Abstract
In the past two decades, the study of oxygen-independent degradation of widely abundant aromatic compounds in anaerobic bacteria has revealed numerous unprecedented enzymatic principles. Surprisingly, the organisms, metabolites and enzymes involved in the degradation of o-phthalate (1,2-dicarboxybenzene), mainly derived from phthalate esters that are annually produced at the million ton scale, are sparsely known. Here, we demonstrate a previously unknown capacity of complete phthalate degradation in established aromatic compound-degrading, denitrifying model organisms of the genera Thauera, Azoarcus and ‘Aromatoleum'. Differential proteome analyses revealed phthalate-induced gene clusters involved in uptake and conversion of phthalate to the central intermediate benzoyl-CoA. Enzyme assays provided in vitro evidence for the formation of phthaloyl-CoA by a succinyl-CoA- and phthalate-specific CoA transferase, which is essential for the subsequent oxygen-sensitive decarboxylation to benzoyl-CoA. The extreme instability of the phthaloyl-CoA intermediate requires highly balanced CoA transferase and decarboxylase activities to avoid its cellular accumulation. Phylogenetic analysis revealed phthaloyl-CoA decarboxylase as a novel member of the UbiD-like, (de)carboxylase enzyme family. Homologs of the encoding gene form a phylogenetic cluster and are found in soil, freshwater and marine bacteria; an ongoing global distribution of a possibly only recently evolved degradation pathway is suggested.
Introduction
Aromatic compounds are highly abundant in nature, mainly as secondary plant metabolites (for example lignin), aromatic amino acids or components of crude oil (Fuchs et al., 2011). Many mono- and polycyclic aromatic compounds are known as environmental pollutants that are often harmful to the environment and toxic or cancerogenic for humans (Satyanarayana and Johri, 2012). The complete mineralization of aromatic compounds to CO2 is only accomplished by microorganisms (Fuchs et al., 2011).
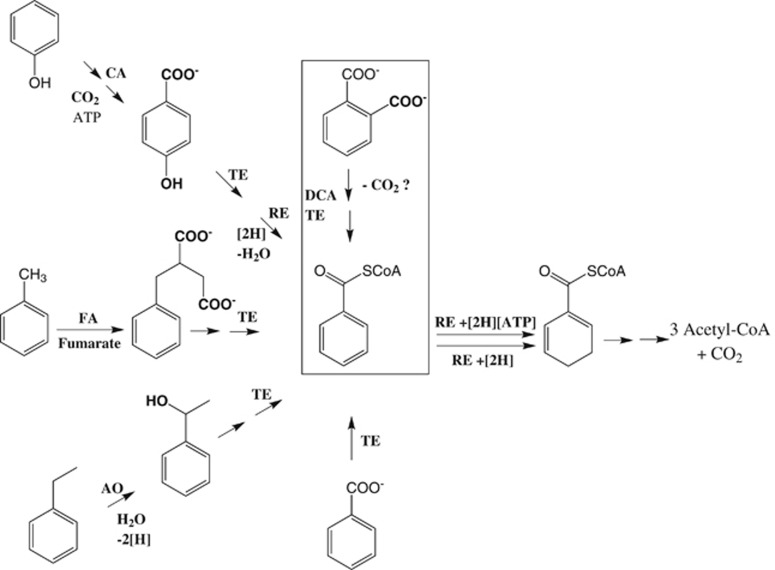
Under oxic conditions, oxygenases play a major role in the degradation of aromatic compounds, and are involved in ring/alkyl side chain hydroxylations, ring dearomatization and cleaving reactions. These processes are no option for bacteria at anoxic sites such as aquifers with a high carbon load or freshwater and marine sediments. In aromatic compounds-degrading anaerobes, most monocyclic aromatic compounds are channeled into the central intermediate benzoyl-coenzyme A (benzoyl-CoA). In the past two decades, the enzymes involved in these reactions have been studied in vitro to some detail, many of which follow unprecedented enzymatic principles (Figure 1; for recent reviews see Carmona et al., 2009; Fuchs et al., 2011; Philipp and Schink, 2012; Boll et al., 2014; Buckel et al., 2014). Benzoyl-CoA is finally dearomatized to a conjugated dienoyl-CoA either by ATP-dependent (Boll and Fuchs, 1995) or ATP-independent (Weinert et al., 2015) benzoyl-CoA reductases. In spite of the insights obtained in the oxygen-independent biomineralization of most environmentally relevant monocyclic aromatics, the anaerobic degradation of phthalate (1,2-dicarboxybenzene) has remained largely unknown.
Figure 1.
Selected characteristic reaction types involved in the anaerobic degradation of monocyclic aromatic compounds. CA=carboxylation, FA=fumarate addition; AO=anoxic oxidation (hydroxylation with water); TE=thioesterification; RE=reduction either for dehydroxylation or dearomatization. The proposed decarboxylation (DCA) of phthalate finally resulting in benzoyl-CoA formation (solid line rectangle) has not been demonstrated so far.
Phthalate predominantly derives from biodegradation of phthalic acid esters (PAE) via sequential hydrolysis by esterases; it is therefore considered as a xenobiotic compound (Engelhardt and Wallnöfer, 1978; Benckiser and Ottow, 1982; Shelton et al., 1984; Vega and Bastide, 2003; Maruyama et al., 2005). PAE are used worldwide as plasticizers to enhance flexibility and other technical properties of crude oil-derived materials (Mersiowsky et al., 2001). Most commonly they are incorporated into high-molecular weight polymers such as polyvinyl chloride but are also found in other consumer products (Latini, 2005). The annual PAE production is in the range of 5–8 million tons (Mackintosh et al., 2006; Net et al., 2015). Several studies of potential adverse health effects of PAE suggest an endocrine-disrupting activity (Caldwell, 2012). PAE are not covalently bound to polymers, and can easily migrate into the environment during manufacturing, use and disposal of plastics (Staples et al., 1997; Mersiowsky et al., 2001; Fujii et al., 2003; Afshari et al., 2004).
Biodegradation by microorganisms is considered to be the main PAE remediation process (Cousins et al., 2003; Huang et al., 2013), and has predominantly been studied in aerobic cultures (Liang et al., 2008; Gao and Wen, 2016). Degradation is initiated by the hydrolysis of PAE to phthalate and alcohols by esterases. Phthalate is subsequently converted by dioxygenases and dehydrogenases either into 3,4-dihydroxyphtalate in Gram-positive bacteria (Eaton and Ribbons, 1982) or into 4,5-dihydroxyphthalate in Gram-negative bacteria (Keyser et al., 1976; Nomura et al., 1992). The formation of a phenolic functionality in ortho or para to a carboxyl-group is essential for the subsequent decarboxylation as it contributes to the stabilization of the negatively charged transition state. Both pathways yield the central intermediate 3,4-dihydroxybenzoate (protocatechuate), which is then cleaved by well-known ring-cleaving dioxygenases (Eaton and Ribbons, 1982; Taylor and Ribbons, 1983).
Under anoxic conditions, the PAE biodegradation rate is largely decreased resulting in PAE accumulation at anoxic sites. The initial steps involving PAE hydrolysis to phthalate and alcohols are identical under oxic and anoxic conditions. However, the anaerobic degradation of phthalate has to follow completely different principles, and is considered as rate-limiting step of anaerobic PAE degradation (Gao and Wen, 2016). To date, only a fermenting Pelotomaculum strain (Qiu et al., 2006) and two nitrate-reducing species (Nozawa and Maruyama, 1988; Junghare et al., 2015) were reported as pure cultures that use phthalate as carbon and energy source. Phthalate degradation was supposed to involve the decarboxylation to benzoate, which is then activated by thioesterification to the central benzoyl-CoA (Eaton and Ribbons, 1982; Taylor and Ribbons, 1983) (Figure 1). However, such a reaction is mechanistically hardly conceivable as decarboxylation of phthalate would result in the formation of a non-stabilized dianionic transition state. The initially suggested reduction of phthalate to a diene system prior to decarboxylation is thermodynamically not feasible with biological electron donors. As an alternative, formation of phthaloyl-CoA by an acyl-CoA synthetase followed by decarboxylation to benzoyl-CoA has been hypothesized (Nozawa and Maruyama, 1988), but never been shown.
To date, the metabolites, genes and enzymes involved in anaerobic phthalate degradation are unknown. All hypothesized phthalate degradation scenarios have in common to channel phthalate to the central intermediate benzoyl-CoA, which is then further metabolized by enzymes of the anaerobic benzoyl-CoA-degradation pathway (Harwood, 1998; Fuchs et al., 2011). In this study, we discovered a previously unknown capacity of various aromatic compound-degrading, denitrifying bacteria including Thauera chlorobenzoica 3CB-1, ‘Aromatoleum aromaticum' EbN1 and Azoarcus evansii KB740 to completely degrade phthalate to CO2. With the biomass obtained from T. chlorobenzoica, we elucidate the previously obscure anaerobic degradation pathway of the industrial xenobiotic.
Materials and methods
Cultivation of bacteria and preparation of cell extracts
T. chlorobenzoica strain 3CB-1 (DSM-18012), Azoarcus evansii strain KB740 (DSM-6898), ‘A. aromaticum' strain EbN1 (Rabus and Widdel, 1995), Azoarcus buckelii strain U120 (DSM-14744), T. aromatica strain K172 (DSM-6984) and Azoarcus sp. CIB (CECT 5669) were grown anaerobically at 30 °C in 0.1–2 l cultivation volume (strain collection reference numbers; DSM: Leibniz-Institut DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; CECT: Spanish Type Culture Collection, Valencia, Spain). Media routinely contained 2–5 mm phthalate or benzoate as sole carbon source and 5–15 mm nitrate as electron acceptor (Rabus and Widdel, 1995; Ebenau-Jehle et al., 2003). If necessary, phthalate or benzoate and nitrate were supplemented several times. The td-values were calculated from three different biological replicates. T. chlorobenzoica was additionally grown in a 200 l bioreactor (Bioengineering AG, Wald, Switzerland), which was operated in a continuous fed-batch mode using a 0.5 m phthalate and 1.8 m NaNO3 stock solution. Under these conditions no accumulation of nitrate or nitrite was observed. Growth was followed by measuring the OD (optical density) at 578 nm. Cells grew with a generation time of 15–35 h owing to a growth limiting feeding rate. Cells were harvested in the exponential growth phase at an OD578 of 3.5. Harvested cells were kept frozen in liquid nitrogen until use. Phthalate concentrations in cell cultures were determined photometrically after acidification with 1 m HCl to pH 2; absorption was measured at 276 nm (ɛ=1200 M−1 cm−1).
Extracts were prepared under anaerobic conditions. Frozen cells of all strains were suspended in twofold volume of 50 mm sodium phosphate buffer (pH 7.5) with or without 20 m potassium chloride and with 0.1 mg ml−1 DNase I. Cell suspensions were passed through a chilled French pressure cell at 9 MPa. The cell lysate was ultracentrifuged (1 h, 200 000 g, 4 °C) and the supernatant was used immediately. Depending on the experiment, cell extracts were desalted with a PD Minitrap G25 column (GE Healthcare, Chalfont St Giles, UK).
Synthesis of CoA thioesters
Benzoyl-CoA, succinyl-CoA and acetyl-CoA were synthesized from the corresponding anhydrides and coenzyme A (Schachter and Taggart, 1953). 2-cyanobenzoyl-CoA, 2-nitrobenzoyl-CoA and phthaloyl-CoA methyl ester were synthesized starting from esterification of the acids with N-hydroxysuccinimide (Blecher, 1981). Synthesis of the CoA esters from succinimdylesters was identical to the syntheses starting from anhydrides. Purity was determined by liquid chromatography (UPLC, Waters Corporation, Milford, MA, USA).
Liquid chromatography analyses
Metabolites were analyzed using a Waters Acquity H-class UPLC system with a Waters BEH 1.7 μm C18-reverse phase column. Proteins were precipitated with 50% (v/v) 1 m HCl and the supernatant was applied onto the column. For CoA esters a gradient of 2–30% acetonitrile in 10 mm potassium phosphate buffer (pH 6.8) was used for separation. For free acids a gradient of 9–45% methanol in 0.1% (v/v) formic acid/water was applied. Metabolites were identified by their retention times and ultraviolet/Vis spectra compared with standards.
Enzyme assays
Succinyl-CoA-dependent product formation was investigated at 30 °C by adding varying volumes of cell extract to 100 mm sodium phosphate buffer pH 7.5 containing 0.15 mm CoA donor (succinyl-CoA or acetyl-CoA) and 0.2 mm phthalate or phthalate analogs. If not otherwise stated, 50 mm potassium chloride was added to the assay. Direct decarboxylation of phthalate to benzoate was measured by adding cell-free extracts to reaction assays containing 50 mm MOPS/KOH pH 7.5, 0.1 mm MnCl2 and 0.5 mm phthalate. Samples taken at different time points were analyzed by UPLC. Ligase reactions were monitored in a coupled, continuous photometric assay (Ziegler et al., 1989); CoA transferase reactions were determined in a discontinuous assay by UPLC analysis of the CoA ester formed at different time points. For inhibition assays, cell extracts were preincubated with the inhibitor for ten minutes at 4 °C. Oxygen sensitivity was determined by incubating cell extracts in air (4 °C) under gentle shaking; anaerobically incubated cell extracts served as a control. Specific enzyme activities of the cell extracts were calculated from metabolite formation or consumption rate.
Proteome profile identification on one-dimensional gels and draft genome of T. chlorobenzoica
Proteins were identified by mass spectrometry (Verónica I. Dumit, Core Facility Proteomics, Center for Biological System Analysis, Freiburg, Germany). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, in-gel proteolytic cleavage using trypsin followed by liquid chromatography mass spectrometric analysis (ESI/Q-TOF, Agilent Technologies, Santa Clara, CA, USA). MassHunter (Agilent Technologies) was used for data acquisition and processing. The Mascot algorithm was used to search MS data of ‘A. aromaticum' EbN1 and A. evansii KB740 against the ‘A. aromaticum' EbN1 protein-coding sequences. MS data of T. chlorobenzoica 3CB-1 was searched against protein translations from a draft of its genome assembled from one 700 bp paired-end and three larger insert mate-paired libraries. Illumina MiSeq reads were processed with the allpaths software (Maccallum et al., 2009), yielding a 3 746 743 bp assembly that was estimated to be 99% complete. The assembly consists in 12 scaffolds, with the largest one measuring 2 452 906 bp, and the three largest ones accounting for 99% of the genomic sequence.
Mass spectrometry-based proteome analyses
For whole proteome analysis ‘A. aromaticum' was cultivated anaerobically in 100 ml bottles on 5 mm phthalate/15 mm nitrate or 5 mm benzoate/15 mm nitrate at 30 °C. Cells were harvested in the mid-exponential phase at OD578 0.2. Cells were washed in 0.1 m Tris/HCl pH 7.5 buffer, followed by resuspension in 2 ml sodium dodecyl sulphate buffer (1.25% sodium dodecyl sulphate, 0.1 m TRIS, 0.3% dithiothreitol). Excised protein band were digested by trypsin (Promega, Madison, WI, USA), and separated by UHPLC (Ultimate 3000, Dionex/Thermo Fisher Scientific, Idstein, Germany). For details see Supplementary Information.
Mass spectrometry was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) with a TriVersa NanoMate (Advion, Ltd., Harlow, UK) source in LC chip-coupling mode. Proteome Discoverer (version 2.1, Thermo Scientific) was used for protein identification and the acquired MS/MS spectra were searched with Sequest HT against the T. chlorobenzoica 3CB-1 database (containing 3519 protein-coding sequences). Only peptides with a false discovery rate<0.01 calculated by Percolator and peptide rank=1 were considered as identified. The abundance of one protein was quantified using the average of the top-3 peptides assigned to this protein. For details of mass spectrometric analyses and protein identification see Supplementary information.
Computational analyses
Gene clusters were compared using Geneious R8 (Kearse et al., 2012). If not previously annotated, genes were predicted using Glimmer 3 (Delcher et al., 2007). Homology of the corresponding gene products was analyzed using the blastp algorithm (Altschul et al., 1990). Phylogenetic analysis of 42 sequences of UbiD-like enzymes was conducted in MEGA6 (Tamura et al., 2013) using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992).
Results
Growth of denitrifying bacteria pure cultures with phthalate
The preliminary knowledge of anaerobic phthalate degradation mainly derives from studies with the non-classified Pseudomonas strain P136 almost three decades ago (Nozawa and Maruyama, 1988), and comprises the description of growth curves, adaptation to carbon source utilization using whole-cell suspensions, analyses of compounds in the culture medium and some carboxylic acid CoA ligase activity measurements. The global occurrence of the industrially produced PAE suggest that anaerobic phthalate-degradation capacity should be highly abundant in anaerobic organisms growing with aromatic compounds. To identify suitable model organisms for unraveling the anaerobic phthalate catabolic pathway, we tested the known aromatic compound-degrading, denitrifying model organisms T. aromatica, T. chlorobenzoica, Azoarcus sp. CIB, A. evansii and A. buckelii as well as the non-classified ‘A. aromaticum' for growth with phthalate (5 mm) and nitrate (15 mm) in mineral salt media under strictly anoxic conditions.
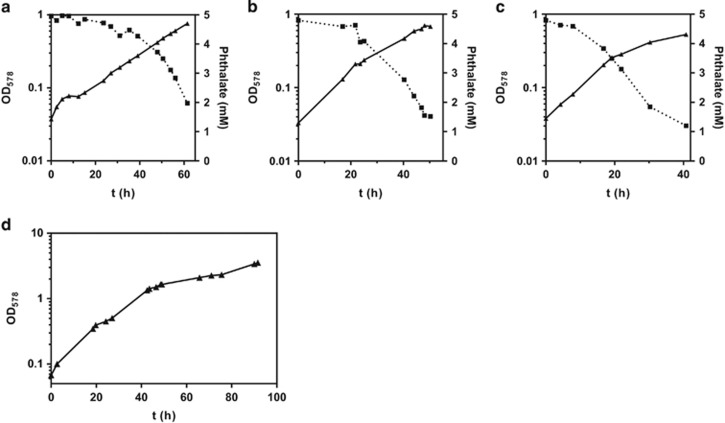
For ‘A. aromaticum', T. chlorobenzoica and A. evansii growth with phthalate/nitrate as sole sources of carbon and energy was observed as indicated by the increase in OD and the complete consumption of phthalate with doubling times between 11 and 15 h (Figures 2a–c). This doubling time is around threefold higher than that determined with benzoate under identical growth conditions (between 3 and 5 h). A comparable growth with phthalate/nitrate was also observed with A. buckelii (not shown), whereas T. aromatica and Azoarcus sp. CIB were not capable of using phthalate/nitrate as carbon/energy source. None of the four phthalate-degrading strains showed growth with isophthalate, terephthalate, diethylphthalate or dibutylphthalate. To gain cell mass for detailed studies of the genes and enzymes involved in phthalate degradation, T. cholorobenzoica was grown in a 200 l fermenter (Figure 2d) with 5 mm phthalate in a fed-batch culture under nitrate-limiting conditions to avoid nitrite accumulation; ~530 g cells (wet mass) were harvested after 4 days (Figure 2d).
Figure 2.
Representative growth curves of (a) ‘A. aromaticum' EbN1 (td=14.7±0.7 h), (b) T. chlorobenzoica 3CB-1 (td=14.0±1.9 h) and (c) A. evansii KB740 (td=11.6±2.9 h) anaerobically cultivated with phthalate as sole carbon source and nitrate as electron acceptor. The td values given represent the mean value±s.d. from td values calculated from three independent growth curves, respectively. (d) Growth curve of T. chlorobenzoica 3CB-1 anaerobically cultivated in a 200-L bioreactor constantly supplied with phthalate and nitrate. Cells were harvested at OD578=3.5; (▲) OD578, (■) phthalate in mM.
In vitro assays for enzymes involved in phthalate degradation
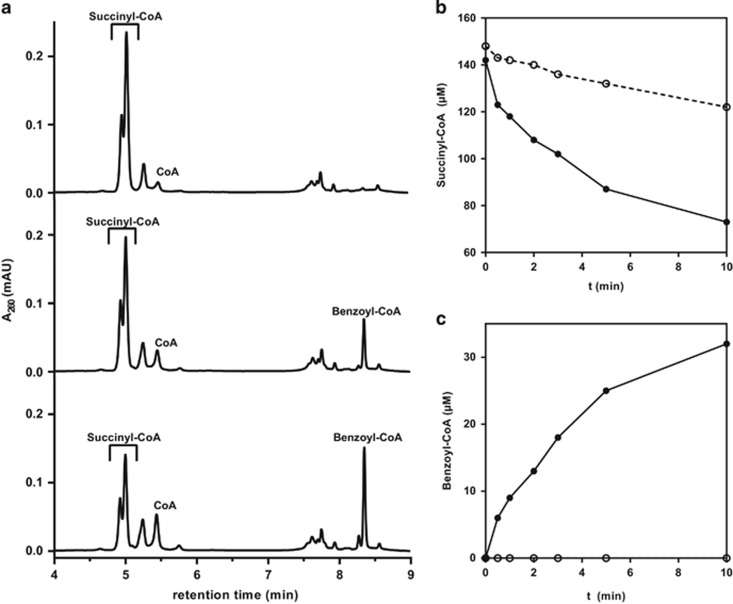
Using cell extracts from T. chlorobenzoica anaerobically grown with phthalate the conversion of phthalate (0.5 mm) to benzoate was observed, however at a rate far too low to explain growth with phthalate (0.3 nmol min−1 mg−1, Table 1). However, the rate of phthalate consumption was more than 100-fold increased when succinyl-CoA was added to the reaction mixture (Table 1). In this case, virtually no benzoate was formed; instead, the time- and protein-dependent formation of benzoyl-CoA, and the concomitant consumption of succinyl-CoA were observed (Figure 3). When phthalate was omitted from the assay, succinyl-CoA was slowly converted to CoA plus succinate, but no benzoyl-CoA formation was observed. This background thioester hydrolyzing activity was in the range of 2–8 nmol min−1 mg−1, and was generally subtracted when the phthalate-dependent succinyl-CoA consumption and CoA formation was determined (Table 1). Most importantly, when phthalate was replaced by benzoate, no formation of benzoyl-CoA was observed excluding that a succinyl-CoA:benzoate CoA transferase was responsible for benzoyl-CoA formation. We interpret the results obtained as the activities of two consecutively acting enzymes: a phthaloyl-CoA forming SCPCT (succinyl-CoA:phthalate CoA transferase) and a benzoyl-CoA forming phthaloyl-CoA decarboxylase. The concomitant formation of CoA is considered to result from partial hydrolysis of the extremely labile phthaloyl-CoA intermediate under the reaction conditions (see below). During phthalate-induced succinyl-CoA consumption, formation of benzoyl-CoA plus CoA accounted to the loss of succinyl-CoA (after subtraction of phthalate-independent hydrolysis). In the course of a typical activity assay, the benzoyl-CoA:CoA ratio was in the range between 3:1 and 2:1.
Table 1. Specific activities of cell extracts from different denitrifying bacteria grown with PA or B.
| Organism | Growth substrate | Substrates in enzyme assay | Succinyl-CoA consumption (nmol min−1 mg−1)a | CoA formation (nmol min−1 mg−1)a | Benzoyl-CoA formation (nmol min−1 mg−1) | Phthalate consumption (nmol min−1 mg−1) |
|---|---|---|---|---|---|---|
| T. chlorobenzoica 3CB-1 | PA | PA | n.d. | n.d. | n.d. | ⩽0.3 |
| T. chlorobenzoica 3CB-1 | PA | Succinyl-CoA PA | 31±9 | 4±1 | 13±2 | 34±9 |
| ‘A. aromaticum' EbN1 | PA | Succinyl-CoA PA | 26±3 | 3±1 | 12±3 | n.d. |
| A. evansii KB740 | PA | Succinyl-CoA PA | 18±3 | 3±1 | 6±1 | n.d. |
| T. chlorobenzoica 3CB-1 | B | Succinyl-CoA PA | n.d. | n.d. | <0.1 | ⩽0.8 |
| T. chlorobenzoica 3CB-1 | PA | Succinyl-CoA B | ⩽2 | ⩽1 | <0.1 | n.d. |
| T. chlorobenzoica 3CB-1 | PA | CoA/MgATP B | n.d. | n.d. | <0.1 | n.d. |
Abbreviations: B, benzoate; n.d., not determined; PA, pthalate. Errors are given as s.d.s of the mean of three biological replicates.
After subtraction of PA-independent succinyl-CoA consumption.
Figure 3.
In vitro assays for anaerobic phthalate degradation in T. chlorobenzoica. (a) Representative LC-chromatogram of the in vitro anaerobic phthalate degradation assay in extracts of cells grown with phthalate/nitrate. Time points are 0 min, 2 min and 6 min (from top to bottom). Under the LC separation conditions used, succinyl-CoA standard eluted as a double peak as indicated. (band c) Time course of succinyl-CoA consumption and benzoyl-CoA formation in the presence and absence of phthalate using cell extracts from T. cholorobenzoica grown with phthalate. (b) ● succinyl-CoA (+phthalate), ○ succinyl-CoA (–phthalate). (c) ● benzoyl-CoA (+succinyl-CoA/+phthalate), ○ benzoyl-CoA (+succinyl-CoA/–phthalate). The individual enzyme activities are presented in Table 1 and derive from three biological replicates.
The observed phthalate- and succinyl-CoA-dependent benzoyl-CoA-forming activities were only observed in T. chlorobenzoica cells grown with phthalate but were negligible in cells grown with benzoate suggesting an upregulation of the genes/enzymes involved in anaerobic phthalate degradation (Table 1). Likewise, extracts from ‘A. aromaticum' and A. evansii grown with phthalate catalyzed the succinyl-CoA- and phthalate-dependent benzoyl-CoA formation, with the rate in A. evansii being 1/3 of that determined in T. chlorobenzoica and ‘A. aromaticum' (Table 1). In all cases, formation of benzoyl-CoA from phthalate was strictly dependent on succinyl-CoA. It could neither be replaced by acetyl-CoA as CoA donor, nor by MgATP plus CoA that could serve as substrates for a potential phthalate activating acyl-CoA synthetase.
The succinyl-CoA- and phthalate-dependent benzoyl-CoA-forming activity was K+-dependent: addition of 50 mm KCl resulted in a reproducible twofold increase of activity (Supplementary Figure S1). In contrast addition of Na+, Mg2+ and Mn2+ had no stimulatory effect. Benzoyl-CoA formation from phthalate and succinyl-CoA was oxygen-sensitive with a half-life time in air of ~20 min (Supplementary Figure S2). In contrast, under aerobic conditions succinyl-CoA consumption rate was not affected, indicating that SCPCT was stable in air for several hours at 4 °C. The CoA-formation rate was even increased in air and was equal to the succinyl-CoA consumption rate. This finding can be rationalized that in case of oxygen-inactivated decarboxylase, phthaloyl-CoA becomes fully hydrolyzed to CoA and phthalic anhydride; the latter spontaneously hydrolyzes to phthalate. Surprisingly, the oxygen sensitivity of phthaloyl-CoA decarboxylation was clearly diminished upon addition of 50 mm KCl (t1/2≈50 min) (Supplementary Figure S2). This observation suggests that K+ ions positively affect both, activity and stability in air of enzymatic phthaloyl-CoA decarboxylation.
Assays for substantiating phthaloyl-CoA as intermediate
During the UPLC-based analyses of phthalate- and succinyl-CoA-dependent benzoyl-CoA formation, no transient accumulation of an intermediate was observed, which may be assigned to phthaloyl-CoA. As the activity of SCPCT was ~1.7–2.4 fold higher than that of the benzoyl-CoA formation (phthaloyl-CoA decarboxylase activity) (Table 1), the failure to detect phthaloyl-CoA cannot be explained by the steady state kinetics of phthaloyl-CoA formation/consumption. Instead, the obvious high tendency of phthaloyl-CoA to intramolecular hydrolysis to phthalic anhydride and free CoA most likely causes CoA release. Indeed, all attempts to chemically synthesize authentic phthaloyl-CoA from phthalic anhydride and CoA using the established protocol by Schachter and Taggart (1953) failed suggesting that under the conditions of chemical CoA ester synthesis (pH 8) the rate of hydrolysis is faster than that of its formation. Notably, the minimal timespan required for sample collection and UPLC separation/detection of CoA esters (>15 min) is far too long to prove whether phthaloyl-CoA stability is sufficient to act as a free intermediate during intracellular phthalate degradation.
To substantiate that phthaloyl-CoA is a cellular intermediate during anaerobic phthalate degradation, the effect of phthaloyl-CoA analogs that are not susceptible to intramolecular hydrolysis was tested. For this purpose succinyl-CoA was reacted with extracts from cells grown with phthalate in the presence of the phthalate analogs 2-nitrobenzoate, 2-cyanobenzoate or mono-methyl phthalate. In no case CoA transfer to the carboxylic acids tested was observed suggesting a high specificity of SCPCT for phthalate. In another attempt, 2-nitrobenzoyl-CoA, 2-cyanobenzoyl-CoA and phthaloyl-CoA methyl ester were chemically synthesized as potential inhibitors of phthaloyl-CoA decarboxylation. Although 2-nitrobenzoyl-CoA and methylphthaloyl-CoA showed only negligible effects on phthalate- and succinyl-CoA dependent benzoyl-CoA formation, 2-cyanobenzoyl-CoA (100 μm, 15 min incubation) was identified as inhibitor of the reaction (Supplementary Table S1). In contrast, 2-cyanobenzoate had no effect on benzoyl-CoA-forming activity. This finding strongly supports that phthaloyl-CoA rather than phthalate is the substrate for enzymatic decarboxylation.
Differential proteome analysis of ‘A. aromaticum'
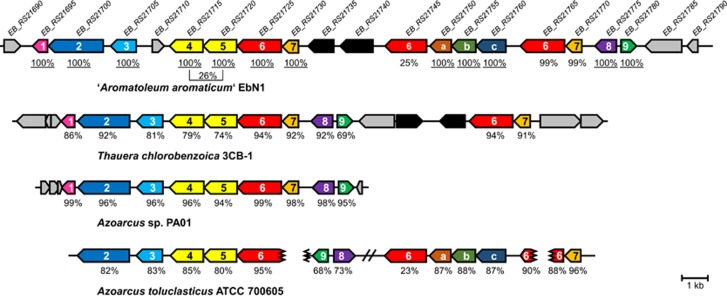
The genome sequence of ‘A. aromaticum' is available (Rabus et al., 2005) and a draft one from T. chlorobenzoica was obtained in this work (see Materials and Methods). To identify phthalate-induced proteins, cells from T. chlorobenzoica, ‘A. aromaticum' and A. evansii were grown with phthalate or benzoate and nitrate, and the proteomes obtained from exponentially grown cells were compared. After one-dimensional separation, a prominent protein band at ~59 kDa was clearly induced in all cells grown with phthalate vs benzoate (Supplementary Figure S3). As a result, UbiD-like carboxylases/decarboxylases were identified. For ‘A. aromaticum' the first two hits were the gene products of EB_RS21765 followed by EB_RS21725 (Figure 5), whereas for T. chlorobenzoica two UbiD-like carboxylases/decarboxylases were identified with equal scores. For A. evansii peptides were matched against the ‘A. aromaticum' proteome and hits were obtained with equal scores for both gene products stated above for ‘A. aromaticum'. UbiD was originally identified as a 3-octaprenyl-4-hydroxybenzoate decarboxylase in Escherichia. coli, a key reaction in ubiquinone biosynthesis (Cox et al., 1969).
Figure 5.
Putative anaerobic phthalate degradation gene clusters. Locus tags are indicated for ‘A. aromaticum' EbN1 that are located on a plasmid. % identities (blastp) of the corresponding gene products are given compared with the products of ‘A. aromaticum' EbN1 genes 1–9 and a to c, respectively. Annotations of ‘A. aromaticum' EbN1 gene products: (1) hypothetical protein (2) TRAP transporter (3) periplasmic binding protein (4) type III CoA transferase subunit, (5) type III CoA transferase subunit, (6) UbiD-like (de)carboxylase, (7) UbiX-like protein, (8) iclR family transcriptional regulator, (9) Nudix family hydrolase; (a) ABC-type transporter periplasmic component, (b) ABC-type transporter ATPase component, (c) ABC-type transporter permease component. Gray-colored genes code for putative transposases/integrases. Black genes have no proposed function in anaerobic phthalate degradation. The end of scaffold is indicated by interrupted lines; //: 28.4 kb genomic separation.
To identify additional phthalate-induced proteins, a global proteome analysis was performed with ‘A. aromaticum' grown with benzoate or phthalate. In order to assess if the proteome data contain significant changes related to the growth substrates, a principle component analysis was calculated (Supplementary Figure S4). There, two investigated groups could be clearly separated based on the data set of 1426 proteins. The first two principle components account for ~62% of the variance in the data set, indicating complex contribution patterns to the global variations among the substrate. Within the whole list of proteins detected (Supplementary Table S2), six were assigned to a function in the proposed anaerobic degradation of phthalate and were more abundant in cultures grown with phthalate compared to benzoate (Table 2).
Table 2. Proteomic analysis of ‘A. aromaticum' grown with phthalate vs benzoate.
| UniProt identifier | Locus tag of gene | Protein description | Fold change (log10) phthalate/benzoate | P-value (adj. BH) a |
|---|---|---|---|---|
| Q5NWI1 | EB_RS21700 | Transmembrane subunit of phthalate transporter (TRAP transporter) | Unique for phthalate (>1b) | n.a. |
| Q5NWI0 | EB_RS21705 | Periplasmatic subunit of phthalate transporter (periplasmic binding protein) | 2.15 | 1.07E−02 |
| Q5NWH8 | EB_RS21715 | Subunit of succinyl-CoA:phthalate CoA transferase (Type III CoA transferase) | 2.07 | 6.56E−03 |
| Q5NWH7 | EB_RS21720 | Subunit of succinyl-CoA:phthalate CoA transferase (Type III CoA transferase) | 2.62 | 1.02E−02 |
| Q5NWH6 | EB_RS21725 | Phthaloyl-CoA decarboxylase (UbiD-like protein) | Unique for phthalate (>2b) | n.a. |
| Q5NWH5 | EB_RS21730 | FMN prenyltransferase for phthaloyl-CoA decarboxylase (UbiX-like protein) | 2.28 | 1.25E−02 |
Only phthalate-induced gene products of the putative phthalate degradation gene cluster are shown. For other proteins see Supplementary Table S2.
Benjamini-Hochberg adjusted P-value, n.a.= not applicable.
Fold changes estimated from protein area (average of the top three peptides) to zero.
Phylogeny and occurrence of phthaloyl-CoA decarboxylases
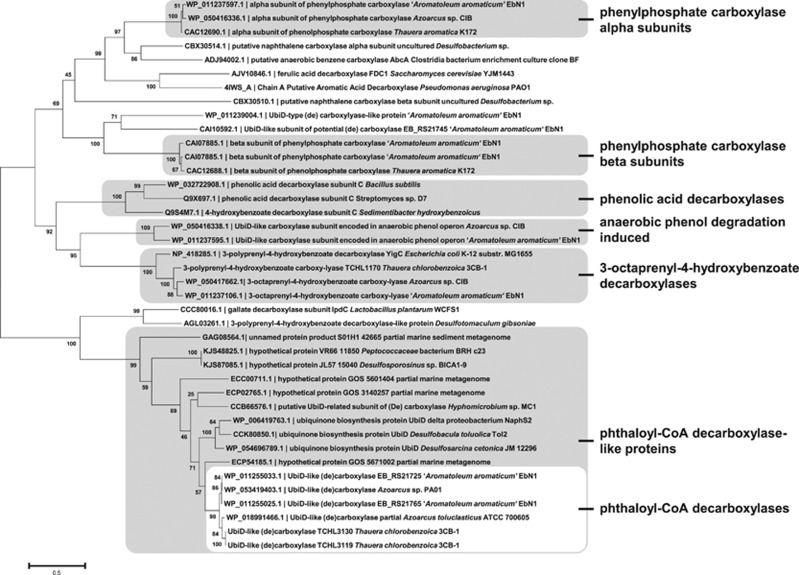
A search using Basic Local Alignment Search Tool identified deduced proteins with amino-acid sequence identities higher than 50% to the UbiD-like phthaloyl-CoA decarboxylase of T. chlorobenzoica that form a phylogenetic subcluster among UbiD-like de(carboxylases) (Figure 4). Deduced proteins of this cluster derive from denitrifying Gram-negative bacteria, Gram-negative and Gram-positive sulfate-reducing bacteria and from a marine metagenome suggesting a wide occurrence of phthaloyl-CoA decarboxylases. More distantly related UbiD-like decarboxylases comprise putative 3-octaprenyl-4-hydroxybenzoate decarboxylases, and experimentally verified or putative carboxylases involved in the degradation of phenol (phenylphosphate carboxylases) (Breinig et al., 2000) as well as other aromatic compounds (Abu Laban et al., 2010; Bergmann et al., 2011; Holmes et al., 2012).
Figure 4.
Phylogenetic analyses of UbiD-like proteins. Protein sequences of the UbiD superfamily pfam01977 in ‘A. aromaticum' EbN1, T. chlorobenzoica 3CB-1, T. aromatica K172 and Azoarcus sp. CIB were considered as well as UbiD-like proteins with previously shown function and proteins homologous (minimal identity/minimal query cover 50%/90% for full length protein sequences) to ‘A. aromaticum' phthaloyl-CoA decarboxylase. Bootstrap values of 1000 replications are given in % for each branch.
Composition of phthalate-induced gene clusters
Based on the phthalate-induced proteins we identified gene clusters of ‘A. aromaticum' and T. chlorobenzoica, and those putatively involved in phthalate degradation in Azoarcus sp. PA01 and Azoarcus toluclasticus (Figure 5). In all cases the gene encoding the phthaloyl-CoA decarboxylase is located next to the two genes putatively encoding a type III CoA transferase, and the genes encoding components of a putative tripartite ATP-independent periplasmic dicarboxylate transporter. They are also directly flanked by an ubiX-like gene (in A. toluclasticus the corresponding scaffold is interrupted in the relevant genomic region). A UbiX-like gene product has previously been shown to catalyse the prenylation of an enzyme-bound flavin mononucleotide using dimethylallylphosphate as cosubstrate (White et al., 2015). In case of a UbiD-like fungal decarboxylase acting on cinnamate, it was demonstrated that the prenylated cofactor formed at the UbiX component is transferred to the UbiD component of the decarboxylase system (Payne et al., 2015). The finding of putative ubiX-like gene products adjacent to the ubiD-like genes in the genomes of phthalate-degrading anaerobes strongly suggests that active phthaloyl-CoA decarboxylase contains a prenylated flavin mononucleotide in the active site.
In ‘A. aromaticum', the genes involved in anaerobic phthalate degradation are located on the plasmid referred to as p2A (Rabus et al., 2005). This plasmid contains the complete set of genes for conjugal DNA transfer with high similarities to the tra and trb gene clusters of the Ti plasmid. We therefore propose that the capacity for anaerobic phthalate degradation can be laterally transferred by conjugation. Notably, with EB_RS21765 and EB_RS21745 there are two additional ubiD-like genes on this plasmid. Although the former appears to be an almost identical duplication of the gene encoding putative phthaloyl-CoA decarboxylase (EB_RS21725), the latter shows only 25% amino-acid sequence identity. This finding along with the presence of adjacent genes encoding an ubiX-like protein and a putative ABC-transporter system suggests that it may be involved in decarboxylation of different, yet unknown carboxylic acid growth substrate. The presence of multiple copies of almost identical ubiD-like genes encoding putative phthaloyl-CoA decarboxylases is also found in the genomes T. chlorobenzoica and A. toluclasticus; most likely they resulted from gene duplication and serve as back-up genes.
Discussion
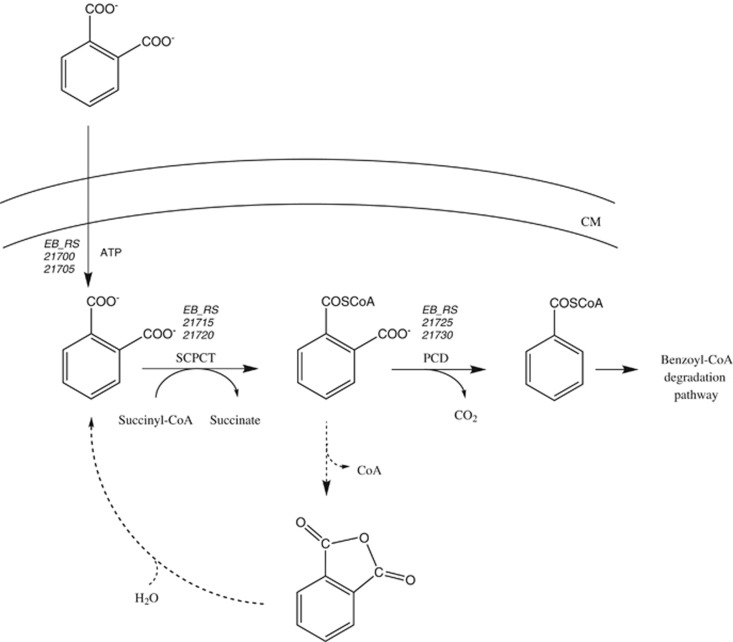
We identified a previously unnoticed capability of a number of aromatic compound-degrading anaerobic model organisms to grow with the environmentally relevant xenobiotic phthalate. First in vitro studies identified a phthalate- and succinyl-CoA-specific CoA transferase together with a UbiD-like phthaloyl-CoA decarboxylase as key enzymes of the newly identified phthalate degradation pathway (Figure 6).
Figure 6.
Genes and enzymes involved in the anaerobic phthalate degradation. The numbers in italics refer to locus tags of the corresponding genes in ‘A. aromaticum'. Solid line arrows refer to putative tripartite phthalate transporter and enzymatic reactions determined in vitro; dotted line reactions refer to putative proton-driven transport and intramolecular chemical hydrolysis of phthaloyl-CoA to phthalic acid and CoA during sample preparation followed by spontaneous hydrolysis of the latter to phthalate.
The predicted phthaloyl-CoA intermediate is probably the most instable CoA thioester in biology because the planar orientation of the carboxyl-group towards the thioester functionality strongly promotes intramolecular hydrolysis (Figure 6). For this reason it was neither possible to chemically synthesize phthaloyl-CoA by conventional organic chemical protocols, nor to identify it by UPLC in samples taken in the steady state of the in vitro conversion of phthalate to benzoyl-CoA. But the following findings of this work strongly support that phthalate degradation indeed proceeds via phthaloyl-CoA: (i) consumption of succinyl-CoA was clearly stimulated by phthalate, (ii) formation of benzoyl-CoA was strictly dependent on both, phthalate and succinyl-CoA; (iii) the rate of succinyl-CoA-independent formation of benzoate from phthalate was only 1% of benzoyl-CoA-formation rate from succinyl-CoA and phthalate; (iv) phthalate and succinyl-CoA dependent benzoyl-CoA formation was clearly induced by phthalate as growth substrate; (v) 2-cyanobenzoyl-CoA, a stable structural analog of phthaloyl-CoA, inhibited formation of benzoyl-CoA from phthalate plus succinyl-CoA, whereas 2-cyanobenzoate had no effect.
In the steady state of phthalate conversion to benzoyl-CoA, 50–80% of the assumed phthaloyl-CoA intermediate was decarboxylated to benzoyl-CoA, whereas the remainder was hydrolyzed to CoA plus phthalate. This finding demonstrates that even under artificial in vitro conditions phthaloyl-CoA appeared to be stable enough to outcompete chemical hydrolysis to a major extent. To minimize chemical hydrolysis of phthaloyl-CoA, the decarboxylase activity has to exceed CoA transferase activity to minimize phthaloyl-CoA accumulation which requires balanced activities of both enzymes. It is conceivable that futile phthaloyl-CoA hydrolysis may occur to a minor extent even in vivo. In this context it is noteworthy that growth rate with phthalate is only 1/3 compared of that with benzoate.
The strategy evolved for anaerobic phthalate degradation represents an unprecedented example for a compromise between kinetic/mechanistic constraints and metabolite stability. The direct decarboxylation of phthalate to benzoate is mechanistically unfavorable, and in the absence of molecular oxygen, the only option is activation of a carboxyl functionality, albeit at the expense of an extremely hydrolysis-sensitive CoA ester intermediate. The carbonyl functionality of phthaloyl-CoA could largely stabilize anionic or radical transition states resulting from decarboxylation of the non-activated carboxyl-group.
The identification of strongly phthalate-induced UbiD-like gene products suggests that phthaloyl-CoA decarboxylase is a novel member of the steadily growing UbiD (de)carboxylase enzyme family. The involvement of a UbiD-like-enzyme in ubiquinone synthesis is known since decades (Cox et al., 1969). However, detailed structural and functional insights have only very recently been obtained with a UbiD-like enzyme catalyzing the decarboxylation of cinnamate to styrene (Payne et al., 2015). Most importantly, the identification of a prenylated flavin mononucleotide-cofactor in UbiD-like enzymes, formed by the UbiX prenyltransferase, shed light into the function of the UbiD/UbiX enzyme systems (White et al., 2015). It explains why the encoding genes are often located adjacent to each other, as it is also the case for the ubiD-like genes coding for putative phthaloyl-CoA decarboxylases identified in this work. UbiD-like enzymes have also been proposed to be involved in the carboxylation of aromatic rings such as benzene or naphthalene in sulfate-reducing or Fe(III)-reducing bacteria and archaea (Abu Laban et al., 2010; Bergmann et al., 2011; Holmes et al., 2012). However, owing to the extremely slow growth of these organisms with non-substituted aromatic hydrocarbons, the limited biomass available largely hampered the biochemical characterization of such enzymatic reactions. As anaerobic growth with phthalate yields & gt;500 g T. chlorobenzoica in a 200-L-fermenter within four days, the UbiD/UbiX components of phthaloyl-CoA decarboxylase from denitrifying bacteria could serve as a model for UbiD-like aromatic ring (de)carboxylases.
Out of the six tested denitrifying pure cultures in this work, four were capable of complete phthalate degradation, suggesting that at least in denitrifying bacteria, anaerobic phthalate degradation should be widely prevalent. Phylogenetic analyses identified genes likely to encode phthaloyl-CoA decarboxylases in other denitrifying organisms with sequenced genomes, but also in genomes of sulfate-reducing pure cultures and marine metagenomes. This finding suggests a global occurrence of the anaerobic phthaloyl-CoA decarboxylation pathway even in microorganisms with an energy metabolism less efficient than that of denitrifyers.
The results obtained in this study suggest that the minimal enzyme inventory for anaerobic phthalate degradation requires four components: a phthalate-specific dicarboxylate transporter, a phthaloyl-CoA forming and a phthaloyl-CoA decarboxylating enzyme, as well as a prenyltransferase for cofactor maturation of the decarboxylase. In ‘A. aromaticum' all genes encoding these proteins are located on a conjugative plasmid; in T. chlorobenzoica and Azoarcus strains they are flanked by transposases and inverted repeats. These observations indicate an ongoing rapid distribution of a xenobiotic degradation pathway by lateral gene transfer. Considering that massive industrial phthalate ester production started in the late 1950's (Graham, 1973), the global emergence of the anaerobic phthalate degradation pathway occurred in a very short timespan.
Acknowledgments
We thank Georg Fuchs (Freiburg) for providing data from initial studies of anaerobic phthalate degradation and many helpful discussions. Moreover, we are grateful to Wolfgang Buckel (Marburg) and Bernard Golding (Newcastle) for helpful advice concerning mechanistic aspects of phthaloyl-CoA decarboxylation. We thank Karine Labadie and Eric Mahieu (Evry) for assistance with Illumina sequencing. This work was partially funded by the EU collaborative network MAGICPAH. Sequences of genes involved in phthalate degradation of T. chlorobenzoica have been deposited at GenBank under the accession KX267662-KX267672.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abu Laban N, Selesi D, Rattei T, Tischler P, Meckenstock RU. (2010). Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ Microbiol 12: 2783–2796. [DOI] [PubMed] [Google Scholar]
- Afshari A, Gunnarsen L, Clausen PA, Hansen V. (2004). Emission of phthalates from PVC and other materials. Indoor Air 14: 120–128. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Benckiser G, Ottow JC. (1982). Metabolism of the plasticizer di-n-butylphthalate by Pseudomonas pseudoalcaligenes under anaerobic conditions, with nitrate as the only electron acceptor. Appl Environ Microbiol 44: 576–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann F, Selesi D, Weinmaier T, Tischler P, Rattei T, Meckenstock RU. (2011). Genomic insights into the metabolic potential of the polycyclic aromatic hydrocarbon degrading sulfate-reducing Deltaproteobacterium N47. Environ Microbiol 13: 1125–1137. [DOI] [PubMed] [Google Scholar]
- Blecher M. (1981). Synthesis of long-chain fatty acyl-coA thioesters using N-hydroxysuccinimide esters. Methods Enzymol 72: 404–408. [DOI] [PubMed] [Google Scholar]
- Boll M, Fuchs G. (1995). Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem 234: 921–933. [DOI] [PubMed] [Google Scholar]
- Boll M, Löffler C, Morris B, Kung JW 2014). Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: organisms, strategies and key enzymes. Environ Microbiol 16: 612–627. [DOI] [PubMed] [Google Scholar]
- Breinig S, Schiltz E, Fuchs G. (2000). Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J Bacteriol 182: 5849–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W, Kung JW, Boll M. (2014). The benzoyl-coenzyme a reductase and 2-hydroxyacyl-coenzyme a dehydratase radical enzyme family. Chembiochem 15: 2188–2194. [DOI] [PubMed] [Google Scholar]
- Caldwell JC. (2012). DEHP: genotoxicity and potential carcinogenic mechanisms-a review. Mutat Res 751: 82–157. [DOI] [PubMed] [Google Scholar]
- Carmona M, Zamarro MT, Blázquez B, Durante-Rodríguez G, Juárez JF, Valderrama JA et al. (2009). Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73: 71–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins IT, Mackay D, Parkerton TF. (2003) Physical-chemical properties and evaluative fate modelling of pphthalate esters. In: Hutzinger O, Staples CA (eds). The Handbook of Environmental Chemistry. Springer Berlin Heidelberg: Berlin, Heidelberg, 57–84. [Google Scholar]
- Cox GB, Young IG, McCann LM, Gibson F. (1969). Biosynthesis of ubiquinone in Escherichia coli K-12: location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenylphenol. J Bacteriol 99: 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23: 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RW, Ribbons DW. (1982). Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B. J Bacteriol 151: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenau-Jehle C, Boll M, Fuchs G. (2003). 2-Oxoglutarate:NADP(+) oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J Bacteriol 185: 6119–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt G, Wallnöfer PR. (1978). Metabolism of Di- and Mono-n-Butyl Phthalate by Soil Bacteria. Appl Environ Microbiol 35: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Boll M, Heider J. (2011). Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9: 803–816. [DOI] [PubMed] [Google Scholar]
- Fujii M, Shinohara N, Lim A, Otake T, Kumagai K, Yanagisawa Y. (2003). A study on emission of phthalate esters from plastic materials using a passive flux sampler. Atmos Environ 37: 5495–5504. [Google Scholar]
- Gao D, Wen Z. (2016). Phthalate esters in the environment: A critical review of their occurrence, biodegradation, and removal during wastewater treatment processes. Sci Total Environ 541: 986–1001. [DOI] [PubMed] [Google Scholar]
- Graham PR. (1973). Phthalate ester plasticizers—-why and how they are used. Environ Health Perspect 3: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. (1998). Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev 22: 439–458. [Google Scholar]
- Holmes DE, Risso C, Smith JA, Lovley DR. (2012). Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J 6: 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Nkrumah PN, Li Y, Appiah-Sefah G. (2013). Chemical behavior of phthalates under abiotic conditions in landfills. Rev Environ Contam Toxicol 224: 39–52. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992). The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Junghare M, Patil Y, Schink B. (2015). Draft genome sequence of a nitrate-reducing, o-phthalate degrading bacterium, Azoarcus sp. strain PA01(T). Stand Genomic Sci 10: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser P, Pujar BG, Eaton RW, Ribbons DW. (1976). Biodegradation of the phthalates and their esters by bacteria. Environ Health Perspect 18: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini G. (2005). Monitoring phthalate exposure in humans. Clin Chim Acta 361: 20–29. [DOI] [PubMed] [Google Scholar]
- Liang D, Zhang T, Fang HHP, He J. (2008). Phthalates biodegradation in the environment. Appl Microbiol Biotechnol 80: 183–198. [DOI] [PubMed] [Google Scholar]
- Maccallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A et al. (2009). ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol 10: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh CE, Maldonado JA, Ikonomou MG, Gobas Frank A P C. (2006). Sorption of phthalate esters and PCBs in a marine ecosystem. Environ Sci Technol 40: 3481–3488. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Akita K, Naitou C, Yoshida M, Kitamura T. (2005). Purification and characterization of an esterase hydrolyzing monoalkyl phthalates from Micrococcus sp. YGJ1. J Biochem 137: 27–32. [DOI] [PubMed] [Google Scholar]
- Mersiowsky I, Weller M, Ejlertsson J. (2001). Fate of plasticised PVC products under landfill conditions: a laboratory-scale landfill simulation reactor study. Water Res 35: 3063–3070. [DOI] [PubMed] [Google Scholar]
- Net S, Sempéré R, Delmont A, Paluselli A, Ouddane B. (2015). Occurrence, fate, behavior and ecotoxicological state of phthalates in different environmental matrices. Environ Sci Technol 49: 4019–4035. [DOI] [PubMed] [Google Scholar]
- Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. (1992). Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng 74: 333–344. [Google Scholar]
- Nozawa T, Maruyama Y. (1988). Anaerobic metabolism of phthalate and other aromatic compounds by a denitrifying bacterium. J Bacteriol 170: 5778–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KAP, White MD, Fisher K, Khara B, Bailey SS, Parker D et al. (2015). New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 522: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp B, Schink B. (2012). Different strategies in anaerobic biodegradation of aromatic compounds: nitrate reducers versus strict anaerobes. Environ Microbiol Rep 4: 469–478. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Sekiguchi Y, Hanada S, Imachi H, Tseng I, Cheng S et al. (2006). Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch Microbiol 185: 172–182. [DOI] [PubMed] [Google Scholar]
- Rabus R, Kube M, Heider J, Beck A, Heitmann K, Widdel F et al. (2005). The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch Microbiol 183: 27–36. [DOI] [PubMed] [Google Scholar]
- Rabus R, Widdel F. (1995). Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol 163: 96–103. [DOI] [PubMed] [Google Scholar]
- Satyanarayana T, Johri BN. (2012) Microorganisms in Environmental Management. Springer: Netherlands: Dordrecht. [Google Scholar]
- Schachter D, Taggart JV. (1953). Benzoyl coenzyme A and hippurate synthesis. J Biol Chem 203: 925–934. [PubMed] [Google Scholar]
- Shelton DR, Boyd SA, Tiedje JM. (1984). Anaerobic biodegradation of phthalic acid esters in sludge. Environ Sci Technol 18: 93–97. [DOI] [PubMed] [Google Scholar]
- Staples CA, Peterson DR, Parkerton TF, Adams WJ. (1997). The environmental fate of phthalate esters. Chemosphere 35: 667–749. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BF, Ribbons DW. (1983). Bacterial decarboxylation of o-phthalic acids. Appl Environ Microbiol 46: 1276–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega D, Bastide J. (2003). Dimethylphthalate hydrolysis by specific microbial esterase. Chemosphere 51: 663–668. [DOI] [PubMed] [Google Scholar]
- Weinert T, Huwiler SG, Kung JW, Weidenweber S, Hellwig P, Stärk H et al. (2015). Structural basis of enzymatic benzene ring reduction. Nat Chem Biol 11: 586–591. [DOI] [PubMed] [Google Scholar]
- White MD, Payne KAP, Fisher K, Marshall SA, Parker D, Rattray NJW et al. (2015). UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K, Buder R, Winter J, Fuchs G. (1989). Activation of aromatic acids and aerobic 2-aminobenzoate metabolism in a denitrifying Pseudomonas strain. Arch Microbiol 151: 171–176. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.