Abstract
Providing ostensibly healthy individuals with personal results from whole-genome sequencing could lead to improved health and well-being via enhanced disease risk prediction, prevention, and diagnosis, but also poses practical and ethical challenges. Understanding how individuals react psychologically and behaviourally will be key in assessing the potential utility of personal whole-genome sequencing. We conducted an exploratory longitudinal cohort study in which quantitative surveys and in-depth qualitative interviews were conducted before and after personal results were returned to individuals who underwent whole-genome sequencing. The participants were offered a range of interpreted results, including Alzheimer's disease, type 2 diabetes, pharmacogenomics, rare disease-associated variants, and ancestry. They were also offered their raw data. Of the 35 participants at baseline, 29 (82.9%) completed the 6-month follow-up. In the quantitative surveys, test-related distress was low, although it was higher at 1-week than 6-month follow-up (Z=2.68, P=0.007). In the 6-month qualitative interviews, most participants felt happy or relieved about their results. A few were concerned, particularly about rare disease-associated variants and Alzheimer's disease results. Two of the 29 participants had sought clinical follow-up as a direct or indirect consequence of rare disease-associated variants results. Several had mentioned their results to their doctors. Some participants felt having their raw data might be medically useful to them in the future. The majority reported positive reactions to having their genomes sequenced, but there were notable exceptions to this. The impact and value of returning personal results from whole-genome sequencing when implemented on a larger scale remains to be seen.
Introduction
Whole-genome and exome sequencing are increasingly used in clinical, research, and commercial contexts. Whole-genome sequencing (sequencing of most of the euchromatin DNA in a genome) and exome sequencing (sequencing of the protein-coding parts of the genome) are used for diagnosing rare diseases,1, 2 large-scale research into rare and common diseases,3 and are available commercially direct to consumers.4 These technologies are more comprehensive and detect more types of DNA variation than targeted genetic tests and single-nucleotide polymorphism (SNP) arrays. However, the huge volume of data produced by whole-genome sequencing in particular raises many practical and ethical challenges. These include issues around informed consent, data analysis, interpretation of rare variants, and return of results and raw data.5, 6, 7
The increasing availability of whole-genome sequencing inside and outside the clinic has prompted debates about its value for ostensibly healthy individuals.8, 9, 10 The value of DNA-based information can be thought of as the ratio of benefits to harms, and has traditionally been conceptualized as ‘clinical utility' (usefulness in the clinic and impact on clinical end points).11 However, genomic information may also have personal and social value, for example, as entertainment, learning, or a way to relate to others.12, 13 Understanding how individuals react to personal results will be key in assessing how they themselves perceive the potential utility or value of whole-genome sequencing.
Although studies have begun exploring issues related to the value of personal genomic sequencing for healthy individuals,14, 15, 16, 17 these largely focus on health-related results. SNP-based studies have similarly focused primarily on health-related results,18, 19, 20 often for a single disease or trait.21, 22, 23 Current evidence from SNP-based tests suggests that DNA-based disease risk information has little emotional impact on people,18 and does not motivate them to improve health behaviours.24, 25 However, SNP-based tests generally do not detect rare variants that may have more important roles in health.
Given that whole-genome sequencing will become the standard in detecting genomic variation in the future, the full scope of potential reactions to the wide range of personal results that may arise from whole-genome sequencing, including rare disease-associated variants, should be explored. Protection motivation theory (PMT)26 may provide a useful theoretical framework for the health-related component of this exploration. PMT is a social cognition model that arose out of fear-arousing communications research, and is widely used as a framework for understanding health behaviours. According to PMT, individuals are motivated to engage in health-protective behaviours (‘protection motivation') when they perceive a threat to their health (‘threat appraisal') and when they feel able to reduce that threat through their actions (‘coping appraisal'). SNP-based tests may not elicit strong behavioural responses because individuals appraise the health threat conveyed by the disease risk information to be low, and so are not motivated to act on it. However, rare disease-associated variants may elicit stronger emotional reactions and thus stronger protection motivation and subsequent behavioural reactions.
We therefore explored how healthy individuals reacted to receiving a range of personal whole-genome sequencing results in the HealthSeq project, using the PMT to guide our conceptual framework relating to the health-related results. HealthSeq is an exploratory longitudinal study designed to explore motivations for whole-genome sequencing, satisfaction, and the impact of personal whole-genome sequencing results. In light of the public fascination with aspects of genetics such as ancestry,27 non-health as well as health-related results were offered. Raw sequence data was also offered, given current discussions around patients' access to their personal clinical/research data.28, 29, 30 As previously reported, most (33/35) HealthSeq participants wanted all the available results, and expressed a range of motivations, including learning personal health-related information, obtaining ancestry information, contributing to research, and satisfying curiosity.31 We have also previously reported on participants' satisfaction with the first genetic counselling session.32
The overarching aim of this study was to explore how people react to receiving personal results from whole-genome sequencing. The Specific Aims were to explore: (i) the emotional impact and perceived value of personal results from whole-genome sequencing; (ii) the behavioural impact; (iii) whether and how people's reactions vary by results categories (eg, ancestry vs type 2 diabetes risk); and (iv) whether and how people's reactions vary according to the personal results they receive within those results categories (eg, increased vs decreased risk of type 2 diabetes).
Methods
This was a mixed-methods longitudinal cohort study in which participants underwent whole-genome sequencing, received personal results, and completed interviews at five time points (T1–T5). Recruitment and T1 procedures have been previously described.31, 32 After results were returned at T2 (see the section ‘Return-of-results appointments and reports' below), telephone follow-up interviews were conducted 1 week (T3), 6 months (T4), and 1 year (T5) later. We used PMT to guide our conceptual framework (Supplementary Figure 1) and inform our selection of constructs and measures relating to the health-related results (Supplementary Table 1). For example, we assessed fear (operationalized quantitatively as ‘test-related distress'), and intended and actual behavioural reactions, in response to personal results from whole-genome sequencing. We are not aware of any existing models that provide a framework for both health-related and non-health-related personal results simultaneously, so our formal framework focuses on responses to health-related results only. However, the qualitative topic guide and analysis plan were designed to also explore responses to the non-health-related results, as well as other potentially important psychological constructs and behaviours (see the section ‘Six-month follow-up interviews' below). This paper describes the return-of-results appointment, and presents findings from the T4 interviews and repeated measures from T1 to T4.
Return-of-results appointments and reports
At the in-person return-of-results appointment (T2), the study genetic counsellor and medical geneticist verified whether the participant still wanted to receive the results categories they had consented to at T1. The personal results were then returned. The participants were given a paper report comprising the following categories of information and interpreted results: sequencing quality; ancestry; physical traits (lactose intolerance, bitter taster type, cilantro taster type, earwax dry/wet type); pharmacogenomics (simvastatin, clopidogrel, warfarin); common polygenic disease risk (type 2 diabetes, coronary artery disease, age-related macular degeneration (AMD)); Alzheimer's disease risk; monogenic disease carrier variants (‘rare carrier variants'); and monogenic disease variants (‘rare disease-associated variants'). The genetic counsellor and medical geneticist systematically reviewed the report with the participant, reviewing each section, providing background and then discussing the meaning of the results. Questions were answered and concerns were addressed for each section before moving to the next. The participants were given the option to receive their raw sequencing data on a portable hard drive. Results sessions lasted approximately 1 h depending on the quantity of results and participant concerns. Immediately after the counselling session, the participants completed the T2 interview with the research coordinator. On completion of the study (ie, after all follow-up interviews had been completed), the results reports were reviewed and all rare variants that had been classified as pathogenic or likely pathogenic were re-interpreted, to examine whether any classifications had changed in the intervening time.
Six-month follow-up interviews
The T4 telephone interview schedule comprised two parts. Part one was a topic guide containing seven broad questions plus suggested prompts. The questions were developed by a multidisciplinary team and informed by PMT and other relevant literature. The questions were designed to explore reactions to personal results from whole-genome sequencing; understanding and recall of results; and satisfaction with procedures and materials. This paper focuses on reactions to personal results from whole-genome sequencing. The relevant questions in the topic guide included: How are you feeling about your decision to get your genome sequenced? How did your genome sequencing results make you feel? Did your results influence your behaviour in any way? If yes, how? Have you shared or intend to share your results with anyone? If yes, with whom and why? Part two of the interview schedule contained valid, reliable measures of depression,33 anxiety,34 quality of life (QoL),35 positive experiences;36 PMT-related measures were fear (test-related distress36) and lifestyle behaviours.37 Interviews were audio-recorded and transcribed verbatim for qualitative analysis. Responses to closed-ended questions were entered into a database for quantitative analysis.
Analyses
A thematic framework approach was developed by SCS and SAS for the analysis of the transcripts from part one of the interviews.38, 39 The initial broad themes were guided by the literature and the overarching objectives, and included two sets of themes: emotional reactions, perceived value, and behavioural reactions; and reactions to results categories and offer of raw data. For the latter set, sub-themes were determined by the categories in the results reports, for example, reactions to ancestry results, reactions to pharmacogenomics results, and so on. For the former set, more differentiated sub-themes were generated in a ‘bottom-up' manner based on an in-depth analysis of 10 interview transcripts: SCS and SAS each analysed five transcripts, individually developed a set of themes and sub-themes based on these analyses, compared the two sets, resolved any differences through discussion, and jointly produced a single codebook. For behavioural reactions, each sub-theme (eg, ‘Communication with family') was further sub-categorized into ‘Action' (has already done this) ‘Intention' (plans to do this), and ‘None' (has not done this and does not plan to). The codebook is available from the authors on request.
SCS used this codebook to code all T4 transcripts using software package NVivo 10. Although not the primary focus here, SCS also read all T2 and T3 transcripts to establish that no key themes were being missed in the T4 transcripts. SAS coded a subset of T4 transcripts and assisted in resolving any uncertainties in coding. A non-exclusive coding approach was used, for example, a text segment could be coded as both ‘negative emotional reaction' and ‘reactions to Alzheimer's disease results'.
To address Specific Aims 1 and 2, the coded text passages were explored and categorized by SCS and SAS into a final characterization of participants' emotional and behavioural reactions overall. To facilitate the analyses for Specific Aims 3 and 4, a tabular thematic framework in Microsoft Excel was used. In this framework, each participant interview corresponded to one row. The columns corresponded to results categories. Each results category (eg, Alzheimer's disease) was represented by two columns. The first column contained the participant's actual result (eg, APOE e3/e4). The second column contained their reaction to the result (eg, if a text segment from that participant's interview had been coded as ‘Reactions to Alzheimer's disease result', then that coded text would be entered here). This thematic framework approach allowed us to explore whether different themes emerged for different categories of genomic results, as well as for different personal results within those categories. It also allowed us to read across the rows, facilitating understanding of individual participants' stories.
For the measures in part two of the interview, variables were described using means, standard deviations, and frequencies. Changes over time were assessed using repeated-measures analysis of variance (ANOVA) and the Wilcoxon signed-rank test. All statistical analyses were conducted using software package SPSS v.20 (IBM, Chicago, IL, USA).
Results
Socio-demographic characteristics
Twenty-nine (82.9%) of the 35 participants completed T4 and were included in the analyses. As Table 1 shows, the participants were 41% female, aged 26–68 years, 79% White non-Hispanic, and 52% had annual household incomes over $150000.
Table 1. Socio-demographic characteristics of participants.
| Characteristic | Participants who completed 6-month follow-up (N=29) |
|---|---|
| Gender | |
| Male | 17 (58.6%) |
| Female | 12 (41.4%) |
| Age (years), mean (SD), range | 48.6 (12.1), 26–68 |
| Age groups | |
| 18–29 years | 3 (10.3%) |
| 30–39 years | 5 (17.2%) |
| 40–49 years | 5 (17.2%) |
| 50–59 years | 9 (31.0%) |
| 60+ years | 7 (24.1%) |
| Race/Ethnicity | |
| African American | 1 (3.4%) |
| Asian | 1 (3.4%) |
| Hispanic/Latino | 2 (6.9%) |
| More than one race | 2 (6.9%) |
| White non-Hispanic | 23 (79.3%) |
| Education level | |
| Less than Bachelor's degree | 0 (0.0%) |
| Bachelor's degree | 10 (34.5%) |
| Master's degree | 12 (41.4%) |
| PhD/MD/JD | 7 (24.1%) |
| Employment status | |
| Employed full-time | 24 (82.8%) |
| Employed part-time | 1 (86.2%) |
| Missing data | 4 (13.8%) |
| Annual household income | |
| Below $20000 | 2 (6.9%) |
| $20 000–$39 000 | 0 (0.0%) |
| $40 000–$59 000 | 4 (13.8%) |
| $60 000–$79 000 | 1 (3.4%) |
| $80 000–$150 000 | 6 (20.7%) |
| Over $150 000 | 15 (51.7%) |
| Chose not to answer | 1 (3.4%) |
| Number of individuals in household (besides oneself) | |
| Mean (SD), range | 1.3 (1.5), 0–5 |
| Number of biological children | |
| Mean (SD), range | 0.79 (0.98), 0–3 |
| No children | 16 (55.2%) |
| 1 child | 4 (13.8%) |
| 2 children | 8 (27.6%) |
| 3 children | 1 (3.4%) |
Note that all the data are expressed as number (%), unless otherwise indicated.
Results returned to participants
Seven participants received results reports that included one or more pathogenic (P) or likely pathogenic (LP) rare disease-associated variants results. Participant 15 was informed he had an LP variant associated with Brugada syndrome and was one of two participants who received APOE e4/e4 results indicating increased Alzheimer's disease risk. Two participants received genetic risk scores (GRSs) indicating a likelihood ratio of AMD greater than 2.0. Most (25/29) were informed they had at least one variant classified as P/LP for carrier status. Table 2 summarizes each participant's results, and shows which P/LP disease-associated variants results were re-classified, for example, as variants of uncertain significance (VUS), after the study had ended.
Table 2. Participant characteristics and personal results from WGS.
| Additional details regarding P/LP disease variants results returned | |||||||
|---|---|---|---|---|---|---|---|
| ID | Gender | Age | Children | Summary of health-related results and raw data returned | Nomenclature | Associated disease | Interpretation |
| 06 | Female | 40–44 years | 0 | 0 P/LP disease; 3 P/LP carrier; e3/e3; T2D 1.45; CAD 0.77; AMD not displayed; raw data Y | — | — | — |
| 07 | Female | 55–59 years | 0 | 3 LP disease; 7 P/LP carrier; e3/e3; T2D 1.59; CAD 1.07; AMD not displayed; raw data Y | NM_000465.2(BARD1):c.1977A>G (p.R659=) | Hereditary breast & ovarian cancer | Likely pathogenic (LP) at time of study; re-classified as VUS after study completion |
| NM_006393.2(NEBL):c.180G>C (p.K60N) | Dilated cardiomyopathy | Likely pathogenic (LP) at time of study; re-classified as VUS after study completion | |||||
| NM_003098.2(SNTA1):c.770C>G (p.A257G) | Long QT syndrome | Likely pathogenic (LP) at time of study; re-classified as VUS after study completion | |||||
| 08 | Female | 60–64 years | 0 | 0 P/LP disease; 1 P/LP carrier; e3/e3; T2D 1.41; CAD 0.94; AMD not displayed; raw data Y | — | — | — |
| 11 | Male | 60–64 years | 1 | 0 P/LP disease; 3 P/LP carrier; e3/e4; T2D 0.14; CAD 1.08; AMD 0.82; raw data Y | — | — | — |
| 12 | Male | 50–54 years | 0 | 1 LP disease; 3 P/LP carrier; e3/e3; T2D 1.32; CAD 0.71; AMD 0.82; raw data Y | NM_000350.2(ABCA4):c.5908C>T (p.L1970F) | Macular degeneration | Likely pathogenic (LP) at time of study; re-classified as VUS after study completion |
| 13 | Male | 50–54 years | 2 | 0 P/LP disease; 3 P/LP carrier; e3/e4; T2D 0.97; CAD 1.26; AMD 1.04; raw data N | — | — | — |
| 14 | Female | 60–64 years | 0 | 0 P/LP disease; 2 P/LP carrier; e3/e4; T2D 0.61; CAD 0.73; AMD 1.00; raw data Y | — | — | — |
| 15 | Male | 25–29 years | 0 | 1 LP disease; 2 P/LP carrier; e4/e4; T2D 1.35; CAD 1.02; AMD 0.45; raw data Y | NM_198056.2(SCN5A):c.5770G>A (p.A1924T) | Brugada syndrome | Likely pathogenic (LP) at time of study; re-classified as VUS after study completion |
| 16 | Female | 35–39 years | 0 | 0 P/LP disease; 2 P/LP carrier; e3/e3; T2D 1.27; CAD 1.11; AMD 0.45; raw data Y | — | — | — |
| 17 | Male | 50–54 years | 2 | 0 P/LP disease; 1 P/LP carrier; e3/e4; T2D 1.21; CAD 1.01; AMD 1.24; raw data N | — | — | — |
| 18 | Male | 60–64 years | 0 | 0 P/LP disease; 0 P/LP carrier; e3/e3; T2D 1.19; CAD 1.17; AMD 1.21; raw data Y | — | — | — |
| 19 | Female | 45–49 years | 2 | 1 P disease; 2 P/LP carrier; e3/e3; T2D 1.47; CAD 0.81; AMD 1.19; raw data Y | NM_002016.1(FLG):c.2282_2285delCAGT (p.S761CfsX36) | Ichthyosis vulgaris, dry skin condition (semi-dominant) | Pathogenic (P) |
| 20 | Male | 25–29 years | 0 | 0 P/LP disease; 5 P/LP carrier; e3/e3; T2D 0.83; CAD 0.87; AMD 2.83; raw data Y | — | — | — |
| 22 | Female | 50–54 years | 2 | 0 P/LP disease; 2 P/LP carrier; e3/e3; T2D 1.34; CAD 0.71; AMD 0.75; raw data N | — | — | — |
| 23 | Male | 45–49 years | 0 | 1 LP disease; 3 P/LP carrier; e3/e3; T2D 1.20; CAD 0.94; AMD 0.39; raw data Y | NM 000041.2(APOE):c.460C>A (p.R154S) | Familial combined hyperlipoproteinemia type III | Likely pathogenic (LP) |
| 24 | Male | 55–59 years | 2 | 0 P/LP disease; 3 P/LP carrier; e3/e3; T2D 0.73; CAD 1.16; AMD 0.39; raw data Y | — | — | — |
| 26 | Male | 35–39 years | 1 | 0 P/LP disease; 0 P/LP carrier; e3/e3; T2D 0.82; CAD 1.14; AMD 8.98; raw data Y | — | — | — |
| 27 | Female | 50–54 years | 2 | 0 P/LP disease; 1 P/LP carrier; e3/e3; T2D 1.20; CAD 0.67; AMD 1.17; raw data Y | — | — | — |
| 29 | Female | 30–34 years | 0 | 0 P/LP disease; 2 P/LP carrier; e3/e3; T2D 0.73; CAD 0.77; AMD 0.44; raw data Y | — | — | — |
| 30 | Female | 55–59 years | 0 | 0 P/LP disease; 4 P/LP carrier; e3/e3; T2D 0.75; CAD 0.85; AMD 1.05; raw data Y | — | — | — |
| 31 | Male | 25–29 years | 0 | 0 P/LP disease; 2 P/LP carrier; e3/e4; T2D 0.65; CAD 1.14; AMD 0.65; raw data Y | — | — | — |
| 32 | Male | 35–39 years | 1 | 0 P/LP disease; 0 P/LP carrier; e4/e4; T2D 1.77; CAD 0.81; AMD 0.39; raw data Y | — | — | — |
| 34 | Male | 65–69 years | 2 | 0 P/LP disease; 2 P/LP carrier; e3/e4; T2D 1.23; CAD 0.92; AMD 0.39; raw data Y | — | — | — |
| 35 | Female | 55–59 years | 0 | 0 P/LP disease; 2 P/LP carrier; e3/e3; T2D 1.24; CAD 1.15; AMD 0.65; raw data Y | — | — | — |
| 36 | Male | 45–49 years | 2 | 0 P/LP disease; 2 P/LP carrier; e3/e3; T2D 1.38; CAD 0.91; AMD 0.46; raw data Y | — | — | — |
| 37 | Female | 30–34 years | 0 | 0 P/LP disease; 3 P/LP carrier; e3/e3; T2D 0.87; CAD 1.66; AMD 0.46; raw data Y | — | — | — |
| 38 | Male | 45–49 years | 1 | 1 P disease; 1 P/LP carrier; e3/e4; T2D 1.03; CAD 0.96; AMD 1.02; raw data Y | M_002016.1(FLG):c.2282_2285delCAGT (p.S761CfsX36) | Ichthyosis vulgaris, dry skin condition (semi-dominant) | Pathogenic (P) |
| 39 | Male | 55–59 years | 3 | 0 P/LP disease; 5 P/LP carrier; e3/e2; T2D 0.69; CAD 1.07; AMD 0.65; raw data Y | — | — | — |
| 40 | Male | 65–69 years | 0 | 1 P disease; 0 P/LP carrier; e3/e3; T2D 0.58; CAD 1.14; AMD 0.15; raw data Y | NM_198578.3(LRRK2):c.6055G>A (p.G2019S) | Parkinson's disease, late-onset (incomplete penetrance) | Pathogenic (P) |
Abbreviations: AMD, age-related macular degeneration genetic risk score; CAD, coronary artery disease genetic risk score; N, no; T2D, type 2 diabetes genetic risk score; Y, yes.
Total n=29. Bold font highlights participants who received P/LP rare disease-associatd variants results, APOE e4/e4 results indicating increased risk of Alzheimer's disease, and/or AMD GRS-based relative risk scores greater than 2.0.
Emotional and behavioural reactions overall: themes and example quotes
Positive emotional reactions
Most participants expressed positive feelings about having had their genomes sequenced. Six major themes emerged: (i) feeling happy or relieved; (ii) it was interesting; (iii) feeling glad to have contributed to research; (iv) the results or data might be useful to them in the future; (v) the results made them feel more connected to the world; (vi) it was fun. Among the participants who felt happy, some felt this way because they were excited by the science, for example, ‘I'm geeked. I'm still geeked. I was geeked at the beginning. I'm geeked now. I love it. I think it's one of the best things I ever did.' [#32, male, 35–39 years]. See Supplementary Table 2 for sub-themes and additional example quotes.
Negative emotional reactions
Although most participants reacted positively, a few expressed negative reactions. Six major themes emerged: (i) concern; (ii) disappointment; (iii) indifference; (iv) confusion; (v) desire for more results; (vi) not relevant to them. Among those who felt concerned, participants talked about the implications for their health, insurance, and their results going into their medical records if they sought medical follow-up. Among those who felt disappointed, for some this was because they had hoped to get information about a specific disease or trait. For others, the results did not meet their expectations and their disappointment was with the general scope of the results, for example, ‘I left the whole process feeling disappointed a little bit in terms of what I actually learned…' [#13, male, 50–54 years]. See Supplementary Table 3 for sub-themes and example quotes.
Behavioural reactions
Many participants had acted on their results. Four major themes emerged: (1) shared results with family and friends; (2) sought further information from family or online; (3) shared results with a healthcare provider; (4) made lifestyle changes. Among those who had shared their results with a healthcare provider, the majority had just mentioned them at a pre-scheduled appointment. One had their results put in their medical record. One had made an appointment and had a consultation, including tests and procedures, with a healthcare provider as a direct consequence, and one as an indirect consequence, of their results (see the ‘Rare disease-associated variants' section below). Many participants talked about why they had not shared their results with a healthcare provider. Only a few had made lifestyle changes because of their results. See Supplementary Table 4 for sub-themes and example quotes.
Reactions to specific results: themes and example quotes
Rare disease-associated variants
Two of the seven participants who received P/LP rare disease-associated variants results were concerned about and acted on their results. In the first case, participant 15 (who received the LP Brugada-associated variant result) said, ‘It was pretty concerning to me because it is a sudden cardiac death mutation, so it's one of those things that, it really can strike at any time, and it's, so I was scared for that reason.' [#15, male, 25–29 years]. The result directly prompted him to make an appointment and have a consultation, including tests and procedures, with a healthcare provider: ‘So I was able to meet with [the cardiologist], and he did the EKG, which didn't find anything pathological… After talking to him, I was reassured…'. He anticipated that the result might be empowering in the future: ‘It was stressful in the short-term… I think going forward I'll be concerned about the risk of sudden cardiac death, but I think hopefully this information will be empowering.' Over time, he had become less concerned with the Brugada-associated variant, and more concerned about his Alzheimer's disease e4/e4 result, saying, ‘I think I heard the APOE4 APOE4 high risk of Alzheimer's, and kind of didn't think much about that at that point in time because my concern was more on the immediate risk of death, but… I've probably thought more and more about the Alzheimer's than about Brugada.' [#15, male, 25–29 years]. In the second case, participant 7 was concerned enough about her two rare cardiac disease-associated variant results to seek further information from relatives about family cardiac health history, which then prompted her to make an appointment and have a consultation with a healthcare provider. See Table 3 for further details.
Table 3. Summary of results returned and participants' reactions by results category: qualitative outcomes.
| Personal genomic results categories | Participants who received each result | Summary of participants' responses in the qualitative 6-month follow-up interviews | |
|---|---|---|---|
| N | % | ||
| Rare disease variants | |||
| Pathogenic variant(s) Likely pathogenic variant(s) Pathogenic and likely pathogenic variant(s)No pathogenic/likely pathogenic variants | 3 4 0 23 | 10.3 13.8 0.0 79.3 | Two of the 7 participants who received ‘pathogenic' or ‘likely pathogenic' (P/LP) rare disease variant results were concerned about and had acted on their results. The first (#15, male, 25–29 years) had made an appointment and had a consultation, including tests and procedures, with a healthcare provider, as a direct consequence of his SCN5A Brugada-associated result; the second (#07, female, 55–59 years) had sought further information from family about her NEBL and SNTA1 results, and this had then prompted her to also have a cardiac consultation with a healthcare provider. Both had concerns about the implications for health and insurance, and had sought further information online. The other 5 participants who received P/LP disease variant results had little emotional or behavioural reactions to these. Participants who did not have any rare disease variants identified in their results reports had little emotional or behavioural reaction to these results. |
| Alzheimer's disease | |||
| APOE e4/e4 APOE e3/e4 APOE e3/e3 APOE e3/e2 | 2 7 19 1 | 6.9 24.1 65.5 3.4 | Both participants who received APOE e4/e4 results were concerned. One (#15, male, 25–29 years) had also received the SCN5A Brugada-associated disease variant: he initially did not focus on the e4/e4 result as he was more concerned about the SCN5A variant; however, over time, he became less focused on this, and more concerned about the Alzheimer's risk. The other (#32, male, 35–39 years) was concerned, although he was happy about the experience in general, and very excited by the science. Neither had discussed their results with a doctor. Although there was little impact of the e3/e4 results on most participants, one was concerned (#11, male, 60–64 years): he had sought further information online, but this had not reduced his confusion and he remained concerned; he had not discussed the result with a doctor, in part due to insurance concerns; he had started to use a brain-training app. Some participants who received e3/e3 or e2/e3 results expressed relief; many did not react particularly to the result. |
| Type 2 diabetes | |||
| Lifetime risk estimate: gender+age | 18.9% to 45.2% | These results had little emotional or behavioural impact on most participants. A few participants made lifestyle changes in response to increased risk results. | |
| 24 variants GRS-based relative risk | 0.58 to 1.77 | ||
| Lifetime risk estimate: gender+age+GRS | 11.0% to 55.5% | ||
| Coronary artery disease | |||
| Lifetime risk estimate: gender+age | 25.0% to 50.0% | These results had little emotional or behavioural impact on most participants. Very few participants made lifestyle changes in response to increased risk results. | |
| 29 variants GRS-based relative risk | 0.67 to 1.66 | ||
| Lifetime risk estimate: gender+age+GRS | 19.2% to 62.7% | ||
| Age-related macular degeneration | |||
| Lifetime risk estimate: gender+age 5 variants GRS-based relative risk Lifetime risk estimate: gender + age + GRS | 8.0% to 25.0% 0.15 to 8.98 1.2% to 71.9% | Most participants received RRs <2.0 and most did not react emotionally or behaviourally to these results. Two participants received RRs > 2.0. One of these participants (#20, male, 25–29 years) had a relevant family history, and had shared their result with their doctor at a pre-scheduled appointment (RR 2.83); the other (#26, male, 35–39 years) planned to share the result with an ophthalmologist but had not done so yet (RR 8.98). | |
| Rare carrier variants | |||
| Pathogenic variant(s) Likely pathogenic variant(s) Pathogenic and likely pathogenic variant(s)None | 16 2 7 4 | 55.2 6.9 24.1 13.8 | There was little impact of P/LP disease carrier results on most participants who received these results. A few said that the information might be useful for their own, or for their children's, reproductive decision-making in the future. Some felt it was too much information and not relevant to them, and that they should perhaps have opted out of this section. Participants with no rare disease carrier variants identified had little emotional or behavioural reaction to these results. |
| Pharmacogenomics | |||
| Clopidogrel (Plavix) | |||
| Extensive metabolizer | 11 | 37.9 | Some participants felt that this information was interesting, and some felt that it might be useful to them in the future. A few would have liked information about more medications, including antidepressants. |
| Ultrarapid metabolizer | 9 | 31.0 | |
| Intermediate metabolizer | 9 | 31.0 | |
| Simvastatin (Zocor) | |||
| Normal activity | 19 | 65.5 | |
| Intermediate activity | 8 | 27.6 | |
| Low activity | 1 | 3.4 | |
| Insufficient information | 1 | 3.4 | |
| Warfarin (Coumadin) | |||
| *** | *** | *** | |
| Physical traits | |||
| Bitter tasting type Bitter taster Not bitter taster Insufficient information | 17 7 5 | 58.6 24.1 17.2 | Most participants did not react to these results. A few said they were interesting or fun. |
| Cilantro tasting type | |||
| Less soapy taste | 15 | 51.7 | |
| More soapy taste | 3 | 10.3 | |
| Least soapy taste | 3 | 10.3 | |
| Insufficient information | 8 | 27.6 | |
| Earwax type | |||
| Wet earwax | 27 | 93.1 | |
| Dry earwax | 2 | 6.9 | |
| Lactose intolerance | |||
| Not lactose intolerant | 12 | 41.4 | |
| Possibly lactose intolerant | 11 | 37.9 | |
| Insufficient information | 6 | 20.7 | |
| Ancestry (largest percentage) | |||
| Ashkenazi Jews Utahn Whites N European Spaniards | 9 6 3 3 | 31.0 20.7 10.3 10.3 | Many participants found these results interesting, and some found them fun. Many shared these results with their family and friends. A few said their results made them feel more connected to the world. |
| French | 2 | 6.9 | |
| Armenians | 1 | 3.4 | |
| African American | 1 | 3.4 | |
| Basque | 1 | 3.4 | |
| Bamoun | 1 | 3.4 | |
| Singapore Chinese | 1 | 3.4 | |
| Maya | 1 | 3.4 | |
| Raw sequence data | |||
| Yes No | 26 3 | 89.7 10.3 | Several participants felt that their raw sequence data might be useful to them clinically in the future if they got sick. |
Alzheimer's disease
Both the participants who received APOE e4/e4 results were concerned. One of the seven participants who received e3/e4 results was confused and unclear about the health implications. See Table 3 for further details.
Multifactorial diseases
The multifactorial (polygenic) type 2 diabetes and coronary artery disease results generally had little impact on participants. Of the two participants who had received GRSs above 2.0 for AMD, one had shared their result with their healthcare provider at a pre-scheduled appointment, and the other intended to make an appointment with an ophthalmologist but had not done so yet.
Rare carrier variants
The rare carrier variant results generally had little impact, although a few participants said they might be useful to them or their children for reproductive decision-making in the future. Some felt these results were not relevant to them, and that they should have opted out of this section.
Pharmacogenomics
The participants found the pharmacogenomics results interesting. Some felt these results might be useful to them clinically in the future. Some had a desire for more results in this category, and would have liked to know about more medications in general, and about antidepressants specifically.
Physical traits
Most participants did not react to the physical traits results, although a few said they were fun.
Ancestry
Many participants found their ancestry results very interesting, and some found them fun. Many said that they had shared these results in particular with family and friends, for example, ‘I was telling my parents about it, some of my friends. Mostly people were interested about the ancestry part.' [#26, male, 35–39 years]. Some said their ancestry results made them feel more connected to the world, for example, ‘…It was surprising, and fascinating, and just enriching of my sense of myself and my place in the world.' [#18, male, 60–64 years].
Raw sequence data
Several participants felt that, even though most could not open their raw sequence data files, their raw data might be useful to them clinically in the future, for example, ‘Or if God forbid I get cancer, myself, then I would probably want to revisit that data.' [#07, female, 55–59 years].
Quantitative outcomes
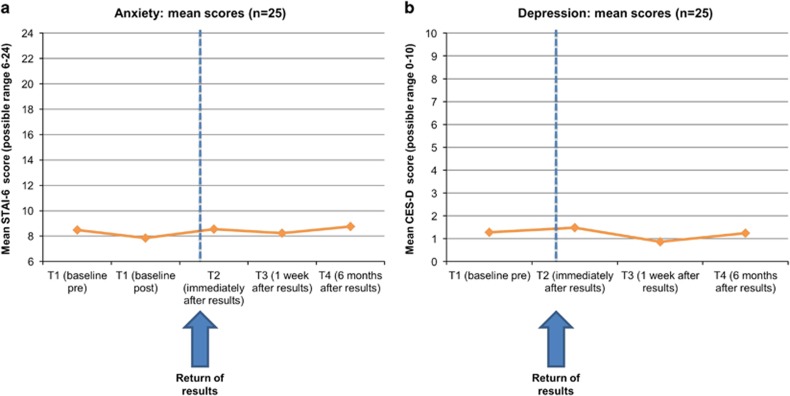
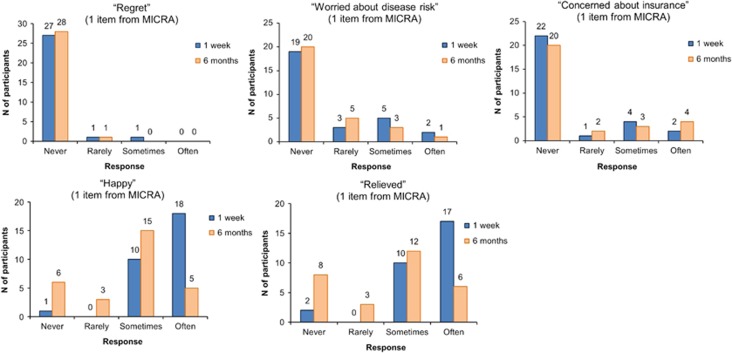
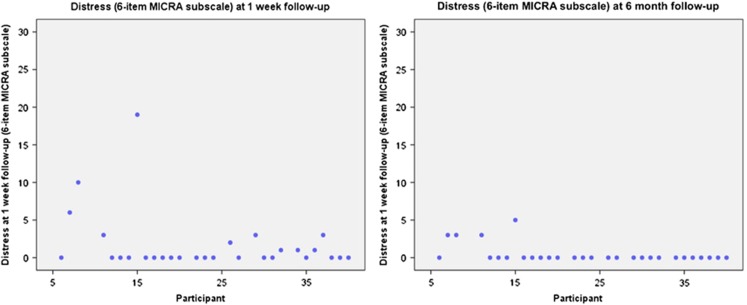
At T1, anxiety and depression were low, and most participants said they were in excellent or good health on the QoL measure: no changes over time were detected in anxiety (F=0.19, P=0.67), depression (F=0.75, P=0.52), or QoL (Z=1.90, P=0.058; see Figure 1 and Supplementary Table 5). MICRA positive experiences scale scores were lower at 1-week than 6-month follow-up (lower scores indicated more positive experiences): mean (SD) scores were 2.55 (3.16) vs 7.45 (6.83), respectively (Z=3.28, P=0.001). On individual MICRA items at T3, most said they were sometimes or often happy (28/29) and relieved (27/29) about their results; again, these numbers were slightly lower at T4 (Figure 2). This likely reflects that the MICRA asks participants how often they have felt this way in the past week. MICRA test-related distress scale scores were low: where 0=low distress and 30=high distress, mean (SD) scores were 1.69 (4.00) at T3 and 0.48 (1.27) at T4. Although low at both time points, the scores were lower at T4 than T3 (Z=2.68, P=0.007). The examination of the scatter plots suggested most participants had low distress but that participant 15 had high distress at T3, which subsided by T4 (Figure 3). As shown in Figure 2, most participants reported they never regretted having their genomes sequenced (27/29 at T3; 28/29 at T4), and never or rarely worried because of their results (22/29 at T3; 25/29 at T4), on individual MICRA items. No changes in lifestyle behaviours were detected (Supplementary Table 5).
Figure 1.
Anxiety and depression: quantitative outcomes. (a) Anxiety; (b) Depression.
Figure 2.
Reactions to personal genomic sequencing results: quantitative outcomes.
Figure 3.
Test-related distress: quantitative outcomes.
Discussion
In this study, most participants had positive emotional reactions to receiving personal results from whole-genome sequencing. Our findings support the suggestion that personal whole-genome sequencing may have value for individuals in ways that go beyond narrow definitions of health-related clinical utility, and that are consistent with broader notions of personal utility or value.11, 12, 13
As observed in previous research using direct-to-consumer and other SNP-based genetic results,18, 25 few participants changed lifestyle behaviours as a consequence of their whole-genome sequencing results. This is consistent with PMT, and supported by our qualitative findings: the small increases in diabetes and heart disease risk conveyed in our study were insufficient to elicit health-related fear or protection motivation among participants. This may also reflect well-known difficulties in facilitating lifestyle changes: even complex interventions explicitly designed to change lifestyles achieve modest at best improvements in lifestyles and related outcomes,40 and HealthSeq did not provide behaviour change interventions.
Although there was little impact of the complex disease risk results on the participants' emotions or lifestyles, several were worried about and/or acted on other results they received, in particular the rare disease variants results. Taken together, these observations are consistent with PMT: individuals who do not feel threatened by their personal results from whole-genome sequencing (or do not feel they can do anything to reduce their risk) may be unlikely to engage in lifestyle change or to pursue clinical follow-up, but individuals who do feel threatened by their results and who believe they might be able to reduce the health threat may engage in subsequent health-protective behaviours.
It is notable that one participant's focus shifted over several months from his Brugada-associated rare variant result to his Alzheimer's disease APOE e4/e4 result, as he processed and dealt with each in turn. This highlights how reactions and counselling needs may change over time, and that counselling protocols may need to reflect this. Ensuring sufficient support is available to people after receiving results is clearly important, but it is not clear yet what the appropriate level of support is nor how to ensure people get the support they need.
A further notable and challenging issue arising in the two cases where ‘likely pathogenic' variants directly or indirectly prompted clinical follow-up, is that these variants were later re-classified as VUSs after study completion. As the knowledge base about rare variants grows, variants are likely to be re-classified over time.41, 42, 43 Studies addressing whether and how often people's sequence data can or should be re-interpreted, and how new interpretations should be returned, are needed.
Several participants were unsure about the significance of their results, and uncertain about whether and what follow-up to pursue. This was despite the results being returned in person by a genetic counsellor and medical geneticist, and despite these participants being early adopters with higher levels of education than the general population. The finding that even under these circumstances some participants struggled to understand their results and their implications underscore that significant efforts will be needed to aid comprehension of results in the future as whole-genome sequencing expands to the wider population.
Several participants had mentioned their results to their regular healthcare providers. This is consistent with previous research reporting that over a third of consumers shared their direct-to-consumer genetic results with a physician.44 These findings further support the need for an educated healthcare workforce that is prepared to interact with patients about their personal genomic information whether or not that information has been obtained within the clinic.45, 46
Recent debates have explored whether research participants have the right to access their own raw data, and whether greater efforts should be made to facilitate this.28, 30, 47 Most HealthSeq participants opted to receive and responded positively to having their raw data. Participants' reasons for wanting their raw data were primarily that they felt having it might be of value to them and their physicians in the future. This novel finding provides early evidence that research participants may value having their raw genomic data because they anticipate it may help in their future clinical care.
Strengths of this study include the qualitative methods and the greater age range compared with previous work.48 Limitations include that participants were self-selected and had high levels of education and income, so the findings are not generalizable to the wider population. This was a small study, although the sample size was not unusual for qualitative research.49, 50 The PMT did not provide a framework for exploring non-health-related results, positive emotional reactions or communication behaviours. A more complete theoretical framework within which to examine the psychological and behavioural effects of results arising from whole-genome sequencing is needed. Development of such a framework may be a particularly valuable focus for future applied psychological research in the genomic sequencing field.
Research participants are key stakeholders in debates about the future directions and value of offering personal results and data from whole-genome sequencing. The findings from this in-depth study with ostensibly healthy individuals suggest the possibility that currently neither the benefits nor harms of personal genome sequencing are significant for most individuals, but that there may be important exceptions to this that warrant further investigation. The impact of returning personal results from whole-genome sequencing results when implemented on a larger scale remains to be seen.
Footnotes
Supplementary Information accompanies this paper on the European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Choi M, Scholl UI, Ji W et al: Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009; 106: 19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranneheim H, Wedell A: Exome and genome sequencing: a revolution for the discovery and diagnosis of monogenic disorders. J Intern Med 2016; 279: 3–15. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Abramowicz M, Al-Mulla F et al: Global implementation of genomic medicine: we are not alone. Genet Med 2015; 7: 290ps213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiec E, Howard HC: Ethical issues in consumer genome sequencing: use of consumers' samples and data. Appl Transl Genom 2016; 8: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, Grove ME, Pan C et al: Clinical interpretation and implications of whole-genome sequencing. JAMA 2014; 311: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinter F: Should we sequence everyone's genome? No. BMJ 2013; 346: f3132. [DOI] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Lupski JR, Gibbs RA: Human genome sequencing in health and disease. Annu Rev Med 2012; 63: 35–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrist M: Personal genomics: where are we now? Appl Transl Genom 2016; 8: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garay ML, McGuire AL, Pereira S, Caskey CT: Personalized genomic disease risk of volunteers. Proc Natl Acad Sci USA 2013; 110: 16957–16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Hu H, Singleton M, De La Vega FM, Reese MG, Yandell M: Global analysis of disease-related DNA sequence variation in 10 healthy individuals: implications for whole genome-based clinical diagnostics. Genet Med 2011; 13: 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse SD, Khoury MJ: What is the clinical utility of genetic testing? Genet Med 2006; 8: 448–450. [DOI] [PubMed] [Google Scholar]
- Foster MW, Mulvihill JJ, Sharp RR: Evaluating the utility of personal genomic information. Genet Med 2009; 11: 570–574. [DOI] [PubMed] [Google Scholar]
- Turrini M, Prainsack B: Beyond clinical utility: the multiple values of DTC genetics. Appl Transl Genom 2016; 8: 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SW, Lee IH, Leshchiner I: Summarizing polygenic risks for complex diseases in a clinical whole-genome report. Genet Med 2015; 17: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Hooker GW, Connors PD: Participant use and communication of findings from exome sequencing: a mixed-methods study. Genet Med 2016; 18: 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Ceyhan-Birsoy O, Christensen KD et al: A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet 2014; 15: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassy JL, Lautenbach DM, McLaughlin HM et al: The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 2014; 15: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss CS, Schork NJ, Topol EJ: Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med 2011; 364: 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RW, O'Brien KE, Waxler JL et al: Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care 2013; 36: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DS, Myers RE, Keenan E et al: Genetic and environmental risk assessment and colorectal cancer screening in an average-risk population: a randomized trial. Ann Intern Med 2014; 161: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsaux CF, Celis-Morales C: Changes in physical activity following a genetic-based internet-delivered personalized intervention: randomized controlled trial (Food4Me). J Med Internet Res 2016; 18: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel SF, Beeken RJ, van Jaarsveld CH, Wardle J: Genetic susceptibility testing and readiness to control weight: results from a randomized controlled trial. Obesity 2015; 23: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson SC, Humphries SE, Hubbart C, Hughes E, Jarvis MJ, Wardle J: Psychological and behavioural impact of genetic testing smokers for lung cancer risk: a phase II exploratory trial. J Health Psychol 2008; 13: 481–494. [DOI] [PubMed] [Google Scholar]
- Hollands GJ, French DP, Griffin SJ et al: The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ 2016; 352: i1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CM, Koehly LM, Sanderson SC, Kaphingst KA: The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annu Rev Public Health 2010; 31: 89–103. [DOI] [PubMed] [Google Scholar]
- Rogers RW: A protection motivation theory of fear appeals and attitude change. J Psychol 1975; 91: 21. [DOI] [PubMed] [Google Scholar]
- Phillips AM: 'Only a click away—DTC genetics for ancestry, health, love... and more: a view of the business and regulatory landscape'. Appl Transl Genom 2016; 8: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunshof JE, Church GM, Prainsack B: Information access. Raw personal data: providing access. Science 2014; 343: 373–374. [DOI] [PubMed] [Google Scholar]
- Middleton A, Wright CF, Morley KI et al: Potential research participants support the return of raw sequence data. J Med Genet 2015; 52: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry SF, Terry PF: Power to the people: participant ownership of clinical trial data. Sci Transl Med 2011; 3: 69cm63. [DOI] [PubMed] [Google Scholar]
- Sanderson SC, Linderman MD, Suckiel SA et al: Motivations, concerns and preferences of personal genome sequencing research participants: baseline findings from the HealthSeq project. Eur J Hum Genet 2016; 24: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckiel SA, Linderman MD, Sanderson SC et al: Impact of genomic counseling on informed decision-making among ostensibly healthy individuals seeking personal genome sequencing: the HealthSeq Project. J Genet Couns 2016; 25: 1044–1053. [DOI] [PubMed] [Google Scholar]
- Kohout FJ, Berkman LF, Evans DA, Cornoni-Huntley J: Two shorter forms of the CES-D (Center for Epidemiological Studies Depression) depression symptoms index. J Aging Health 1993; 5: 179–193. [DOI] [PubMed] [Google Scholar]
- Marteau TM, Bekker H: The development of a six-item short-form of the state scale of the Spielberger State-Trait Anxiety Inventory (STAI). Br J Clin Psychol 1992; 31 (Pt 3): 301–306. [DOI] [PubMed] [Google Scholar]
- Gandek B, Ware JE Jr, Aaronson NK et al: Tests of data quality, scaling assumptions, and reliability of the SF-36 in eleven countries: results from the IQOLA Project. International Quality of Life Assessment. J Clin Epidemiol 1998; 51: 1149–1158. [DOI] [PubMed] [Google Scholar]
- Cella D, Hughes C, Peterman A et al: A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol 2002; 21: 564–572. [PubMed] [Google Scholar]
- (CDC) CfDCaP: Behavioral Risk Factor Surveillance System Survey Questionnaire. Atlanta, GA, USA: US Department of Health and Human Services, CDC, 2006.
- Braun VC: Using Thematic Analysis in Psychology. Qual Res Psychol 2006; 3: 34. [Google Scholar]
- Ritchie JL: Qualitative Research Practice. A Guide for Social Science Students and Researchers. New Delhi/London: Sage/Thousand Oaks, 2003. [Google Scholar]
- Dansinger ML, Gleason JA, Griffith JL, Selker HP, Schaefer EJ: Comparison of the Atkins, Ornish, Weight Watchers, and Zone diets for weight loss and heart disease risk reduction: a randomized trial. JAMA 2005; 293: 43–53. [DOI] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL et al: Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016; 375: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driest SL, Wells QS, Stallings S et al: Association of arrhythmia-related genetic variants with phenotypes documented in electronic medical records. JAMA 2016; 351: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh RT, Thomson KL, Ware JS et al: Reassessment of Mendelian Gene pathogenicity using 7855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2016, e-pub ahead of print 17 August 2016 doi:10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed]
- Bloss CS, Schork NJ, Topol EJ: Direct-to-consumer pharmacogenomic testing is associated with increased physician utilisation. J Med Genet 2014; 51: 83–89. [DOI] [PubMed] [Google Scholar]
- Feero WG, Green ED: Genomics education for health care professionals in the 21st century. JAMA 2011; 306: 989–990. [DOI] [PubMed] [Google Scholar]
- Korf BR, Berry AB, Limson M et al: Framework for development of physician competencies in genomic medicine: report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet Med 2014; 16: 804–809. [DOI] [PubMed] [Google Scholar]
- Schadt EE: The changing privacy landscape in the era of big data. Mol Syst Biol 2012; 8: 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Mullikin JC, Facio FM et al: The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res 2009; 19: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowswell G, Ismail T, Greenfield S, Clifford S, Hancock B, Wilson S: Men's experience of erectile dysfunction after treatment for colorectal cancer: qualitative interview study. BMJ 2011; 343: d5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch J, Jansen J, Barratt A et al: Women's views on overdiagnosis in breast cancer screening: a qualitative study. BMJ 2013; 346: f158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.