Abstract
We aimed to identify the genetic cause of the devastating neurodegenerative disease amyotrophic lateral sclerosis (ALS) in a German family with two affected individuals, and to assess the prevalence of variants in the identified risk gene, FIG4, in a central European ALS cohort. Whole-exome sequencing (WES) and an overlapping data analysis strategy were performed in an ALS family with autosomal dominant inheritance and incomplete penetrance. Additionally, 200 central European ALS patients were analyzed using whole-exome or targeted sequencing. All patients were subjected to clinical, electrophysiological, and neuroradiological characterization to explore genotype–phenotype relationships. WES analysis of the ALS family identified the rare heterozygous frameshift variant FIG4:c.759delG, p.(F254Sfs*8) predicted to delete the catalytic domain and active center from the encoded phosphoinositide 5-phosphatase with a key role in endosomal vesicle trafficking. Additionally, novel or rare heterozygous FIG4 missense variants predicted to be deleterious were detected in five sporadic ALS patients revealing an overall FIG4 variant frequency of 3% in our cohort. Four of six variants identified were previously associated with ALS or the motor and sensory neuropathy Charcot-Marie-Tooth disease type 4J (CMT4J), whereas two variants were novel. In FIG4 variant carriers, disease duration was longer and upper motor neuron predominance was significantly more frequent compared with ALS patients without FIG4 variants. Our study provides evidence for FIG4 as an ALS risk gene in a central European cohort, adds new variants to the mutational spectrum, links ALS to CMT4J on a genetic level, and describes a distinctive ALS phenotype for FIG4 variant carriers.
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease characterized by progressive degeneration of upper motor neurons (UMNs) and lower motor neurons (LMNs), leading to paralysis of voluntary muscles and ultimately death due to respiratory failure within 3–5 years from disease onset.1, 2 While 5–10% of ALS patients have a positive family history (fALS), the majority of patients are sporadic cases (sALS).3 The distinction between fALS and sALS, however, is currently being questioned by the awareness that apparently sALS can be inherited.4 Since 1993, when the superoxide dismutase 1 (SOD1) gene was found to be mutated in fALS patients, an increasing number of disease genes, designated ALS 1–22, have been reported.5
In 2009, ALS11 was identified as a rare autosomal dominant form of ALS associated with heterozygous deleterious variants of FIG4 in North American patients.6 FIG4 encodes a phosphoinositide 5-phosphatase regulating phosphatidylinositol-3,5-bisphosphate, an intracellular signaling lipid with a key role in endosomal vesicle trafficking.7, 8 A loss of function Fig4 variant was found to cause neuronal degeneration in the central nervous system including spinal motor neurons, and peripheral neuropathy in the ‘pale tremor' mouse.9 Biallelically mutated FIG4 was detected in patients with (i) Charcot-Marie-Tooth disease type 4J (CMT4J),9 a recessively inherited form of hereditary motor and sensory neuropathy, (ii) Yunis-Varón syndrome,10 an autosomal recessive disorder with skeletal anomalies and severe neurological involvement, and (iii) familial epilepsy with polymicrogyria.11
Here, we present known and novel heterozygous deleterious FIG4 variants in a cohort of 201 central European fALS and sALS patients, and provide clinical, electrophysiological, and neuroradiological information on FIG4 variant carriers.
Subjects and methods
Patients
The study was approved by the Ethics Board of Hannover Medical School. Each patient and family member provided informed consent for participation in the study. All patients were examined at least once at the motor neuron disease outpatient clinic of Hannover Medical School by a neurologist specialized in ALS. Extensive clinical workup including magnetic resonance imaging (MRI), cerebral spinal fluid analysis, electromyography (EMG), and nerve conduction studies (NCS) was performed to exclude ALS-mimicking conditions. Longitudinal information over a number of years was available for most individuals. A total of 124 male and 77 female patients of almost exclusively central European origin were enrolled in this study, including 6 fALS and 195 sALS cases. The mean age of onset was 58.6±12.5 years (range 19–84 years). Initial symptoms occurred in the bulbar region in 44 patients (21.9%) and the spinal cord in 157 patients (78.1%). UMN and LMN involvement was assessed in all patients as described previously.12 Increased muscle tone, jaw jerk, increased deep tendon reflexes, spread of reflexes, clonus, and Babinski's sign were classified as signs of UMN affection. Muscle atrophy, fasciculations, and weakness were classified as LMN signs. Regarding bulbar involvement, brisk masseter reflex and tongue spasticity were interpreted as UMN signs, whereas tongue atrophy and tongue fasciculations were considered to be LMN signs. The extent and distribution of clinically inapparent LMN affection was also assessed by EMG in all patients. Muscle strength, muscle tone, reflexes, and the degree of atrophy and fasciculations were graded as detailed in Körner et al.12 Using these criteria, 13 patients (6.5%) had a predominant UMN phenotype, eight of which were diagnosed with primary lateral sclerosis (PLS). In the index patient, FamALS006-01, the following genetic tests had been performed before this study: (1) sequence analysis of the genes SOD1, FUS, and TARDBP using conventional chain termination protocols, (2) analysis of the hexanucleotide repeat in the C9orf72 gene by repeat prime PCR and Southern blot analysis.
Genetic analysis
Whole-exome sequencing (WES) was performed on peripheral blood from the index patient, father and mother of ALS family FamALS006, 22 sALS patients, and 48 controls not affected by ALS using Agilent SureSelect Human All Exon v4 Target Enrichment System on an Illumina HiSeq 2000 (Oxford Gene Technology, Begbroke, UK) as described previously.13 All samples were sequenced to a mean target coverage of >50 ×. WES data from ALS family FamALS006 were analyzed using INGENUITY Variant Analysis (INGENUITY Systems; QIAGEN, Redwood City, CA, USA) and our in-house NGS data analysis workflow as described in Results, Supplementary Table S1, and Supplementary Table S2. In 22 sALS patients, FIG4 variants were retrieved from WES data. To verify selected FIG4 variants identified by WES and to screen the coding exons (1–23) and adjacent intronic regions of FIG4 (NG_007977.1 and NM_014845.5) for variants in 178 further ALS patients, conventional chain termination protocols were used (for primer sequences see Supplementary Table S3). Minor allele frequency (MAF) was retrieved from Variant Analysis (INGENUITY Systems) and Exome Aggregation Consortium (ExAC) Browser Beta (http://exac.broadinstitute.org/; http://biorxiv.org/content/early/2015/10/30/030338). For prediction of variant deleteriousness, the tools SIFT and PROVEAN (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and MutationTaster (http://www.mutationtaster.org) were used. The six rare non-silent FIG4 variants predicted to be deleterious identified in ALS patients were submitted to ClinVar (SCV accession nos SCV000299294–SCV000299299; http://www.ncbi.nlm.nih.gov/clinvar/).
Results
Analysis of WES data revealed a rare heterozygous FIG4 frameshift variant shared by patient and father of FamALS006
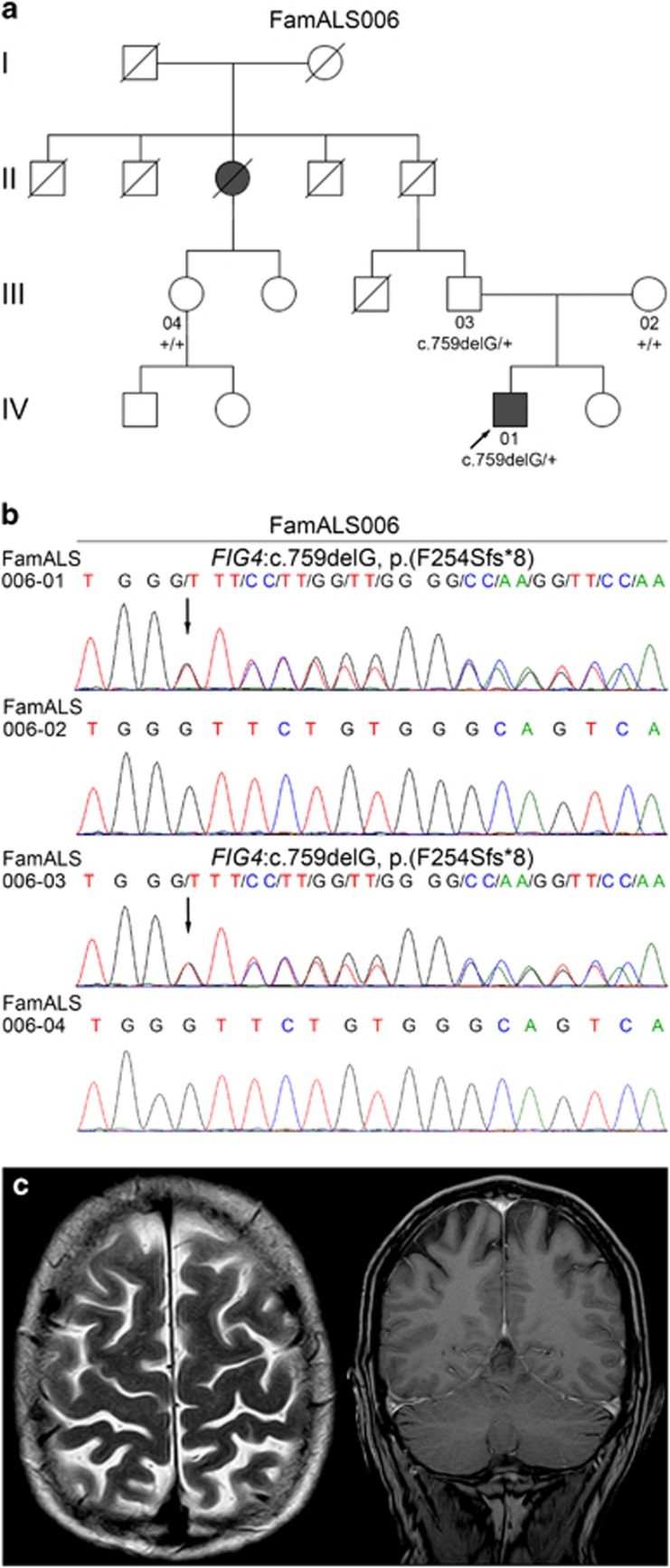
WES was performed in ALS family FamALS006 comprising the index patient and his affected paternal great-aunt, thus displaying autosomal dominant inheritance with incomplete penetrance (Figure 1a). Before WES, analysis of the most common fALS genes (SOD1, FUS, TARDBP, and C9orf72) in the index patient had revealed no pathogenic aberrations. For WES data analysis (Supplementary Table S1), we used an overlapping strategy to detect variants shared by the patient and his father because the affected great-aunt was related on the father's side, and DNA from her or the clinically asymptomatic grandfather was not available. Of 1861 coding variants shared by the index patient and his father and not present in the healthy mother, 1425 variants were non-silent (ie, canonical±1 or 2 splice site, frameshift, in-frame indels, stop gained/lost, and missense variants), and of these 96 variants were rare (MAF of <1%) and not present in 48 in-house controls (Supplementary Table S2). To focus on known ALS-associated genes, we then extracted only variants in genes (n=126) listed in the ALS Online Genetics Database (http://alsod.iop.kcl.ac.uk/),14 resulting in one variant. This variant, a deletion of a single nucleotide in exon 7 of the FIG4 gene, FIG4:c.759delG, p.(F254Sfs*8), causes a frameshift predicted to result in a truncated protein lacking the catalytic domain including the active center. This variant was not found in WES data from 48 neurologically unaffected German controls and is very rare (MAF of 0.00009001) in Europeans (non-Finnish) according to the ExAC database. By Sanger sequencing, the variant was confirmed in the index patient and his father, but not detected in the 62-year-old healthy daughter of the affected great-aunt or the mother of the index patient (Figure 1b). The affected paternal great-aunt and the paternal grandfather have passed away, and the sister of the index patient was also not available for genetic testing, thereby limiting segregation analysis. Therefore, it cannot be excluded that the FIG4 frameshift variant was inherited from the paternal grandmother of the index patient.
Figure 1.
A rare heterozygous FIG4 frameshift variant was identified in family FamALS006 by WES and overlapping strategy. (a) Pedigree of family FamALS006 with two ALS cases, that is, the index patient (designated by an arrow) and his paternal great-aunt, and segregation of the identified heterozygous frameshift variant FIG4:c.759delG, p.(F254Sfs*8). 01, index patient; 02, mother of index patient; 03, father of index patient; 04, daughter of paternal great-aunt of index patient; filled symbol, ALS patient; +, wild-type allele. (b) Electropherograms obtained by Sanger sequencing demonstrating the heterozygous frameshift variant FIG4:c.759delG, p.(F254Sfs*8) in the index patient FamALS006-01 and his father FamALS006-03 (the first affected nucleotide is designated by an arrow), but not in the healthy 62-year-old daughter of the affected paternal great-aunt of the index patient (FamALS006-04) or in his mother FamALS006-02. This variant is predicted to result in a substantially shortened protein lacking the catalytic domain including the active center (see Figure 2c). (c) Cranial MRI findings in 41-year-old fALS patient FamALS006-01 carrying the heterozygous frameshift variant FIG4:c.759delG, p.(F254Sfs*8) demonstrating pronounced frontoparietal atrophy on transverse T2-weighted (left) and coronal T1-weighted scans (right), whereas the temporal lobe was not affected.
Sequence analysis of FIG4 in 200 additional ALS cases revealed five rare heterozygous FIG4 missense variants predicted to be deleterious
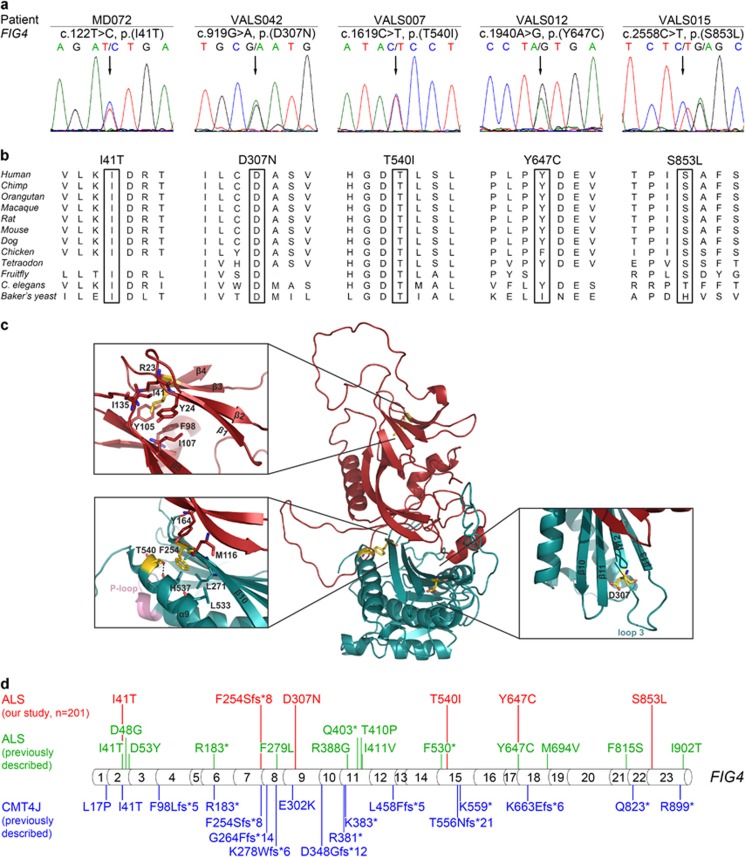
Next, we assessed the frequency of FIG4 variants in an almost exclusively central European cohort of 200 unrelated ALS patients using WES and targeted sequencing. Thereby, we identified novel or rare heterozygous missense variants in the coding region of FIG4 in five additional ALS patients (Table 1 and Figure 2a). None of these patients had a family history of neurodegenerative disorders, and no parental DNA was available for segregation analysis. All five affected amino acids were highly or very highly conserved across species (Figure 2b), and the variants were predicted to have a deleterious effect on protein structure or function by at least two of four prediction tools, that is MutationTaster, PROVEAN, SIFT, or PolyPhen-2 (Table 1). The affected amino acids were located in the N-terminal SAC domain (n=1), the catalytic domain (n=2), or in regions of FIG4 not covered by structural data (n=2; Figure 2c). Three of the missense variants had previously been described in ALS or CMT patients, whereas two variants have not been associated with these disorders (Table 1). The latter is true for the novel variant FIG4:c.1619C>T, p.(T540I) that is neither listed in the single-nucleotide polymorphism database nor in the ExAC browser, and for the very rare variant FIG4:c.919G>A, p.(D307N) with a MAF of 0 in non-Finnish Europeans according to ExAC. The six rare deleterious FIG4 variants we identified in total in 201 ALS patients were distributed throughout the gene without any specific clustering region (Figure 2d). All FIG4 variants detected in our ALS cohort are summarized in Supplementary Table S4, including common, noncoding or silent variants that were not considered deleterious.
Table 1. Rare or novel heterozygous non-silent variants in the FIG4 gene predicted to be deleterious identified in 201 central European ALS patients.
| Prediction | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Chromosomal position according to GRCh37 | Exon | Nucleotide change | Amino-acid change | Reference SNP number | MAF | MutationTaster | PROVEAN | SIFT | PolyPhen-2 | Diagnosis | Previously described in |
| MD072 | 6:110 036 336 | 2 | c.122T>C | p.(I41T) | Rs121908287 | 0.001572 | Disease causing | Deleterious −4.59 | Damaging 0.001 | Probably damaging 1.000 | PLS | ALS19 CMT4J9 |
| FamALS006-01 FamALS006-03 | 6:110 059 638 | 7 | c.759delG | p.(F254Sfs*8) | Rs764717219 | 0.00009001 | Disease causing | — | — | — | fALS — | CMT4J9 CMT16 |
| VALS042 | 6:110 064 355 | 9 | c.919G>A | p.(D307N) | Rs573441014 | 0 | Disease causing | Deleterious −3.71 | Damaging 0.003 | Probably damaging 0.997 | sALS | — |
| VALS007 | 6:110 087 967 | 15 | c.1619C>T | p.(T540I) | — | — | Disease causing | Deleterious −4.75 | Damaging 0.000 | Probably damaging 0.998 | PLS | — |
| VALS012 | 6:110 106 223 | 17 | c.1940A>G | p.(Y647C) | Rs150301327 | 0.000135 | Disease causing | Neutral −2.17 | Damaging 0.031 | Benign 0.008 | sALS | ALS6 CMT25 |
| VALS015 | 6:110 146 302 | 23 | c.2558C>T | p.(S853L) | Rs774805375 | 0.00003 | Disease causing | Deleterious −2.77 | Damaging 0.004 | Probably damaging 0.998 | sALS | CMT16 |
Abbreviations: ALS, amyotrophic lateral sclerosis; CMT, Charcot-Marie-Tooth disease; CMT4J, Charcot-Marie-Tooth disease type 4J; f, familial; MAF, minor allele frequency in European (non-Finnish) population according to Exome Aggregation Consortium Browser (Beta); PLS, primary lateral sclerosis; s, sporadic; SNP, single-nucleotide polymorphism.
PROVEAN uses a score threshold of −2.282, whereby neutral >−2.282 and deleterious <−2.282; SIFT prediction scores range from 1 (=tolerated) to 0 (=damaging); PolyPhen-2 prediction scores range from 0 (=benign) to 1 (=probably damaging).
Reference sequences used: NG_007977.1 and NM_014845.5.
Figure 2.
Rare heterozygous deleterious FIG4 missense variants in five sALS patients detected by whole-exome and targeted sequencing. (a) Electropherograms demonstrating rare heterozygous FIG4 missense variants in five sALS patients (affected nucleotides are designated by an arrow). All variants are predicted to be deleterious by at least two prediction tools. Parents were not available for segregation analysis. (b) Missense variants detected in FIG4 affect highly or very highly conserved amino acids (data taken from Alamut Visual 2.6.1., Interactive Biosoftware, Rouen, France). (c) Mapping of the amino-acid residues affected by the ALS-associated variants detected here on a structural model of human phosphoinositide 5-phosphatase FIG4. One of the affected residues (p.I41) is located in the N-terminal SAC domain (red), and three affected residues (p.F254; p.D307; p.T540) are situated in the catalytic domain (cyan). The identified frameshift variant, FIG4:c.759delG, p.(F254Sfs*8), is predicted to result in a truncated protein lacking the catalytic domain (cyan) including the P-loop (active center). The insets show close-up views of the corresponding regions. The structural model was generated using MODELLER32 based on the crystal structure of SAC1 (PDB: 3LWT).20 There are no structural data covering the regions encompassing the residues p.Y647 and p.S853, which are, therefore, missing in the model. (d) Summary of FIG4 variants detected in ALS patients in this or previous studies or previously in CMT4J patients. Variants found in ALS patients from our cohort are indicated in red; variants previously described in ALS patients6, 19, 33 are indicated in green; variants previously described in CMT4J patients9, 16 are indicated in blue. Protein sequence variants are given according to the Human Genome Variation Society recommendation v2.0; the exon structure of the human FIG4 gene (NG_007977.1, NM_014845.5) was based on Alamut Visual 2.6.1.
When comparing the FIG4 variant frequency in our 201 central European ALS patients with ethnicity-matched controls (non-Finnish Europeans from the ExAC database, n=33 370 individuals), 6 of 201 ALS cases (3%) carried rare (MAF<1%) non-silent deleterious (all loss of function variants, that is, frameshift, stop gained/lost, canonical ±1 or 2 splice site, and initiation codon variants, as well as missense variants predicted to be deleterious by either SIFT or PolyPhen-2 prediction tools) FIG4 variants versus 441 of 33 370 controls (1.32%). This difference is statistically significant (two-tailed P-value<0.05, χ2 test), whereby it should be considered that the two cohorts do not match in size and may not match in age and gender distribution.
Phenotypic spectrum of six ALS patients with deleterious variants of FIG4: disease duration was longer and UMN predominance was significantly more frequent in FIG4 variant versus non-variant carriers
Clinical, electrophysiological, and neuroradiological characteristics of ALS patients carrying deleterious FIG4 variants are summarized in Table 2. The index patient, FamALS006-01, carrying the frameshift variant FIG4:c.759delG, p.(F254Sfs*8) developed ALS at 40 years of age, with atrophic paresis of the left upper limb and increased deep tendon reflexes as initial symptoms. Within 3 years, atrophies and weakness progressed to the right upper and both lower extremities. No marked respiratory deficiency was observed. Cranial MRI at the age of 41 years showed pronounced frontoparietal atrophy (Figure 1c), whereas the Edinburgh Cognitive and Behavioral ALS Screen15 revealed no abnormalities. FamALS006-03, the index patient's father, also carrying FIG4:c.759delG, p.(F254Sfs*8) did not develop neurological symptoms until he died of cardiovascular disease at 75 years of age several months after the genetic test. The great-aunt of the index patient, who was diagnosed with ALS, had slowly progressive atrophic weakness of upper and lower limbs with dysphagia and died ∼30 years after symptom onset. Patients MD072 carrying FIG4:c.122T>C, p.(I41T) and VALS007 carrying FIG4:c.1619C>T, p.(T540I) had initially been diagnosed with UMN predominant ALS. In both cases, the diagnosis later changed to PLS because of slow disease progression over 3 years, no clinical signs of LMN involvement, and only mild LMN signs detectable by needle EMG in one body region in patient VALS007. Similarly, patient VALS042 carrying FIG4:c.919G>A, p.(D307N) had an UMN predominant clinical phenotype, but in contrast to MD072 and VALS007, progressive LMN signs developed after 3 years of stable disease. Comparable to MD072, VALS007, and VALS042, patient VALS012 carrying FIG4:c.1940A>G, p.(Y647C) had a relatively long disease duration (5 years) before he died of a traumatic subdural hemorrhage. He had been diagnosed with flail arm syndrome with no UMN signs. In contrast, patient VALS015 carrying FIG4:c.2558C>T, p.(S853L) suffered from classical Charcot-type ALS with rapid disease progression and death from respiratory failure within 12 months. Remarkable neurological non-motor neuron symptoms in the FIG4 variant carriers were mild cognitive impairment in VALS007 and sensory deficits in three of six patients. By initial neurophysiological studies, no major slowing of nerve conduction velocity indicating peripheral demyelination, as seen in CMT4J, was detected in the six FIG4 variant carriers. However, amplitude reduction or lack of detectable response of the sural nerve was observed in three patients indicating sensory axonal neuropathy in addition to the ALS-related motor axonal impairment. Varying degrees of brain atrophy, particularly in the frontoparietal region, were found in all FIG4 variant carriers by cranial MRI.
Table 2. Clinical, electrophysiological, and neuroradiological characteristics of ALS patients carrying heterozygous deleterious FIG4 variants.
| Patient | Amino-acid change | Inheri-tance | Gender | Country of origin | Age of onset (years) | Site of onset | Diagnosis | El Escorial | Diseaseduration (years)a | Neurological non-motor neuron symptoms | EMG | NCS | Cranial MRI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MD072 | p.(I41T) | Sporadic | F | Italy | 43 | Spinal | PLS | Possible ALS | 12.25 | Pathological laughing and crying | No acute/chronic denervation | Normal | Precentral gyrus thinning, mild frontoparietal atrophy, no ventricular enlargement, no T2 hyperintensity in the CST |
| FamALS006–01 | p.(F254Sfs*8) | Familial | M | Germany | 40 | Spinal | ALS | Clinically probable ALS | 2.67 | None | Acute/chronic denervation UE, LE, thoracic | Reduction of CMAP and prolongation of DML | Severe frontoparietal atrophy, no ventricular enlargement, no T2 hyperintensity in the CST |
| VALS042 | p.(D307N) | Sporadic | M | Germany | 78 | Bulbar | ALS-UMN | Clinically probable ALS | 5.25 | Sensory impairment for vibration, light touch | Chronic denervation UE, LE | Motor-sensory axonal neuropathy | Age-related symmetric atrophy and ventricular enlargement, no T2 hyperintensity in the CST |
| VALS007 | p.(T540I) | Sporadic | F | Germany | 72 | Bulbar | PLS | Possible ALS | 3.25 | Mild cognitive impairment, sensory impairment for vibration | Sparse and stable signs of acute/chronic denervation (LE only) | Reduction of CMAP and prolongation of DML | Moderate frontoparietal atrophy and ventricular enlargement, no T2 hyperintensity in the CST |
| VALS012 | p.(Y647C) | Sporadic | M | Germany | 66 | Spinal | ALS-flail arm | — | 5.25 | None | Acute/chronic denervation UE, LE | Motor-sensory axonal neuropathy | Mild frontoparietal atrophy and ventricular enlargement, no T2 hyperintensity in the CST |
| VALS015 | p.(S853L) | Sporadic | F | Germany | 48 | Spinal | ALS | Clinically probable ALS | 0.92 | Minimal sensory impairment for vibration | Acute/chronic denervation | Mild motor–sensory axonal neuropathy | Mild frontoparietal atrophy, no ventricular enlargement, no T2 hyperintensity in the CST |
Abbreviations: ALS, amyotrophic lateral sclerosis; CMAP, compound motor action potential; CST, corticospinal tract; DML, distal motor latency; EMG, electromyography; F, female; LE, lower extremity; M, male; MRI, magnetic resonance imaging; NCS, nerve conduction study; PLS, primary lateral sclerosis; UE, upper extremity; UMN, upper motor neuron.
Reference sequences used: NG_007977.1 and NM_014845.5.
Until last follow-up (MD072, FamALS006-01, VALS042, VALS007) or death (VALS012: due to traumatic subdural hemorrhage; VALS015: due to respiratory failure caused by ALS).
When comparing the phenotype in ALS patients from our cohort with (n=6) versus without (n=195) deleterious FIG4 variants, predominance of UMN signs was significantly more frequent in FIG4 variant carriers (3/6: 50%) compared with non-FIG4 variant carriers (10/195: 5% P=0.004, Fisher's exact test, two-sided). Bulbar onset was not significantly more frequent in patients with (2/6: 33%) versus without (42/195: 21.5%) FIG4 variants (P=0.614, Fisher's exact test, two-sided). Disease duration was compared in the subset of 189 patients with disease onset between 2004 and 2014. Mean disease duration was longer in the six FIG4 variant carriers (4.93±3.94 years) compared with that in the 183 non-FIG4 variant carriers (3.18±2.21 years), although this difference did not reach statistical significance (P=0.212, Mann–Whitney U-test, two-tailed).
Discussion
Heterozygous deleterious variants in the FIG4 gene have first been implicated in sALS and fALS in 2009.6 To date, at least 14 rare nonsynonymous FIG4 variants have been detected in ALS cases (Figure 2d). However, the contribution of FIG4 variants to ALS pathogenesis has been debated because in smaller cohorts no deleterious FIG4 variants were found, and some non-penetrant FIG4 variant carriers have been described.16, 17, 18 Our sequence analysis of the FIG4 gene in 201 almost exclusively central European ALS patients confirms known and identifies novel or rare heterozygous variants.
Several lines of evidence suggest that the six FIG4 variants detected in our cohort are pathogenic and associated with an ALS phenotype. The frameshift variant, FIG4:c.759delG, p.(F254Sfs*8), was identified by WES in the fALS patient and his father, which is consistent with the fact that the other ALS patient in the family, the great-aunt of the index patient who was not available for genetic testing, was in the paternal line. This frameshift variant is very rare and predicted to truncate the protein at the C-terminal end of the β10-strand. Therefore, the frameshift variant could result in a substantially shortened protein lacking the catalytic domain including the P-loop, which corresponds to the active center, or in the loss of expression at the mRNA level because of nonsense-mediated decay. The variant FIG4:c.122T>C, p.(I41T) that we detected in sALS patient MD072 was reported previously in one of 349 sALS patients.19 According to Manford et al.,20 it may affect local folding or protein stability. Isoleucine at position 41 is located at the C-terminal end of the β3-strand, is surrounded by hydrophobic residues of the adjacent β-strands, and might stabilize folding by hydrophobic interactions. Using a yeast two-hybrid system, the p.I41T variant impaired interaction of mutant FIG4 with the scaffold protein VAC14, causing an unstable protein and low protein levels in an animal model and patient fibroblasts.21 The very rare variant FIG4:c.919G>A, p.(D307N) was identified in sALS patient VALS042. Aspartic acid at position 307 is located at the C-terminal end of the β12-strand, directly upstream of the protruding loop 3. The residue is surface exposed and, therefore, might affect protein folding and stability. In a previous study, a missense variant in close proximity, p.E302K, was shown to act as a functional null allele of FIG4.16 The substitution by lysine did not rescue the phenotype of Fig4p-null yeast, possibly due to reduced protein stability.16 The novel variant FIG4:c.1619C>T, p.(T540I) was detected in sALS patient VALS007. Threonine at position 540 is part of the N-terminal turn of the α9-helix. The residue stabilizes the α-helix by a hydrogen bond of the hydroxyl side chain with the backbone carbonyl oxygen of H537 (Figure 2c). The change of threonine at position 540 to a nonpolar residue prevents this important interaction and might destabilize the initial α-turn of the helix, which is in close vicinity of the catalytic P-loop, thereby directly affecting the active site. The very rare variant FIG4:c.1940A>G, p.(Y647C) identified in sALS patient VALS012 was previously reported in one of 473 ALS patients.6 Taken together, the frequency of rare non-silent deleterious FIG4 variants identified here in central European ALS patients (3%) is significantly higher compared with that in ethnicity-matched controls from the ExAC database (1.32%), whereas there is no lower-than-expected number of FIG4 LoF, that is, stop-gained and essential splice site, variants in the ExAC cohort. Our findings are in line with previous results reporting FIG4 gene variants in 2% of North American ALS patients of European ancestry.6 Comparably, variants in FIG4 were identified as ALS risk factors in two recent large-scale sequencing studies.19, 22 However, there is incomplete penetrance in the ALS family described here, which includes an unaffected father carrying a FIG4 frameshift variant, suggesting that FIG4 variants are not causative alone, but that ALS patients may need to carry rare variants in multiple genes to show disease. This notion is corroborated by the finding that at least 30 genes are implicated in ALS pathogenesis with a low percentage of ALS cases explained by variants in each of these.22 In addition, there is direct evidence for an oligogenic basis of ALS, for example, 7 of 14 ALS patients with TARDBP variants carried variants in 4 other ALS genes sequenced,23 and 15 of 391 (3.8%) ALS cases had variants in more than one of 17 analyzed ALS genes.19
Our data provide evidence for a possible link between the motor neuron disease ALS and the hereditary motor and sensory neuropathy CMT4J at the genetic and phenotypic level, comparable to the detection of deleterious variants in the DYNC1H1 (dynein, cytoplasmic 1, heavy chain 1) gene previously associated with classical CMT, in patients with UMN syndromes.24 Deleterious biallelic variants of FIG4 were initially identified in CMT4J,9 which is an autosomal recessive form of CMT with severe combined axonal and demyelinating peripheral neuropathy and potentially severe muscle weakness and wasting.16 Four of the FIG4 variants identified here in ALS patients were either previously found in a compound heterozygous state with other FIG4 variants in CMT4J patients (c.122T>C and c.759delG),9 or as heterozygous variants in patients from CMT disease cohorts (c.1940A>G and c.2558C>T).16, 25 In contrast to ALS, the typical CMT phenotype involves a sensory deficit. We, therefore, examined FIG4 variant carriers with respect to sensory impairment. Clinical and electrophysiological examination of the six FIG4 variant carriers revealed four cases with sensory impairment or sensory–motor axonal neuropathy, whereby two patients were older than 70 years. Although lower extremity sensory nerve response can be difficult to elicit in the elderly, our findings suggest that ALS patients carrying FIG4 variants may show sensory deficits as a sign of a clinical overlap with CMT, however, without neurophysiological evidence of demyelination as in CMT4J.16
Interestingly, Fig4 loss in the ‘pale tremor' mouse results in both neuronal degeneration in the central nervous system including spinal motor neurons and in sensory and autonomic ganglia, as well as in electrophysiological and pathological abnormalities in peripheral nerves.9 While a peripheral neuronopathy is detectable in the ‘pale tremor' mouse, an ALS phenotype in later life cannot be excluded because Fig4 loss causes juvenile lethality.9 Similar to neuronal loss in the cerebral cortex and ventricular enlargement in the ‘pale tremor' mouse, all ALS patients with FIG4 variants studied here showed varying degrees of brain atrophy of the frontal or parietal cortex, some combined with enlarged ventricles upon neuroradiological evaluation. Frontoparietal atrophy was particularly severe in the fALS patient carrying the FIG4 frameshift variant, while the temporal lobe was unaffected. Comparably, frontal lobe atrophy was described in two siblings with a homozygous FIG4 frameshift variant and Yunis-Varón syndrome,10 whereby extramotor, in particular frontotemporal, affection occurs in the majority of fALS as well as sALS cases.2, 26, 27
By clinical and electrophysiological assessment, we found that UMN predominance was significantly more frequent in patients with versus without FIG4 variants of our cohort. Similarly, very prominent corticospinal tract findings were previously reported in four of nine ALS patients carrying heterozygous FIG4 variants.6 Among these, two of nine FIG4 variant carriers were diagnosed with PLS.6 Consistently, in our study, two of six patients carrying heterozygous FIG4 variants exhibited a PLS phenotype. PLS involves UMNs only, represents 1–4% of cases with motor neuron disease,28 and is now considered a restricted phenotype of ALS in the most recent revision of the El Escorial diagnostic criteria.29 As PLS is associated with a better prognosis than other ALS forms,2, 30 we compared disease duration in FIG4 variant versus non-variant carriers. In our ALS cohort, mean disease duration was longer in patients carrying deleterious FIG4 variants compared with non-FIG4 variant carriers (4.9 versus 3.2 years). Although not statistically significant, this difference in mean disease duration is particularly striking because one of six variant carriers died prematurely because of an accident and two others did not show a PLS/UMN predominant phenotype. Mean disease duration of our ALS patients with deleterious FIG4 variants was also longer than median survival from onset in a recent epidemiologic study of 1217 Dutch ALS patients (4.9 versus 2.9 years).31 This finding is corroborated by the even longer mean disease duration, that is, 9.1 years, reported by Chow et al.6 for eight ALS patients with heterozygous deleterious variants of FIG4. From this and previous studies, the notion emerges that ALS patients carrying FIG4 variants may often have a distinctive phenotype, although our six patients do include cases with rapid progression and mainly LMN involvement.
In summary, our data demonstrate heterozygous deleterious FIG4 variants in 3% of ALS patients from a central European cohort providing further evidence that variants of FIG4 are associated with ALS and suggesting that it may be worthwhile to perform FIG4 genetic testing in ALS patients, particularly in patients with a predominant UMN phenotype. If identified in ALS patients, FIG4 variants may serve as markers for a relatively good prognosis.
Acknowledgments
We thank the patients and their families for participating in this study, and are indebted to Christopher Baum, Reinhard Dengler, and Brigitte Schlegelberger for generous support and helpful discussions. Isolde Rangnau received a scholarship from the Klin-StrucMed program of Hannover Medical School funded by the Else Kröner-Fresenius-Stiftung. Anne Kosfeld and Ruthild G Weber received research support from the Else Kröner-Fresenius-Stiftung (Grant No. 2014_A234). Alma Osmanovic, Susanne Petri, and Ruthild G Weber received research support from the Petermax-Müller-Stiftung.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Robberecht W, Philips T: The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci 2013; 14: 248–264. [DOI] [PubMed] [Google Scholar]
- Bäumer D, Talbot K, Turner MR: Advances in motor neurone disease. J R Soc Med 2014; 107: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Walsh C, Lynch C et al: Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 2011; 82: 623–627. [DOI] [PubMed] [Google Scholar]
- Turner MR, Hardiman O, Benatar M et al: Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol 2013; 12: 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, Wu ZY: Genotype–phenotype correlations of amyotrophic lateral sclerosis. Transl Neurodegener 2016; 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK et al: Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet 2009; 84: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford AC, Traer C, Wassmer T et al: The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci 2006; 119: 3944–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fu Z et al: Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem 2007; 282: 23878–23891. [DOI] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ et al: Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 2007; 448: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Lenk GM, Lu JT et al: Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet 2013; 92: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S, Lenk GM, Dufresnois B et al: Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology 2014; 82: 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner S, Kollewe K, Fahlbusch M et al: Onset and spreading patterns of upper and lower motor neuron symptoms in amyotrophic lateral sclerosis. Muscle Nerve 2011; 43: 636–642. [DOI] [PubMed] [Google Scholar]
- Classen CF, Riehmer V, Landwehr C et al: Dissecting the genotype in syndromic intellectual disability using whole exome sequencing in addition to genome-wide copy number analysis. Hum Genet 2013; 132: 825–841. [DOI] [PubMed] [Google Scholar]
- Abel O, Powell JF, Andersen PM, Al-Chalabi A: ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat 2012; 33: 1345–1351. [DOI] [PubMed] [Google Scholar]
- Lulé D, Burkhardt C, Abdulla S et al: The Edinburgh Cognitive and Behavioural Amyotrophic Lateral Sclerosis Screen: a cross-sectional comparison of established screening tools in a German-Swiss population. Amyotroph Lateral Scler Frontotemporal Degener 2015; 16: 16–23. [DOI] [PubMed] [Google Scholar]
- Nicholson G, Lenk GM, Reddel SW et al: Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain 2011; 134: 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CP, Soong BW, Lin KP, Tu PH, Lin JL, Lee YC: FUS, TARDBP, and SOD1 mutations in a Taiwanese cohort with familial ALS. Neurobiol Aging 2011; 32: 553e13–553e21. [DOI] [PubMed] [Google Scholar]
- Verdiani S, Origone P, Geroldi A et al: The FIG4 gene does not play a major role in causing ALS in Italian patients. Amyotroph Lateral Scler Frontotemporal Degener 2013; 14: 228–229. [DOI] [PubMed] [Google Scholar]
- Cady J, Allred P, Bali T et al: Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol 2015; 77: 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK et al: Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J 2010; 29: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk GM, Ferguson CJ, Chow CY et al: Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet 2011; 7: e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S et al: Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015; 347: 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blitterswijk M, van Es MA, Hennekam EA et al: Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet 2012; 21: 3776–3784. [DOI] [PubMed] [Google Scholar]
- Strickland AV, Schabhüttl M, Offenbacher H et al: Mutation screen reveals novel variants and expands the phenotypes associated with DYNC1H1. J Neurol 2015; 262: 2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiVincenzo C, Elzinga CD, Medeiros AC et al: The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med 2014; 2: 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Pagani M, Agosta F, Calvo A, Cistaro A, Filippi M: Neuroimaging in amyotrophic lateral sclerosis: insights into structural and functional changes. Lancet Neurol 2014; 13: 1228–1240. [DOI] [PubMed] [Google Scholar]
- Swinnen B, Robberecht W: The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 2014; 10: 661–670. [DOI] [PubMed] [Google Scholar]
- Statland JM, Barohn RJ, Dimachkie MM, Floeter MK, Mitsumoto H: Primary lateral sclerosis. Neurol Clin 2015; 33: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludolph A, Drory V, Hardiman O et al: A revision of the El Escorial criteria—2015. Amyotroph Lateral Scler Frontotemporal Degener 2015; 16: 291–292. [DOI] [PubMed] [Google Scholar]
- Pringle CE, Hudson AJ, Munoz DG, Kiernan JA, Brown WF, Ebers GC: Primary lateral sclerosis. Clinical features, neuropathology and diagnostic criteria. Brain 1992; 115: 495–520. [DOI] [PubMed] [Google Scholar]
- Huisman MH, de Jong SW, van Doormaal PT et al: Population based epidemiology of amyotrophic lateral sclerosis using capture–recapture methodology. J Neurol Neurosurg Psychiatry 2011; 82: 1165–1170. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL: Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993; 234: 779–815. [DOI] [PubMed] [Google Scholar]
- Couthouis J, Raphael AR, Daneshjou R, Gitler AD: Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet 2014; 10: e1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.