Abstract
Hearing impairment (HI) is genetically heterogeneous which hampers genetic counseling and molecular diagnosis. Testing of several single HI-related genes is laborious and expensive. In this study, we evaluate the diagnostic utility of whole-exome sequencing (WES) targeting a panel of HI-related genes. Two hundred index patients, mostly of Dutch origin, with presumed hereditary HI underwent WES followed by targeted analysis of an HI gene panel of 120 genes. We found causative variants underlying the HI in 67 of 200 patients (33.5%). Eight of these patients have a large homozygous deletion involving STRC, OTOA or USH2A, which could only be identified by copy number variation detection. Variants of uncertain significance were found in 10 patients (5.0%). In the remaining 123 cases, no potentially causative variants were detected (61.5%). In our patient cohort, causative variants in GJB2, USH2A, MYO15A and STRC, and in MYO6 were the leading causes for autosomal recessive and dominant HI, respectively. Segregation analysis and functional analyses of variants of uncertain significance will probably further increase the diagnostic yield of WES.
Introduction
DNA diagnostics of any genetically heterogeneous disease based on single gene testing is highly inefficient, laborious and expensive. High-throughput sequencing technologies such as whole-exome sequencing (WES) have coped with these disadvantages allowing the analysis of all protein-coding exons in a single cost-effective attempt.1
Hearing impairment (HI) is the most common sensory disorder with an incidence of one in 750 newborns (>40 dB hearing loss) in developed countries.2 About half of the cases are attributed to genetic factors with more than a hundred syndromic and non-syndromic genes known to date (http://hereditaryhearingloss.org/). HI is most frequently manifested as non-syndromic (NSHI) accounting for about 70% of the hereditary cases. About 77% of hereditary NSHI cases exhibit autosomal recessive inheritance (arNSHI) with 62 genes and 92 loci known to date, whereas it is dominant (adNSHI) in about 22% of the cases with 36 genes and 58 loci known. The remaining 1% shows an X-linked, a Y-linked or a mitochondrial type of inheritance pattern with seven loci and six genes known to date (http://hereditaryhearingloss.org/). Thus, NSHI is a perfect example of a genetically heterogeneous disorder. Many genes have been described to be involved in only one or a few families with HI.3, 4, 5 Some exceptions are known, for example, mutations in GJB2 followed by mutations in STRC and MYO15A are the most common causes for arNSHI worldwide.6, 7, 8, 9
Since many genes contribute to hereditary HI, targeting all or a selection of protein-coding exons in a single experiment, as in WES, might currently be the best option for a comprehensive genetic analysis of HI individuals. The implementation of WES in a diagnostic setting has been much slower than in scientific research due to the relatively low sensitivity of this method in detection of genetic variation in some exonic regions, for example, extremely GC-rich regions, as compared with Sanger sequencing.10 Despite this fact, the diagnostic yield in hereditary HI obtained by WES is expected to be higher compared with the approach of phenotype-based pretesting of one or two genes by Sanger sequencing.11 Therefore, the aim of this study is to evaluate the diagnostic utility of WES targeting a panel of HI-related genes in a group of 200 Dutch index patients with presumed hereditary HI.
Patients and methods
Patients
A retrospective cohort study was performed in 200 patients with HI, mainly of Dutch origin, who underwent WES in diagnostics in the period of 2011–2014. Only index cases were included. Non-genetic causes of the HI were considered to be unlikely on basis of medical history and ENT examination. Type of inheritance, age of onset, and phenotype were based on family history, medical history and available audiogram(s) (Supplementary Tables S2–S4). The inheritance pattern indicated by the referring clinician was autosomal dominant in 66 cases, autosomal recessive in 31 cases, X-linked in one case and the remaining 102 cases were isolated. In the majority of subjects, the age of onset of HI was congenital (n=79) or in the first decade of life (n=60). Patients with both nonsyndromic and syndromic HI were included. The audiometric phenotype was assessed according to the recommendations by the GENDEAF study group.12 Thirty-six patients have been previously reported in a study on the utility of WES,11 and one patient has been reported before in a publication on novel and recurrent CIB2 variants.13 Prior to WES, causative variants in one or more genes involved in HI had been excluded by Sanger sequencing in 137 patients. Prescreening was performed in these patients because WES was not available at that time or because the clinician had a high clinical suspicion for mutations in a specific gene. Diagnostic WES was approved by the medical ethics committee of the Radboud university medical center, Nijmegen, The Netherlands (registration number 2011–188). For all patients, written informed consent for WES was obtained after counseling by a clinical geneticist.
WES and bioinformatics
Before sequencing, genomic DNA fragments of all patients were enriched for exome sequences using the Agilent (Santa Clara, CA, USA) SureSelectXT Human All Exon 50 Mb kit (n=30) or the version 4 (V4) kit (n=170) (Agilent Technologies, Santa Clara, CA, USA). For 44 patients (30 cases Agilent 50 Mb, 14 cases Agilent V4), WES was performed with a 5500xl SOLiD system (Life Technologies, Carlsbad, CA, USA) at the department of Human Genetics, Radboudumc Nijmegen, and data were analyzed using LifeScopeTM software as previously reported.11 For the remaining patients (n=156, Agilent V4) WES was performed at BGI-Europe (Copenhagen, Denmark), employing an Illumina HiSeq2000TM machine (Illumina, San Diego, CA, USA). For these samples, ‘read alignment' using BWA and ‘variant calling' with GATK were performed at BGI.14
For all patients, variants were annotated with an in-house developed annotation and prioritization pipeline.11 Variants in genes associated with HI were selected and analyzed. In the first 44 patients, a panel of 98 HI genes was analyzed (gene list DGD07092012).11 In 2014, the gene panel was updated to 120 genes: four genes were deleted from the gene list, because proof for the involvement of the gene in HI is questionable, and 26 published novel HI-related genes were added (gene list DGD20062014). The remaining 156 patients were analyzed with the updated list of 120 HI genes. In retrospect, the WES data of the first 44 patients were analyzed for variants in the 26 genes added to gene list DGD20062014. Detailed information on both gene lists can be found in Supplementary Table S1.
The coverage was determined for the HI-related genes. The targeted genes enriched with the Agilent 50 Mb and Agilent V4 were compared with the longest RefSeq transcript to identify untargeted exons (Supplementary Table S1). These exons were omitted from coverage calculations. The coverage was calculated per sample on a base pair resolution, using the coverage function of BEDtools (v2.19.1; PMID 20110278). Subsequently, the mean percentage of base pairs with at least 20 reads (≥20 × coverage) was determined per sample, for each gene and technological WES condition. Finally, the median ≥20 × coverage was calculated per gene and technological WES condition.
For all patients ‘copy number variant calling' was carried out using CoNIFER 0.2.0,15 and variant annotation was performed using an in-house developed strategy.16
Interpretation and classification of variants
To systematically predict their pathogenicity, variants were classified according to the existing guidelines from the American College of Medical Genetics and Genomics: benign (class 1), likely benign (class 2), uncertain significance (class 3), likely pathogenic (class 4) and pathogenic (class 5).17 Patients were grouped based on the variant classification, segregation analysis and associated phenotype known from literature. Three groups were distinguished: (1) patients with causative variants, that is, (likely) pathogenic variant(s) matching the phenotype and segregating with the HI in the family; (2) patients with variants of uncertain significance, that is, variants that could not be further classified by segregation analysis or not matching the phenotype; and (3) patients without detected causative variants. All detected variants were submitted to the Leiden Open Variant Database (LOVD, http://databases.lovd.nl/shared/genes, patient IDs 79876, 79998, 80001-80064, 80136 and 80138-80147).
Validation of selected variants
All reported sequence variants have been validated by Sanger sequencing (primer sequences and PCR conditions are available upon request). Copy number variants (CNVs) were validated by MLPA (STRC, homemade MLPA kit s139; USH2A, MRC-Holland kits P361A1 and P362A2) or deletion-specific PCR (OTOA, kindly provided by Guney Bademci, MD).
Results
The exomes of 200 individuals with presumed hereditary HI, mostly of Dutch origin, were sequenced in this study. Subsequently, targeted analysis of WES data was performed for a panel of 120 genes (DGD20062014) associated with HI. A median coverage of at least 20 × was reached for 72.0% and 97.8% of the targeted genes with the SOLiD system (n=44), for the Agilent 50 Mb and V4 enrichment kits, respectively (Supplementary Figure S1a and b, Supplementary Table S1). The median coverage was 97.5% with the HiSeq system (n=156; Supplementary Figure S1c). The percentage of identified causative variants did not increase with improvement of the technological WES conditions. In 31.8% (n=14) of the samples performed with the SOLiD system causative variants were identified, compared with 34.0% (n=53) of the samples performed with the Illumina HiSeq2000TM (Supplementary Table S3).
Diagnostic yield with WES in HI patients
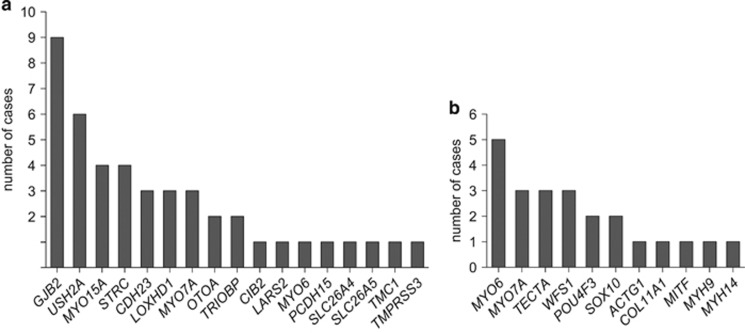
We identified causative variants in 33.5% (67 cases) out of 200 cases with presumed hereditary HI (Table 1, Supplementary Tables S2 and S3). In 44 of these patients, homozygous or compound heterozygous variants in genes associated with autosomal recessive HI (arHI) were detected, being large homozygous deletions of several exons or complete genes in eight of these cases. No causative heterozygous CNVs were identified. GJB2 was found to be the most frequently mutated gene (13.4% of positive cases), followed by USH2A, MYO15A and STRC, together accounting for 34.3% of the positive cases (Figure 1a).
Table 1. List of patients with causative variants in HI-related genes.
| Patient | Gene, Ref Seq transcript ID | Variant 1 nucleotide change (protein change) | Variant 2 nucleotide change (protein change) | Zygositya | Segregating with HIb |
|---|---|---|---|---|---|
| AD1c | MYO6 NM_004999.3 | c.1546+1G>T (p.?) | - | het | Yes |
| AD2 | WFS1 NM_006005.3 | c.2051C>T (p.(Ala684Val))c | - | het | ND |
| AD3 | TECTA ENST00000392793.1 | c.6002G>T (p.(Cys2001Phe)) | - | het | de novo |
| AD4 | MYO7A NM_000260.3 | c.1373A>T (p.(Asn458Ile))c | - | het | Yes |
| AD5c | MYO6 NM_004999.3 | c.1211del (p.(Gly404Glufsa4)) | - | het | Yes |
| AD6c | MYH9 NM_002473.5 | c.2507C>T (p.(Pro836Leu)) | - | het | Yes |
| AD7 | POU4F3 NM_002700.2 | c.828_829insT (p.(Lys277a)) | - | het | ND |
| AD8 | MYO7A NM_000260.3 | c.652G>A (p.(Asp218Asn)) | - | het | Yes |
| AD9 | POU4F3 NM_002700.2 | c.668T>C (p.(Leu223Pro))c | - | het | ND |
| AD10 | MYO6 NM_004999.3 | c.3395del (p.(Lys1132Serfsa12)) | - | het | Yes |
| AD11 | SOX10 NM_006941.3 | c.1195C>T (p.(Gln399a)) | - | het | ND |
| AD12 | MITF NM_000248.3 | c.649del (p.(Arg217Aspfsa4))c | - | het | Yes |
| AD13 | MYO7A NM_000260.3 | c.2617C>T (p.(Arg873Trp))c | - | het | Yes |
| AD14 | TECTA ENST00000392793.1 | c.5794A>C (p.(Thr1932Pro)) | - | het | Yes |
| AD15 | MYH14 NM_001145809.1 | c.5176C>T (p.(Arg1726Trp)) | - | het | Yes |
| AD16 | WFS1 NM_006005.3 | c.2032T>C (p.(Trp678Arg)) | - | het | Yes |
| AD17 | MYO6 NM_004999.3 | c.584C>A (p.(Ala195Glu)) | - | het | Yes |
| AD18 | TECTA ENST00000392793.1 | c.5597C>T (p.(Thr1866Met))c | - | het | Yes |
| AR1c | USH2A NM_206933.2 | c.(9371+1_9372-1)_(9570+1_9571-1)del (p.?) | c.(9371+1_9372-1)_(9570+1_9571-1)del (p.?) | hom | ND |
| AR2 | OTOA ENST00000388958 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | ND |
| AR3c | CIB2 NM_006383.3 | c.97C>T (p.(Arg33a)) | c.196C>T (p.(Arg66Trp)) | compound het | Yes |
| AR4 | STRC NM_153700.2 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | Yes |
| AR5 | USH2A NM_206933.2 | c.(8845+1_8846-1)_(9371+1_9372-1)del (p.?) | c.(8845+1_8846-1)_(9371+1_9372-1)del (p.?) | hom | Yes |
| AR6 | TMC1 NM_138691.2 | c.646del (p.(Leu216Serfsa54)) | c.790C>T (p.(Arg264a)) | ND | ND |
| AR7 | CDH23 NM_022124.5 | c.6442G>A (p.(Asp2148Asn))c | c.1545_1547del (p.(Ile515del))c | compound het | ND |
| AR8 | USH2A NM_206933.2 | c.1606T>C (p.(Cys536Arg))c | c.9815C>T (p.(Pro3272Leu))c | compound het | Yes |
| AR9 | GJB2 NM_004004.5 | c.35del (p.(Gly12Valfsa2))c | c.71G>A (p.Trp24a))c | ND | ND |
| AR10 | GJB2 NM_004004.5 | c.35del (p.(Gly12Valfsa2))c | c.35del (p.(Gly12Valfsa2))c | hom | ND |
| AR11 | CDH23 NM_022124.5 | c.2096A>G (p.(Asp699Gly)) | c.4562A>G (p.(Asn1521Ser)) | compound het | Yes |
| AR12 | GJB2 NM_004004.5 | c.-23+1G>A (p.?)c | c.35del (p.(Gly12Valfsa2))c | compound het | ND |
| AR13 | MYO7A NM_000260.3 | c.3289C>T (p.(Gln1097a)) | c.3862G>C (p.(Ala1288Pro))c | compound het | ND |
| c.5227C>T (p.Arg1743Trp)c | |||||
| AR14 | STRC NM_153700.2 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | ND |
| AR15 | PCDH15 NM_033056.3 | c.3374-2A>G (p.?) | c.4127C>A (p.(Ala1376Asp)) | compound het | Yes |
| AR16 | STRC NM_153700.2 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | ND |
| AR17 | LOXHD1 NM_144612.6 | c.1618dup (p.(Thr540Asnfsa24)) | c.1730T>G (p.(Leu577Arg)) | compound het | Yes |
| AR18 | USH2A NM_206933.2 | c.5018T>C (p.(Leu1673Pro)) | c.2299del (p.(Glu767Serfsa21)) | compound het | ND |
| c.7871C>T (p.(Pro2624Leu)) | |||||
| ISO1c | MYO15A NM_016239.3 | c.625G>T (p.(Glu209a)) | c.1137del (p.(Tyr380Metfsa64))c | ND | ND |
| ISO2c | USH2A NM_206933.2 | c.5385T>A (p.(Tyr1795a)) | c.6846_6849dup (p.(His2284Asnfsa48)) | ND | ND |
| ISO3 | CDH23 NM_022124.5 | c.8480_8481del (p.(Leu2827Hisfsa23)) | c.8480_8481del (p.(Leu2827Hisfsa23)) | hom | ND |
| ISO4c | MYO15A NM_016239.3 | c.6764+2T>A (p.?) | c.3844C>T (p.(Arg1282Trp) | ND | ND |
| c.5287C>T (p.(Arg1763Trp)) | |||||
| ISO5 | GJB2 NM_004004.5 | c.101T>C (p.(Met34Thr))17 | c.109G>A (p.(Val37Ile))17 | compound het | ND |
| ISO6 | WFS1 NM_006005.3 | c.2051C>T (p.(Ala684Val))2 | - | het | ND |
| ISO7 | MYO15A NM_016239.3 | c.6787G>A (p.(Gly2263Ser)) | c.7893+1G>A (p.?) | compound het | ND |
| ISO8 | TRIOBP NM_001039141.2 | c.2653del (p.(Arg885Alafsa120)) | c.5014G>T (p.(Gly1672a)) | compound het | ND |
| ISO9 | USH2A NM_206933.2 | c.2299del (p.(Glu767Serfsa21)) | c.920_923dup (p.(His308Glnfsa16)) | compound het | ND |
| ISO10 | MYO7A NM_000260.3 | c.3476G>T (p.(Gly1159Val))c | c.5560G>A (p.Val1854Met))c | compound het | ND |
| ISO11 | GJB2 NM_004004.5 | c.250G>C(p.(Val84Leu)) | c.269T>C (p.(Leu90Pro))c | ND | ND |
| ISO12 | GJB2 NM_004004.5 | c.109G>A (p.(Val37Ile))c | c.109G>A (p.(Val37Ile))c | hom | ND |
| ISO13 | TMPRSS3 NM_024022.2 | c.916G>A (p.(Ala306Thr))c | c.1276G>A (p.(Ala426Thr))c | ND | ND |
| ISO14 | STRC NM_153700.2 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | ND |
| ISO15 | GJB2 NM_004004.5 | c.35del (p.(Gly12Valfsa2))c | c.101T>C (p.(Met34Thr))c | compound het | ND |
| ISO16 | ACTG1 NM_001199954.1 | c.773C>T (p.(Pro258Leu)) | - | het | de novo |
| ISO17 | GJB2 NM_004004.5 | c.35del (p.(Gly12Valfsa2))c | c.508_511dup p.Ala171Glufsa40))c | compound het | ND |
| ISO18 | MYO15A NM_016239.3 | c.3311dup (p.(Glu1105a)) | c.3311dup (p.(Glu1105a)) | hom | ND |
| ISO19 | TRIOBP NM_001039141.2 | c.3460_3461del (p.(Leu1154Alafsa29)) | c.3232dup (p.(Arg1078Profsa6))c | compound het | ND |
| ISO20 | MYO7A NM_000260.3 | c.5618G>A (p.(Arg1873Gln))c | c.6028G>A (p.(Asp2010Asn))c | compound het | ND |
| ISO21 | MYO6 NM_004999.3 | c.3610C>T (p.(Arg1204Trp))c | - | het | de novo |
| ISO22 | SLC26A5 NM_198999.2 | c.355C>T (p.(Pro119Ser)) | c.355C>T (p.(Pro119Ser)) | hom | ND |
| ISO23 | GJB2 NM_004004.5 | c.101T>C (p.(Met34Thr))c | c.109G>A (p.(Val37Ile))c | compound het | ND |
| ISO24 | SOX10 NM_006941.3 | c.482G>A (p.(Arg161His))c | - | het | de novo |
| ISO25 | MYO6 NM_004999.3 | c.3335A>G (p.(Tyr1112Cys)) | c.1897del (p.(Gln633Lysfsa19)) | compound het | ND |
| ISO26 | LARS2 NM_015340.3 | c.683G>A (p.(Arg228His)) | c.880G>A (p.(Glu294Lys)) | compound het | ND |
| ISO27 | OTOA ENST00000388958 | c.(?_-1)_(a1_?)delc | c.(?_-1)_(a1_?)delc | hom | ND |
| ISO28c | COL11A1 NM_001854.3 | c.1630-2del (p.?) | - | het | ND |
| ISO29 | LOXHD1 NM_144612.6 | c.3061+1G>A (p.?) | c.6353G>A (p.(Gly2118Glu)) | ND | ND |
| ISO30 | LOXHD1 NM_144612.6 | c.3061C>T (p.(Arg1021a)) | c.5885C>T (p.(Thr1962Met)) | compound het | ND |
| ISO31 | SLC26A4 NM_000441.1 | c.505del (p.(Thr169Leufsa3)) | c.1334T>G (p.(Leu445Trp))c | compound het | ND |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; het, heterozygous; HI, hearing impairment; hom, homozygous; ISO, isolated; ND, not determined or not conclusive.
Zygosity, determined on basis of segregation analysis in the parents.
Segregating with HI, determined on basis of segregation analysis in affected and/or unaffected family members.
Mutations that have been described in literature before, the corresponding references are provided in Supplementary Table S2.
Figure 1.
Overview of HI genes and number of cases in which causative variant(s) were identified in these genes. (a) Cases with arHI. (b) Cases with adHI.
In the remaining 23 cases, heterozygous causative variants in 11 different genes associated with autosomal dominant HI (adHI) were found (Supplementary Table S2, Figure 1b). In four cases, the heterozygous variants were de novo. Causative variants in MYO6 were the leading cause in this cohort of presumed adHI cases (Figure 1b).
The diagnostic yield was related to the type of inheritance and the age of onset of HI in the patients. For patients with suspected arHI, the diagnostic yield was 58.1%, of which 16.7% (n=3) caused by variants in GJB2 (Table 1, Supplementary Table S3). In 30.4% of the cases without a (known) family history of HI (isolated cases), the molecular etiology could be identified, the majority harboring causative variants in genes associated with arHI. In 19.4% (n=6) of these cases causative variants in GJB2 were found (Table 1,Supplementary Table S3). For adHI, causative variants were found in 27.3% of the cases (Table 1, Supplementary Table S3). Causative variant(s) could be identified in 49.4% and 36.7% of the subjects with congenital and first decade onset HI, respectively (Supplementary Table S3, Supplementary Figure S2). This percentage strongly declines with the increase in age of onset.
We identified variants of uncertain significance in 10 cases (5.0% Supplementary Tables S3 and S4). In eight of these, segregation analysis of the variant(s) could not be performed, because DNA of family members was unavailable. Segregation analysis is essential in order to classify these variants as causative or non-causative. In the remaining two cases, segregation analysis was performed, but the variants could neither be classified as the cause of the HI, nor could they be discarded as not disease causing. In one case (AD19), the reported phenotype (unilateral HI) is different from the type of HI known to be associated with mutations in the gene (MYO7A). In another patient (AR19), two class 3 variants in USH2A were found. The patient was 14 years old and had no signs of retinitis pigmentosa. The significance of the (previously unreported) USH2A missense variants in this patient is unclear and they cannot be definitely classified as causative for the HI in this patient, although they are located in trans and co-segregate with the HI.
In our study, 61.5% of the 200 exomes (123 cases) did not reveal causative or putative causative variants. In the majority of them (95 cases), no putative causative variants remained after the data-filtering procedure. In 24 cases, the variants were not segregating with the HI in the family. In three cases, only a single variant was identified in a gene known to underlie arHI. In two of these patients (ISO41 and ISO45), the genes were analyzed with Sanger sequencing and/or MLPA, which did not reveal a second variant. In the third patient (AD65), a heterozygous variant c.1322C>T (p.(Ser441Leu)) was found in SLC26A5. As the patient had autosomal dominant, profound, asymmetric HI with an onset in the sixth decade, the phenotype was not compatible with DFNB61. Therefore, the SLC26A5 gene was not further analyzed. Finally, in one case (ISO87), a class 3 variant was identified in NLRP3, a gene known to underlie autosomal dominant cryporin-associated periodic syndromes, for example, Muckle–Wells syndrome.18 Further clinical evaluation in the patient revealed, however, no evidence for this syndrome and the variant was therefore considered not to be the cause of the HI.
Prescreening of single genes
In 137 patients (68.5%), one or more HI-related genes had been prescreened with Sanger sequencing prior to WES (Tables 2a and b). On average, 1.5 genes were pretested per individual. The numbers of individuals with pretests and the number of pretested genes were comparable between the groups of inheritance and between the groups of patients with causative variants, variants of uncertain significance and without detected causative variants. GJB2 was most frequently tested (80 times), followed by TECTA (36 times). Although prescreening in these 137 patients was negative and therefore WES was performed, it is known that patients with a specific phenotype associated with mutations in one or a few genes (eg, Pendred syndrome) are quite often solved by targeted testing.19
Table 2a. Single gene tests before WES per category.
| Categories | Subcategories | No. of individuals with single gene test(s) (%) | No. of single gene tests (per individual) | No. of individuals with GJB2/GJB6 test (%) |
|---|---|---|---|---|
| Total | 137 (68.5%) | 306 (1.5) | 80 (40%) | |
| WES result | Causative variant(s) | 45 (67.2%) | 101 (1.5) | 32 (47.8%) |
| Variant(s) of uncertain significance | 8 (80.0%) | 17 (1.7) | 4 (36.4%) | |
| No detected causative variant(s) | 84 (68.3%) | 188 (1.5) | 44 (35.8%) | |
| Inheritance | adHI | 45 (68.2%) | 106 (1.6) | 11 (16.7%) |
| arHI | 24 (77.4%) | 43 (1.4) | 17 (54.8%) | |
| Isolated HI | 67 (55.8%) | 154 (1.5) | 51 (50.0%) | |
| X-linked HI | 1 (100.0%) | 3 (3.0) | 1 (100.0%) |
Table 2b. Single gene tests prior to WES per gene.
| Gene | No. of requests |
|---|---|
| Total | 306 |
| GJB2/GJB6 | 80 |
| TECTA | 36 |
| KCNQ4 | 18 |
| COCH | 16 |
| COL11A2 | 15 |
| SLC26A4 | 15 |
| TMPRSS3 | 14 |
| MYO6 | 10 |
| POU4F3 | 10 |
| ACTG1 | 9 |
| WFS1 | 8 |
| Asper array | 6 |
| EYA4 | 6 |
| USH2A | 6 |
| MITF | 5 |
| MYO7A | 5 |
| DFNA5 | 4 |
| MYO15A | 4 |
| PAX3 | 4 |
| DIAPH1 | 3 |
| OTOF | 3 |
| Array | 2 |
| COLL11A2 | 2 |
| POUF4F3 | 2 |
| PTPRQ | 2 |
| TMC1 | 2 |
| CDH23 | 1 |
| DFNB59 | 1 |
| EYA | 1 |
| EYA1 | 1 |
| FMR1 | 1 |
| KCNE1 | 1 |
| KCNQ | 1 |
| KCNQ1 | 1 |
| POLR1C | 1 |
| POLR1D | 1 |
| POU3F4 | 1 |
| RP-R | 1 |
| SIX1 | 1 |
| SIX5 | 1 |
| SMPX | 1 |
| SNAI2 | 1 |
| SOS1 | 1 |
| TCOF1 | 1 |
| TIMM8A | 1 |
Abbreviations: adHI, autosomal dominant hearing impairment; arHI, autosomal recessive hearing impairment.
To evaluate the utility of prescreening in individuals with HI, we made an overview of all in-house gene analysis requests for HI in 2013–2014 and the diagnostic yield (Supplementary Table S5). The vast majority of these tests were performed in patients of Dutch origin. The three genes with the highest diagnostic yield were COCH (36.8%), KCNQ4 (15.4%) and GJB2 (7.2%). For these three genes, founder or hotspot mutations occur in the Dutch population explaining the high incidence of mutations found in DNA diagnostics.20, 21, 22 The diagnostic yield for COL11A1, DFNA5, EYA1, MYO7A, NDP, OTOF, SLC26A4 and USH2A was higher than 10%, but the number of requests was less than 10 times. Therefore, the diagnostic yield for these genes is not reliable.
Discussion
In this study, we aimed to evaluate the diagnostic yield of WES-based targeted analysis of genes involved in HI. WES technology allowed the efficient identification of single-nucleotide variants, small insertions or deletions (indels) and large deletions that affect the protein coding regions of HI genes in a single experiment.11 Our study underlines the great genetic heterogeneity of HI, as causative variants were found in 26 different genes (Table 1, Figure 1).
In 61.5% of the cases, no causative variants were identified in the targeted HI-related genes. A part of these cases might be explained by variants that are not identified due to insufficient enrichment or coverage.23 Although coverage has greatly improved over time (Supplementary Figure S1, Supplementary Table S1), the identification rate of causative variants has remained stable (Supplementary Table S3). This implies that part of the causative variants in known HI genes cannot be identified by WES, for example deep intronic variants affecting splicing, variants in non-coding exons, repeat regions and regulatory regions. Another part of the cases without detected causative variants may be explained by mutations in yet undiscovered genes. There are tens of HI-related loci known, of which the causative gene is still not identified (http://www.hereditaryhearingloss.org/). As de novo variants in known adNSHI genes were identified in four of the cases in our study, we hypothesize that novel genes for adNSHI can be identified by a de novo strategy (ie, sequencing affected individuals and their unaffected parents).24 For the isolated cases, comprising about half of the subjects in this study, involvement of non-genetic causes cannot be fully excluded. Also, in subjects with late-onset HI non-genetic causes or a combination of (multiple) genetic and non-genetic factors cannot be discarded, despite the thorough patient evaluation. This could well explain why no variants were identified in the 16 subjects with an age of onset in the fifth or sixth decade (Supplementary Table S3, Supplementary Figure S2).
Variants of uncertain significance were mainly reported for patients without a family history of HI (isolated cases) or presumed adHI (Supplementary Tables S3 and S4). As in most of these cases no family members were available for segregation analysis, the causality of these variants remained unclear. This highlights the importance of taking an accurate family history and collecting clinical data and DNA samples of family members. In addition, it is essential to provide a thorough description of the phenotype of the patient in order to evaluate whether the gene with the identified variant has previously been associated with this specific phenotype.
A subset of patients (36 cases) in the present study was previously reported by Neveling et al.11 In 16 out of these 36 cases likely causative variants were identified, leading to a diagnostic yield of 44.4% for WES in HI. However, in nine of these families, segregation analysis was still needed to confirm the genetic diagnosis. This analysis was performed in the current study and in seven families the variants did not segregate with the HI. This lowers the diagnostic yield of the cases included in the study by Neveling et al to 22%, which is comparable to the yield in our study and again underlines the importance of segregation analysis.
The wide use of WES in routine diagnostics and research is producing large amounts of data on sequence variants in HI. Variants that have initially been reported as causative, based on the knowledge at that time, might be reclassified as benign due to increasing availability of allele frequency data.25 This highlights the importance of population-based allele frequency data to evaluate the causality of variants. However, rare variants can still be difficult to classify. We identified novel missense variants in USH2A (AR19) and classified these as variants of uncertain significance, despite the fact that (1) they are predicted to be damaging, (2) they were not reported in any public database so far and (3) they segregated with the hearing impairment in the corresponding family. The patient did not show symptoms of retinitis pigmentosa at the age of 14 years, but visual symptoms in Usher syndrome type 2 normally start in the second decade of life.26 Without support from functional studies, the pathogenicity of these missense variants will remain uncertain, since Petrovski et al.27 calculated a residual variation score of 4.18 for USH2A (75th percentile of scored genes, frequency data based on NHLBI Exome Sequencing Project) suggestive of a great tolerance of this gene to genetic variation. This is casting some doubt on the extensive variation in USH2A reported as likely pathogenic in public databases such as the LOVD. Importantly, these uncertainties are extremely difficult for genetic counseling, as parents have to be informed about the possible development of Usher syndrome in their children.
CNV detection in our cohort could identify large homozygous deletions in 4% of the cases, which is comparable to the 1.5–7.3% presented in the literature.28, 29, 30, 31, 32, 33, 34 A relatively high frequency of STRC deletions was found in our Dutch population (2%), as has also been reported in other populations.8, 9
In one case (ISO31), we found causative variants in a gene that is associated with an identifiable phenotype and segregating with a recessive inheritance pattern. This patient had an incomplete partition of the cochlea and mutations in SLC26A4.35 We did not find other cases with an identifiable phenotype such as progressive HI with a downsloping audiogram caused by TMPRSS3 mutations,7, 36 and the stable HI with a cookie-bite audiogram configuration caused by mutations in TECTA.7 This is most likely due to the fact that these genes are generally pretested in patients with these identifiable phenotypes.
In our cohort, the diagnostic yield of WES targeting a panel of HI-related genes is 33.5%. Other studies using massively parallel sequencing have reported similar overall diagnostic rates, despite of using different technologies and testing different populations.32, 33, 34, 37, 38 We found that causative variants in GJB2, USH2A, MYO6, STRC and MYO15A underlie HI in 14.0% of the cases in our cohort. This is in agreement with the previously published studies on the involvement of HI genes in other populations.6, 7, 8, 9, 32, 33, 34, 36, 38, 39, 40 The diagnostic yield of WES targeting a panel of HI-related genes is generally higher than that of single gene testing. Therefore, we recommend to reduce prescreening of single genes to a minimum. As the utility and yield of prescreening of single genes prior to WES is population specific, our recommendations apply in particular for the Dutch population. We suggest that for nonsyndromic congenital or first decade onset HI it would be cost-effective to prescreen GJB2, because of its relatively frequent association with HI. For recognizable phenotypes (such as Pendred syndrome, Waardenburg syndrome and Usher syndrome) or for genes with a relatively common founder mutation in a specific population (such as mutations in COCH in the Dutch and Belgian population)20 prescreening of specific genes might still be useful. This is supported by the relatively high diagnostic yield of targeted sequencing of GJB2 (7.2%) and COCH (36.8%). In all other cases, we recommend to perform WES targeting a panel of HI-related genes as a first diagnostic test.
Acknowledgments
We are grateful to all participating patients and their families. We also thank Kim van der Donk and Jaap Oostrik for expert technical assistance, and Rick de Reuver and Nienke Wieskamp for their support in bioinformatics analysis. We are thankful to all clinicians for collecting and referring patients. This work was financially supported by grants from the Netherlands Organization for Scientific Research (016.136.088, to MS), the Fonds NutsOhra foundation (1204-076, to HK) and the Heinsius Houbolt foundation (to HK).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Shearer AE, DeLuca AP, Hildebrand MS et al: Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA 2010; 107: 21104–21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE: Newborn hearing screening—a silent revolution. N Engl J Med 2006; 354: 2151–2164. [DOI] [PubMed] [Google Scholar]
- Seco CZ, Oonk AM, Dominguez-Ruiz M et al: Progressive hearing loss and vestibular dysfunction caused by a homozygous nonsense mutation in CLIC5. Eur J Hum Genet 2015; 23: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, Ruiz-Palmero L, Kalay E et al: Mutations of the gene encoding otogelin are a cause of autosomal-recessive nonsyndromic moderate hearing impairment. Am J Hum Genet 2012; 91: 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariz KO, Duman D, Seco CZ et al: Mutations in OTOGL, encoding the inner ear protein otogelin-like, cause moderate sensorineural hearing loss. Am J Human Genet 2012; 91: 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M, Nishio SY, Hattori M et al: Mutations in the MYO15A gene are a significant cause of nonsyndromic hearing loss: massively parallel DNA sequencing-based analysis. Ann Otol Rhinol Laryngol 2015; 124: 158S–168S. [DOI] [PubMed] [Google Scholar]
- Hoefsloot LH, Feenstra I, Kunst HP, Kremer H: Genotype phenotype correlations for hearing impairment: approaches to management. Clin Genet 2014; 85: 514–523. [DOI] [PubMed] [Google Scholar]
- Francey LJ, Conlin LK, Kadesch HE et al: Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A 2012; 158A: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B, Hofrichter MA, Neuner C et al: DFNB16 is a frequent cause of congenital hearing impairment: implementation of STRC mutation analysis in routine diagnostics. Clin Genet 2015; 87: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA: Disease gene identification strategies for exome sequencing. Eur J Human Genet 2012; 20: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C et al: A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013; 34: 1721–1726. [DOI] [PubMed] [Google Scholar]
- Mazzoli M, Van Camp G, Newton V, Giarbini N, Declau F, Parving A: Recommendations for the description of genetic and audiological data for families with nonsyndromic hereditary hearing impairment. Audiol Med 2003; 1: 148–150. [Google Scholar]
- Seco CZ, Giese AP, Shafique S et al: Novel and recurrent CIB2 variants, associated with nonsyndromic deafness, do not affect calcium buffering and localization in hair cells. Eur J Hum Genet 2015; 24: 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C et al: From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013; 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A et al: Copy number variation detection and genotyping from exome sequence data. Genome Res 2012; 22: 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfundt R, del Rosario M, Vissers LE et al: Detection of clinically relevant copy number variants by exome sequencing in a large cohort of genetic disorders. Genet Med 2016, e-pub ahead of print 27 October 2016 doi:10.1038/gim.2016.163. [DOI] [PMC free article] [PubMed]
- Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrabay G, Grandemange S, Touitou I: Diagnosis of cryopyrin-associated periodic syndrome: challenges, recommendations and emerging concepts. Expert Rev Clin Immunol 2015; 11: 827–835. [DOI] [PubMed] [Google Scholar]
- Dai P, Stewart AK, Chebib F et al: Distinct and novel SLC26A4/Pendrin mutations in Chinese and U.S. patients with nonsyndromic hearing loss. Physiol Genomics 2009; 38: 281–290. [DOI] [PubMed] [Google Scholar]
- Fransen E, Verstreken M, Bom SJ et al: A common ancestor for COCH related cochleovestibular (DFNA9) patients in Belgium and The Netherlands bearing the P51S mutation. J Med Genet 2001; 38: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini P, Rabionet R, Barbujani G et al: High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet 2000; 8: 19–23. [DOI] [PubMed] [Google Scholar]
- Topsakal V, Pennings RJ, te Brinke H et al: Phenotype determination guides swift genotyping of a DFNA2/KCNQ4 family with a hot spot mutation (W276S). Otol Neurotol 2005; 26: 52–58. [DOI] [PubMed] [Google Scholar]
- Lelieveld SH, Spielmann M, Mundlos S, Veltman JA, Gilissen C: Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum Mutat 2015; 36: 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112. [DOI] [PubMed] [Google Scholar]
- Shearer AE, Eppsteiner RW, Booth KT et al: Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet 2014; 95: 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Berlin CI, Hejtmancik JF et al: Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet 1994; 50: 32–38. [DOI] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB: Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013; 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai EA, Berman MA, Conlin LK et al: PECONPI: a novel software for uncovering pathogenic copy number variations in non-syndromic sensorineural hearing loss and other genetically heterogeneous disorders. Am J Med Genet A 2013; 161A: 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Diaz-Horta O, Guo S et al: Identification of copy number variants through whole-exome sequencing in autosomal recessive nonsyndromic hearing loss. Genetic Test Mol Biomarkers 2014; 18: 658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Kolbe DL, Azaiez H et al: Copy number variants are a common cause of non-syndromic hearing loss. Genome Med 2014; 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Lu J, Wang J, Li H, Lin X: Combined examination of sequence and copy number variations in human deafness genes improves diagnosis for cases of genetic deafness. BMC Ear Nose Throat Disord 2014; 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommen M, Schrauwen I, Vandeweyer G et al: DNA diagnostics of hereditary hearing loss: a targeted resequencing approach combined with a mutation classification system. Hum Mutat 2016; 37: 812–819. [DOI] [PubMed] [Google Scholar]
- Yan D, Tekin D, Bademci G et al: Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet 2016; 135: 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE et al: Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 2016; 135: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps PD, Coffey RA, Trembath RC et al: Radiological malformations of the ear in Pendred syndrome. Clin Radiol 1998; 53: 268–273. [DOI] [PubMed] [Google Scholar]
- Bademci G, Foster J 2nd, Mahdieh N et al: Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med 2015; 18: 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Smith RJ: Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol Head Neck Surg 2015; 153: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Black-Ziegelbein EA, Hildebrand MS et al: Advancing genetic testing for deafness with genomic technology. J Med Genet 2013; 50: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atik T, Onay H, Aykut A et al: Comprehensive analysis of deafness genes in families with autosomal recessive nonsyndromic hearing loss. PLoS One 2015; 10: e0142154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Babanejad M, Beheshtian M et al: Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet 2015; 52: 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.