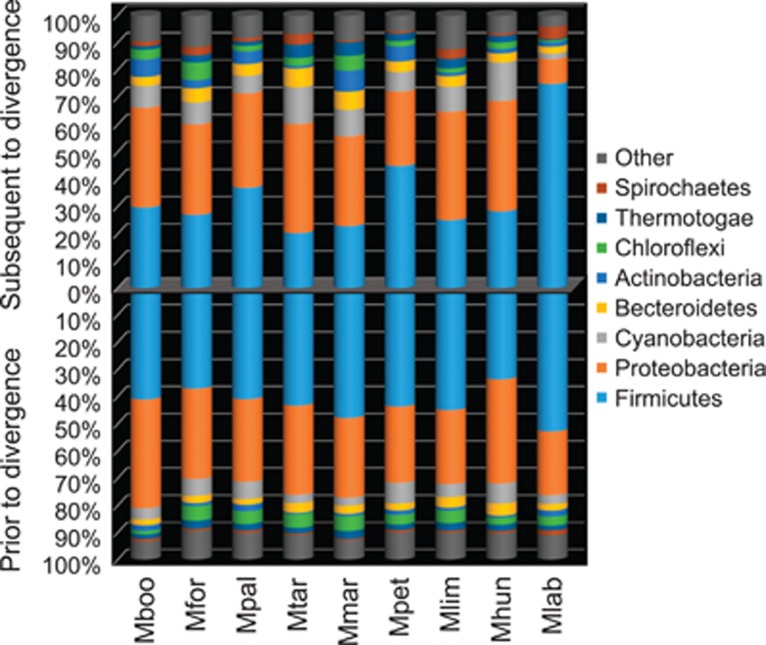
This test was coupled with the screening for HGT candidates, which is a key for its role in encoding novel products into recipients' genomes. The three methodologies used in the HGT screening showed significant variation (Supplementary Figure 3). This reflects the fact that screening for phylogenetically atypical proteins can detect acquisitions of different ages while sequence composition biases may smooth out over time following acquisition (Lawrence and Ochman, 1997). The strains putatively acquired between 19.2% and 31.9% of their coding sequences via HGT, primarily from groups within the Proteobacteria and Firmicutes (Figure 3). Despite gene flow dynamics evidencing genome reduction in M. labreanum, this strain had the highest fraction of HGT candidates relative to its genome's size (Supplementary Figure 4). M. labreanum also showed a different profile of putative HGT donors, with gene gains from the Firmicutes representing a larger fraction than those from the Proteobacteria (Figure 3).
Figure 3.
Phyla affiliations of most probable HGT donors. The phyla affiliations of putative bacterial HGT donors as inferred by DarkHorse to have occurred prior and subsequent to the divergence of the Methanomicrobiales order are presented. The numbers of HGT candidates are scaled to 100%. Abbreviations as in Figure 1.
Assessment for positive selection in the protein coding core genome and differential content analysis in variable genome
Patterns of positive selection in the 699 core protein clusters present as single orthologs only were assessed on the various branches within the phylogeny presented in (Figure 1a). In all, 269 of the 699 core orthologous protein clusters had evidence of positive selection at some point with the false positive detection rate controlled to 1% (Supplementary Data 2). Of these, 106 were not localized to only one branch of Figure 1a. There were between 13 and 39 instances of protein clusters showing evidence of positive selection on only one of each of the individual branches.
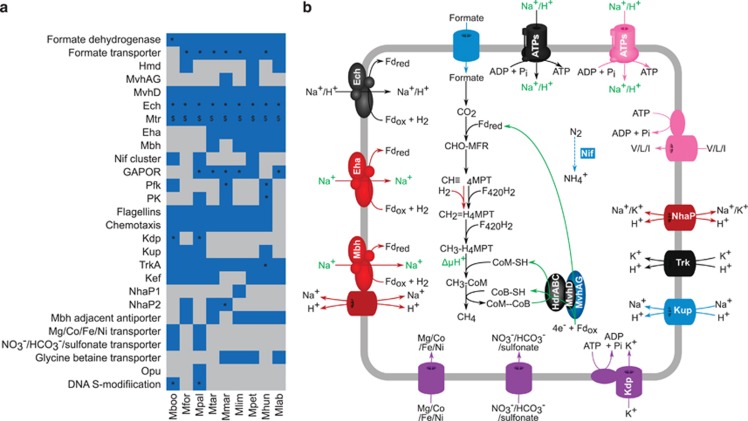
The differential content analysis of non-core protein clusters (Supplementary Data 1) revealed 60 clusters were found only in both peat-dwelling strains, 13 were only found in strains tolerant of greater than 0.5 m NaCl and 11 clusters were present only in strains isolated from bioreactors (Supplementary Table 2). Functional annotations of these genes suggested contrasting properties (Figure 4), although also a significant fraction did not present an annotation (57.4%) for better interpretation. The differential content analysis also revealed that M. labreanum lacks 96 protein clusters present in all other strains (Supplementary Table 3), while the other strains lacked only between 2 and 15 of such clusters. Notably, 14 of the 96 clusters absent in M. labreanum code for archaellum (archaeal flagellum) and chemotaxis proteins in the other order members, suggesting the loss of these functions.
Figure 4.
Adaptations of Methanomicrobiales to contrasting habitats. (a) Heatmap showing presence (blue) and absence (gray) patterns of various processes in the nine Methanomicrobiales isolates. ‘*' denotes HGT and ‘$' denotes positive selection evidence (in at least one subunit of multisubunit systems). (b) Cellular overview highlighting many of the processes evident in part (a). Features in black are present in all strains. Those in blue are present in some, but not all, strains with no relation to habitat. Features in purple are present in only M. boonei and M. palustris and are potentially adaptive to peat dwelling. Features in cyan are found only in bioreactor-dwelling strains. Processes in green require biochemical and/or genetic evidence to determine the nature of the electron source or translocated ion. ATPs, ATP synthase; V/L/I, valine/leucine/isoleucine; CHO-MFR, formyl-methanofuran; CH≡H4MPT, methenyl-tetrahydromethanopterin; CH2=MPT, methylene-H4MPT; CH3-H4MPT, methyl-tetrahydromethanopterin; Hmd, H2-dependent methylene-H4MPT dehydrogenase; Mtr, methyl-tetrahydromethanopterin S-methyltransferase; Mvh, F420-nonreducing hydrogenase; Hdr, heterodisulfide reductase; Ech, energy conserving hydrogenase; Eha, energy harvesting hydrogenase; Mbh, membrane-bound hydrogenase; Nif, nitrogen fixation; GAPOR, glyceraldehyde-3-phosphate ferredoxin oxidoreductase; Pfk, phosphofructokinase; PK, pyruvate kinase; Kdp, ABC-type K+ transporter; Kup, K+ uptake protein; TrkA, K+ uptake protein; Kef, K+ efflux transporter; NhaP, Na+/H+ antiporter; Opu, outer membrane porin; Fdrex/ox, reduced/oxidized ferredoxin; Pi, inorganic phosphate. Strain name abbreviations are defined in Figure 1.
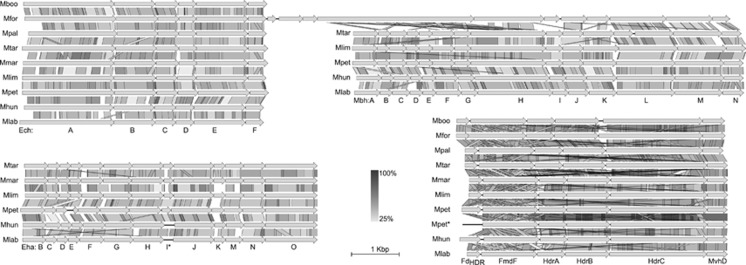
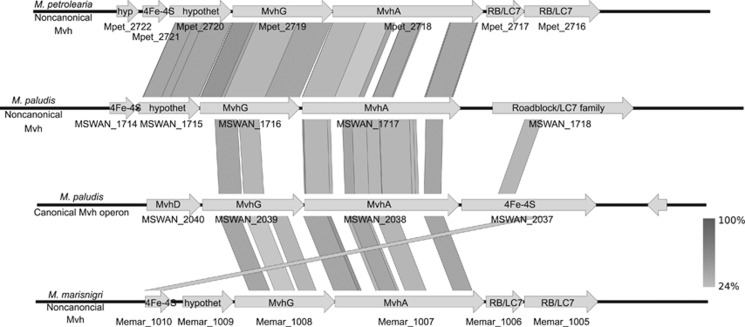
We also analyzed the distribution of central metabolism genes including methanogenesis, energy conservation and electron trafficking (Figure 4, Clusters ‘S' in Supplementary Data 1). Among these, three different membrane-bound hydrogenases, Ech (cluster Sα), Eha (cluster Sβ) and Mbh (cluster Sχ), were found. Only Ech was present in all strains (core) whereas Mbh and Eha were found only in six and five strains, respectively (Figures 4 and 5). A synteny analysis was performed on these membrane-bound hydrogenases and adjacent multisubunit Na+/H+ antiporters, if present, and on the HdrED genes that were all adjacent to genes encoding the D-subunit of the F420-nonreducing hydrogenase (Mvh) (cluster Sκ) (Figure 5). Furthermore, synteny analysis was also performed on the loci encoding MvhAG subunits (cluster Sλ) (Figure 6) present only in two strains, M. marisnigri and M. petrolearia (Figure 4). The results of these analyses (discussed below) support hypotheses on the use of H+- rather than Na+-transmembrane chemiosmotic gradients, and the potential for differences in electron bifurcation steps in the central metabolism of members of this order.
Figure 5.
Synteny of membrane-bound hydrogenases and HdrED. Ech and Mbh subunits are not encoded adjacent to each other, except in the instance shown for M. formicica. *M. petrolearia possesses two adjacent loci encoding the HdrED, with one of these loci lacking a ferredoxin encoding gene (FdHDR). The scale bars show the length corresponding to 1 kb and percent identity calculated from tblastx searches in grayscale. Abbreviations of strains are defined in Figure 1.
Figure 6.
Synteny of F420-nonreducing hydrogenases. The loci encoding Mvh subunits A and G from M. marisnigri and M. petrolearia are shown in comparison with canonical and non-canonical Mvh loci from Methanobacterium paludis. Locus tags are shown below the genes represented. Percent identities between loci were calculated using tblastx and are presented in grayscale. hyp, hypothetical protein; hypothet, hypothetical protein; RB/LC7, Roadblock/LC7 family protein.
Discussion
Pangenome and phylogenomics of the Methanomicrobiales
The Methanomicrobiales core' genome (832 genes) was found to be smaller than Methanocellales (1187 genes) (Lyu and Lu, 2015) but larger than Methanomassiliicoccales (658 genes) (Borrel et al., 2014). However, an in-depth analysis of order-level core genome across methanogenic Archaea is limited by the uneven sampling of taxonomic diversity and number of genomes within groups, plus variation in criteria when delineating clusters. Meanwhile, inference from core genome content allowed assessing the phylogenetic structure of the strains in this study, which highlight some discrepancies previously noted elsewhere. For instance, analysis of SSU rRNA by neighbor-joining places Methanospirillum hungatei near but outside the Methanoregulaceae family while likelihood-based analyses place M. hungatei within such family (Cadillo-Quiroz et al., 2009; Brambilla et al., 2010; Bräuer et al., 2011; Yashiro et al., 2011). It is known that when applying phylogenomics to distantly related organisms, the phylogenetic signal may become corrupted by long branch attraction artifacts (a particular issue for parsimony-based methods), through recombination and through substitution saturation (Philippe et al., 2011). Our reconstructions indicate that M. labreanum and M. hungatei had long terminal branch lengths to nodes with relatively short internal branch lengths (Figure 1), indicating that the placement of these strains is not fully resolved yet and requires a few more related genomes to reduce this possible source of phylogenetic incongruence. HGT effects on core genome could be considered as an additional source of incongruence. However, concatenated approaches and Bayesian reconstruction, as done here, have been proposed as robust to low HGT frequencies (Lapierre et al., 2014; Davidson et al., 2015). Nevertheless, the incongruent branching order of the families between different data sets and methods indicates that future revisions of phylogenetic relationships between these families are required.
Gene flow dynamics evidence a different life history for M. labreanum
Assessment of expansion and/or streamlining of a genome can reveal putative adaptations (Ochman et al., 2000; Lee and Marx, 2012). For instance, M. labreanum was the only strain in this study to evidence net gene loss since the divergence of the order. M. labreanum also evidenced differences in HGT, both in terms of acquisition rates (Supplementary Figure 4) and profile of HGT donor affiliations (Figure 3). These differences in M. labreanum's HGT history cannot be accounted for by a lack of Proteobacteria in its habitat as Proteobacteria members are present in tar pits (Kim and Crowley, 2007). Notably, the absence of archaellum and chemotaxis protein coding genes in M. labreanum is supported by phenotypic evidence showing the lack of motility or archaella under the microscope (Anderson et al., 2009b). More generally, the genome of M. labreanum was deficient in genes associated with COG categories V (defense mechanisms) and T (signal transduction mechanisms) compared with the other strains (Supplementary Figure 5). This loss of secondary functions suggests genome downsizing leading to niche specialization (Lee and Marx, 2012). However, M. labreanum's putative specialization is still unclear. For instance in other microbes, genome downsizing can sometimes indicate specialization to low substrate availability (Giovannoni et al., 2005) but M. labreanum's relatively rapid doubling time (t1/2=12.5 h) in pure culture indicates that it is capable of taking advantage of abundant substrate (H2) supply.
Adaptations to peatlands
Methanoregula boonei and M. palustris were isolated as representatives of dominant methanogenic clades of contrasting peatlands, from an ombrotrophic bog (Bräuer et al., 2006) and a minerotrophic fen (Cadillo-Quiroz et al., 2008), respectively. Both strains must cope with low levels of inorganic nutrients and low pH (Dettling et al., 2007), especially in the bog. Among the 60 clusters found only in peat-dwelling strains (Supplementary Table 2), an ATP-energized K+-transport system (Kdp) and two transport systems (one for cations and one for anions) of unknown specificity were evidenced. These three transport systems, along with a complete nitrogen fixation (nif) cluster (cluster Sδ, Supplementary Data 1) shared with M. petrolearia, suggest that adaptation to low availability of inorganic nutrients is important for peat dwelling. Other protein clusters unique to the two peat-dwelling organisms include various metabolism-related proteins, putative heat-shock proteins/chaperones and a DNA sulfur modification system (possibly a defense-oriented host-specific restriction system; Xu et al., 2010). Both peat-dwelling strains also appeared to show specializations in their iron uptake systems. While the other seven strains have gene clusters encoding several (two to nine) ABC-type iron transporters, M. boonei and M. palustris possess only one each. The fact that the only one type of substrate-binding protein is seen in the iron transporters of each peatland strain, while all other strains possess two to eight different substrate-binding domains in their iron transporters suggest that M. boonei and M. palustris have specialized iron acquisition systems.
Additionally, Kdp was identified as an HGT candidate of high importance for adaptation to peatland environments in M. boonei (Bräuer et al., 2015; Cadillo-Quiroz et al., 2015) and also in M. palustris (Bräuer et al., 2015; Cadillo-Quiroz et al., 2015). In peat soils, K+ concentrations are low (3–25 μm) (Dettling et al., 2007), but high levels of K+ are required intracellularly (up to 154 mm in M. hungatei GP 1; Sprott and Jarrell, 1981) for roles in pH homeostasis, cell turgor and membrane potential (Epstein, 2003). The two sets in M. boonei and one set of Kdp operons in M. palustris likely allow both organisms to overcome low K+ concentrations present in oligotrophic and minerotrophic peats. Kdp has a high affinity for K+ (Km=2 μm in Escherichia coli K-12, with 37–63% protein identity to Kdp subunits of M. boonei and M. palustris) and has been shown to concentrate K+ in the cytoplasm when extracellular concentrations were as low as 50 nm (Ballal et al., 2007). Our analyses of gene flow dynamics indicated that Kdp was likely acquired subsequent to the divergence of the Methanoregulaceae family, in which the presence of Kdp operons in two peat-dwelling strains, M. boonei and M. palustris, and its absence in two bioreactor-dwelling ones, Methanoregula formicica and M. tarda, suggest that peatlands likely favor methanogens that acquired Kdp. As for the remaining strains in this study, all nine possess Trk- and some have Kup-type lower affinity K+-transport systems (Figure 4), both of which have Km values around the order of 1 mm (Trchounian and Kobayashi, 1999; Epstein, 2003).
Our analyses indicated that the peatland methanogens, which are known to be highly sensitive to Na+ (Cadillo-Quiroz et al., 2009; Bräuer et al., 2011), are the only ones of this Methanomicrobiales study to not encode Na+-transporters. All other strains have a NhaP-type Na+/H+ antiporter and/or a multisubunit Na+/H+ antiporter adjacent to a membrane-bound hydrogenase (Figure 4). This corroborates the hypothesis that M. boonei has adapted to the low Na+/high H+ challenges of peat dwelling by using H+ rather than Na+ transmembrane chemiosmotic gradients (Bräuer et al., 2015). Up to this point, based on studies of Methanobacteriales and Methanococcales methanogens, it was thought that all cytochrome-deficient hydrogenotrophic methanogens conserve energy using a Na+ transmembrane chemiosmotic gradient, which is generated by tetrahydromethanopterin S-methyltransferase (Mtr) and consumed by a Na+-translocating ATP synthase (Schlegel and Müller, 2013). According to the new hypothesis, Methanomicrobiales members are expected to generate a H+ gradient using Mtr (as supported by previous analyses of the MtrE subunit; Bräuer et al., 2015), and consume the H+ gradient, thus generating ATP, using a H+/Na+ promiscuous ATP synthase (Schlegel et al., 2012; Bräuer et al., 2015).
Adaptations to bioreactors and saline environments
The strains from bioreactors, M. tarda, M. formicicum and M. hungatei, all possessed another ATP synthase (cluster Sɛ, in Supplementary Data 1) (in addition to the one core to the order), a branched-chain amino-acid transporter (cluster Sφ), as well as a universal stress protein (cluster Sγ) not seen in any of the other strains. M. tarda and M. hungatei shared yet another ATP synthase (cluster Sη) with M. limicola. Although all ATP synthases found in this order are predicted as Na+/H+ promiscuous type (Grüber et al., 2014), alternative ATP synthases can have adaptive value for different growth rates or for different concentrations of Na+ and H+. The presence of a branched-chain amino-acid transporter indicates an ability to take advantage of substrates likely more available in eutrophic habitats, or environments with high microbial cell density and, possibly, significant dead microbial mass-turnover as in bioreactors.
All three Methanomicrobiaceae strains were isolated from saline environments (Romesser et al., 1979; Wildgruber et al., 1982; Ollivier et al., 1997) and are also the most saline tolerant strains (0.69–0.86 M maximum NaCl tolerance) in this study. M. labreanum is also moderately NaCl tolerant (up to 0.51 m NaCl), while bioreactor strains are less tolerant to NaCl (up to 0.26 m NaCl). The peatland strains are the most NaCl sensitive of the strains in this study, tolerating no >0.1 m NaCl (Bräuer et al., 2011). Microorganisms must maintain an osmotic pressure at least equivalent to their surroundings, which may be achieved by (i) the accumulation of salts, usually K+ and Cl− and known as the ‘salt-in strategy' or (ii) by the accumulation of organic osmotic solutes (Oren, 2015). Since all strains possess various K+-transporters, regardless of their NaCl tolerances (Figure 4), it would seem that the accumulation of K+ is not necessarily the means by which Methanomicrobiales strains tolerate more or less NaCl. A putative glycine-betaine transporter was found in all three Methanomicrobiaceae strains and also in M. labreanum (cluster Sϕ) and not in any of the other strains (Figure 4). This organic solute transporter is proposed as a candidate for mediating NaCl/osmotic stress tolerance in the Methanomicrobiales. As the media in which the maximum NaCl tolerances were determined for the three Methanomicrobiaceae strains and M. labreanum contained complex ingredients (for example, yeast extract and peptone hydrolysate) it is possible that these media contained suitable osmotic solutes for cytoplasmic osmolarity homeostasis. However, de novo synthesis of osmotically active solutes cannot be discounted.
It is important to consider that the genome availability of NaCl tolerant Methanomicrobiales is rather limited, and no genome sequences are currently available for strains within the most NaCl tolerant family of the order, the Methanocalculaceae, or for Methanomicrobiaceae strains isolated from wetlands or bioreactors (non-marine environments), hence it is likely that further genomic strategies to cope with NaCl/osmotic stress may yet become apparent within the Methanomicrobiales.
Analysis of primary metabolic genes reveals different lifestyles within the Methanomicrobiales
In hydrogenotrophic methanogens membrane-bound hydrogenases, such as Ech, Eha and Mbh, would be expected to catalyze the endergonic reduction of ferredoxin with H2 at the expense of a transmembrane chemiosmotic gradient. Considering (i) our hypothesis that M. boonei and M. palustris utilize H+- rather than Na+-transmembrane chemiosmotic gradients, (ii) that M. boonei and M. palustris are the only strains in this study that appear to lack Na+/H+ antiporters and (iii) that Ech is the only membrane-bound hydrogenase apparent in the genomes of M. boonei and M. palustris, it could be expected that Ech is capable of translocating H+. Our results indicate that multisubunit Na+/H+ antiporters are adjacent to Mbh (where present) and adjacent to Ech in M. formicica (Figure 5). This suggests that some of the Methanomicrobiales could concurrently use Na+ and H+ transmembrane chemiosmotic gradients as proposed in the Methanosarcinales (Schlegel and Müller, 2013). Ultimately, experimental investigations into the nature of ions translocated by the membrane-bound hydrogenases (Ech, Eha and Mbh), Mtr and ATP synthases will be required to understand the energy conservation and transduction mechanisms in the Methanomicrobiales. As a consequence of having only one membrane-bound hydrogenase, it would be expected that M. boonei and M. palustris are incapable of using separate membrane-bound hydrogenases to discriminate between anabolic and anaplerotic pools of ferredoxin as seen in Methanococcus maripaludis (Lie et al., 2012), though they may possess another means of such discrimination.
In addition to differences in energy conservation pathways, these analyses also evidenced putative differences in the pathways of electron flow. Balancing cellular pools of reducing equivalents (H2, co-factor F420 (F420/F420H2) and ferredoxin) is important as these reducing equivalents are required during several steps of methanogenesis and anabolic metabolism, and moreover, maintaining a suitably low H2 concentration is important to the thermodynamic constraints of syntrophic metabolism (Schink, 1997; Walker et al., 2012). An F420-reducing alcohol dehydrogenase (MMPG_2660) was found in M. marisnigri, M. petroleariapetrolearius and M. tarda. The former two are verified to be able to utilize secondary alcohols (Maestrojuán et al., 1990; Ollivier et al., 1997), while M. tarda was unable to utilize 2-propanol and 2-butanol (Imachi et al., 2008). All strains had a cytoplasmic F420-reducing hydrogenase, but they showed differential gene content at other steps of methanogenesis. In distantly related orders of cytochrome-deficient methanogens, the Methanobacteriales and Methanococcales, it was shown, during hydrogenotrophic methanogenesis, that two pairs of electrons are bifurcated in a process whereby the exergonic reduction of the coenzyme M-coenzyme B heterodisulfide is energetically coupled to the endergonic production of formyl-methanofuran, in a protein complex that incorporates cytoplasmic heterodisulfide reductase (HdrABC), formyl-methanofuran dehydrogenase, subunit D of Mvh and one of either formate dehydrogenase (Fdh), MvhABG, or an F420-reducing hydrogenase (Costa et al., 2010) (Figure 4). The proximity of MvhD to HdrABC (Figure 5) suggests that this subunit could interface with HdrABC during electron bifurcation as previously demonstrated in the Methanococcales order (Costa et al., 2013). However, the organization of Mvh subunits (Figure 6) and lack of homologs to MvhB differs to what has been observed in M. maripaludis (Costa et al., 2010) where experimental evidence showed that proximally encoded MvhABGD subunits (canonical enzyme system) mediated electron bifurcation. Overall, this indicates that there are profound differences in the mechanisms of electron bifurcation (particularly in the generation of low potential electrons) within the Methanomicrobiales and between the Methanomicrobiales and other orders of cytochrome-deficient methanogens. The implications of these differences in MvhABG content are not clear, as the pathways of electron bifurcation in hydrogenotrophic methanogens are still being elucidated and have been studied only in Methanobacteriales and Methanococcales. However, it may be of particular interest that Fdh increases in relative abundance in electron bifurcating complexes at the expense of MvhABG in M. maripaludis when hydrogen is in limiting supply (Costa et al., 2013). This is interesting because Fdh is encoded in the Methanomicrobiales' core genome, including M. boonei, which is the only strain to lack a formate transporter and be unable to utilize formate as a methanogenic substrate (Bräuer et al., 2011). This suggests a possible role for Fdh in electron bifurcation in the Methanomicrobiales, although experimental evidence is still pending.
The genome of M. palustris encodes a putative alternative membrane-bound HdrED, a key enzyme complex of the cytochrome containing Methanosarcinales (Buan and Metcalf, 2010). Homology search results indicate that M. palustris is the only archaeon outside the Methanosarcinales to possess a putative HdrE. HGT and maximum likelihood analyses (Supplementary Figure 6) indicate that these genes could have been horizontally acquired from a Methanosaeta lineage, although more HdrED-containing genomes will improve this test. In terms of potential for functionality, there did not appear to be any means of synthesizing the methanophenazine electron carriers required by HdrED encoded in the genome of M. palustris, suggesting that HdrED may not have a canonical role in methanogenesis of M. palustris. However, the role and evolutionary history of HdrED, or HdrD alone, across methanogens is intriguing in light of this, plus new findings of HdrD presence (Borrel et al., 2014) and active functioning (in the absence of HdrE) as HdrED for energy conservation in Methanomassiliicoccus (Kröninger et al., 2016).
Two methylene-tetrahydromethanopterin (methylene-H4MPT) dehydrogenases, the F420-dependent Mtd and the H2-dependent Hmd, catalyze the same step of methanogenesis, the reduction of methenyl-H4MPT to methylene-H4MPT. Expression studies have shown that one of Mtd or Hmd takes on quantitatively greater importance in response to H2 limitation, rapid growth rate and syntrophic growth (Hendrickson et al., 2007; Kato et al., 2008; Enoki et al., 2011; Walker et al., 2012). In this study, Mtd was found in all strains, while Hmd was only found in three (Figure 4). This suggests that the adaptation to low H2 concentrations afforded by Mtd is highly relevant for Methanomicrobiales under syntrophic lifestyles. Moreover, codon alignments of five F420-dependent genes (Mtd, methylene-H4MPT reductase, and three out of four subunits of the cytoplasmic F420-reducing hydrogenase) and three subunits of the energy conserving Mtr evidenced positive selection (Supplementary Data 2). This indicates that electron trafficking and energy conservation systems are subject to selection in these organisms. When also taking into account that many genes involved in central anabolic pathways such as acetate assimilation, amino-acid biosynthesis and gluconeogenesis are also subject to positive selection (Supplementary Data 2), it would appear that both energy metabolism and growth-related systems are under selective pressure. Such selective pressure can derive from syntrophic interactions, which can be subject to tight thermodynamic constraints. It is known that methanogens undergo gene regulation programs, impacting greatly on methanogenesis (energy production) and anabolic reactions (growth), to optimize syntrophic interactions (Shimoyama et al., 2009; Walker et al., 2012).
Conclusions
Here, we presented predicted differences in energy conservation, electron trafficking and other metabolic functions from the genomic make up of nine organisms within the Methanomicrobiales. Although different evidence lines converge in several of such putative predictions (Figure 4a), they require future examination via biochemical/genetic studies to understand the physiology of Methanomicrobiales as distinct from other methanogens. The importance of this point is highlighted by the fact that seemingly subtle differences in energy conservation and electron trafficking pathways can be central to different lifestyles in contrasting environments, as seen, for example, in freshwater versus marine Methanosarcina spp. (Ferry and Lessner, 2008; Kulkarni et al., 2009).
Comparative genomic analyses at the order level have allowed the proposition of a number of possible adaptations and differences across Methanomicrobiales strains from distinct taxonomic families and environments. The results of differential gene content, HGT, selection signatures and gene flow dynamics allow elaborating several hypotheses to further our understanding of the adaptations of methanogens to vastly different ecosystems.
Acknowledgments
The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported under Contract No. DE-AC02-05CH11231. We thank the three anonymous reviewers for their important comments, suggestion and assessment for the betterment of this manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD et al. (2009. a). Genomic characterization of methanomicrobiales reveals three classes of methanogens. PLoS One 4: e5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson IJ, Sieprawska-Lupa M, Goltsman E, Lapidus A, Copeland A, Glavina Del Rio T et al. (2009. b). Complete genome sequence of Methanocorpusculum labreanum type strain Z. Stand Genomic Sci 1: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet J-C, Barberan A, Casamayor EO. (2009). Global ecological patterns in uncultured Archaea. ISME J 4: 182–190. [DOI] [PubMed] [Google Scholar]
- Ballal A, Basu B, Apte S. (2007). The Kdp-ATPase system and its regulation. J Biosci 32: 559–568. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. [Google Scholar]
- Bialek K, Cysneiros D, O'Flaherty V. (2013). Low-temperature (10 °C) anaerobic digestion of dilute dairy wastewater in an EGSB bioreactor: Microbial community structure, population dynamics, and kinetics of methanogenic populations. Archaea 2013: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla E, Djao ODN, Daligault H, Lapidus A, Lucas S, Hammon N et al. (2010). Complete genome sequence of Methanoplanus petrolearius type strain (SEBR 4847). Stand Genomic Sci 3: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer S, Cadillo-Quiroz H, Yashiro E, Yavitt JB, Zinder SH. (2006). Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442: 192–194. [DOI] [PubMed] [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. (2011). Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol 61: 45–52. [DOI] [PubMed] [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Kyrpides N, Woyke T, Goodwin L, Detter C et al. (2015). Genome of Methanoregula boonei 6A8 reveals adaptations to oligotrophic peatland environments. Microbiology 161: 1572–1581. [DOI] [PubMed] [Google Scholar]
- Brugère J-F, Borrel G, Gaci N, Tottey W, O'Toole PW, Malpuech-Brugère C. (2014). Archaebiotics. Gut Microbes 5: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buan NR, Metcalf WW. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol Microbiol 75: 843–853. [DOI] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Yashiro E, Yavitt JB, Zinder SH. (2008). Characterization of the archaeal community in a minerotrophic fen and terminal restriction fragment length polymorphism-directed isolation of a novel hydrogenotrophic methanogen. Appl Environ Microbiol 74: 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Yavitt JB, Zinder SH. (2009). Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. Int J Syst Evol Microbiol 59: 928–935. [DOI] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Browne P, Kyrpides N, Woyke T, Goodwin L, Detter C et al. (2015). Complete genome sequence of Methanosphaerula palustris E1-9CT, a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. Genome Announc 3: e01280–01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA 107: 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa KC, Lie TJ, Xia Q, Leigh JA. (2013). VhuD facilitates electron flow from H2 or formate to heterodisulfide reductase in Methanococcus maripaludis. J Bacteriol 195: 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurös M. (2010). Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26: 1910–1912. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R, Vachaspati P, Mirarab S, Warnow T. (2015). Phylogenomic species tree estimation in the presence of incomplete lineage sorting and horizontal gene transfer. BMC Genomics 16: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettling MD, Yavitt JB, Cadillo-Quiroz H, Sun C, Zinder SH. (2007). Soil-methanogen interactions in two peatlands (Bog, Fen) in central New York State. Geomicrobiol J 24: 247–259. [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2: 414–424. [DOI] [PubMed] [Google Scholar]
- Enoki M, Shinzato N, Sato H, Nakamura K, Kamagata Y. (2011). Comparative proteomic analysis of Methanothermobacter themautotrophicus ΔH in pure culture and in co-culture with a butyrate-oxidizing bacterium. PLoS One 6: e24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein W. (2003). The roles and regulation of potassium in bacteria. Progress in Nucleic Acid Research and Molecular Biology. Academic Press, pp 293–320. [DOI] [PubMed] [Google Scholar]
- Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350: 434–438. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). Phylip—phylogeny inference package. Cladistics 5: 164–166. [Google Scholar]
- Ferry JG, Smith PH, Wolfe RS. (1974). Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int J Syst Evol Micr 24: 465–469. [Google Scholar]
- Ferry JG, Lessner DJ. (2008). Methanogenesis in marine sediments. Ann NY Acad Sci 1125: 147–157. [DOI] [PubMed] [Google Scholar]
- Ferry JG. (2011). Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr Opin Biotechnol 22: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 188: 642–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J-L, Ollivier B, Whitman W. (2006) The order Methanomicrobiales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds), The Prokaryotes. Springer: New York, USA, pp 208–230. [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Grüber G, Manimekalai MSS, Mayer F, Müller V. (2014). ATP synthases from archaea: The beauty of a molecular motor. Biochim Biophys Acta 1837: 940–952. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Haydock AK, Moore BC, Whitman WB, Leigh JA. (2007). Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc Natl Acad Sci USA 104: 8930–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W, Wan I, Jones SJ, Brinkman FSL. (2003). IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19: 418–420. [DOI] [PubMed] [Google Scholar]
- Imachi H, Sakai S, Sekiguchi Y, Hanada S, Kamagata Y, Ohashi et al. (2008). Methanolinea tarda gen. nov., sp. nov., a methane-producing archaeon isolated from a methanogenic digester sludge. Int J Syst Evol Microbiol 58: 294–301. [DOI] [PubMed] [Google Scholar]
- Kato S, Kosaka T, Watanabe K. (2008). Comparative transcriptome analysis of responses of Methanothermobacter thermautotrophicus to different environmental stimuli. Environ Microbiol 10: 893–905. [DOI] [PubMed] [Google Scholar]
- Kim J-S, Crowley DE. (2007). Microbial diversity in natural asphalts of the Rancho La Brea Tar Pits. Appl Environ Microbiol 73: 4579–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröninger L, Berger S, Welte C, Deppenmeier U. (2016). Evidence for the involvement of two heterodisulfide reductases in the energy-conserving system of Methanomassiliicoccus luminyensis. FEBS J 283: 472–483. [DOI] [PubMed] [Google Scholar]
- Kulkarni G, Kridelbaugh DM, Guss AM, Metcalf WW. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc Natl Acad Sci USA 106: 15915–15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre P, Lasek-Nesselquist E, Gogarten JP. (2014). The impact of HGT on phylogenomic reconstruction methods. Brief Bioinform 15: 79–90. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. (1997). Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol 44: 383–397. [DOI] [PubMed] [Google Scholar]
- Lee M-C, Marx CJ. (2012). Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet 8: e1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. (2012). Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci USA 109: 15473–15478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Csuros M, Hughes AL, Moran MA. (2013). Evolution of divergent life history strategies in marine alphaproteobacteria. MBio 4: e00373–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Z, Lu Y. (2015). Comparative genomics of three Methanocellales strains reveal novel taxonomic and metabolic features. Environ Microbiol Rep 7: 526–537. [DOI] [PubMed] [Google Scholar]
- Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS et al. (2006). The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within Methanosarcinal Genomes. J Bacteriol 188: 7922–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrojuán GM, Boone DR, Xun L, Mah RA, Zhang L. (1990). Transfer of Methanogenium bourgenseMethanogenium marisnigriMethanogenium olentangyi, and Methanogenium thermophilicum to the genus Methanoculleus gen. nov., Emendation of Methanoculleus marisnigri and Methanogenium, and description of new strains of Methanoculleus bourgense and Methanoculleus marisnigri. Int J Syst Bacteriol 40: 117–122. [Google Scholar]
- Mihajlovski A, Alric M, Brugère J-F. (2008). A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res Microbiol 159: 516–521. [DOI] [PubMed] [Google Scholar]
- Nobu MK, Narihiro T, Kuroda K, Mei R, Liu W-T. (2016). Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J e-pub ahead of print 4 March 2016 doi:doi:10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed]
- Notredame C, Higgins DG, Heringa J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollivier B, Cayol JL, Patel BKC, Magot M, Fardeau ML, Garcia JL. (1997). Methanoplanus petrolearius sp. nov., a novel methanogenic bacterium from an oil-producing well. FEMS Microbiol Lett 147: 51–56. [DOI] [PubMed] [Google Scholar]
- Oren A. (2015). Halophilic microorganisms and adaptation to life at high salt concentrations—evolutionary aspects. In: Bakermans C (ed), Microbial Evolution under Extreme Conditions. De Gruyter: Berlin, Boston, pp 97–110. [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G et al. (2011). Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol 9: e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T. (2007). DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol 8: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwantini E, Torto-Alalibo T, Lomax J, Setubal JC, Tyler BM, Mukhopadhyay B. (2014). Genetic resources for methane production from biomass described with the Gene Ontology. Front Microbiol 5: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romesser JA, Wolfe RS, Mayer F, Spiess E, Walther-Mauruschat A. (1979). Methanogenium, a new genus of marine methanogenic bacteria, and characterization of Methanogenium cariaci sp. nov. and Methanogenium marisnigri sp. nov. Arch Microbiol 121: 147–153. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Imachi H, Hanada S, Ohashi A, Harada H, Kamagata Y. (2008). Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I', and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol 58: 929–936. [DOI] [PubMed] [Google Scholar]
- Schink B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Reviews 61: 262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel K, Leone V, Faraldo-Gómez JD, Müller V. (2012). Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci USA 109: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel K, Müller V. (2013). Evolution of Na+ and H+ bioenergetics in methanogenic archaea. Biochem Soc Trans 41: 421–426. [DOI] [PubMed] [Google Scholar]
- Shimoyama T, Kato S, Si Ishii, Watanabe K. (2009). Flagellum mediates symbiosis. Science 323: 1574. [DOI] [PubMed] [Google Scholar]
- Sprott GD, Jarrell KF. (1981). K+, Na+, and Mg2+ content and permeability of Methanospirillum hungatei and Methanobacterium thermoautotrophicum. Can J Microbiol 27: 444–451. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591. [DOI] [PubMed] [Google Scholar]
- Trchounian A, Kobayashi H. (1999). Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett 447: 144–148. [DOI] [PubMed] [Google Scholar]
- van Dongen SM. (2000) Graph Clustering by Flow Simulation. University of Utrecht, Netherlands. [Google Scholar]
- Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF et al. (2006). Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He Z, Hendrickson EL et al. (2012). Functional responses of methanogenic archaea to syntrophic growth. ISME J 6: 2045–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildgruber G, Thomm M, König H, Ober K, Richiuto T, Stetter K. (1982). Methanoplanus limicola, a plate-shaped methanogen representing a novel family, the methanoplanaceae. Arch Microbiol 132: 31–36. [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV. (2012). Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. (2003). An index of substitution saturation and its application. Mol Phylogenet Evol 26: 1–7. [DOI] [PubMed] [Google Scholar]
- Xia X, Lemey P. (2009) Assessing substitution saturation with DAMBE. In: Lemey P, Salemi M, Vandamme A-M (eds), The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. Cambridge University Press, Cambridge. [Google Scholar]
- Xia X. (2013). DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol 30: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yao F, Zhou X, Deng Z, You D. (2010). A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res 38: 7133–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen H, Zhou R. (2011). Genome-wide evidence for positive selection and recombination in Actinobacillus pleuropneumoniae. BMC Evol Biol 11: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yashiro Y, Sakai S, Ehara M, Miyazaki M, Yamaguchi T, Imachi H. (2011). Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int J Syst Evol Microbiol 61: 53–59. [DOI] [PubMed] [Google Scholar]
- Youngblut ND, Wirth JS, Henriksen JR, Smith M, Simon H, Metcalf WW et al. (2015). Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment. ISME J 9: 2191–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Boone DR, Mah RA, Boone JE, Xun L. (1989). Isolation and characterization of Methanocorpusculum labreanurn sp. nov. from the LaBrea tar pits. Int J Syst Evol Micr 39: 10–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.