Abstract
Huntington disease (HD) is a dominant neurodegenerative disorder caused by a CAG repeat expansion in the Huntingtin (HTT) gene. HD occurs worldwide, but the causative mutation is found on different HTT haplotypes in distinct ethnic groups. In Latin America, HD is thought to have European origins, but indigenous Amerindian ancestry has not been investigated. Here, we report dense HTT haplotypes in 62 mestizo Peruvian HD families, 17 HD families from across Latin America, and 42 controls of defined Peruvian Amerindian ethnicity to determine the origin of HD in populations of admixed Amerindian and European descent. HD in Peru occurs most frequently on the A1 HTT haplotype (73%), as in Europe, but on an unexpected indigenous variant also found in Amerindian controls. This Amerindian A1 HTT haplotype predominates over the European A1 variant among geographically disparate Latin American controls and in HD families from across Latin America, supporting an indigenous origin of the HD mutation in mestizo American populations. We also show that a proportion of HD mutations in Peru occur on a C1 HTT haplotype of putative Amerindian origin (14%). The majority of HD mutations in Latin America may therefore occur on haplotypes of Amerindian ancestry rather than on haplotypes resulting from European admixture. Despite the distinct ethnic ancestry of Amerindian and European A1 HTT, alleles on the parent A1 HTT haplotype allow for development of identical antisense molecules to selectively silence the HD mutation in the greatest proportion of patients in both Latin American and European populations.
Introduction
Huntington disease (HD, (MIM 143100)) is an autosomal dominant neurodegenerative disorder characterized by choreic movements, cognitive impairment, and behavioural disturbances.1 HD is caused by an expanded CAG trinucleotide repeat in the first exon of HTT, encoding expanded polyglutamine residues with pathogenic effects.2 HD occurs worldwide, but shows marked geographic differences in prevalence. Studies using defined clinical and genetic diagnostic standards report 10.6–13.7 cases per 100 000 inhabitants in Western populations, with higher rates among subpopulations of European ancestry.3 In contrast, Black African and East Asian populations are reported to have the lowest rates worldwide at 0.25–0.7 cases per 100 000 inhabitants.4, 5 In populations of European ancestry where prevalence is highest, the expanded CAG repeat occurs preferentially on haplotypes A1, A2, and A3a, whereas in African and East Asian patients the HD mutation occurs on a heterogeneous mix of local haplotypes.6, 7, 8, 9 European HD haplotypes A1 and A2 do not occur in African and East Asian populations, suggesting that these haplotypes may account in part for higher HD prevalence rates in populations of European ancestry.4, 7
HD has been reported in many countries of Latin America, but detailed genetic investigations are lacking.10 Early clinical descriptions were reported in Cuba and Brazil, and subsequently in Argentina, Mexico, Peru, Chile, and Colombia.11 No population-based prevalence estimates of HD are available in Latin America, but a major focus of HD in Latin America and the world is in the state of Zulia, surrounding Lake Maracaibo in Venezuela.12 Across Latin America, the HD mutation is believed to originate from European founders with subsequent admixture in indigenous populations.10, 12 Limited haplotyping studies of HD in Latin America suggest that the HD mutation occurs on specific haplotypes, although not necessarily from European founders.10 HTT haplotypes in Venezuelan patients, including the CCG repeat in exon 1 (GRCh37 chr4:g.3076673_3076675[7]) and Δ2642 codon deletion in exon 58 (GRCh37 chr4:g.3230411_3230413delGAG),13 suggest a common origin of the HD mutation among affected families of Venezuela, but without definitive data of European ancestry.14 (HTT exon numbering follows Ensembl transcript HTT-001/ENST00000355072.9, NCBI reference mRNA NM_002111.8, and exon numbering originally described in Ambrose et al.13)
The Cañete Valley of southern Lima has been reported as a demographic focus of HD in Peru,15 and the HD mutation in Peru has been hypothesized to originate in Cañete, possibly from European sources.16 Today HD is known to be widespread in Lima and across northern and southern regions of Peru, with additional reports of HD within native communities of the Peruvian Amazon.17 Peru is mainly composed of a mestizo population with predominant Amerindian genetic ancestry and heterogeneous levels of non-indigenous ancestry, principally from Europe.18 The term ‘mestizo' is used to refer to these Peruvian individuals of mixed ethnic ancestry.19 Genetic data from mestizo Peruvians are consistent with historically documented post-Columbian immigration and admixture from European sources, with minor contributions of African and East Asian immigrants.20, 21 Relative European and indigenous ancestry of HD is unknown in Peru and elsewhere in Latin America. In the absence of detailed haplotype data for the HD mutation in Latin America, the potential to therapeutically target HTT SNPs for allele-specific gene silencing in Latin American patients also remains unclear.22
To illuminate the origins of HD in Peru and other Latin American populations, here we comprehensively haplotype the HTT gene region in a mestizo Peruvian HD patient cohort, in a collection of HD families originating from across Latin America, and in a control population of defined Amerindian ancestry. We uncover a surprising indigenous variant of the A1 HTT haplotype in ethnically distinct Amerindian controls, which is strongly associated with the HD mutation in Peruvian patients and in HD families of diverse Latin American origins. In contrast, this Amerindian A1 haplotype is rare in Caucasian European patients and controls. We propose that the parent A1 HTT haplotype may be the most frequent haplotype of the HD mutation in Latin America, and show that distinct A1 variant haplotypes of European and Amerindian origin are present in contemporary mestizo populations. Alleles shared by European A1 and Amerindian A1 haplotypes may offer optimal targets for the development of allele-specific therapies in ethnically diverse patient populations across North America, South America, and Europe.
Materials and methods
HD patient and control cohorts
Mestizo Peruvian HD patients have been genetically diagnosed at the Neurogenetics Research Center (NRC) at the Instituto Nacional de Ciencias Neurológicas (INCN) in Lima, Peru since 2000. DNA samples from Peruvian patients and controls were collected and stored for diagnostic and research purposes at INCN. IRB approval for this study was obtained from the Ethical Committee at INCN. HD and control donors gave informed consent that allowed for further studies on HD. Amerindian Peruvian control samples were originally collected as part of a genetic study of Parkinson disease, and were granted proper authorization for use in further studies related to neurodegenerative diseases.23 All Peruvian samples were de-identified at the NRC before shipment to the Centre for Molecular Medicine and Therapeutics (CMMT) in Vancouver. Additional Latin American HD families were identified in the UBC HD BioBank from samples collected through the Centre for HD at the University of British Columbia. Latin American HD families were defined by confirmed origin of the proband or both parents from Mexico, Colombia, Ecuador, El Salvador, Argentina or Venezuela. Additional HD patients of European ancestry were screened from the UBC HD BioBank under existing research ethics protocols (UBC C&W CREB H06-70467 and H05-70532).
CAG and CCG Repeat Sizing
CAG repeat genotyping of 265 Peruvian HD and 42 Amerindian control samples was performed at both the NRC and CMMT. CAG and CCG repeat lengths were confirmed in Peruvian, Amerindian, and Latin American individuals against controls of known CAG and CCG length at the CMMT. CAG and CCG repeat sizes were determined as previously described9 using fluorescently labelled primers flanking the CAG repeat (HD344F, 5′-HEX-CCTTCGAGTCCCTCAAGTCCTTC-3′ and HD450R, 5′-GGCGGCGGTGGCGGCTGTTG-3′), the CCG repeat (HD419F, 5′-AGCAGCAGCAGCAACAGCC-3′ and HD482R, 5′-6FAM-GGCTGAGGAAGCTGAGGAG-3′), and both CAG and CCG repeats (HD344F, 5′-HEX-CCTTCGAGTCCCTCAAGTCCTTC-3′ and HD482R, 5′-GGCTGAGGAAGCTGAGGAG-3′) following established HD diagnostic testing guidelines.24
HTT SNP genotyping and haplotype reconstruction
Overlapping custom Illumina GoldenGate arrays of 92 SNPs and 77 SNPs spanning the HTT gene region (Illumina, San Diego, CA, USA) were directly genotyped in 142 mestizo Peruvian, 42 Amerindian Peruvian, and 63 Latin American individuals.7 Curated genotype clusters for each SNP were exported as genotype calls using GenomeStudio (Illumina, San Diego, CA, USA) and formatted for haplotype inference with PHASE v2.1 for each assay design. Accuracy of reconstructed haplotypes was verified where possible by pedigree analysis of genotyped family members. Haplotypes of all Peruvian HD alleles were confirmed by familial segregation. Haplotypes of Latin American HD alleles from the CMMT were confirmed by familial segregation and by CCG repeat association as previously described.7 Following phasing and comparison of haplotype sequences across all genotyped individuals, common haplotypes consisting of 125 SNPs could be inferred from individual 92 SNP or 77 SNP data, of which 79 SNPs had informative heterozygosity in our sample populations (Supplementary Table S1). Genotype and haplotype data from our 62 mestizo Peruvian HD probands have been submitted to the Leiden Open Variation Database (www.LOVD.nl/HTT; individuals 81358-81419).
Identification and genotyping of Amerindian A1 SNPs
Chromosomal Native American ancestry annotations from 1000 Genomes were examined from previously published data.25 Among 25 reference Latin American controls from 1000 Genomes Project Phase 3 with homozygous Native American ancestry across HTT, one was homozygous for the A1 HTT haplotype previously defined by rs72239206 (GRCh37 chr4:g.3142661_3142664delACTT), rs149109767 (GRCh37 chr4:g.3230411_3230413delGAG), and rs362307 (GRCh37 chr4:g.3241845C>T).7 Three additional HTT SNPs in this individual had homozygous variant alleles exclusive to the A1 haplotype among all 1000 Genomes Project samples: rs12508079 (GRCh37 chr4:g.3080238T>C), rs188072823 (GRCh37 chr4:g.3188616C>T), and rs186719032 (GRCh37 chr4:g.3255302G>A). SNP rs12508079:T>C was genotyped in all Peruvian HD chromosomes, all Amerindian control chromosomes, and all Latin American HD chromosomes by TaqMan assay (C___2480924_10, Applied Biosystems, Foster City, CA, USA). SNPs rs188072823:C>T and rs186719032:G>A were genotyped by direct amplicon sequencing (rs188072823: F 5′-TCGTCACTCCAAACACAATGG-3′, R 5′-ACATAAGTCACAGCTGAAGAAAAA-3′, rs186719032: F 5′-GGCCGCTTCGAGGATGAT-3′, R 5′-AACTTTCCGTGCAGCTCAA-3′).
Ancestry analysis of mestizo Peruvian and Amerindian individuals
Twelve Amerindian control samples (six individuals with Amerindian A1 or derivative crossover haplotype and six individuals without A1) and three mestizo Peruvian HD probands with C1 HD haplotypes were selected for ancestry analysis using 1000 Genomes Project Phase 3 samples as a reference. Selected samples were genotyped for 7907 markers using a custom Infinium ADME Core Panel (Illumina), an approach that has been shown to accurately infer genetic ancestry.26, 27 Genotypes were generated in GenomeStudio (Illumina) following standard quality control protocols. PLINK (v1.07) was then employed to remove markers with <95% call rate and deviations from Hardy–Weinberg equilibrium (P<0.001) as well as ensure that all samples had a >95% call rate. The intersection of markers between the genotyped samples and the 1000 Genomes Project samples was subsequently examined. Markers with minor allele frequencies of >1% were subjected to linkage disequilibrium pruning in PLINK (pairwise genotypic correlation: window 50, step 5, r2 0.2). This pruned data set was then assessed using principal component analysis (PCA) with EIGENSOFT (v5.0) and ADMIXTURE (v1.2) employing cross validation to determine the optimal K-value.
Results
HD in Peru occurs predominantly on the A1 HTT haplotype
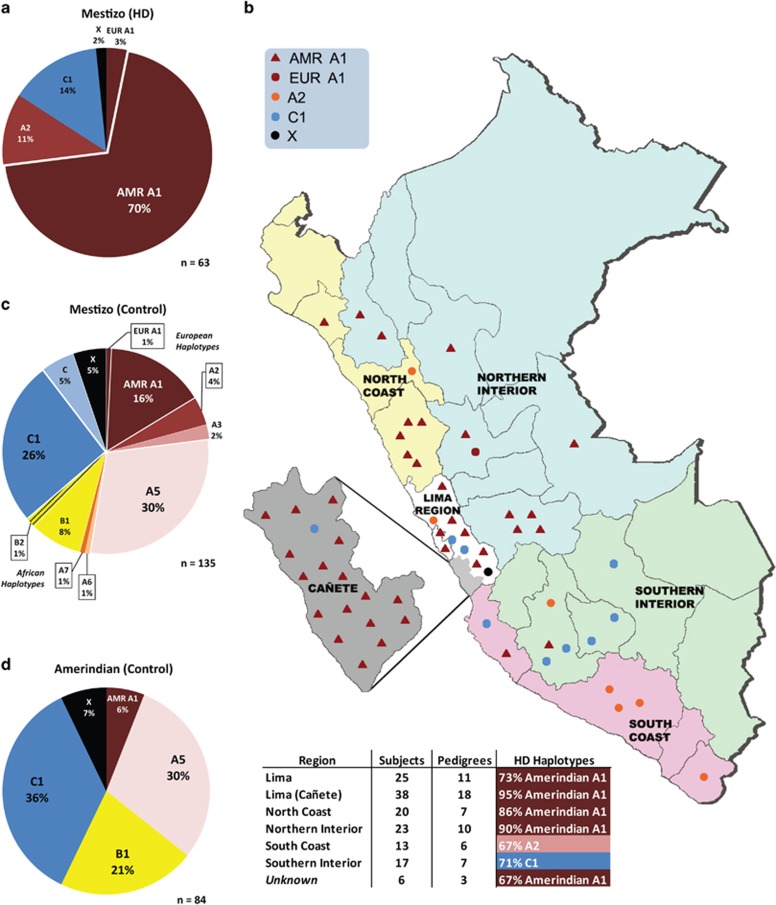
To determine the genetic ancestry of the expanded CAG repeat in Peruvian HD patients, we genotyped 142 individuals from 62 unrelated mestizo Peruvian HD families at either 92 SNPs or 77 SNPs spanning the HTT gene, as previously genotyped in HD patients of European ancestry.7 These mestizo Peruvian HD families spanned five broad geographic regions of the country: Lima, Northern Interior, North Coast, Southern Interior, and South Coast. One HD homozygote was found in one family, and was counted as two independently inherited HD chromosomes. In total, gene-spanning HTT haplotypes of 79 informative SNPs (Supplementary Tables S1 and S2) were fully phased to CAG repeat length in 63 unrelated HD chromosomes and 135 unrelated control chromosomes from mestizo Peruvian patient families.
Forty-six out of 63 (73.0%) Peruvian HD mutations were found on the A1 HTT haplotype (Figure 1a), previously shown to be the most common gene-spanning HTT haplotype in HD chromosomes of European ancestry.7 A1 HD haplotypes occur at an even higher frequency in Peruvian patients than in patients of European ancestry (P=0.0008, χ2, 73% versus 48% in Peru and the UBC BioBank, respectively). Seven of 63 (11.1%) Peruvian HD mutations were found on A2, previously shown to be the second most common HD haplotype in patients of European ancestry. However, Peruvian HD mutations also occurred on the C1 haplotype at similar frequency to A2 (9/63, 14.3%), whereas in European patients the HD mutation on C1 is rare (3.2–4.5%). In East Asian and Southeast Asian patient populations, the HD mutation has also been shown to occur frequently on C1.8, 28 Among black South African patients, the HD mutation occurs on a heterogenous collection of A, B, and C haplotypes, usually informative of African origin.9 In our mestizo Peruvian HD families, no HD haplotypes indicative of African origin were observed.
Figure 1.
HTT haplotype distributions in the Peruvian population. (a) The HD mutation in Peru occurs predominantly on the A1 haplotype (73%), of which nearly all are the Amerindian A1 variant haplotype. (b) Haplotypes of the HD mutation differ by geographic region of Peru. HD occurs most frequently on the A1 haplotype in Lima (including Cañete), North Coast, and the Northern Interior regions, but is more frequent on A2 in South Coast, and C1 in the Southern Interior. (c) Control HTT haplotypes in Mestizo Peruvians reflect indigenous ancestry with major European admixture and minor African contributions. (d) Control Amerindian HTT haplotypes reflect low genetic diversity in this subpopulation relative to Mestizo controls, and reveal the Amerindian A1 haplotype in the indigenous population of Peru.
There are significant regional differences in the frequency of HD haplotypes across Peru (Figure 1b). A1 predominates in Lima, the North Coast, and the Northern Interior, whereas A2 and C1 are more common in the South Coast and Southern Interior, respectively. In Cañete, where a historical focus of HD is reported, HD occurs almost exclusively on A1.
Control chromosomes from Peruvian HD pedigrees represent diverse ethnic origins, including typically European, East Asian, and African HTT haplotypes (Figure 1c). Interestingly, A1 occurs at a higher frequency in mestizo Peruvian controls (16.3%) than in European controls (8.0–9.7%), whereas this haplotype is absent in East Asian populations.7 A5 also occurs at a higher frequency in mestizo Peruvian controls (29.6%) than in European controls (6.1–6.9%), but at a rate comparable to East Asian controls (27%).8
A1 haplotype occurs in the indigenous Amerindian population of Peru
In order to investigate indigenous Amerindian contributions to the ancestry of HD in mestizo Peruvian patients, we additionally haplotyped 42 control Amerindian individuals with our 77 SNP panel (Figure 1d). Haplotype diversity among the Amerindian control individuals was low, with only 5 distinct gene-spanning haplotypes revealed by our SNP panel versus 21 in mestizo Peruvians (Supplementary Table S2), reflecting prior findings of low genetic diversity and serial founder effects in Native American populations.18, 29 Unexpectedly, the A1 haplotype defined by rs72239206 (GRCh37 chr4:g.3142661_3142664delACTT), rs149109767 (GRCh37 chr4:g.3230411_3230413delGAG), and rs362307 (GRCh37 chr4:g.3241845C>T), known to be associated with HD in European populations, was found in multiple Amerindian control chromosomes (5/84, 6.0%), whereas it is entirely absent in East Asian reference populations to which indigenous Americans are more closely related. Other common HTT haplotypes that would indicate European ancestry, such as A2, are absent among our Amerindian control individuals, arguing against European admixture as the source of A1 in this population.
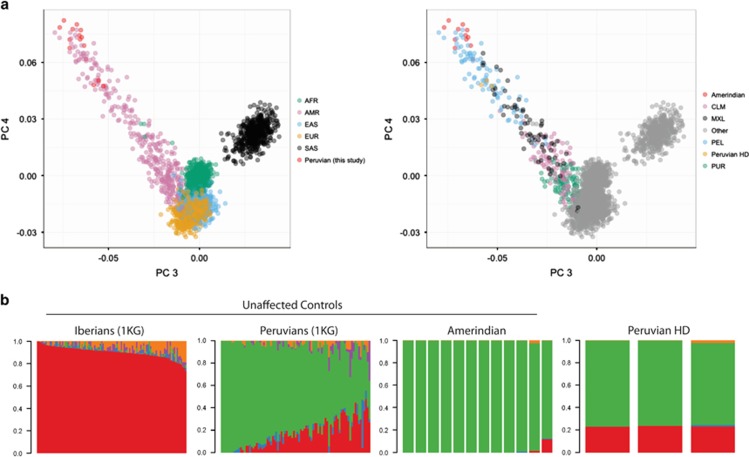
To examine the possibility of cryptic admixture in our Amerindian controls, which could support a European origin of A1 in these individuals, we genotyped 7907 SNPs in a subset of 12 Amerindian controls and three mestizo HD patients using a custom genetic diversity panel. All five Amerindian A1 haplotype carriers were included, in addition to one putative A1 crossover haplotype. It has been shown by our group and others that genotypes from the ADME panel enable identification of individuals with misattributed ancestry by principal component analysis (PCA).26 We extracted genotypes representing the ADME panel from Phase 3 of the 1000 Genomes Project and performed PCA and ADMIXTURE analysis with our Amerindian and mestizo Peruvian HD samples. PCA revealed that the Amerindian samples formed an extreme genetic cluster closest to Peruvian reference samples (PEL) from the 1000 Genomes Project (Figure 2a). There was no difference in the clustering of Amerindian A1 carriers versus Amerindian individuals without A1. Our three selected mestizo Peruvian HD patients, each with HD on C1, were intermediate between the Amerindian cluster and individuals of European ancestry, and overlapped with reference Peruvian samples. ADMIXTURE analysis confirmed the predominant indigenous ancestry of our Amerindian controls, in contrast to varying degrees of European admixture in reference Peruvian individuals and the mestizo Peruvian HD probands (Figure 2b; Supplementary Figure S1). These data strongly suggest that our Amerindian controls are of uniform indigenous descent despite the presence of the A1 HTT haplotype typical of Europeans. From these data, we hypothesized that the A1 haplotype in Amerindians and in some mestizo Peruvians may be of ancient Amerindian origin rather than recent European origin via admixture.
Figure 2.
Genome-wide admixture analysis of selected Amerindian controls and mestizo HD patients from Peru relative to the 1000 Genomes Project reference samples. (a) Plotting of principal components 3 and 4 from Amerindian Peruvian individuals reveals extreme clustering by ethnicity and partial overlap with reference Peruvians from the 1000 Genomes Project. Amerindian Peruvian controls are most similar to reference Peruvians, reflecting known heterogeneity of indigenous genetic ancestry across Latin America. (b) ADMIXTURE analysis suggests a nearly exclusive indigenous ancestry among Amerindian Peruvian controls, including all Amerindian individuals with the A1 haplotype. Mestizo Peruvian HD probands show similar levels of admixture as reference Peruvians relative to Europeans from Spain and Portugal (K=5, see Supplementary Figure 1).
Amerindian A1 haplotype is marked by specific SNPs and represents the majority of HD mutations in Peru
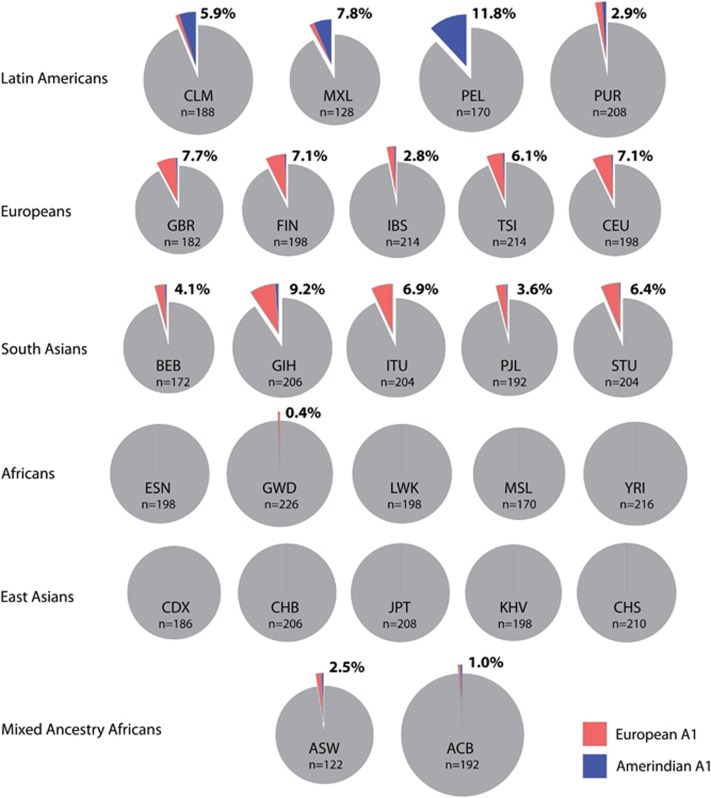
To determine if additional genetic variants could distinguish A1 haplotypes of Amerindian origin from A1 haplotypes of European origin, we examined HTT variants in A1 haplotypes of admixed Latin American individuals from the 1000 Genome Project. Specific chromosomal segments from these individuals have been previously identified as Native American, European, or African by local genomic ancestry inference.25 Among 25 reference Latin American individuals from 1000 Genomes Project Phase 3 with homozygous Native American ancestry across HTT, one was homozygous for the A1 HTT haplotype defined by rs72239206 (GRCh37 chr4:g.3142661_3142664delACTT), rs149109767 (GRCh37 chr4:g.3230411_3230413delGAG), and rs362307 (GRCh37 chr4:g.3241845C>T). This individual was found to be homozygous for three additional SNP variants specific to the A1 haplotype: rs12508079:T>C in HTT intron 1, rs188072823:C>T, and rs186719032:G>A. In the 1000 Genomes Project Phase 3 data set, the rs12508079:T>C variant allele is highly enriched in A1 from Latin American individuals (Figure 3 and Supplementary Figure S2; Supplementary Table S3). A1 with rs12508079:T>C represents 9/10 (90.0%), 8/10 (80.0%), and 22/22 (100%) of Colombian, Mexican, and Peruvian A1 control haplotypes, respectively. In contrast, the rs12508079:T>C variant allele is rare among A1 HTT control chromosomes from European and South Asian reference populations, where the A1 haplotype is commonly found. The rs12508079:T>C variant allele is never found in individuals of exclusive African or East Asian ancestry, in agreement with the previously established absence of A1 in these populations.7, 8, 9
Figure 3.
European and Amerindian A1 HTT haplotype frequencies across all reference populations of the 1000 Genomes Project. Latin American reference populations have the highest frequency of the Amerindian A1 variant haplotype, constituting the majority of all A1 haplotypes among Colombians (CLM), Mexicans (MXL), and Peruvians (PEL). Amerindian A1 also constitutes a third of all A1 haplotypes among Puerto Ricans (PUR). In contrast, Amerindian A1 is rare in European and South Asian populations where the parent A1 haplotype is common: British (GBR), Finnish (FIN), Iberian (IBS), Toscani Italian (TSI), Utah Caucasian (CEU), Bengali (BEB), Gujarati (GIH), Telugu (ITU), Punjabi (PJL), and Sri Lankan Tamil (STU). The parent A1 haplotype is entirely absent in Black African and East Asian populations, except for one instance in a Gambian individual. European and Amerindian A1 are both found among admixed Africans from the United States (ASW) and Barbados (ACB).
Variant alleles of rs186719032:G>A and rs188072823:C>T were found on 20/22 (91.0%) A1 control haplotypes in the Peruvian reference population, but only half of Mexican and Colombian A1 control haplotypes, suggesting a diversity of Amerindian subtypes among A1 haplotypes bearing rs12508079:T>C. We thus define the Amerindian A1 haplotype as any HTT haplotype carrying the rs12508079:T>C variant allele in addition to the canonical A1 variant alleles at rs72239206, rs149109767, and rs362307. Some, but not all Amerindian A1 haplotypes also bear rs186719032:G>A and rs188072823:C>T variant alleles, representing Amerindian A1 subtypes.
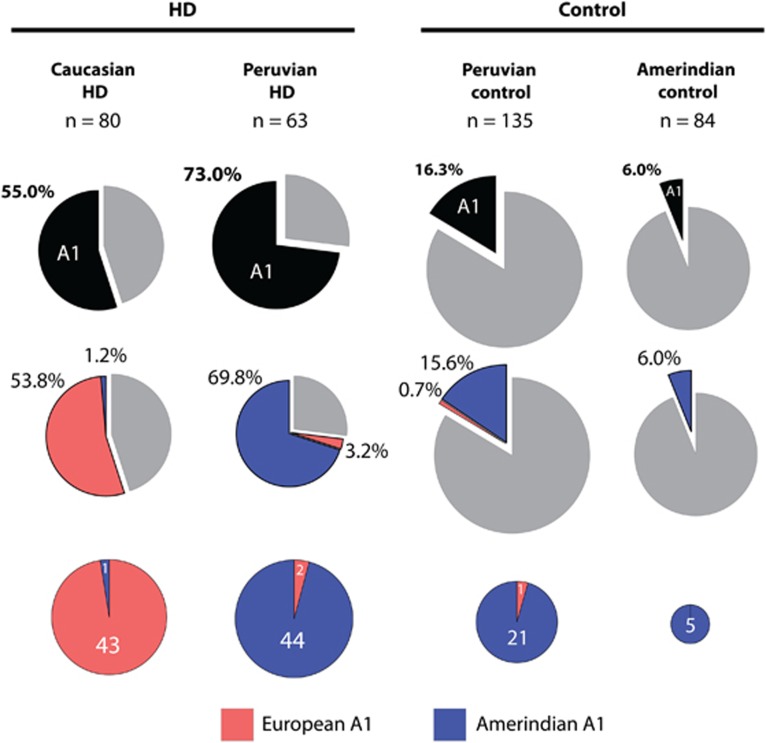
We directly genotyped rs12508079:T>C, rs186719032:G>A, and rs188072823:C>T in all Amerindian control samples and all Peruvian HD patients, then phased to 79-SNP HTT haplotypes and CAG repeat length by familial segregation (Figures 1 and 4; Supplementary Tables S2 and S4). All five Amerindian control chromosomes with A1 variant alleles at rs72239206, rs149109767, and rs362307 also carried variant alleles at rs12508079, rs186719032, and rs188072823, suggesting a common Amerindian A1 haplotype by descent. Strikingly, 41/45 (91.1%) mestizo Peruvian A1 HD chromosomes occur on the Amerindian A1 haplotype bearing all 3 Amerindian A1-specific alleles, and not on the European A1 haplotype lacking these 3 derived Amerindian variants, supporting a pre-Columbian origin of HD in this population rather than a proximate European origin as previously supposed. Specific Amerindian A1 variant alleles at rs12508079:T>C, rs186719032:G>A and rs188072823:C>T did not occur on any other haplotypes than A1 in direct genotyping of our Peruvian HD patients and Amerindian controls.
Figure 4.
The frequency of European and Amerindian A1 HTT haplotypes differs dramatically between HD chromosomes from Caucasian Canadian patients and mestizo Peruvian patients. The A1 HTT haplotype in mestizo Peruvian HD and control chromosomes is almost exclusively the Amerindian A1 variant. In Amerindian Peruvian controls, the A1 HTT haplotype is exclusively the Amerindian A1 variant. In contrast, the Amerindian A1 haplotype variant occurs rarely in Caucasian Canadian HD chromosomes. The A1 HTT haplotype is defined by variant alleles at rs72239206 (GRCh37 chr4:g.3142661_3142664delACTT), rs149109767 (GRCh37 chr4:g.3230411_3230413delGAG), and rs362307 (GRCh37 chr4:g.3241845C>T). The Amerindian A1 variant haplotype is additionally defined by the C allele at rs12508079 (GRCh37 chr4:g.3080238T>C).
Additionally, we genotyped 80 unrelated HD chromosomes of European ancestry for rs12508079:T>C, 44 (55%) of which were previously found to have the A1 HTT haplotype defined by variant alleles at rs72239206, rs149109767, and rs362307 (Figure 4). In stark contrast to Peruvian HD chromosomes with the A1 haplotype, only 1/44 European HD chromosome with the A1 haplotype carried the Amerindian A1-defining allele of rs12508079:T>C. Fully 43/44 (97.7%) European A1 HD chromosomes lack the Amerindian A1-defining variant allele, in agreement with the rarity of rs12508079:T>C in A1 chromosomes from European controls in the 1000 Genomes Project.
Amerindian A1 is the most common HD haplotype in HD families of Latin American origin
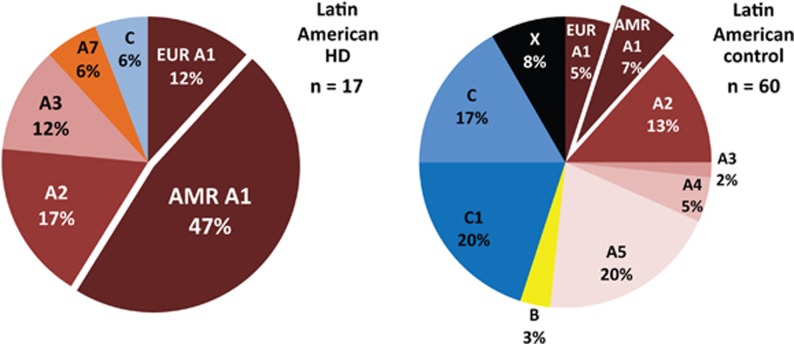
To extend our findings of an Amerindian-specific A1 variant to HD chromosomes from other Latin American populations, we haplotyped 63 subjects from 17 HD families of diverse Latin American origins in the UBC HD BioBank (Figure 5). Ten out of 17 (59%) Latin American HD chromosomes were found on the A1 HTT haplotype marked by variant alleles at rs72239206, rs149109767, and rs362307. Eight of these 10 Latin American A1 HD chromosomes (80%) also carried the rs12508079:T>C variant allele, similar to the enrichment for Amerindian A1 observed in Peruvian HD chromosomes. One HD mutation was observed on the A7 haplotype, suggesting African ancestry, and one on the C4 haplotype found in both European and African patients. Among seven unrelated A1 control chromosomes from our Latin American HD families, four carried the Amerindian-specific variant allele of rs12508079:T>C, similar to the enrichment for Amerindian A1 in Latin American controls from the 1000 Genomes Project. Control chromosomes from Latin American HD families were also enriched for A5 (20%) relative to European controls, as in mestizo Peruvian (30%) and Amerindian controls (30%). A1 and A2 of European ancestry were observed in our admixed Latin American controls but few haplotypes of definitive African ancestry, suggesting predominant Amerindian and European ancestry in these families.
Figure 5.
HTT haplotype distribution of HD and control chromosomes in Latin American HD families from the UBC HD BioBank. Amerindian A1 is the most frequent HD haplotype in a collection of patients from across Latin America (47%). The parent A1 haplotype, which could be therapeutically targeted for allele-specific HTT silencing in European patient populations, constitutes the majority (59%) of HD chromosomes in Latin American patients. Latin American control haplotypes reflect indigenous Amerindian and European admixture.
The occurrence of the HD mutation on the parent A1 haplotype in both European and Latin American patients suggests that common defining A1 variant alleles could represent shared targets for allele-specific silencing in both populations. In European patients, A1 and A2 allele targets in combination allow allele-specific treatment of the most HD patients.7 We find that defining A1 alleles at rs72239206 (GRCh37 chr4:g.3142661_3142664delACTT), rs149109767 (GRCh37 chr4:g.3230411_3230413delGAG), and rs362307 (GRCh37 chr4:g.3241845C>T) allow allele-specific treatment of the most Peruvian HD patients given one allele target (35/62, 56.5%), and allow treatment of the most Peruvian HD patients overall in combination with A2 (42/62, 67.7%). A1 and A2 alleles in combination also allow treatment of 71% (12/17) of Latin American HD probands among our UBC BioBank families. These data suggest that A1 and A2 allele targets may represent optimal targets for allele-specific therapy in Latin American HD patient populations, as in patient populations of predominant European ancestry.
Discussion
To date, the origin of HD in Latin America has remained unclear in the absence of clear genetic ancestry markers within HTT. To our knowledge, this is the first analysis of dense haplotypes of the HD mutation in Latin America, allowing description of the ancestry of the HD mutation in Peru and other patients of Latin American origin. We show that HD in Latin America occurs most frequently on the A1 haplotype, as in European HD patients. Remarkably, a subtype of the A1 haplotype occurs in individuals of confirmed Amerindian ancestry, and represents the most frequent HD haplotype in patients from Peru and elsewhere in Latin America. HD in Latin American patients is also found on haplotypes of European ancestry and in rare instances on haplotypes suggestive of African origin, reflecting extensive admixture in Latin America.30 HD mutations on C1 may represent either an indigenous origin or a recent founder effect resulting from East Asian immigrants that arrived in Peru over the past two centuries.21 However, ADMIXTURE analysis shows minimal East Asian ancestry among Peruvian patients with the HD mutation on C1, supporting an indigenous origin of the C1 disease haplotype in these families. Peruvian HD chromosomes on A2, frequent in the South Coast, may represent recent admixture from European sources.
Ancestry admixture in Latin America is known to occur in distinct proportions by country and region. Interestingly, our HTT haplotype data and admixture analysis of individuals from the Peruvian population reflect the known demographic and genetic history of Peru. For example, the Peruvian population has one of the highest Amerindian ancestry components (83%) among all evaluated groups, with admixed Spanish European ancestry and marginal African and Asian contributions.18 Indeed, our HTT haplotyping data of Peruvian HD patients shows a predominant Amerindian ancestry at this locus, with European admixture and isolated African contributions among HD and control chromosomes. Interestingly, a recent epidemiological study of the incidence of HD among American Indians across the United States revealed an absence of the disease among the Navajo.31 In addition to admixture differences, there may be considerable genetic heterogeneity of HD among different Native American subpopulations.
The presence of the A1 haplotype in Amerindian individuals with no discernable European admixture, and the identification of ancestry-specific A1 alleles that are frequent in Latin American individuals but rare in Europeans, suggests that the parent A1 haplotype has deep common ancestry between European and Native American populations. This is further supported by the presence of the A1 haplotype in all reference populations of South Asian ancestry. Shared ancestry of the parent A1 haplotype between Europeans and Native Americans is unexpected given the absence of A1 among contemporary East Asian individuals to which Native Americans are believed to be related. Recent reports have suggested that Native American populations may have closer ancestry to indigenous Siberian and Central Asian populations than to contemporary East Asians, and that gene flow between Eurasian populations and Native Americans may have occurred. Nuclear DNA extracted from 24 000-year-old remnants of a boy in south-central Siberia indicated a shared ancestry between contemporary Native Americans and Europeans but divergent from modern East Asians.32 We hypothesize that the A1 haplotype was present in a similar ancient population of Central Asia, distinct from the ancestors of contemporary East Asians. Alternatively, A1 may have been lost in contemporary East Asians by bottleneck effects and genetic drift. The defining Amerindian A1 alleles at rs12508079:T>C, rs186719032:G>A, and rs188072823:C>T are observed at low frequency outside Latin American populations, and these alleles may represent reverse admixture from New World sources. Alternatively, these alleles may have been present in ancestral Indo-European populations and become enriched in Amerindian populations by serial founder events. Detailed examination of Amerindian A1 alleles in HD and control cohorts from other defined indigenous populations would be necessary to clarify and test different migration models of this haplotype.
Regardless of the ancestral origin of A1 worldwide, the common occurrence of HD on the parent A1 haplotype in both Europe and Latin America suggests that allele-specific therapies targeting A1 could be designed for patients in both populations. We previously demonstrated that ~40% of HD patients of European ancestry could be treated with antisense reagents selective for the A1 HTT haplotype.7 In our Peruvian HD cohort the percentage of treatable patients is higher than in European populations, with 57% of Peruvian HD probands heterozygous for A1 and phased to the HD mutation. Among our Latin American HD patients, 53% would be treatable with A1 targets, also exceeding the heterozygosity observed in European patients. This suggests that antisense reagents developed for allele-specific suppression of the A1 haplotype (ie, against the deleted allele of rs72239206, the deleted allele of rs149109767, or the T allele of rs362307) may have similar or greater utility in treatment of Latin American HD patients as in patients of major European descent. Further, as in European patients, alleles specific to the A1 and A2 haplotypes allow treatment of the greatest number of Peruvian patients in combination (68%) among all possible combinations of two allele targets, suggesting that A1 and A2 may also represent prioritized panels of primary and secondary allele targets in Latin American patient populations. In conclusion, our study shows that the targetable A1 HD haplotype is common in Latin America, but that this haplotype has genetically distinct Amerindian and European origins in contemporary mestizo American populations.
Acknowledgments
We thank the many patients cared for by the INCN and the UBC Centre for HD for their donation of DNA for this study. We also thank Simon Warby for initial communications that led to the collaboration between INCN and the CMMT at UBC. We acknowledge Michelle Higginson and Nasim Massah for their assistance in genotyping. CK is supported by a Doctoral Research Award from the Canadian Institutes of Health Research. Funding for this study was provided by the Canadian Institutes of Health Research (CIHR: MOP-84438).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Bates GP, Dorsey R, Gusella JF et al: Huntington disease. Nat Rev Dis Primers 2015; 1: 15005. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group: A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993; 72: 971–983. [DOI] [PubMed] [Google Scholar]
- Fisher ER, Hayden MR: Multisource ascertainment of Huntington disease in Canada: prevalence and population at risk. Mov Disord 2014; 29: 105–114. [DOI] [PubMed] [Google Scholar]
- Kay C, Fisher ER, Hayden M: Epidemiology (in Huntington's Disease), 4th ed. Oxford University Press: New York, NY, USA, 2014. [Google Scholar]
- Baine FK, Krause A, Greenberg LJ: The frequency of Huntington disease and Huntington disease-like 2 in the South African population. Neuroepidemiology 2016; 46: 198–202. [DOI] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR et al: CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet 2009; 84: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay C, Collins JA, Skotte NH et al: Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol Ther 2015; 23: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Visscher H, Collins JA et al: HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet 2011; 19: 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baine FK, Kay C, Ketelaar ME et al: Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur J Hum Genet 2013; 21: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilhos RM, Augustin MC, Santos JA, Perandones C, Saraiva-Pereira ML, Jardim LB: Genetic aspects of Huntington's disease in Latin America. A systematic review. Clin Genet 2015; 89: 295–303. [DOI] [PubMed] [Google Scholar]
- Torres Ramirez L, Cosentino Esquerre C, Mori Quispe N: Actualización sobre la enfermedad de Huntington y experiencia de 30 años en el Instituto Nacional de Ciencias Neurológicas. Diagnostico 2008; 47: 65–69. [Google Scholar]
- Wexler NS, Lorimer J, Porter J et al: Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci USA 2004; 101: 3498–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Duyao M, Barnes G et al: Structure and expression of the Huntington's disease gene: Evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet 1994; 20: 27–38. [DOI] [PubMed] [Google Scholar]
- Paradisi I, Hernandez A, Arias S: Huntington disease mutation in Venezuela: age of onset, haplotype analyses and geographic aggregation. J Hum Genet 2008; 53: 127–135. [DOI] [PubMed] [Google Scholar]
- Cuba JM: [A focus of Huntington's chorea in Peru]. Rev Neurol (Paris) 1986; 142: 151–153. [PubMed] [Google Scholar]
- Cornejo-Olivas MR, Inca-Martinez MA, Espinoza-Huertas K et al: Clinical and molecular features of late onset Huntington disease in a Peruvian cohort. J Huntingtons Dis 2015; 4: 99–105. [PubMed] [Google Scholar]
- Cornejo-Olivas M, Mori N, Alva I, Zunt J P M. Huntington's Disease in an Indigenous Village in the Peruvian Amazon Jungle. 16th International Congress of Parkinson's Disease and Movement Disorders. Dublin, Ireland, 2012, vol 17-21.
- Sandoval JR, Salazar-Granara A, Acosta O et al: Tracing the genomic ancestry of Peruvians reveals a major legacy of pre-Columbian ancestors. J Hum Genet 2013; 58: 627–634. [DOI] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W et al: Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet 2008; 4: e1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García LN: Historia de las Américas. Madrid: Editorial Alhambra Universidad de Sevilla, 1991. [Google Scholar]
- Herrera I: Los Inmigrantes Chinos en la Amazonia Peruana. Bull Inst Français d'Etudes Andines 1986; 15: 49–60. [Google Scholar]
- Kay C, Skotte NH, Southwell AL, Hayden MR: Personalized gene silencing therapeutics for Huntington disease. Clin Genet 2014; 86: 29–36. [DOI] [PubMed] [Google Scholar]
- Cornejo-Olivas M, Mata I, Dorschner M, Inca-Martinez M, Medina A, Shetty AC. , (LARGE-PD) LARCotGoPsD: Target sequencing analysis of Parkinson's disease genes in a healthy Amerindian population from Puno-Peru. 64th Annual Meeting of the American Society of Human Genetics, San Diego, CA, USA, 2014.
- Bean L, Bayrak-Toydemir P: American College of Medical Genetics and Genomics Standards and Guidelines for Clinical Genetics Laboratories, 2014 edition: technical standards and guidelines for Huntington disease. Genet Med 2014; 16: e2. [DOI] [PubMed] [Google Scholar]
- Gravel S, Zakharia F, Moreno-Estrada A et al: Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet 2014; 9: e1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher H, Ross CJ, Dube MP et al: Application of principal component analysis to pharmacogenomic studies in Canada. Pharmacogenomics J 2009; 9: 362–372. [DOI] [PubMed] [Google Scholar]
- Jackson JN, Long KM, He Y, Motsinger-Reif AA, McLeod HL, Jack J: A comparison of DMET Plus microarray and genome-wide technologies by assessing population substructure. Pharmacogenet Genomics 2016; 26: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkes T, Papsing C, Wattanapokayakit S, Mahasirimongkol S: CAG-expansion haplotype analysis in a population with a low prevalence of Huntington's disease. J Clin Neurol 2014; 10: 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M et al: Genetic variation and population structure in native Americans. PLoS Genet 2007; 3: e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Linares A, Adhikari K, Acuna-Alonzo V et al: Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet 2014; 10: e1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PH, Mehal JM, Rowland AS, Cheek JE, Bartholomew ML: Huntington disease among the Navajo: A population-based study in the Navajo Nation. Neurology 2016; 86: 1552–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE et al: Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 2014; 505: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.