Abstract
Alzheimer's disease (AD) is the most frequent cause of dementia. Misfolded protein pathological hallmarks of AD are brain deposits of amyloid-β (Aβ) plaques and phosphorylated tau neurofibrillary tangles. However, doubts about the role of Aβ in AD pathology have been raised as Aβ is a common component of extracellular brain deposits found, also by in vivo imaging, in non-demented aged individuals. It has been suggested that some individuals are more prone to Aβ neurotoxicity and hence more likely to develop AD when aging brains start accumulating Aβ plaques. Here, we applied genome-wide transcriptomic profiling of lymphoblastoid cells lines (LCLs) from healthy individuals and AD patients for identifying genes that predict sensitivity to Aβ. Real-time PCR validation identified 3.78-fold lower expression of RGS2 (regulator of G-protein signaling 2; P=0.0085) in LCLs from healthy individuals exhibiting high vs low Aβ sensitivity. Furthermore, RGS2 showed 3.3-fold lower expression (P=0.0008) in AD LCLs compared with controls. Notably, RGS2 expression in AD LCLs correlated with the patients' cognitive function. Lower RGS2 expression levels were also discovered in published expression data sets from postmortem AD brain tissues as well as in mild cognitive impairment and AD blood samples compared with controls. In conclusion, Aβ sensitivity phenotyping followed by transcriptomic profiling and published patient data mining identified reduced peripheral and brain expression levels of RGS2, a key regulator of G-protein-coupled receptor signaling and neuronal plasticity. RGS2 is suggested as a novel AD biomarker (alongside other genes) toward early AD detection and future disease modifying therapeutics.
Introduction
Alzheimer's disease (AD), a progressive neurodegenerative disorder, is the most frequent cause of dementia. Old age is a major AD risk factor: the annual AD incidence increases from 1% between ages of 60 and 70 years to 6–8% at the age of 85 or older.1, 2 AD is characterized by misfolded protein pathological brain hallmarks: extracellular deposits of amyloid-β (Aβ) plaques and accumulation of phosphorylated tau neurofibrillary tangles. The Aβ1–42 peptide aggregates are predominant in AD brain plaques and considered the most neurotoxic Aβ form.3, 4, 5, 6, 7 However, there are individuals who exhibit Aβ plaques in the absence of dementia symptoms.1, 8, 9, 10 Mild cognitive impairment (MCI) is a state when there is mild loss of memory, considered normal for old age. Fifty percent of MCI patients will progress to AD over 4 years.1
Efforts have been made for identifying early AD biomarkers that may detect high-risk individuals so that they are prioritized for disease-modifying drugs that are being developed.11, 12 Imaging techniques based on in vivo measurements of brain Aβ have been disappointing,13 and indeed one of the biggest mysteries in AD pathophysiology is that some aged individuals show, upon brain imaging, large quantities of brain Aβ deposits without showing clinical AD signs and while maintaining good cognitive skills into their 80s.13 This has recently led to strong doubts about the validity of the ‘amyloid cascade hypothesis' that assumes a central role for Aβ in AD pathology.14, 15 It has been proposed that some individuals could be more prone to Aβ-mediated neurotoxicity, while Aβ brain deposition per se may represent part of the normal brain aging process.13, 16
To further understand the pathophysiology of AD toward potential prevention and disease-modifying treatments, disease biomarkers may prove beneficial. One approach is the candidate gene approach, which we (IG) recently took, finding correlation between serum activity-dependent neuroprotective protein (ADNP) and intelligence test scores of elderly individuals, coupled with lower ADNP messenger RNA (mRNA) in blood cells correlated with increased Aβ deposits and significant deregulation of activity-dependent neuroprotective protein mRNA expression in AD lymphocytes.17 Another approach entails proteomic screening.18, 19 In our present work, we applied a third approach, namely, genome-wide transcriptomics of human lymphoblastoid cell lines (LCLs) from unrelated healthy individuals and AD patients for searching gene expression levels that are correlated with in vitro Aβ sensitivity. We report several genes, most notably RGS2 (regulator of G-protein signaling 2) and DLGAP1 (disks, large (Drosophila) homolog-associated protein 1) with low expression correlated with higher Aβ sensitivity in LCLs from healthy individuals and lower expression in LCLs from AD patients, as well as in postmortem AD brain tissues and both AD and MCI peripheral blood.
Materials and methods
Human LCLs and materials
LCLs from adult donors were obtained from the National Laboratory for the Genetics of Israeli Populations (NLGIP; http://nlgip.tau.ac.il) at Tel Aviv University, Israel (23 LCLs of healthy controls) and from The University of Cagliari, Italy (28 AD patients and 16 healthy controls). Detailed demographic data and cognitive scores of the AD patients and controls are presented in Supplementary Table 1. The cell lines were generated from peripheral blood lymphocytes donated by consenting patients and healthy controls. The cells were maintained in optimal growth conditions as described.20 Tissue-culture reagents were purchased from Biological Industries (Beit-Haemek, Israel). Amyloid-β1–42 (Aβ1–42) peptide was purchased from Genemed Synthesis (San Antonio, TX, USA). Aβ1–42 peptide was dissolved in sterile tissue-culture grade water (1 mg ml−1) and stored (as 100 μl aliquots) at −20 °C. Before the experiments, an aliquot of Aβ1–42 in water was preincubated at 37 °C for 3 days21, 22 for assuring the generation of Aβ fibrils.23, 24
Cell proliferation assay
Growth inhibition of LCLs was examined by exposure to 8 μm Aβ1–42 fibrils for 3 days (unless otherwise indicated). LCLs were first washed in phosphate-buffered saline and suspended with serum-free RPMI medium containing the commercial serum supplement 4% BIOGRO-2 (Biological Industries). This BIOGRO-2 concentration was optimal for long-term serum-free growth of LCLs.25 The serum-free conditions are essential for observing Aβ1–42 mediated growth inhibition. The cells were counted and diluted in the same media to a concentration of 250 000 cells ml−1, followed by plating 100 μl per well in 96-well plates (Corning, Corning, NY, USA). The LCLs from healthy controls or AD patients were similarly assayed. The XTT cell proliferation assay (Biological Industries) was carried out after 3 days, as earlier described.26 Each cell line was tested for the effect of Aβ1–42 in three independent experiments.
RNA extraction
RNA extraction was performed from cells incubated in upright T-25 flasks under optimal growth conditions in serum-containing media at a cell density of 0.5 × 106–1 × 106 cells ml−1 as previously described.20 RNA was quantified using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, USA), with both 260/280 nm and 260/230 nm parameters >2.0. RNA quality was confirmed using 1% agarose gels.
Gene expression microarrays
The RNA samples (N=16) from optimally growing LCLs, exhibiting high or low sensitivities to Aβ (8 each) were chosen for genome-wide expression profiling. The RNA samples (250 ng) were prepared and hybridized to Affymetrix Human Gene 2.1 ST arrays as described in the Affymetrix website. Microarray analysis was performed on CEL files using Partek Genomics Suite (Partek, Chesterfield, MO, USA). Genes of interest that were differentially expressed in the two phenotypic groups of the LCLs (fold-difference cutoff >1.5 and P<0.05) were obtained.
Real-time PCR
Real-time PCR was performed to validate the microarray expression patterns of selected genes using the same RNA samples used for the microarray experiment. The complementary DNA (cDNA) samples were prepared from 1 μg RNA samples using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA) containing 10 × RT buffer, 10 × RT random primers, 25 × dNTP mix, RNAse inhibitor and MultiScribe Reverse transcriptase. Reverse transcription was performed using a thermal cycler over three steps (25 °C for 10 min, followed by 37 °C for 120 min and 85 °C for 5 min). Real-time PCR experiments were done with 20 μl mixtures containing 20 ng of cDNA, Absolute Blue qPCR ROX mix (Thermo Scientific, Waltham, MA, USA) and Primers (TaqMan Gene Expression Assay; Applied Biosystems). GUSB (glucuronidase, beta) was used as reference gene as recommended for transcriptomic analysis of LCLs.27 TaqMan Gene Expression Assay IDs are listed below:
PCR reactions were performed using ABI Step One (Applied Biosystems) and the cycle protocol was as follows: 50 °C for 2 min, 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Comparative critical threshold (Ct) values were used for analyzing relative gene expression in selected sample groups according to 2−ΔCт (ΔCт=Ct target Gene—Ct reference gene). For SNORD116-13 and for the reference gene GUSB, real-time PCR was done using the SYBR Green (Kapa SYBR, Wilmington, MA, USA) technique. Primers (shown below) were purchased from IDT (Coralville, IA, USA).

GEO data mining
The NCBI Gene Expression Omnibus (GEO) was searched for expression data sets of human AD and MCI blood and postmortem brain tissues. Data sets GSE5281 (ref. 28) from six postmortem brain regions (87 AD and 74 controls), and GSE63060 (ref. 29) from whole blood (145 AD, 80 MCI and 104 controls) were identified as the largest cohorts. GEO files were downloaded using the R package GEO. The five selected candidate genes were tested for differential expression between AD, MCI and controls using the R package Limma.
Results
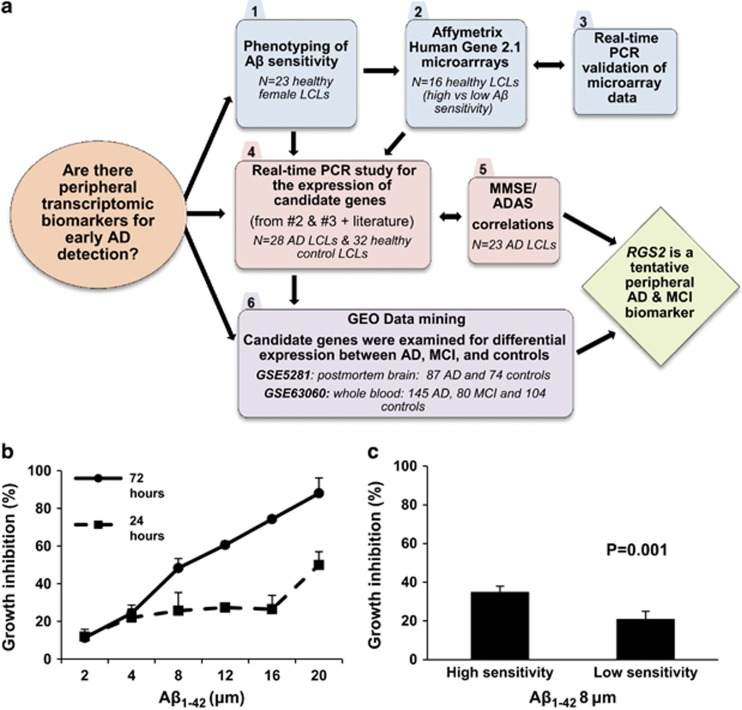
A flowchart outlining our study design is presented in Figure 1a. As a preparatory step for genome-wide transcriptomic search for genes implicated in Aβ1–42 sensitivity of cells from unrelated individuals, we initially screened human LCLs from healthy female donors for growth inhibition following incubation with several concentrations of Aβ1–42 (range 1 to 20 μm) for 24 or 72 h in serum-free medium (see the Materials and methods' section). This first phase included exclusively female LCLs, as sex was shown to affect gene expression by human LCLs.30 Confirming previous reports, Aβ1–42 did not grossly affect cell growth or survival in serum-containing media22 (Figure 1b). Thus, the concentration of 8 μm Aβ1–42 and the incubation period of 72 h in serum-free medium were selected for phenotyping Aβ-mediated growth inhibition in a panel of LCLs from 23 unrelated healthy female donors using XTT cell proliferation assay (see the 'Materials and methods' section). Three repeat experiments were performed (each in triplicate) for each cell line. Eight LCLs exhibiting the highest Aβ1–42 sensitivity (35±3% growth inhibition) and eight LCLs exhibiting the lowest Aβ1–42 sensitivity (21±4% growth inhibition; Figure 1c) were selected for comparative genome-wide expression profiling (see the 'Materials and methods' section; step #1, Figure 1a). Average donor ages were similar for the high and low Aβ1–42 sensitivity groups (38±8 and 58±10 years, respectively; P=0.142).
Figure 1.
(a) A flowchart presenting the study design. Aβ1–42 sensitivity determined in lymphoblastoid cells lines (LCLs) of healthy female donors. (b) Lymphoblastoid cells from a healthy female donor were plated in 96-well plates (25 000 cells per well) and incubated with the indicated concentrations of Aβ1–42 for 24 or 72 h followed by determination of viable cell numbers with the XTT color reagent (see the 'Materials and methods' section). Data are from a representative experiment, with similar observations obtained in a repeat experiment. (c) Aβ1–42 sensitivity (8 μm, 72 h) is shown for two LCL groups (eight unrelated donors each) from healthy female donors selected for the microarray experiment based on their different Aβ1–42 sensitivity phenotypes. Average growth inhibition values were 35±4 and 21±3 in the high- and low-sensitivity groups, respectively (P=0.001; Mann–Whitney U-test). See the 'Materials and methods' section for experimental protocol. AD, Alzheimer's disease; ADAS, Alzheimer's Disease Assessment Scale; MCI, mild cognitive impairment; MMSE, Mini Mental State Examination.
Genome-wide microarray expression and RT-PCR validation
The RNA samples were prepared from the 16 selected healthy female LCLs growing under optimal conditions in serum-containing medium (see the 'Materials and methods' section). Genome-wide expression profiles were compared in the healthy donor LCLs exhibiting high or low Aβ1–42 sensitivity (n=8 per group) using Affymetrix Human Gene 2.1 ST arrays (See the 'Materials and methods' section; step #2, Figure 1a). Table 1 shows 27 transcripts found to exhibit >1.5 fold difference (P<0.05) in basal expression levels comparing healthy female LCLs exhibiting high vs low Aβ1–42 sensitivity.
Table 1. Genome-wide transcriptomic profiling comparing individual LCLs with high vs low Aβ1 –42 sensitivities (eight LCLs in each group; Affymetrix GeneChip Human Gene 2.1 ST arrays).
| Gene/transcript | Full name | Fold difference (high vs low) | P-value |
|---|---|---|---|
| DNASE1L3 | Deoxyribonuclease I-like 3 | −2.40 | 0.003 |
| ABHD6 | Abhydrolase domain containing 6 | −1.62 | 0.006 |
| MERTK | c-mer proto-oncogene tyrosine kinase | −1.64 | 0.008 |
| PEX5L | Peroxisomal biogenesis factor 5-like | −1.54 | 0.010 |
| FARP1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 | 1.69 | 0.010 |
| LOC728419 | Ubiquitin carboxyl-terminal hydrolase 17-like | −1.63 | 0.012 |
| OR5K4 | Olfactory receptor, family 5, subfamily K, member 4 | −1.88 | 0.017 |
| FAH | Fumarylacetoacetate hydrolase | 1.51 | 0.021 |
| ZNF804A | Zinc finger protein 804A | −1.77 | 0.023 |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | −1.81 | 0.027 |
| RNU6-55 | RNA, U6 small nuclear 55 | −1.64 | 0.029 |
| OR5H14 | Olfactory receptor, family 5, subfamily H, member 14 | −2.40 | 0.031 |
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | −2.14 | 0.035 |
| KDM5B | Lysine (K)-specific demethylase 5B | −2.10 | 0.035 |
| SNORD116-13 | Small nucleolar RNA, C/D box 116-13 | 1.57 | 0.035 |
| CCL28 | Chemokine (C-C motif) ligand 28 | −1.51 | 0.036 |
| INPP4B | Inositol polyphosphate-4-phosphatase, type II, 105 kDa | −2.28 | 0.038 |
| PTPN14 | Protein tyrosine phosphatase, non-receptor type 14 | −2.17 | 0.038 |
| TRNAU2 | Transfer RNA selenocysteine 2 | 1.64 | 0.038 |
| SNORD45C | Small nucleolar RNA, C/D box 45C | 1.58 | 0.041 |
| PHYHIPL | Phytanoyl-CoA 2-hydroxylase interacting protein-like | −1.61 | 0.043 |
| DLGAP1 | Disks, large (Drosophila) homolog-associated protein 1 | −2.10 | 0.044 |
| ANKRD20A11P | Ankyrin repeat domain 20 family, member A11, pseudogene | −1.68 | 0.044 |
| PAG1 | Phosphoprotein associated with glycosphingolipid microdomain | −1.73 | 0.045 |
| SNORD116-18 | Small nucleolar RNA, C/D box 116-18 | 1.75 | 0.045 |
| GLIPR2 | GLI pathogenesis-related 2 | −1.53 | 0.046 |
| BCHE | Butyrylcholinesterase | −1.82 | 0.049 |
Abbreviations: Aβ, amyloid-β LCL, lymphoblastoid cells line.
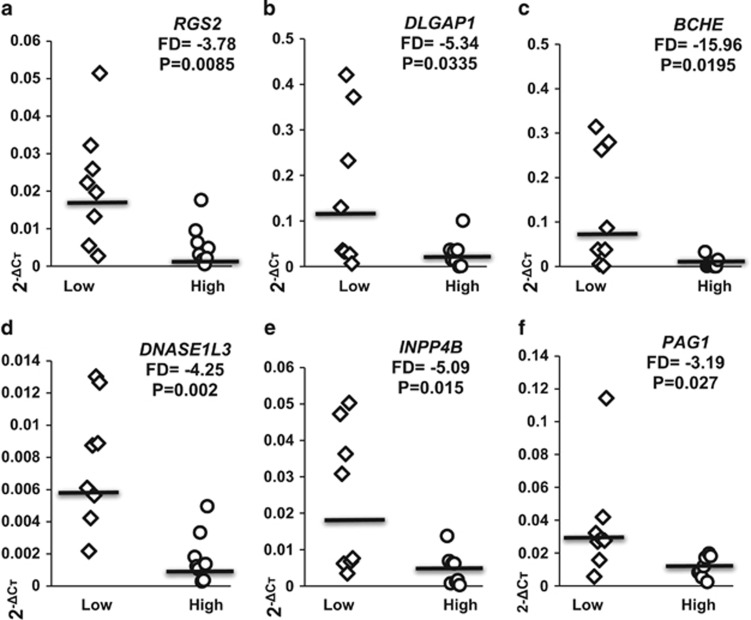
The 27 listed transcripts differed by >1.5-fold with P<0.05 in eight LCLs exhibiting high Aβ1–42 sensitivity compared with eight LCLs exhibiting low Aβ1–42 sensitivity (as shown in Figure 1c). Genes are arranged by increasing P-values. The expression differences for eight selected genes (in bold font) were tested in the same RNA samples by real-time PCR experiments (Figures 2a–f) and further tested in Alzheimer's disease LCLs (Figures 3a–e; Supplementary Table 2).
The same RNA samples from the LCLs exhibiting high or low Aβ1–42 sensitivity (eight in each group) were converted to cDNA. Eight genes from the 27 found as differentially expressed were selected for validation by real-time PCR based on their high expression in human brain tissues as well as their relevance for neuronal function. The expression levels of BCHE, DLGAP1, INPP4B, DNASE1L3, RGS2 and PAG1 (Table 1) are presented as scatter plots (Figures 2a–f). The expression level differences between the two Aβ1–42 sensitivity groups of control LCLs are clearly evident.
Figure 2.
Differences in the expression levels of RGS2, DLGAP1, BCHE, DNASE1L3, INPP4B, and PAG1 as determined by real-time PCR in two groups of healthy female lymphoblastoid cells lines (LCLs). Fold-difference (FD) values for real-time PCR validation of the microarray experiment are shown for LCLs exhibiting high vs low Aβ1–42 sensitivity (eight in each group; Figure 1) FD and P-values are shown for (a) RGS2; (b) DLGAP1; (c) BCHE; (d) DNASE1L3; (e) INPP4B and (f) PAG1. Note: SNORD116-13 and FARP1, two genes from the eight selected for real-time PCR validation (from those depicted in bold font in Table 1) showed the expected trend, however, with P>0.05 and their expression data are not displayed.
Expression levels of candidate genes in AD vs healthy control LCLs
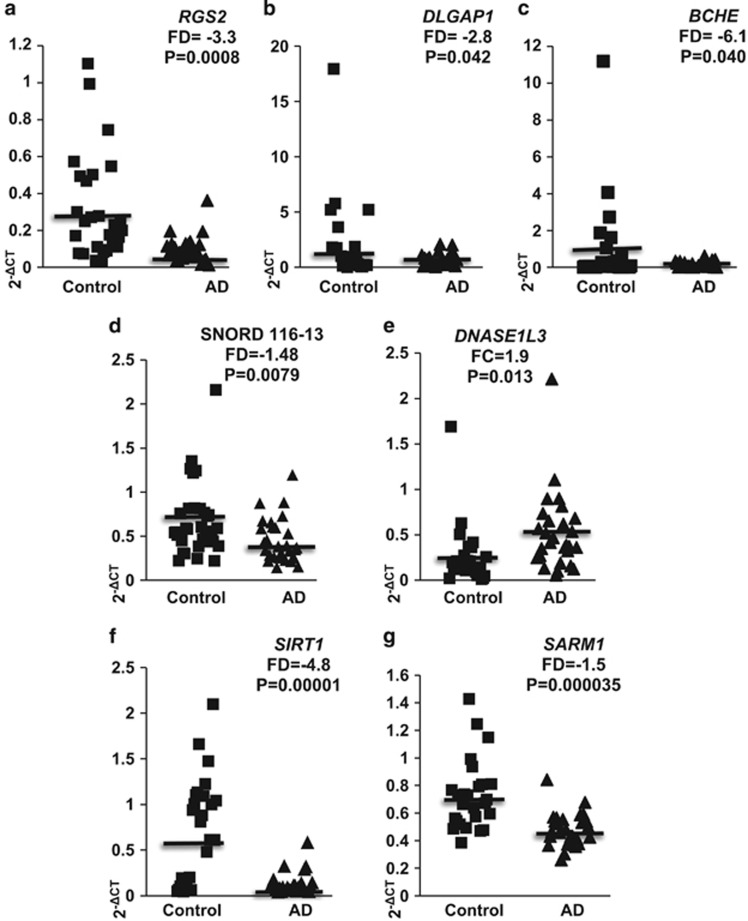
Next, the expression levels of selected genes found to be differentially expressed in healthy control LCLs with high vs low Aβ1–42 sensitivity were determined by real-time PCR in 28 AD and 32 healthy control LCLs growing under optimal conditions (see the 'Materials and methods' section). The expression levels of two additional genes, SIRT1 and SARM1, albeit not found in our genome-wide transcriptomic experiment, were also analyzed in the same AD and healthy control LCLs, as both have been implicated in AD,31, 32, 33, 34, 35 and SIRT1 expression was reduced in postmortem AD parietal cortex.36 The RNA samples were extracted and converted to cDNA for determining the expression levels of selected genes by real-time PCR (Supplementary Table 2). The expression levels of RGS2, DLGAP1, BCHE, SNORD116-13, DNASE1L3, SIRT1 and SARM1 are also presented as scatter plots (Figures 3a–g). No correlations were observed between expression levels of RGS2, DLGAP1 or BCHE and between control or AD patient ages, or between ages and growth inhibition by 8 μm Aβ1–42 in individual LCLs (Supplementary Figure 1). However, a correlation was observed between the expression levels of RGS2 and growth inhibition by Aβ1–42 in 26 individual healthy control LCLs (R=−0.565; P=0.003) but not in 32 AD LCLs (Supplementary Figure 2). In addition, a correlation (R=0.688; P=0.000000006) was found between the expression levels of DLGAP1 and BCHE in individual LCLs from pooled 55 control and AD LCLs (Supplementary Figure 3). Correlations were also found between the expression levels of SIRT1 and the expression of RGS2 or SARM1 in individual LCLs (Supplementary Figure 4).
Figure 3.
Expression levels of RGS2, DLGAP1, BCHE, SNORD116-13, DNASE1L3, SIRT1 and SARM1 in lymphoblastoid cells lines (LCLs) of Alzheimer's disease (AD) patients and healthy controls. The values were determined by real-time PCR; fold-difference (FD) values are shown for 2−ΔCT (see the 'Materials and methods' section). (a) RSG2: AD (N=28) and control (N=32). (b) DLGAP1: AD (N=24) and control (N=28). (c) BCHE: AD (N=27) and control (N=28). (d) SNORD116-13: AD (N=28) and control (N=32). (e) DNASE1L3: AD (N=28) and control (N=32). (f) SIRT1: AD (N=28) and control (N=32). (g) SARM1: AD (N=28) and control (N=32).
RGS2 expression levels in LCLs correlate with dementia scores
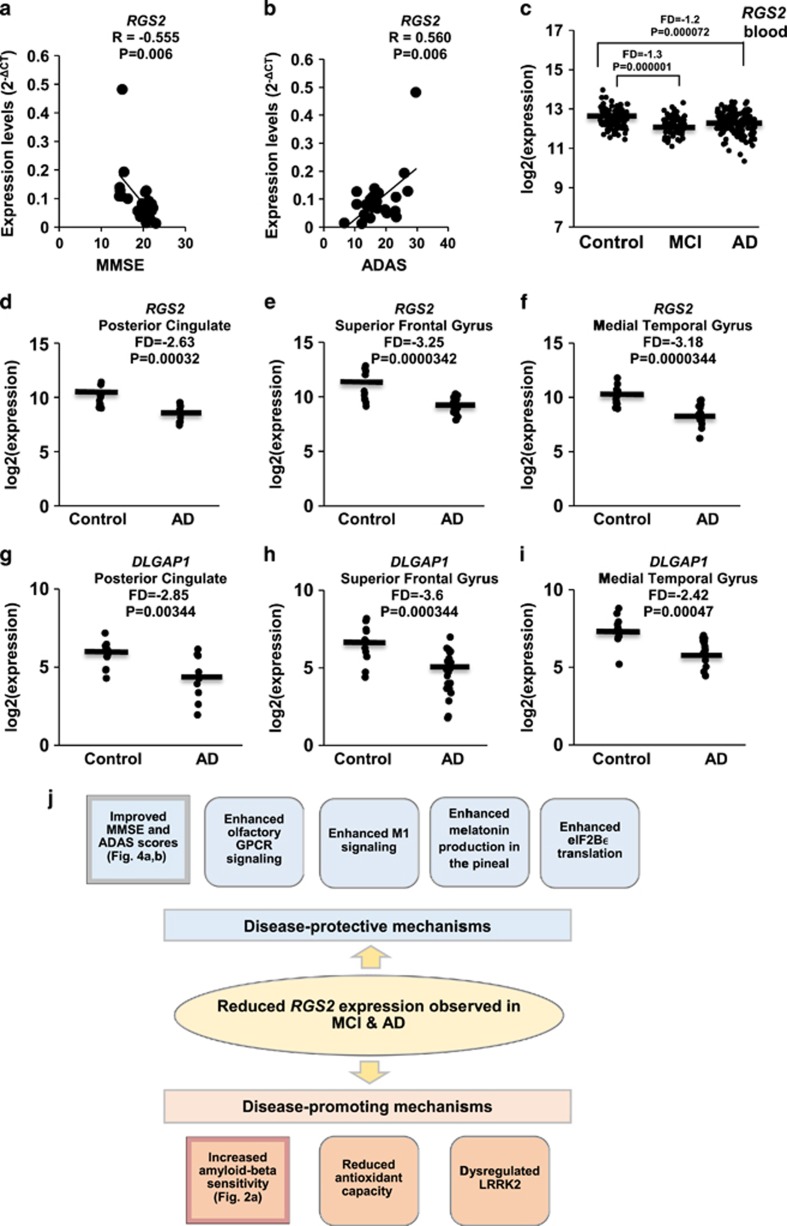
The expression levels of RGS2 in LCLs from AD patients for whom cognitive scores were available (n=23) were examined for correlations with these scores (see Supplementary Table 1). A significant correlation (R=−0.555; P=0.006) was observed between the MMSE (Mini Mental State Examination) scores of AD patients and the RGS2 expression levels in their LCLs (Figure 4a). Moreover, a significant correlation (R=0.560; P=0.006) was observed between RGS2 expression levels and ADAS (Alzheimer's Disease Assessment Scale) scores (Figure 4b).
Figure 4.
(a and b) Correlations between the expression levels of RGS2 in lymphoblastoid cells lines (LCLs) from 23 individual Alzheimer's disease (AD) patients (Mini Mental State Examination (MMSE) <24) and their cognitive test scores as determined at the time of blood withdrawal for LCL generation. Note that higher MMSE scores reflect better cognition, whereas it is the opposite for Alzheimer's Disease Assessment Scale (ADAS) scores (a) MMSE: a negative correlation of R=−0.555 (P=0.006) was observed (b) ADAS: a positive correlation of R=0.560 (P=0.006) was observed. Note: MMSE<24 scores were determined for 23 out of 28 AD patients (Supplementary Table 2). (c) Expression levels of RGS2 in 80 mild cognitive impairment (MCI), 104 healthy controls and 145 AD patients from whole blood (Data set GSE63060) (d–i) Expression levels of RGS2 and DLGAP1 in postmortem brains (Data set GSE5281) from AD patients and age-matched non-demented controls. (d and g) Posterior cingulate: AD (N=9) and control (N=13). (e and h) Superior frontal gyrus: AD (N=23) and control (N=11). (f and i) Medial temporal gyrus: AD (N=16) and control (N=12). (j) A scheme summarizing tentative disease-protective and disease-promoting events associated with reduced RGS2 expression in AD brain tissues. Squares on the left summarize our observations; adjacent squares summarize tentative related consquences (see the 'Discussion' section).
GEO data mining indicates reduced RGS2 expression in AD brain and blood
Data mining was performed for GEO data sets GSE5281 (ref. 28) from six postmortem brain regions (87 AD and 74 controls) and GSE63060 (ref. 29) from whole blood (145 AD, 80 MCI and 104 controls)—two data sets identified as the largest AD cohorts deposited on the NCBI Gene Expression Omnibus (GEO; see the 'Materials and methods' section). Both RGS2 and DLGAP1 exhibited significantly lower expression in postmortem AD brain tissues compared with matched controls of data set GSE5281.28 Moreover, RGS2 also exhibited lower expression in whole blood data set GSE63060 (ref. 29) for both AD and MCI patients compared with healthy controls (FD=−1.2 and −1.3; P=0.000072 and 0.0000012, respectively; Figure 4c), suggesting that its low blood expression may serve as a peripheral MCI and AD biomarker.
Discussion
Research on risk genes for late-onset AD (LOAD), the most common cause of dementia in the elderly, has been largely focused on the role of the ApoE4 genotype, the most notable genetic variation contributing to AD risk, whereas relatively few other genetic clues for this disease have been established. Yet, only about half of LOAD patients are ApoE4 carriers,37 suggesting that further genomic or epigenomic variations contribute to this neurodegenerative disease. These may include DNA sequence variations, gene or ncRNA transcripts, or epigenomic modifications that affect the sensitivity of brain cells to Aβ and/or tau. Our present study was aimed at discovering transcriptomic correlates for Aβ sensitivity as first step toward identifying LOAD risk genes.
The failure of genome-wide association studies to find major LOAD risk alleles besides ApoE4 suggests that transcriptomic and proteomic studies should be performed as, unlike genome-wide association studies, transcriptomic and proteomic studies also capture effects of microdeletions and individual epigenomic variations. In the present study, we have applied a microarray-based genome-wide transcriptomic approach, starting with LCLs from unrelated individual female donors, for searching gene expression variations that may correlate with LOAD risk due to more pronounced individual Aβ toxicity, and thereby, presumably, to LOAD. Notably, it has recently been demonstrated that LCLs capture life-course environmental epigenomic signatures.38 This genome-wide phase was followed by gene validation in a cohort of 28 AD patient LCLs compared with 32 LCLS from non-demented controls. We report 27 transcripts with expression levels associated with Aβ sensitivity of control LCLs (Table 1). Four of these genes, RGS2, DLGAP1, BCHE and SNORD116-13 exhibited significantly lower expression in the AD LCL cohort compared with healthy controls, whereas DNASE1L3 exhibited higher expression in the latter (Figures 3a–e). Among these genes, only the expression of RGS2 correlated with patient MMSE and ADAS cognitive scores (Figures 4a and b). Both RGS2 and DLGAP1 exhibited, in addition, lower expression in published gene expression data set (GSE5281; ref. 28) of postmortem AD compared with control brain tissues, and RGS2 exhibited lower expression also in a whole blood data set (GSE63060; ref. 29) of AD and MCI patients compared with control (Figures 4c–i). Below, we discuss the LOAD risk relevance of these genes.
RGS2 expression
Our genome-wide transcriptomic profiling detected 2.1-fold reduced RGS2 expression in a group of healthy donors LCLs exhibiting high Aβ sensitivity (P=0.035; Table 1). Next, we observed a 3.3-fold reduced RGS2 expression in AD LCLs compared with matched controls (P=0.0008; Figure 3a). To our knowledge, this is the first report on reduced RGS2 expression in AD cells. RGS proteins, comprising a family with 20 members, have key roles in synaptic signaling and neuronal plasticity: these proteins function as negative regulators of G-protein-coupled receptors (GPCR) signaling, acting as GTPase activating proteins for Gα subunits, thereby accelerating the turnoff of GPCR signaling.39 RGS2 has widespread brain expression,40 and its altered expression has been implicated in several neurodegenerative and psychiatric diseases.41, 42, 43, 44, 45, 46, 47 Unlike other RGS family members, RGS2 is an immediate early gene, rapidly upregulated in response to stimuli evoking brain plasticity39 such as high frequency stimulation.48 Notably, RGS2 expression was induced in rat hippocampus 2 h following acute electroconvulsive shock,47 and in human astrocytoma cells following heat shock or oxidative stress.49
RGS2 was identified as key regulator of LRRK2 (leucine-rich repeat kinase 2; also known as PARK8). LRRK2 mutations cause shortening of the dendritic tree and are among the primary genetic causes of Parkinson's disease.41, 45 Reduced RGS2 expression was observed in the striata of LRRK2-mutated and sporadic Parkinson's disease patients.45 In addition, RGS2 rs4606 polymorphism is a risk allele for schizophrenia43 and is associated with antipsychotic-induced parkinsonism.42 RGS2 has also been suggested to be implicated in antioxidant defense.50
Decreased striatal RGS2 expression has been suggested to be neuroprotective in Huntington's disease (HD).44 A similar compensatory response may underlie the lower RGS2 expression observed in our AD LCLs (Figure 3a) and in postmortem AD brain tissues (Figures 4d–f). Notably, the decreased blood RGS2 expression is already apparent at the MCI stage (Figure 4c) and prevails in AD blood samples. This may partly explain our observations that lower RGS2 expression levels were correlated with better MMSE and ADAS scores (Figures 4a and b). Whatever the explanation, our data suggest that RGS2 expression levels are implicated in AD pathology, either as causative or as disease-triggered protective mechanism, as has been suggested for its reduced expression in HD brains.44
Genes coding for GPCRs comprise the largest family in the human genome, with 791 different genes (~4% of the human exome), half coding for olfactory receptors.51 The activity of the olfactory receptors is tightly regulated by RGS family proteins, including RGS2.52, 53 Reduced olfactory sensing is a common feature in AD, observed already in some MCI patients.54, 55, 56 It is accompanied by reduced neuronal stem cell renewal in the olfactory epithelium, a tissue of central origin,57 owing to impaired neuronal stem cell migration and proliferation, possibly secondary to amyloid-β accumulation.58 Thus, it is plausible that reduced RGS2 expression in MCI and AD patients represents a compensatory mechanism aimed at improving a deteriorating olfactory capacity.
Dysregulation of acetylcholine receptors, in particular the M1 muscarinic receptor, has received considerable interest in AD research, as this GPCR is implicated in memory consolidation59, 60, 61 and as acetylcholinesterase (AChE) inhibitors remain among first-line AD therapeutics. Decreased levels M1 muscarinic receptors have been demonstrated in several AD postmortem brain regions including CA1, temporal cortex and occipital cortex.62, 63, 64 Yet, M1 muscarinic signaling capacity was shown to be preserved in AD brain tissues.65 Notably, RGS2 has been shown to bind directly and selectively to the M1 muscarinic acetylcholine receptor (via the receptor's third intracellular loop) and modulate Gq/11alpha signaling66 resulting in suppression of M1 muscarinic receptor-mediated activation of KCNQ channels that in turn regulate neuronal excitability.67 It is therefore plausible that preserved M1 G-protein coupling capacity persists in AD brain tissues in spite of compromised acetylcholine levels in part owing to reduced RGS2 expression that allows enhanced M1 muscarinic receptor signaling. This tentative scenario agrees with the above suggestion for a compensatory neuroprotective role of reduced brain RGS2 expression, as also proposed for HD.44
Melatonin treatment has been suggested to ameliorate AD pathology and cognitive decline in animal models.68, 69, 70 Notably, melatonin production in the rat pineal gland was reduced following Rgs2 transfection.71 Lower RGS2 expression in AD LCLs, blood and brain (Figure 3a, Figures 4c–f) may indicate enhanced melatonin production. Moreover, melatonin treatment of multiple sclerosis patients upregulated SIRT1 expression in their blood cells72 and reduced sepsis-induced brain injury through upregulation of Sirt1 and Bcl-2 in mice.73 Thus, lower RGS2 expression in AD may allow higher pineal melatonin production and in turn improve neuroprotection.
Last, RGS2 has been reported as the only RGS family member that inhibits the mRNA translation into protein of eIF2Bε (eukaryotic initiation factor 2B ε subunit),74 a protein crucial for correct protein folding, a process dysfunctional in neurodegenerative disorders including HD, AD and prion diseases, and mutations in which cause childhood ataxia.75 This novel role of RGS2 supports its postulated defensive mechanism in both HD and AD, whereby reduced RGS2 expression reflects an attempt to protect cells from misfolded protein accumulation by enhancing eIF2Bε translation.76
GPCRs are the largest gene family in the human genome (~800 members) and ~40% of current therapeutics are GPCR ligands.77 Our findings of the GPCR regulator RGS2 as deregulated in AD LCLs (Figure 3a), and that its expression was correlated with AD patients' MMSE and ADAS scores (Figures 4a and b), are intriguing. Moreover, RGS2 was found to be downregulated in published GEO data sets from postmortem AD brain tissues (Figures 4d–f), as well as in both AD and MCI peripheral blood (Figure 4c). A scheme summarizing tentative disease-protective and disease-promoting events associated with reduced RGS2 expression is shown in Figure 4j. These observations attest to the complexity of the disease, with fundamental pathways led astray. It further highlights the need for innovative approaches to AD therapeutics.
DLGAP1 expression
DLGAP1 expression was 2.1-fold lower in a group of healthy donors LCLs exhibiting high Aβ sensitivity (P=0.044; Table 1). We subsequently observed 2.8-fold reduced DLGAP1 expression in AD LCLs compared with matched controls (P=0.042; Figure 3b). The proteins encoded by DLGAP1 (also known as GKAP) along with DLC2 take part in neuronal N-methyl-d-aspartate (NMDA)-receptor-associated scaffolding complex. NMDA glutamate receptors are strongly implicated in neurodegenerative diseases,78 and comprise the drug target of memantine, the first non-cholinesterase inhibitor FDA-approved AD drug.79 Interference of the DLGAP1–DLC2 interaction inhibits NMDA receptor activity in dendritic spines.80 In turn, synaptic activity-induced DLGAP1–DLC2 interaction in dendritic spines stabilizes the scaffolding complex and enhances the NMDA currents.81, 82
Of note, the NMDA receptor GluN1 subunit was increased 6-fold in postmortem AD frontal cortex compared with controls,83 supporting a key role for elevated NMDA receptor activity in glutamate-mediated neurodegeneration.78 Moreover, Aβ was shown to induce degradation of GKAP, the protein encoded by DLGAP1.84 Further studies are needed for clarifying how the latter observation is related to the reduced DLGAP1 expression observed in our AD LCLs (Figure 3b).
The reduced DLGAP1 expression in AD LCLs may represent, similarly to our above suggestions for RGS2, a compensatory mechanism for protecting against NMDA-mediated neuronal cell death. This tentative explanation needs further exploration, as the function of NMDA receptors in immune cells, although apparent, remains little studied.85
BCHE expression
The expression of BCHE, coding for BChE, was 1.82-fold lower in the group of high Aβ sensitivity LCLs (Table 1). We subsequently observed 6.1-fold lower BCHE expression in AD LCLs compared with healthy controls (P=0.04; Figure 3c). BChE, along with AChE, comprise the targets of the first-generation AD drug rivastigmine and the (discontinued) first AD drug Tacrine. BChE was shown to prevent Aβ fibril formation,86 an observation that may explain the increased AD risk in carriers of BChE K, a variant with reduced enzymatic activity87 and found by a recent meta-analysis to pose increased AD risk.88 In support of our observations, significantly lower plasma BChE activity levels were reported in AD plasma samples compared with controls, and were associated with faster disease progression.89 Thus, our findings on reduced BCHE expression in control LCLs showing higher Aβ sensitivity (Figure 2c), as well as in AD LCLs (Figure 3c) seem to fit a putative protective role of BChE against Aβ toxicity, while questioning the benefit of mixed AChE/BChE inhibitors (such as rivastigmine) as AD therapeutics. Perhaps the benefit from inhibiting acetylcholine hydrolysis by AChE outweighs the disadvantage of BChE inhibition by such drugs. The impact of reduced BCHE expression on AD risk and pathology as well as potential clinical implications for choosing selective AChE inhibitors vs mixed AChE/BChE inhibitors in AD treatment should be further explored.
SNORD116 transcripts
Two SNORD116 transcripts, SNORD116-13 and SNORD116-18, exhibited higher expression in the LCL group having higher Aβ sensitivity in the genome-wide expression profiling microarrays. The SNORD116-13 microarray data were validated by real-time PCR, albeit only with a trend for significance (P=0.07), indicating 1.70-fold higher expression levels in LCLs exhibiting high Aβ sensitivity. The same SNORD116-13 transcript showed 1.48-fold lower expression in AD LCLs vs healthy controls (P=0.0079; Figure 3d).
SNORD116 deletions cause Prader–Willi syndrome, a neurodevelopmental genetic disorder manifested in cognitive and behavioral deficits.90 SNORD transcripts are noncoding nucleolar RNAs acting similarly to transcription factors. SNORD116 was shown to be developmentally regulated in maturing neurons91 and its overexpression affects the expression of over 200 genes.92 SNORD116 transfection increased the expression of MAP2 (microtubule-associated protein 2, an axonal marker) and TUBB4 (tubulin beta-4 A chain), both important for microtubule assembly. The expression of both MAP2 and TUBB4 were decreased in postmortem posterior hypothalamus from Prader–Willi syndrome.92 Our findings on decreased SNORD116-13 expression in AD LCLs compared with controls, and increased expression in LCLs exhibiting higher Aβ sensitivity, suggest that some genes regulated by SNORD116 may be implicated in neurodegeneration, possibly by modifying cellular responses to chronic Aβ exposure.
Strengths and constraints
Our observations suggest that the protein products of the genes discussed above may be implicated in the pathophysiology of sporadic AD. The correlations we have observed between their lower expression levels and higher Aβ sensitivity in healthy female donors LCLs suggest that their low expression may be among the causes rather than consequences for sporadic AD. Yet, considering reports of a compensatory neuroprotective role for reduced RGS2 levels in HD, it may well be that the reduced RGS2 expression levels that we observed in AD LCLs and postmortem brain reflect a similar compensatory mechanism in AD.
A key limitation of our study is that transcriptomic profiling assays were conducted in blood-derived cells, namely LCLs, rather than in neurons. Nonetheless, neuroimmune interactions have a key role in neurodegenerative diseases including AD,93, 94, 95 and the recent demonstration of a functional meningeal lymphatic system that drains cerebrospinal fluid to deep cervical lymph nodes96 emphasizes the relevance of neuroimmune interactions in neurodegenerative diseases. In favor of applying LCLs transcriptomic profiling for AD research are our observations on reduced SIRT1 and SARM1 expression in AD personal LCLs (Figures 3f–g), moreover, SIRT1 expression was reduced in AD brains.97
Our hypothesis-free findings on lower expression of RGS2 and DLGAP1 in AD LCLs are supported by analysis of published gene expression data sets of postmortem AD brain tissues. RGS2 expression levels were also lower in AD and MCI patients' blood (Figures 4c–i). Personal LCLs may thus serve, in the absence of neuronal tissues, as surrogate for brain cells, and may point to altered transcriptomic profiles that could be implicated in AD pathology.
Conclusions
Our findings, based on a genome-wide transcriptomic search for genes implicated in Aβ sensitivity, show lower expression levels of several key regulatory genes. In particular, lower expression levels of RGS2, DLGAP1 and BCHE are implicated in the higher Aβ sensitivity of LCLs from some individuals. Furthermore, lower expression levels of RGS2 and DLGAP1 were also found in LCLs of AD patients compared with non-demented control donors, as well as in two published gene expression data sets (GSE5281 and GSE63060) of postmortem AD brain tissues and in MCI and AD patients' blood. Taken together, we suggest the involvement of lower expression of RGS2 and DLGAP1 in AD pathophysiology. In particular, the potential diagnostic value of blood RGS2 expression levels should be explored, as this reduction is already noticeable in blood samples of MCI patients. Further studies are required for elaborating the roles of these genes and their protein products, till now not implicated in AD, in the disease pathophysiology, as well as the potential of their expression levels as early AD biomarkers, and tentative utility as AD drug targets.
Acknowledgments
This study was supported by the Israel Science Foundation to IG and DG (Grant #1424/14). IG is supported by the AMN Foundation, the Dr Diana and Zelman Elton (Elbaum) Laboratory for Molecular Neuroendocrinology and the Lily and Avraham Gildor Chair for the Investigation of Growth Factors at Tel Aviv University. DG is supported by the Yoran Institute for Human Genome Research at Tel Aviv University. EM was supported by a postdoctoral fellowship from the Shabbetai Donnolo Fellowships between Italy and Israel. PM is supported by grants from Charles University in Prague (P24/LF1/3, UNCE 204011/2012) and from MZ CR (RVO-VFN 64165/2012). NS is supported by the I-CORE Program of the Planning and Budgeting Committee, Israel. We thank the anonymous donors of the NLGIP Biobank at Tel Aviv University, Israel, whose altruism and trust in biomedical research have made this study possible. We thank Professor Nechama Kosower (Tel Aviv University) for helpful discussions. This study is in partial fulfillment for the graduate study requirements for AH at the Dr Miriam and Sheldon G. Adelson Graduate School of Medicine, Sackler Faculty of Medicine, Tel Aviv University.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
The authors declare no conflict of interest.
Supplementary Material
References
- Ruiz-Ruiz FJ. Early Alzheimer's disease. N Engl J Med 2004; 350: 80–82, author reply 80–82. [PubMed] [Google Scholar]
- Alzheimer's Association. 2011 Alzheimer's disease facts and figures. Alzheimers Dement 2011; 7: 208–244. [DOI] [PubMed] [Google Scholar]
- Mann DM, Iwatsubo T, Ihara Y, Cairns NJ, Lantos PL, Bogdanovic N et al. Predominant deposition of amyloid-beta 42(43) in plaques in cases of Alzheimer's disease and hereditary cerebral hemorrhage associated with mutations in the amyloid precursor protein gene. Am J Pathol 1996; 148: 1257–1266. [PMC free article] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43). Neuron 1994; 13: 45–53. [DOI] [PubMed] [Google Scholar]
- Gozes I, Divinski I, Piltzer I. NAP and D-SAL: neuroprotection against the beta amyloid peptide (1-42). BMC Neurosci 2008; 9(Suppl 3): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike CJ, Burdick D, Walencewicz AJ, Glabe CG, Cotman CW. Neurodegeneration induced by beta-amyloid peptides in vitro: the role of peptide assembly state. J Neurosci 1993; 13: 1676–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Kowall NW, Ferrante RJ. Neurotoxicity and oxidative damage of beta amyloid 1-42 versus beta amyloid 1-40 in the mouse cerebral cortex. Ann N Y Acad Sci 1999; 893: 314–320. [DOI] [PubMed] [Google Scholar]
- Kawas CH, Kim RC, Sonnen JA, Bullain SS, Trieu T, Corrada MM. Multiple pathologies are common and related to dementia in the oldest-old: the 90+ Study. Neurology 2015; 85: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp KV, Mormino EC, Amariglio RE, Munro C, Dagley A, Schultz AP et al. Biomarker validation of a decline in semantic processing in preclinical Alzheimer's disease. Neuropsychology 2015; 30: 624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T, Schultz AP, Rieckmann A, Mormino EC, Johnson KA, Sperling RA et al. Multiple brain markers are linked to age-related variation in cognition. Cereb Cortex 2016; 26: 1388–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal M, Biswas A. Molecular diagnostics of neurodegenerative disorders. Front Mol Biosci 2015; 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol Psychiatry 2011; 16: 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat G, La Joie R, Villain N, Perrotin A, de La Sayette V, Eustache F et al. Amyloid imaging in cognitively normal individuals, at-risk populations and preclinical Alzheimer's disease. Neuroimage Clin 2013; 2: 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. The case for rejecting the amyloid cascade hypothesis. Nat Neurosci 2015; 18: 794–799. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Karran E. The cellular phase of Alzheimer's disease. Cell 2016; 164: 603–615. [DOI] [PubMed] [Google Scholar]
- Harrison JR, Owen MJ. Alzheimer's disease: the amyloid hypothesis on trial. Br J Psychiatry 2016; 208: 1–3. [DOI] [PubMed] [Google Scholar]
- Malishkevich A, Marshall GA, Schultz AP, Sperling RA, Aharon-Peretz J, Gozes I. Blood-borne activity-dependent neuroprotective protein (ADNP) is correlated with premorbid intelligence, clinical stage, and Alzheimer's disease biomarkers. J Alzheimers Dis 2015; 50: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med 2007; 13: 1359–1362. [DOI] [PubMed] [Google Scholar]
- Baird AL, Westwood S, Lovestone S. Blood-based proteomic biomarkers of Alzheimer's disease pathology. Front Neurol 2015; 6: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved K, Morag A, Pasmanik-Chor M, Rehavi M, Shomron N, Gurwitz D. Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl Psychiatry 2013; 3: e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y, Gotoh Y, Zieg J, Barrett T, Takano H, Flavell R et al. Beta-amyloid induces neuronal apoptosis via a mechanism that involves the c-Jun N-terminal kinase pathway and the induction of Fas ligand. J Neurosci 2001; 21: 7551–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind E, Vaisid T, Kornspan JD, Barnoy S, Rottem S, Kosower NS. Neuroprotective effects of Mycoplasma hyorhinis against amyloid-beta-peptide toxicity in SH-SY5Y human neuroblastoma cells are mediated by calpastatin upregulation in the mycoplasma-infected cells. Neurochem Int 2011; 58: 497–503. [DOI] [PubMed] [Google Scholar]
- Soreghan B, Kosmoski J, Glabe C. Surfactant properties of Alzheimer's A beta peptides and the mechanism of amyloid aggregation. J Biol Chem 1994; 269: 28551–28554. [PubMed] [Google Scholar]
- Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB et al. Amyloid beta-protein: monomer structure and early aggregation states of Abeta42 and its Pro19 alloform. J Am Chem Soc 2005; 127: 2075–2084. [DOI] [PubMed] [Google Scholar]
- Milanesi E, Hadar A, Maffioletti E, Werner H, Shomron N, Gennarelli M et al. Insulin-like growth factor 1 differentially affects lithium sensitivity of lymphoblastoid cell lines from lithium responder and non-responder bipolar disorder patients. J Mol Neurosci 2015; 56: 681–687. [DOI] [PubMed] [Google Scholar]
- Morag A, Pasmanik-Chor M, Oron-Karni V, Rehavi M, Stingl JC, Gurwitz D. Genome-wide expression profiling of human lymphoblastoid cell lines identifies CHL1 as a putative SSRI antidepressant response biomarker. Pharmacogenomics 2011; 12: 171–184. [DOI] [PubMed] [Google Scholar]
- de Brouwer AP, van Bokhoven H, Kremer H. Comparison of 12 reference genes for normalization of gene expression levels in Epstein-Barr virus-transformed lymphoblastoid cell lines and fibroblasts. Mol Diagn Ther 2006; 10: 197–204. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics 2007; 28: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S, Gallagher IJ, Lunnon K, Rullman E, Keohane A, Crossland H et al. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol 2015; 16: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, Matigian NA, Vadlamudi L, Mulley JC, Mowry B, Martin NG et al. Replicated effects of sex and genotype on gene expression in human lymphoblastoid cell lines. Hum Mol Genet 2007; 16: 364–373. [DOI] [PubMed] [Google Scholar]
- Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD(+) destruction. Science 2015; 348: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, DiAntonio A. Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. J Neurosci 2014; 34: 9338–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy JA, Zolezzi JM, Braidy N, Inestrosa NC. Role of Sirt1 during the ageing process: relevance to protection of synapses in the brain. Mol Neurobiol 2014; 50: 744–756. [DOI] [PubMed] [Google Scholar]
- Theendakara V, Peters-Libeu CA, Spilman P. Direct transcriptional effects of apolipoprotein E. J Neurosci 2016; 36: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 2014; 32: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien C, Tremblay C, Emond V, Lebbadi M, Salem N Jr., Bennett DA et al. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J Neuropathol Exp Neurol 2009; 68: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993; 261: 921–923. [DOI] [PubMed] [Google Scholar]
- Suderman M, Pappas JJ, Borghol N, Buxton JL, McArdle WL, Ring SM et al. Lymphoblastoid cell lines reveal associations of adult DNA methylation with childhood and current adversity that are distinct from whole blood associations. Int J Epidemiol 2015; 44: 1331–1340. [DOI] [PubMed] [Google Scholar]
- Gerber KJ, Squires KE, Hepler JR. Roles for regulator of G protein signaling proteins in synaptic signaling and plasticity. Mol Pharmacol 2016; 89: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstein-Dunn E, Young KH, Cockett MI, Khawaja XZ. Regional distribution of regulators of G-protein signaling (RGS) 1, 2, 13, 14, 16, and GAIP messenger ribonucleic acids by in situ hybridization in rat brain. Mol Brain Res 2001; 88: 113–123. [DOI] [PubMed] [Google Scholar]
- Geurts M, Maloteaux JM, Hermans E. Altered expression of regulators of G-protein signaling (RGS) mRNAs in the striatum of rats undergoing dopamine depletion. Biochem Pharmacol 2003; 66: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Smith RC, Rigbi A, Strous R, Teltsh O, Kanyas K et al. Further evidence for association of the RGS2 gene with antipsychotic-induced parkinsonism: protective role of a functional polymorphism in the 3'-untranslated region. Pharmacogenomics J 2009; 9: 103–110. [DOI] [PubMed] [Google Scholar]
- Higa M, Ohnuma T, Maeshima H, Hatano T, Hanzawa R, Shibata N et al. Association analysis between functional polymorphism of the rs4606 SNP in the RGS2 gene and antipsychotic-induced Parkinsonism in Japanese patients with schizophrenia: results from the Juntendo University Schizophrenia Projects (JUSP). Neurosci Lett 2010; 469: 55–59. [DOI] [PubMed] [Google Scholar]
- Seredenina T, Gokce O, Luthi-Carter R. Decreased striatal RGS2 expression is neuroprotective in Huntington's disease (HD) and exemplifies a compensatory aspect of HD-induced gene regulation. PLoS One 2011; 6: e22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusonchet J, Li H, Guillily M, Liu M, Stafa K, Derada Troletti C et al. A Parkinson's disease gene regulatory network identifies the signaling protein RGS2 as a modulator of LRRK2 activity and neuronal toxicity. Hum Mol Genet 2014; 23: 4887–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francelle L, Galvan L, Brouillet E. Possible involvement of self-defense mechanisms in the preferential vulnerability of the striatum in Huntington's disease. Front Cell Neurosci 2014; 8: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Heifets BD, Pudiak CM, Potts BW, Nestler EJ. Regulation of regulators of G protein signaling mRNA expression in rat brain by acute and chronic electroconvulsive seizures. J Neurochem 2002; 82: 828–838. [DOI] [PubMed] [Google Scholar]
- Ingi T, Krumins AM, Chidiac P, Brothers GM, Chung S, Snow BE et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J Neurosci 1998; 18: 7178–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS. Oxidative stress and heat shock stimulate RGS2 expression in 1321N1 astrocytoma cells. Arch Biochem Biophys 2001; 392: 192–196. [DOI] [PubMed] [Google Scholar]
- Salim S, Asghar M, Taneja M, Hovatta I, Wu YL, Saha K et al. Novel role of RGS2 in regulation of antioxidant homeostasis in neuronal cells. FEBS Lett 2011; 585: 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M, Munger SD. Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem 2009; 109: 1570–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature 2001; 409: 1051–1055. [DOI] [PubMed] [Google Scholar]
- Norlin EM, Berghard A. Spatially restricted expression of regulators of G-protein signaling in primary olfactory neurons. Mol Cell Neurosci 2001; 17: 872–882. [DOI] [PubMed] [Google Scholar]
- Mesholam RI, Moberg PJ, Mahr RN, Doty RL. Olfaction in neurodegenerative disease: a meta-analysis of olfactory functioning in Alzheimer's and Parkinson's diseases. Arch Neurol 1998; 55: 84–90. [DOI] [PubMed] [Google Scholar]
- Masurkar AV, Devanand DP. Olfactory dysfunction in the elderly: basic circuitry and alterations with normal aging and Alzheimer's disease. Curr Geriatr Rep 2014; 3: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks KH, Chuah MI, King AE, Vickers JC. Connectivity of pathology: the olfactory system as a model for network-driven mechanisms of Alzheimer's disease pathogenesis. Front Aging Neurosci 2015; 7: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamo BR, Rudel R, Kosik KS, Lee VM, Neff S, Adelman L et al. Pathological changes in olfactory neurons in patients with Alzheimer's disease. Nature 1989; 337: 736–739. [DOI] [PubMed] [Google Scholar]
- Ayala-Grosso CA, Pieruzzini R, Diaz-Solano D, Wittig O, Abrante L, Vargas L et al. Amyloid-abeta Peptide in olfactory mucosa and mesenchymal stromal cells of mild cognitive impairment and Alzheimer's disease patients. Brain Pathol 2015; 25: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan G, Bolanos-Badillo E, Gonzalez-Sanchez H, Quirarte GL, Prado-Alcala RA. Selective M1 muscarinic receptor antagonists disrupt memory consolidation of inhibitory avoidance in rats. Neurosci Lett 1997; 230: 93–96. [DOI] [PubMed] [Google Scholar]
- Ferreira AR, Furstenau L, Blanco C, Kornisiuk E, Sanchez G, Daroit D et al. Role of hippocampal M1 and M4 muscarinic receptor subtypes in memory consolidation in the rat. Pharmacol Biochem Behav 2003; 74: 411–415. [DOI] [PubMed] [Google Scholar]
- Young MB, Thomas SA. M1-muscarinic receptors promote fear memory consolidation via phospholipase C and the M-current. J Neurosci 2014; 34: 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Kito S, Yamamura Y, Tahara E, Kanazawa I. Alteration of muscarinic receptor subtypes in CA1 field of hippocampus in senile dementia of Alzheimer type: an autoradiographic study. Hiroshima J Med Sci 1990; 39: 119–124. [PubMed] [Google Scholar]
- Nordberg A, Alafuzoff I, Winblad B. Nicotinic and muscarinic subtypes in the human brain: changes with aging and dementia. J Neurosci Res 1992; 31: 103–111. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Zhu SZ, Mash DC, el-Fakahany EE. Comparison of the concentration of messenger RNA encoding four muscarinic receptor subtypes in control and Alzheimer brains. Brain Res Mol Brain Res 1992; 16: 64–70. [DOI] [PubMed] [Google Scholar]
- Pearce BD, Potter LT. Coupling of m1 muscarinic receptors to G protein in Alzheimer disease. Alzheimer Dis Asso Disord 1991; 5: 163–172. [DOI] [PubMed] [Google Scholar]
- Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI et al. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J Biol Chem 2004; 279: 21248–21256. [DOI] [PubMed] [Google Scholar]
- Suh BC, Horowitz LF, Hirdes W, Mackie K, Hille B. Regulation of KCNQ2/KCNQ3 current by G protein cycling: the kinetics of receptor-mediated signaling by Gq. J Gen Physiol 2004; 123: 663–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Dong W, Huang F. Anti-amyloidogenic and anti-apoptotic role of melatonin in Alzheimer disease. Curr Neuropharmacol 2010; 8: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Fu Z, Li Y, Feng J, Luo J et al. Melatonin ameliorates Alzheimer-like pathological changes and spatial memory retention impairment induced by calyculin A. J Psychopharmacol 2011; 25: 1118–1125. [DOI] [PubMed] [Google Scholar]
- Olcese JM, Cao C, Mori T, Mamcarz MB, Maxwell A, Runfeldt MJ et al. Protection against cognitive deficits and markers of neurodegeneration by long-term oral administration of melatonin in a transgenic model of Alzheimer disease. J Pineal Res 2009; 47: 82–96. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Coon SL, Klein DC. RGS2 is a feedback inhibitor of melatonin production in the pineal gland. FEBS Lett 2013; 587: 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamgholipour S, Hossein-Nezhad A, Sahraian MA, Askarisadr F, Ansari M. Evidence for possible role of melatonin in reducing oxidative stress in multiple sclerosis through its effect on SIRT1 and antioxidant enzymes. Life Sci 2016; 145: 34–41. [DOI] [PubMed] [Google Scholar]
- Zhao L, An R, Yang Y, Yang X, Liu H, Yue L et al. Melatonin alleviates brain injury in mice subjected to cecal ligation and puncture via attenuating inflammation, apoptosis, and oxidative stress: the role of SIRT1 signaling. J Pineal Res 2015; 59: 230–239. [DOI] [PubMed] [Google Scholar]
- Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR et al. Translational control by RGS2. J Cell Biol 2009; 186: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP, Mohammad SS, Pavitt GD. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol Cell Biol 2004; 24: 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L, Ohno M. PERK mediates eIF2alpha phosphorylation responsible for BACE1 elevation, CREB dysfunction and neurodegeneration in a mouse model of Alzheimer's disease. Neurobiol Aging 2014; 35: 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG. Thematic minireview series: new directions in G protein-coupled receptor pharmacology. J Biol Chem 2015; 290: 19469–19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Mechanisms of excitotoxicity in neurologic diseases. FASEB J 1992; 6: 3338–3344. [PubMed] [Google Scholar]
- Matsunaga S, Kishi T, Iwata N. Memantine monotherapy for Alzheimer's disease: a systematic review and meta-analysis. PLoS One 2015; 10: e0123289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutin E, Compan V, Raynaud F, Clerte C, Bouquier N, Labesse G et al. The stoichiometry of scaffold complexes in living neurons - DLC2 functions as a dimerization engine for GKAP. J Cell Sci 2014; 127(Pt 16): 3451–3462. [DOI] [PubMed] [Google Scholar]
- Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M et al. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci 2012; 15: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroy J, Moutin E. Scaffold remodeling in space and time controls synaptic transmission. Bioarchitecture 2012; 2: 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba G, Vernay A, Kraftsik R, Tardif E, Riederer BM, Savioz A. Pathological reorganization of NMDA receptors subunits and postsynaptic protein PSD-95 distribution in Alzheimer's disease. Curr Alzheimer Res 2014; 11: 86–96. [DOI] [PubMed] [Google Scholar]
- Roselli F, Livrea P, Almeida OF. CDK5 is essential for soluble amyloid beta-induced degradation of GKAP and remodeling of the synaptic actin cytoskeleton. PLoS One 2011; 6: e23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldyrev AA, Carpenter DO, Johnson P. Emerging evidence for a similar role of glutamate receptors in the nervous and immune systems. J Neurochem 2005; 95: 913–918. [DOI] [PubMed] [Google Scholar]
- Diamant S, Podoly E, Friedler A, Ligumsky H, Livnah O, Soreq H. Butyrylcholinesterase attenuates amyloid fibril formation in vitro. Proc Natl Acad Sci USA 2006; 103: 8628–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podoly E, Shalev DE, Shenhar-Tsarfaty S, Bennett ER, Ben Assayag E, Wilgus H et al. The butyrylcholinesterase K variant confers structurally derived risks for Alzheimer pathology. J Biol Chem 2009; 284: 17170–17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiang Y, Wang X, Du Y, Xiao D, Deng Y et al. Butyrylcholinesterase K variant and Alzheimer's disease risk: a meta-analysis. Med Sci Monit 2015; 21: 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono GF, Simao-Silva DP, Batistela MS, Josviak ND, Dias PF, Nascimento GA et al. Butyrylcholinesterase: K variant, plasma activity, molecular forms and rivastigmine treatment in Alzheimer's disease in a Southern Brazilian population. Neurochem Int 2015; 81: 57–62. [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med 2011; 14: 10–26. [DOI] [PubMed] [Google Scholar]
- Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum Mol Genet 2009; 18: 4227–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva M, Surface J, Shen M, de la Grange P, Stamm S. SNORD116 and SNORD115 change expression of multiple genes and modify each other's activity. Gene 2015; 572: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MA. The impact of neuroimmune changes on development of amyloid pathology; relevance to Alzheimer's disease. Immunology 2014; 141: 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H, Elgueta D, Montoya A, Pacheco R. Neuroimmune regulation of microglial activity involved in neuroinflammation and neurodegenerative diseases. J Neuroimmunol 2014; 274: 1–13. [DOI] [PubMed] [Google Scholar]
- Grant JL, Ghosn EE, Axtell RC, Herges K, Kuipers HF, Woodling NS et al. Reversal of paralysis and reduced inflammation from peripheral administration of beta-amyloid in TH1 and TH17 versions of experimental autoimmune encephalomyelitis. Sci Transl Med 2012; 4: 145ra105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau A, Harris TH, Kipnis J. Revisiting the mechanisms of CNS immune privilege. Trends Immunol 2015; 36: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalla R, Donmez G. The role of sirtuins in Alzheimer's disease. Front Aging Neurosci 2013; 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.