Abstract
Local concentrations of mutations are well-known in human cancers. However, their 3-dimensional (3D) spatial relationships have yet to be systematically explored. We developed a computational tool, HotSpot3D, to identify such spatial hotspots (clusters) and to interpret the potential function of variants within them. We applied HotSpot3D to >4,400 TCGA tumors across 19 cancer types, discovering >6,000 intra- and inter-molecular clusters, some of which showed tumor/tissue specificity. In addition, we identified 369 rare mutations from genes including TP53, PTEN, VHL, EGFR, and FBXW7 and 99 medium recurrence mutations from genes such as RUNX1, MTOR, CA3, PI3, and PTPN11, all residing within clusters having potential functional implications. As a proof of concept, we validated our predictions in EGFR using high throughput phosphorylation data and cell-line based experimental evaluation. Finally, drug-mutation cluster/network analysis predicted over 800 promising candidates of druggable mutations, raising new possibilities for designing personalized treatments for patients carrying specific mutations.
Introduction
With tens of thousands of tumor-normal pairs already sequenced, accumulation of cancer genomic data continues to accelerate. The vast majority of mutations are incidental with no discernable role in tumor development. Various computational approaches1–6 have been developed to winnow mutation lists down to the drivers, including searching for genes or pathways having mutation rates higher than that explained by chance, genes having either mutually exclusive or co-occurring mutations, or those having neighboring mutations on the linear DNA/protein sequences.
Mutational impact on protein structure has not yet been systematically analyzed, but recent developments are moving in this direction. For example, MuPIT7, an extension of LS-SNP/PDB8, maps sequence variants onto protein structures, Interactome3D9 annotates protein-protein interactions with structural details, other web tools10–12 map and visualize variants on protein structures, SpacePAC13 identifies mutation clusters via simulation, CLUMPS14 clusters cancer genes and examines protein-protein interactions where at least one protein is known to be cancer related, and Mechismo identifies interaction sites contributing to the binding forces between proteins and other peptides15. However, no system yet provides comprehensive analysis for understanding mutational consequences or implications for drug delivery.
Here we present a novel computational tool, HotSpot3D, which identifies mutation-mutation and mutation-drug clusters using three-dimensional structures and correlates these clusters with known or potentially interacting functional variants, domains, and proteins. We describe its testing and subsequent application to more than 4,000 TCGA tumors across 19 cancer types. Over 6,000 interacting cluster discoveries are identified, many of which are likely undetectable by conventional approaches, with a subset supported by high throughput phosphorylation data and cell-line based experimental evaluation included in this study as well as accumulated experimental evidence16–18. We also report 800 promising candidate, druggable mutations, generally characterized by complex, multi-dimensional interactions between drugs and mutations. The list furnishes substantial possibilities for future therapeutics.
Results
Intra- and inter-mutation clusters across 19 cancer types
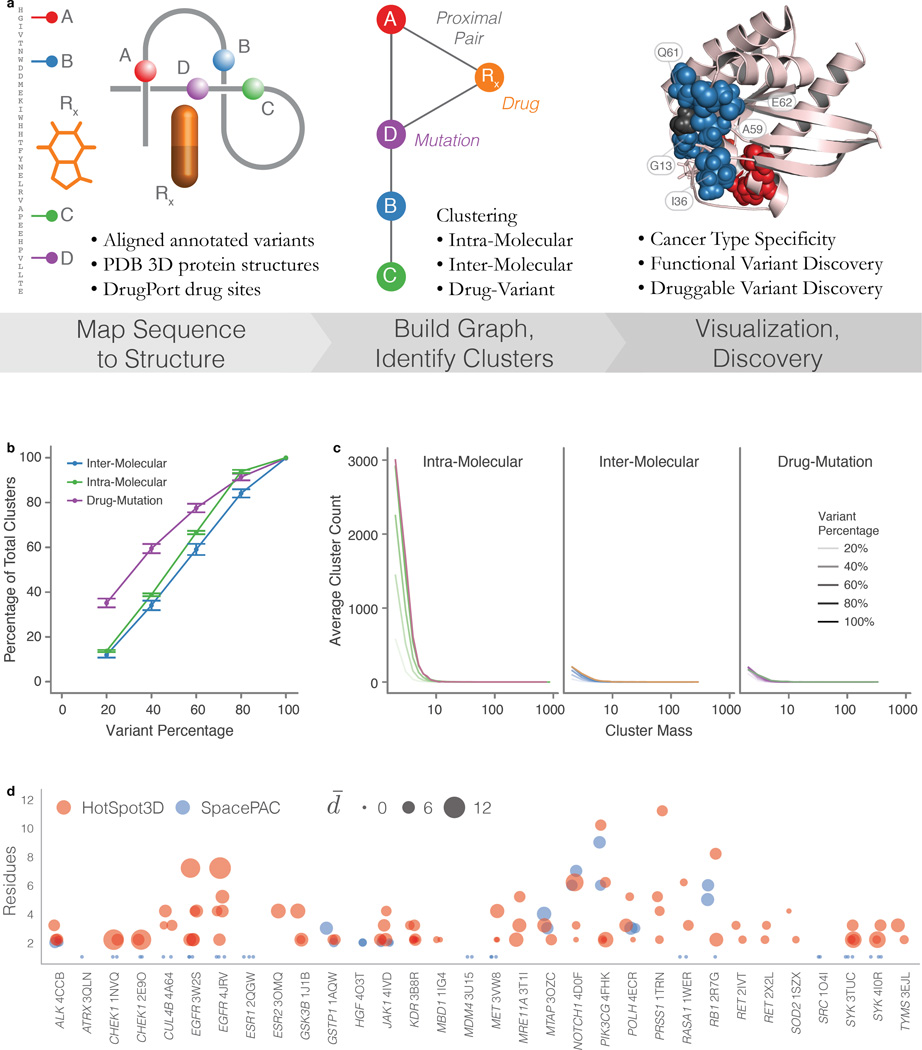
HotSpot3D is a multifaceted tool that integrates sequence mutations with three dimensional protein structures (Methods and Supplementary Note). It identifies significant spatial mutation and mutation-drug clusters in the form of novel or rare mutations co-clustering with known hotspot residues, medium recurrent mutations that collectively exhibit enrichment, cancer type-specific mutation clusters within and between proteins, and mutations potentially interacting with cancer drugs. HotSpot3D utilizes structures from the Protein Data Bank (PDB)19 and mutation/drug co-structures from DrugPort (Methods and Figure 1a). We evaluated HotSpot3D clustering performance and compared it to existing tools to demonstrate its advancement for mutation cluster analysis (Figure 1a–d and Supplementary Note).
Figure 1. HotSpot3D workflow, robustness simulations, and comparison to SpacePAC.
a) HotSpot3D work-flow can be grouped to three processing steps, (from left to right), Data Preprocessing, Structural Analysis, and Post Processing. First, annotation resources from several databases are used to contextualize input datasets, including user-defined DNA variants. Variants are then annotated and mapped onto appropriate PDB structures. DrugPort annotations are used to map pharmaceutical/nutraceuticals onto PDB molecules as a part of the drug module. Mutation pairwise calculations are performed and users can perform clustering of the paired mutations. Users can then visualize mutation clusters along with annotated information. Analyses by users can then lead to in silico discoveries for functional validation hypotheses. b) Robustness simulations show a steady reduction in the percentage of clusters found relative to the percentage of the variant set used. Error bars represent one standard deviation from the mean over 50 random trials. c) Cluster mass distributions show steady decline in clusters of all sizes. Each variant percentage curve (below 100%) is an average over the random trials represented in panel b. d) Significant mutation clusters (P ≤ 0.05) are shown as circles found by HotSpot3D (red) and SpacePAC (blue). The number of residues in each cluster is shown for each structure, labeled by HUGO Symbol and PDB ID. Centers are slightly offset from each residue number, with SpacePAC on the left and HotSpot3D on the right. For all structures, molecule chain A was used. The size of each circle indicates the average inner cluster distance.
We applied HotSpot3D to somatic non-truncational mutations (549,295 unique missense mutations and 4,201 in frame indels) in 4,405 samples from 19 major cancer types (Methods). To identify potential intra-molecular (within a single protein), inter-molecular (between proteins in a complex), and drug-mutation interactions (e.g. near drug binding pocket), we focused on detecting pairs within the typical protein interaction range of 10Å20. We applied Hotspot3D to specifically target intra-molecular mutation pairs separated by at least 20 amino acids (Methods). Clustering was performed on pairs within significant proximity (P < 0.05) and ultimately compared to a known cancer gene list of 624 genes (Supplementary Table 1).
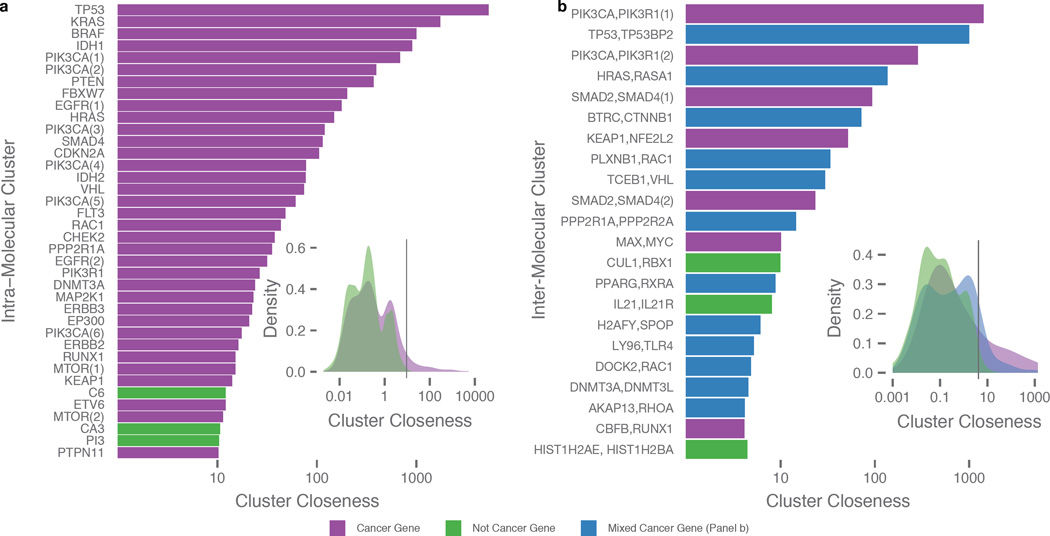
Among the 5,822 intra-molecular clusters identified, 698 clusters are from 244 known cancer genes and 5,124 clusters are from 2,275 non-cancer genes (Supplementary Table 2 and 3). 38 clusters (35 “cancer genes” and 3 “non-cancer genes”) were above the cluster closeness (Cc) threshold (Cc > 10.3, see Methods). The top 5 cancer genes exhibiting high cluster closeness are TP53, KRAS, BRAF, IDH1, and PIK3CA, as expected and due largely to their high mutation rates in cancer (Figure 2a). TP53 has the highest cluster closeness, a result of both numerous mutations in close proximity (192 unique mutations) and mutation recurrence (38 hotspot residues) throughout the gene. We observed a shift towards higher cluster closeness for mutation clusters in cancer genes as compared to non-cancer genes (P ≈ 5.3e-13) (Figure 2a inset) (Methods).
Figure 2. Significant spatial clusters.
Panels are divided into intra-molecular (a) and inter-molecular (b) results and purple and green shading denoting gene type, i.e. cancer and non-cancer genes, respectively. a) List of intra-molecular clusters having the highest cluster closeness as defined by the same type of threshold procedure on cluster closeness distribution (inset). b) List of inter-molecular clusters having the highest cluster closeness, with threshold set at top 20% (inset). Here, inter-molecular clusters are divided into 3 groups: clusters of strictly cancer genes (purple), clusters with at least one cancer gene (blue), and cluster composed solely of non-cancer genes (green) and axis labels only include the top two genes contributing the most number of mutations. Multiple clusters within a single protein or protein complex are differentiated with a numerical suffix in parentheses.
Clustering analysis of protein complexes resulted in 488 clusters, of which 34 were comprised only of cancer genes, 122 contained at least one cancer gene, and 332 contained no cancer genes (Supplementary Table 2 and 4). Similar to the intra-molecular analysis, we selected top inter-molecular clusters (Cc > 4.1, see Methods) for downstream analyses (Figure 2b). Of the 22 clusters that passed the threshold, clusters containing cancer genes exhibit significantly higher cluster closeness than those having no cancer genes (Figure 2b inset).
Oncogenes and tumor suppressor genes (TSGs) have distinct mutation signatures, the former characterized by recurrent mutations at activating sites and the latter having higher abundances of truncations scattered across their sequences21. However, the mutational patterns of non-truncational mutations in TSGs have not been intensively studied. Using 64 oncogenes and 74 TSGs classified by Vogelstein et al.21, we observed 124 and 89 intra-molecular clusters in 36 oncogenes and 38 TSGs, respectively (Supplementary Fig. 1 and Supplementary Tables 5 and 6). Nine oncogenes (HRAS, KRAS, IDH1, IDH2, BRAF, PPP2R1A, SPOP, PIK3CA, and MAP2K1) and five TSGs (TP53, CDKN2A, B2M, FBXW7, and MAP2K4) account for >50% of non-truncational mutations included in clusters; Difference between oncogene/TSG in the number of genes with a majority of mutations in clusters is not significant (P ≈ 0.4). Clusters in both categories tend to correlate with known functional domains, suggesting functional implications (Supplementary Tables 7 and 8).
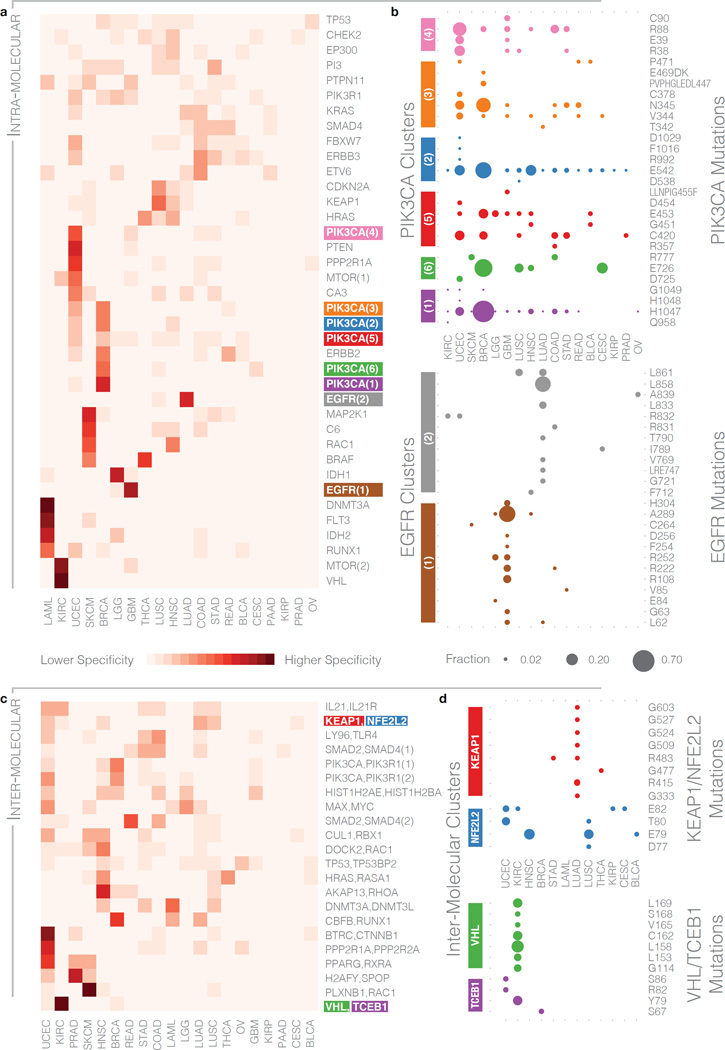
Significant mutation clusters with cancer type specificity
To explore cancer type specificities within significant clusters, we performed unsupervised clustering of cancers with the 38 intra-molecular clusters (Cc > 10.3) and 22 inter-molecular clusters (Cc > 4.1) (Methods and Figure 3a,c). Non-specific intra-molecular clusters included those from TP53, PIK3R1, and KRAS (Figure 3a). We further identified 18 intra-molecular clusters that were at least 50% specific to one cancer type (Supplementary Table 9), suggesting diverse roles in different cell types. High specificity is associated with VHL and MTOR (Supplementary Fig. 2a), having 95% and 86% of their respective mutation clusters specific to KIRC, and DNMT3A with 91% specificity to AML. High-specificity clusters can be the result of a hotspot site having most of its mutations in one cancer type, as is the case with DNMT3A residue Arg882. Conversely, VHL and MTOR show distribution across multiple residues.
Figure 3. Cancer type specificity of intra-molecular and inter-molecular clusters.
a) Cancer specificity heat map of intra-molecular clusters exceeding the threshold defined in Figure 1b. Each row represents a cluster, with intensity of shading indicating the proportion of mutations across all samples in a cluster observed in a particular cancer type. b) Distribution of cancer type specificities of 6 PIK3CA (purple, green, blue, red, orange, and pink) and 2 EGFR (brown and gray) clusters at the residue level. Bubble sizes indicate the fraction of mutations in the cluster that occur at specific residues (labeled on y-axis) for each of the 19 cancer types (x-axis). Bubble color indicates corresponding clusters on the heat map in panel (a), with a trailing suffix in parenthesis to distinguish multiple clusters within same gene. c) Cancer specificity heat map of the inter-molecular clusters exceeding the threshold defined in Figure 1d. d) Distribution of cancer type specificities of the KEAP1/NFE2L2 (red and blue, respectively) and VHL/TCEB1 (green and purple, respectively) clusters at a residue level. Here, colors correspond to the specific genes that make up the cluster.
PIK3CA has 6 top-scoring, distinct clusters, exhibiting both UCEC and BRCA specificity (Figure 3b and Supplementary Fig. 2b). The PIK3CA(4) cluster at centroid Arg88 is primarily UCEC specific (54% of its mutations) and is distributed among three different residues (Arg38, Glu39, and Arg88) that show little BRCA specificity. Conversely, the PIK3CA(1) cluster is primarily BRCA specific (69% of its mutations), and the His1047 centroid is primarily responsible for the overall BRCA specificity. Finally, the PIK3CA(5) cluster with centroid Cys420 shows distribution across multiple cancer types. We found mild GBM specificity in PIK3CA across 4 residues (Arg38, Glu39, Arg88, and Cys90) in the PIK3CA(4) cluster and CESC specificity at Glu726 in the PIK3CA(6) cluster. EGFR also has two different clusters that contribute to different cancer types (Figure 3b): an extracellular cluster, EGFR(1), with centroid at Ala289 enriched in LGG/GBM and the kinase domain cluster, EGFR(2), with centroid at Leu858 enriched in LUSC/LUAD.
Several inter-molecular clusters also showed tumor specificity, with 8 clusters >50% specific to one cancer type, including well-known oncogenic protein complexes ASB9/SOCS4/TCEB1/VHL (KIRC), BTRC/CTNNB1 (UCEC), AKAP13/ARHGEF12/RHOA (HNSC), PPP2R1A/PPP2R2A (UCEC), and CBFB/RUNX1 (BRCA) (Figure 3c and Supplementary Table 10). KEAP1/NFE2L2 showed mutual exclusivity, with KEAP1 mutations in adenocarcinomas LUAD and STAD and NFE2L2 mutations in multiple other cancer types (Figure 3d). Two of the residues, Arg415 and Arg483 from KEAP1, have been experimentally validated and shown both to be in the KEAP1 binding pocket and to play a major role in the stability of the KEAP1/NFE2L2 complex22. We also identified 4 TCEB1 residues, Arg82, Ser67, Ser86, and Tyr79 in UCEC, BRCA, UCEC, and KIRC, respectively, clustering with 7 VHL residues, Cys162, Leu153, Leu158, Leu169, Ser168, Gly114, and Val165 in KIRC; Tyr79 has been experimentally validated to disrupt the TCEB1/VHL complex16 (Figure 3d and Supplementary Table 11).
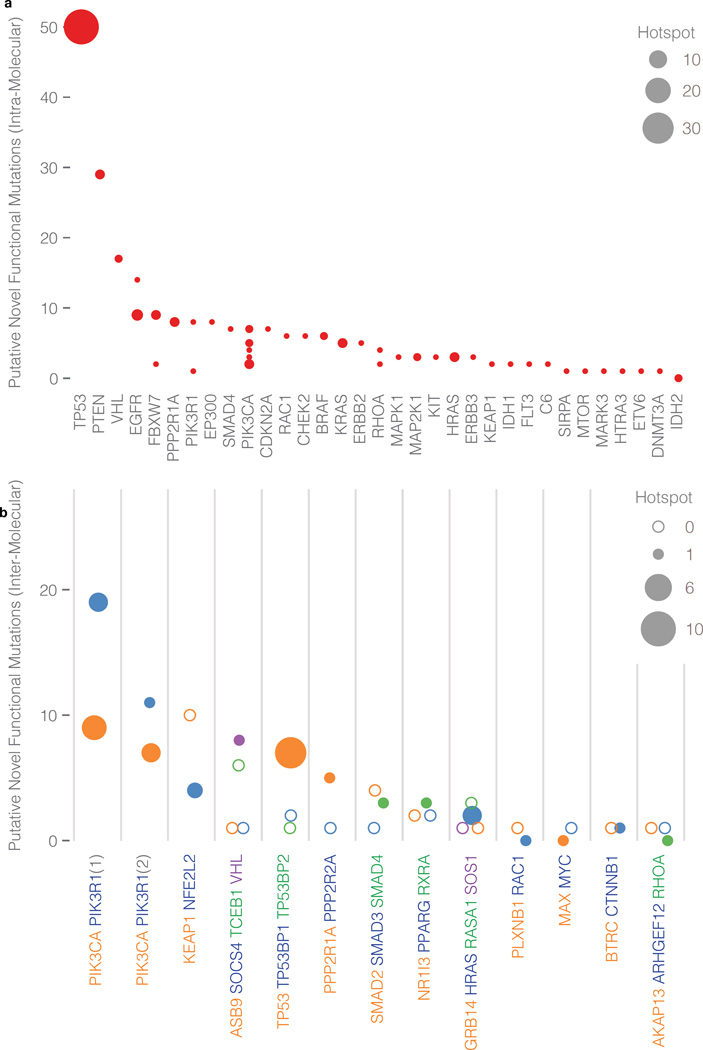
Rare and medium recurrence functional mutation discovery
Rare and medium recurrent drivers are often missed by frequency-based approaches1, 2. We define hotspot residues as those mutated in at least 5 different patient samples, regardless of the amino acid change. Mutations that fall in the same cluster as the hotspot residues are considered potential novel functional mutations.
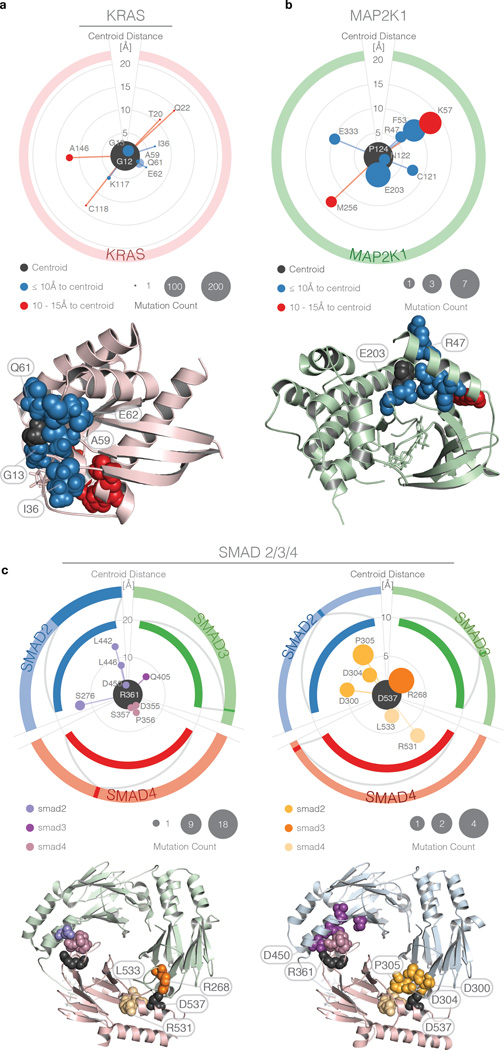
We found 100 hotspot residues and 249 potentially novel functional mutations (Supplementary Table 12 and Figure 4a) clustered with hotspot residues from intra-molecular analysis. TP53, PTEN, VHL, EGFR, and FBXW7 contain the top 5 clusters contributing the most novel functional mutations. A KRAS cluster had the second highest cluster closeness across all clusters, which is a consequence of the high frequency of mutations at the centroid and nearby hotspots. The centroid is at Gly12 (found in 198 patient samples) and has multiple amino acid changes (Gly12Cys/Asp/Ser/Val/Ala/Phe). For this particular cluster, we have 3 hotspot residues Gly12, Gly13, and Gln61 (Figure 5a). Additional possible functional mutations outside of hotspot residues are Ile36M, Ala59Glu/Gly/Thr (each in one sample), and Glu62Lys. Importantly, mutations Ala59Glu/Gly/Thr have a geodesic length of only 3Å from the highly mutated centroid Gly12 in 3D space, even though they are 47 amino acids away in the linear sequence. Ala59 has a higher closeness centrality than expected due to its close proximity to highly mutated residues (Gln61, Gly12, and Gly13). Likewise, Ile36Met is more than 20 amino acids away from all other hotspot residues in the cluster, but has a geodesic distance of only 5.8Å from Gly12. These 5 potential novel functional mutations could be good candidates for subsequent functional validation. Another interesting observation is a MAP2K1 cluster with centroid at Pro124, which is recurrently mutated in 7 patient samples. Additionally, it contained another hotspot at Glu203, mutated 5 times (Figure 5b). Other potential functional candidates in this cluster are Arg47Gln (mutated only once, but having geodesic length of 5.9 Å from the centroid) and Asn122Asp and Glu333Ala (likewise mutated once, but geodesics within 10 Å of centroid). Experimental evidence exists for our prediction that rare mutation Arg47Gln is functional in cancer (Supplementary Table 11). Arg47Gln led to increased phosphorylation of downstream kinases ERK1/2, supporting the activating potential of the mutation17.
Figure 4. Intra-molecular and inter-molecular clusters with unique hotspot mutations and novel mutations.
Numbers of unique hotspot and novel mutations are indicated by bubble area and y-axis position, respectively. a) Intra-molecular clusters: Proteins are labeled on the x-axis and each bubble denotes a cluster from each protein. b) Inter-molecular clusters: Clusters are labeled on the x-axis and bubble colors correspond to member proteins (multiple clusters involving the same proteins are designated in parenthesis). Hollow bubbles indicate that a protein has novel unique mutations but does not have a hotspot.
Figure 5. Polar plots showing rare/medium recurrent functional mutation discovery in intra-molecular and inter-molecular clusters.
Centroids (black) and mutations are represented by bubbles. The latter are ordered clockwise according to primary sequence position, with the radial extent proportional to centroid-mutation spatial distance (rather than geodesics used for clustering). Bubble area indicates number of samples in which the mutations are found. Outer and inner rings represent, respectively, the entire protein linear sequence and a subsection within which the mutations are found. Corresponding clusters on the 3D protein structure are shown below each polar plot. Although there is a linear limit of 20 peptides between paired mutations (Methods), clusters represent networks with edge lengths as the pairwise distance, thus picking up mutations between linearly limited mutations through chaining mutations. a) KRAS Gly12 cluster, with colors indicating mutation distance from the centroid, and corresponding 3D protein structure. b) MAP2K1 Pro124 cluster with same scaling as panel (a) and corresponding 3D structure. c) SMAD2/3/4 clusters with centroid located at SMAD4 Arg361 (top left) and SMAD4 Asp537 (top right). The three proteins are distinguished on the polar plots by differing colors of the outer and inner rings (which correspond to protein backbone color on 3D structure) and slight variation in hue for the bubbles. SMAD3/SMAD4 complex 3D structure on bottom left shows SMAD4 Arg361 (purple) and SMAD4 Asp537 (orange). SMAD2/SMAD4 complex 3D structure is on bottom right with same color key.
Similarly, we can uncover potentially novel, functional variants from inter-molecular clusters. We found 33 hotspot residues and 120 potentially novel functional variants, 4 of which were already observed in intra-molecular clusters (Supplementary Table 13 and Figure 4b). Notable examples are the SMAD2, SMAD3, and SMAD4 complexes. Two separate inter-molecular clusters (Figure 5c) account for 28.6% of the SMAD2/SMAD3/SMAD4 missense mutations and in-frame indels. For one of the complexes (purple cluster, Figure 5c), we were able to identify 7 rare variants, each mutated only once from SMAD2 (Leu442Val, Leu446Val, Ser276Leu), SMAD3 (Gln405Leu), and SMAD4 (Asp355Gly, Pro356Leu, Ser357Pro) and all in close spatial proximity with the SMAD4 Arg361 hotspot (Arg361Cys/His/Pro/Ser). In addition, Asp450Asn in SMAD2 is mutated only once and is the closest spatially (2.6Å) to the SMAD4 hotspot residue, making it another functional candidate. Recent work confirms our prediction that mutations (Asp450 and Ser276 from SMAD2) in close proximity to the Arg361 hotspot on SMAD4 destabilize the SMAD2/4 and SMAD3/4 complexes18 (Supplementary Table 11).
Our analysis also identified five such intra-molecular cases above the cluster closeness threshold involving RUNX1, MTOR, CA3, PI3, and PTPN11. None have hotspot residues, but all contain mutations having medium recurrence or rare variants that are spatially dense. All of the mutations in each of the five clusters collectively contribute to the high cluster closeness and could all be novel functional mutations (Supplementary Table 14). For example, the cluster in RUNX1 contains Arg162 recurrently mutated 4 times, Pro113 mutated twice, and four other singleton mutations (Leu161Pro, Val118Ala, Asp160Gly, and Ala134Pro). In terms of inter-molecular cases, there are 9 clusters with significant cluster closeness, but no hotspot residues (Supplementary Table 15). The other SMAD2/3/4 cluster (orange cluster, Figure 5c) contains Asp537 (SMAD4) mutated 4 times, Arg268 (SMAD3) mutated 3 times, Pro305 (SMAD2) mutated twice, and four singletons (Arg531 and Leu533 from SMAD4, Asp304 and Asp300 from SMAD2). Additionally, RBX1, CUL1 and GLMN form a cluster, but none are on the cancer gene list. This cluster contains Arg506, Gly543, and Glu758 from CUL1 and Met50 from RBX1, which are all mutated twice, and 6 remaining mutations that are singletons (Supplementary Table 16).
Validation by protein array and functional experiment
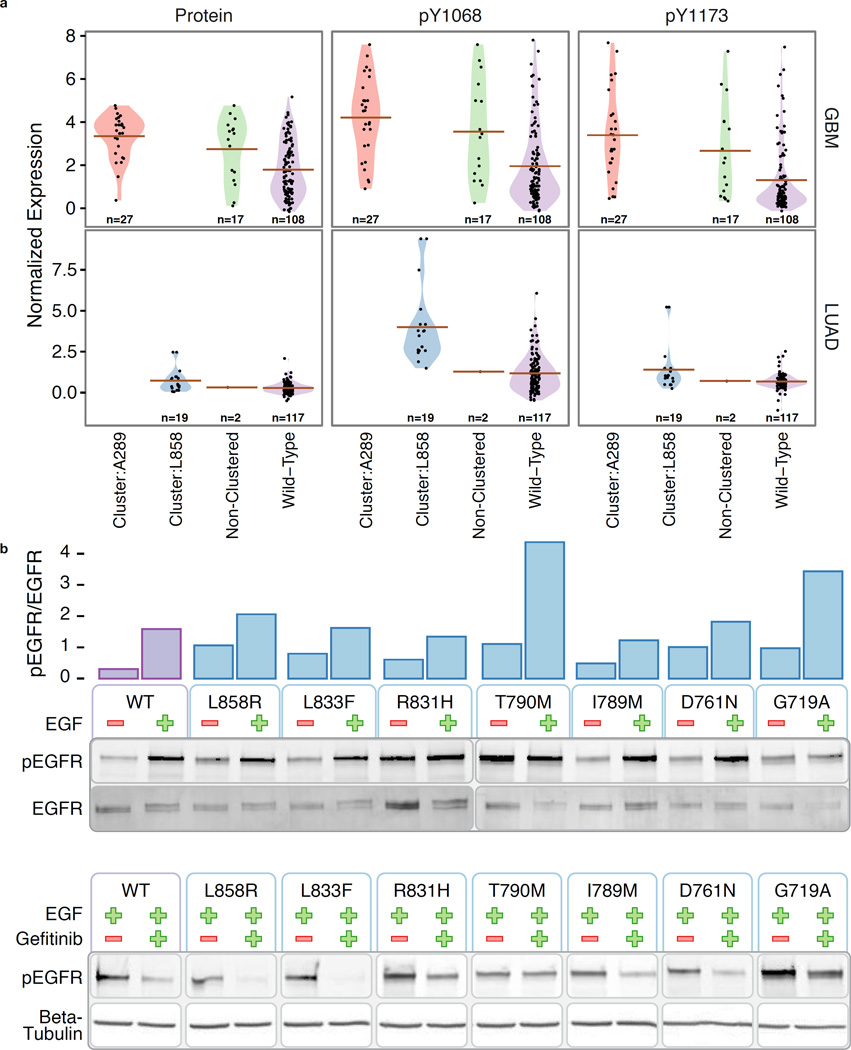
In cancer, mutations within extracellular and kinase domains of Receptor Tyrosine Kinases (RTKs) can cause ligand-independent activation, leading to autophosphorylation. We keyed on this phenomenon to validate the performance of HotSpot3D for identifying functional variants. Specifically, we first conducted validation using Reverse Phase Protein Array (RPPA) expression data to assess whether predicted clusters in EGFR actually have higher levels of protein expression/autophosphorylation than either the wild type or mutations outside clusters. EGFR is an excellent test case because of the high number of mutations found across multiple patient samples and the two most significant clusters being highly cancer specific. The latter is important because RPPA varies by cancer type. We used the RPPA values to examine EGFR protein expression and site-specific phosphorylation at major autophosphorylation sites pTyr1173 and pTyr1068.
We validated the two clusters in EGFR that exceeded the Cc threshold, one specific to GBM with centroid at Ala289 from the extracellular domain and the other specific to LUAD with centroid at Leu858 from the kinase domain. The mean protein and phosphoprotein (pTyr1173 and pTyr1068) levels were significantly higher in GBM samples with mutations from the Ala289 cluster as compared to wild type EGFR, P=2.3e-8, P=1.9e-5, P=1.5e-6, respectively (Figure 6a) Means were also higher than for samples with EGFR mutations outside of any cluster, but there were insufficient data to establish this observation as statistically significant. Almost all of the mutations for LUAD in the kinase domain are from the L858 cluster, so here we focus on comparing it to the wild type. Mean protein and phosphoprotein (pTyr1173 and pTyr1068) levels were again significantly higher for samples containing a mutation in the Leu858 cluster, P=0.01, P=0.04, P=4.6e-5, respectively (Figure 6a). We also conducted validation on one ERBB2 cluster in the kinase domain having its centroid at Val842Ile using RPPA data for ERBB2 protein expression and autophosphorylation site pTyr1248. This cluster exhibited the same trend as the two EGFR clusters; the mean protein and phosphoprotein (pTyr1248) levels were the highest for samples having mutations in the Val842Ile cluster (Methods).
Figure 6. Functional assessment using phosphorylation data and experimental validation.
a) Protein and phosphoprotein (pTyr1068 and pTyr1173) levels in GBM and LUAD samples with mutations in EGFR from the Ala289 cluster (red), the Leu858 cluster (green), non-clustered (blue), and wild type (purple). b) Ligand-independent activity of the mutant EGFR. Bar plot shows normalized relative intensities of pEGFR/EGFR from the western blots below. NIH3T3 clone2.2 cells were transiently transfected with wild type (WT) or mutant EGFR constructs were cultured in 0.5% calf serum for 24h before stimulating with EGF (50ng/ml) for 10 minutes. EGFR autophosphorylation was analyzed by quantifying phosphorylated EGFR (pEGFR, phospho Tyr1068). Tyrosine 1068 of mature EGFR is equivalent to Tyrosine 1092 of uncleaved EGFR. c) NIH3T3 clone2.2 cells were transiently transfected with wild type or mutant EGFR constructs were cultured in 0.5% calf serum for 21h. A 3h gefitinib (1uM) treatment was started at this time and it was followed by a 10-minute EGF stimulation.
We also performed EGFR phosphorylation experiments on mutations from the EGFR Leu858Arg cluster in cultured NIH3T3 cells to more conclusively assess functional predictions from HotSpot3D. This cluster included well-known mutations such as Leu858Arg, Gly719Ala, and Thr790Met. Additional rare mutations, having no available direct evidence of autophosphorylation consequence, include Asp761Asn, Ile789Met, Arg831His, and Leu833Phe, although a few reports suggested weak/partial response to tyrosine kinase inhibitors in samples with other known druggable mutations23–25. Our phosphorylation experiment targeting autophosphorylation site pTyr1068 showed a low level of pTyr1068 phosphorylated EGFR (pEGFR, 0.21, normalized by the total EGFR) in the wild type without EGF treatment (Figure 6b). Leu858Arg, Gly719Ala, and Thr790Met have higher levels of normalized pEGFR (0.79, 0.89, and 1.08, respectively), indicating ligand-independent activation. Asp761Asn, Ile789Met, Arg831His, and Leu833Phe also yielded higher levels of normalized pEGFR (0.78, 0.38, 0.32, and 0.55, respectively), suggesting potential ligand-independent activation as well (Figure 6b). In addition, similar to Thr790Met and Gly719Ala, Asp761Asn shows a much higher normalized pEGFR level (1.76) when compared to the wild type (1.08) under EGF stimulation. These observations demonstrate that some of the variants do not just have ligand-independent activation; their levels of autophosphorylation upon EGF stimulation can be higher than that of the wild type (Figure 6b and Supplementary Table 17). Furthermore, we performed an experiment examining sensitivity of the EGFR variants to gefitinib. We found that Thr790Met is resistant to gefitinib, consistent with previous reports26. The other 6 variants are all sensitive to gefitinib (Figure 6c). In aggregate, these results furnish convincing evidence of the HotSpot3D approach.
Mutation-drug networks and clinical implications
The HotSpot3D drug module targets mutations in spatial proximity to actionable sites for pharmaceuticals and nutraceuticals derived from DrugPort (Methods). We identified 394 significant drug-mutation clusters involving 153 drugs and 359 genes (Supplementary Table 2, 18). Top HGNC gene families and drug classes are in Supplementary Note (Figure 7a and Supplementary Tables 19, 20, 21). While we have obtained drug-mutation relationships from multiple databases (Methods), only 14 unique mutations (with different amino acid position and/or change) in the clusters have been reported in these sources, implying the remaining 844 unique mutations are potentially novel drug interacting candidates.
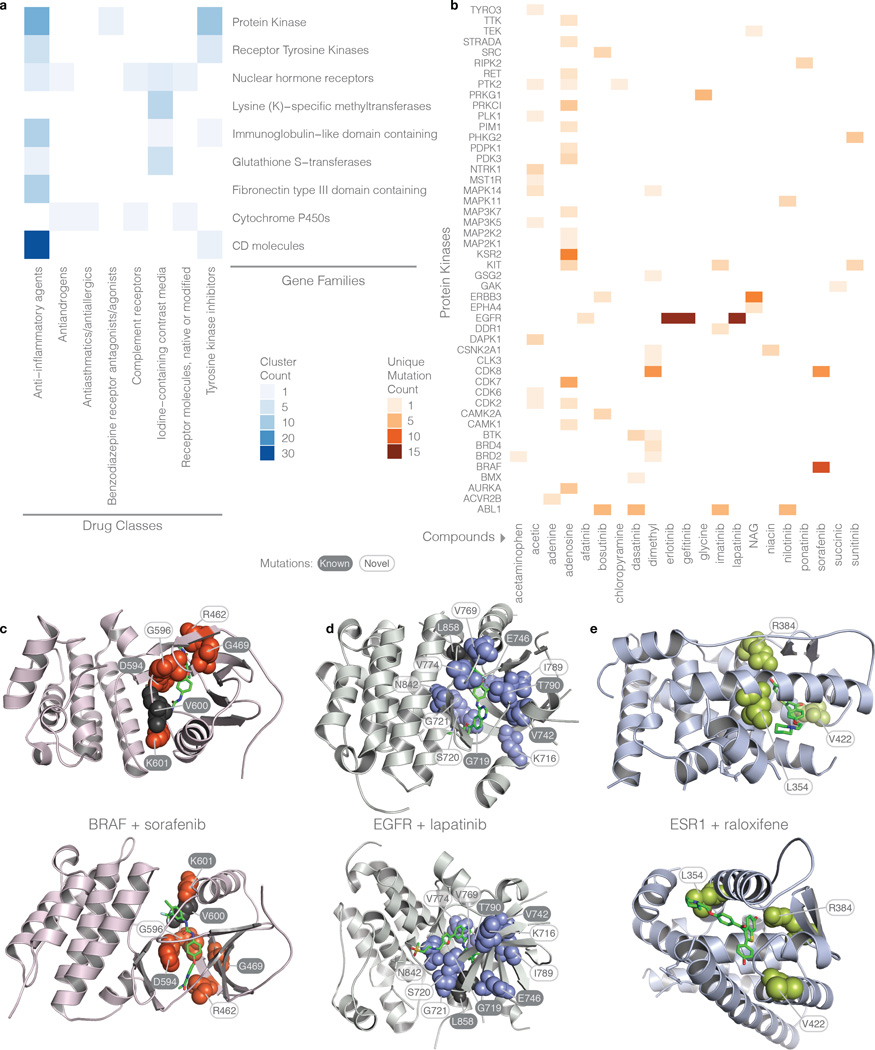
Figure 7. Drug-mutation interaction heat maps and structures.
a) Number of clusters across gene families and drug classes. Gene families and protein kinases are determined by the HUGO Gene Nomenclature Committee (HGNC) and the Gene Ontology (GO) databases, respectively. Protein kinase family is a superset of the receptor tyrosine kinase family. b) Number of unique mutations involving specific protein kinases and drugs. c) 3D structures displaying drug-mutation clusters for BRAF, EGFR, and ESR1 with sorafenib, lapatinib, and raloxifene, respectively. Mutations are depicted as spheres while drugs are represented as green stick models. Black residues represent the centroids; however, for the ESR1 cluster, the drug is the centroid. Two views are shown at different rotations.
Of particular interest, we have detected 48 protein kinases, interacting with 21 drugs (Figure 7b) with strong mutation-drug clusters found in EGFR, BRAF, KSR2, ERBB3, CDK7/8, and ABL1. Our analysis also showed that 24 out of the 394 mutation-drug clusters have cluster closeness scores greater than 2.5 (Table 1), including several protein kinases (BRAF, ERBB3, EGFR, PDK3, and NTRK1), nuclear hormone receptors (ESR1 and PPARD), CD molecules (ACE, CD40LG, and ITGAX), as well as tumor suppressors (TP53 and VHL). Among the kinase-drug clusters, BRAF (a serine/threonine kinase) with sorafenib (a tyrosine kinase inhibitor) tops the list due to hotspots at Val600 and Lys601. Interestingly, there are 8 unique BRAF mutations in this cluster: Arg462Lys, Gly469Ala/Arg, Asp594Gly/His/Asn, Gly596Asp, and Val600Arg that are each observed in one or two samples. Three of these mutations (Arg462Lys, Gly469Arg, Gly596Asp) are not in the current releases of MyCancerGenome (MCG), CancerDR (CDR), Personalized Cancer Therapy (PCT), or Gene-Drug Knowledge Database (GDKD), and eight (Gly469Ala, Asp594Gly/His/Asn, Val600Glu/Lys/Arg, and Lys601Glu) are present in at least one or more of these databases, but have unknown effects on drug binding affinity. Our analysis lends weight to the potential druggability of the 3 functionally unknown, unique BRAF mutations (Figure 7c). We also found two drug-mutation clusters of ERBB3 in which 8 of the 9 unique mutations were not catalogued in these databases (from the extracellular domain cluster: Val104Leu/Met, Ala245Val, Gly284Arg in GDKD, Lys329Glu/Thr, R103H, and R388Q and from the kinase cluster: L792V) and V104 is the centroid mutated in 11 samples. The larger ERBB3 cluster evidently interacts with 4 n-acetyl-d-glucosamine (NAG) molecules throughout the extracellular domain spanning both receptor L domains and the Furin-like cysteine rich region. The second ERBB3 cluster involves bosutinib, a tyrosine kinase inhibitor. Two EGFR drug-mutation clusters were found in which 11 out of 16 unique mutations are novel (Figure 7d). None of the three mutations of the PDK3 drug-mutation cluster have been reported in the four druggable mutation databases (Arg299Cys/Ser and Phe324Leu). The three mutations of NTRK1 were likewise not found in these databases (Arg649Leu/Trp and Arg702Cys) and are observed with an acetic ion binding in the C-terminal lobe adjacent to the binding pocket and DFG motif (within 10Å).
Table 1.
Top (cluster closeness > 2.5) drug-mutation clusters with HGNC gene families and drug classifications from NIH and DrugBank.
| Cluster Closeness |
Mutations (in databases) |
Unique Mutations |
Genes and Drugs / Compounds |
HGNC Families and GO Protein Kinases |
DrugBank Classifications |
NIH Classifications |
|---|---|---|---|---|---|---|
| 1007.764 | 337 (324) | 11 | BRAF; sorafenib | Protein Kinase | Antineoplastic Agents | Unclassified |
| 110.854 | 38 | 4 | TP53; acetic | Unclassified | Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 24.591 | 19 (1) | 8 | ERBB3; n-acetyl-d- glucosamine |
Protein Kinase; Receptor Tyrosine Kinases |
Dietary Supplements; Micronutrients; Supplements |
Anti-inflammatory agents (acetic acid derivatives) |
| 20.353 | 23 (14) | 14 | EGFR; erlotinib; gefitinib; lapatinib |
Protein Kinase; Receptor Tyrosine Kinases |
Unclassified | Tyrosine kinase inhibitors; Unclassified |
| 15.109 | 12 | 8 | KEAP1; acetic | BTB (POZ) domain containing; Kelch- like |
Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 8.189 | 15 | 14 | ACE; acetic | CD molecules | Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 5.049 | 10 | 9 | PLG; acetic; aminocaproic |
Unclassified | Unclassified | Anti-inflammatory agents (acetic acid derivatives); Unclassified |
| 4.612 | 5 | 5 | ESR1; diethylstilbestrol; estradiol; estriol; estrone; raloxifene |
Nuclear hormone receptors |
Anti-menopausal Agents; Antihypocalcemic Agents; Bone Density Conservation Agents; Carcinogens; Contraceptive Agents; Estrogen Antagonists; Estrogens; Estrogens, Non-Steroidal; Selective Estrogen Receptor Modulators; Unclassified |
Iodine-containing contrast media; Unclassified |
| 4.49 | 4 (3) | 3 | VHL; acetic | Unclassified | Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 4.257 | 4 | 3 | PDK3; adenosine | Protein Kinase | Analgesics; Anti- Arrhythmia Agents; Cardiovascular Agents; Vasodilator Agents |
Unclassified |
| 4.106 | 4 | 3 | NTRK1; acetic | Immunoglobulin- like domain containing; Protein Kinase; Receptor Tyrosine Kinases |
Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 4.092 | 3 | 3 | FDPS; alendronate; ibandronate; pamidronate; risedronate; zoledronate |
Unclassified | Antihypocalcemic Agents; Antiresorptives; Bisphosphonates; Bone Density Conservation Agents; Calcium Channel Blockers |
Androgens; Calcium metabolism regulators |
| 3.935 | 7 | 7 | CD40LG; n-acetyl- d-glucosamine |
CD molecules; Endogenous ligands; Tumor necrosis factor (ligand) superfamily |
Dietary Supplements; Micronutrients; Supplements |
Anti-inflammatory agents (acetic acid derivatives) |
| 3.174 | 7 | 4 | LRP6; n-acetyl-d- glucosamine |
Low density lipoprotein receptors |
Dietary Supplements; Micronutrients; Supplements |
Anti-inflammatory agents (acetic acid derivatives) |
| 2.954 | 6 | 6 | REN; acetic; remikiren |
Unclassified | Unclassified | Anti-inflammatory agents (acetic acid derivatives); Unclassified |
| 2.954 | 5 (1) | 3 | ALB; diazepam; diflunisal |
Unclassified | Unclassified | Unclassified |
| 2.937 | 3 | 3 | CA2; acetazolamide; brinzolamide; dichlorphenamide; dorzolamide; ethoxzolamide; furosemide; topiramate |
Carbonic anhydrases |
Anticonvulsants; Carbonic Anhydrase Inhibitors; Diuretics; Sodium Potassium Chloride Symporter Inhibitors; Unclassified |
Anti-inflammatory agents (acetic acid derivatives); Carbonic anhydrase inhibitors; Diuretics (furosemide type); Unclassified |
| 2.811 | 3 | 3 | ITGAX; n-acetyl-d- glucosamine |
CD molecules; Complement system; Integrins |
Dietary Supplements; Micronutrients; Supplements |
Anti-inflammatory agents (acetic acid derivatives) |
| 2.8 | 5 | 5 | GSTA1; ethacrynic; glutathione |
Glutathione S- transferases |
Dietary Supplements; Micronutrients; Supplements; Unclassified |
Anti-inflammatory agents (acetic acid derivatives); Iodine-containing contrast media |
| 2.787 | 6 | 6 | HMGCR; atorvastatin; fluvastatin; rosuvastatin |
Unclassified | Anticholesteremic Agents; Hydroxymethylglutaryl- CoA Reductase Inhibitors |
Antiasthmatics/antiallergics (not acting primarily as antihistamines, leukotriene biosynthesis inhibitors) |
| 2.649 | 5 | 5 | NCAM2; n-acetyl-d- glucosamine |
Fibronectin type III domain containing; I-set domain containing |
Dietary Supplements; Micronutrients; Supplements |
Anti-inflammatory agents (acetic acid derivatives) |
| 2.608 | 9 | 7 | ADH7; acetic | Alcohol dehydrogenases |
Unclassified | Anti-inflammatory agents (acetic acid derivatives) |
| 2.608 | 2 | 2 | PPARD; icosapent | Nuclear hormone receptors |
Dietary Supplements; Micronutrients; Supplements |
Receptor molecules, native or modified; complement receptors |
| 2.572 | 6 | 4 | BCAT1; gabapentin | Unclassified | Analgesics; Anti-Anxiety Agents; Anticonvulsants; Antimanic Agents; Antiparkinson Agents; Calcium Channel Blockers; Excitatory Amino Acid Antagonists |
Gabamimetics |
ESR1, PPARD, and PPARG top the nuclear hormone receptor family of mutation-drug clusters (Supplementary Table 18). The ESR1 cluster with Cc = 4.6, has 4 unique mutations interacting with 5 different compounds: raloxifene, estradiol, estrone, estriol, and diethylstilbestrol (Figure 7e). Raloxifene is a FDA-approved estrogen receptor modulator for reducing the risk of invasive breast cancer27, while estradiol, estrone, and estriol are estrogenic hormones functioning through ESR1. Arg394His/Leu mutations in ESR1 form significant pairs with all 5 compounds and could potentially affect their responses (Figure 7e). HotSpot3D analysis suggests multiple putative therapeutic options for one mutation, but functional validation will still be required for confirmation and to determine which drug is most appropriate. Peroxisome proliferator-activated receptor delta (PPARD) is found with 2 unique mutations, His287Arg and His287Tyr, adjacent to icosapent, a micronutrient which has been used to treat a variety of symptoms and diseases and most notably has been suggested to improve chemotherapy response28. Another PPAR drug-mutation cluster involves 6 unique PPARG mutations that are associated with 4 drugs (indomethacin, pioglitazone, rosiglitazone, and telmisartan) (Supplementary Table 18). The action site for indomethacin, a non-steroidal anti-inflammatory drug (NSAID), neighbors all 6 mutations of the cluster, while the sites for pioglitazone and rosiglitazone (anti-diabetic drugs) and telmisartan (an angiotensin II receptor antagonist (ARB)) neighbor two (Ile277Asn and Ile290Met), three (Ile290Met, Arg316Cys, and His494Tyr), and two (Arg316Cys and E352K) mutations, respectively. It is significant that, although none of these drugs has any previously known use in treating cancer, their action sites have all been found near a frequently mutated binding pocket in cancer. Both clusters of ESR1 and PPARG exist in the hormone receptor domain, suggesting that drug binding in this region may be affected by cancer mutations.
The drug-module in HotSpot3D allows users to identify mutation-drug clusters involving multiple drugs, independent of on-label (drug approved for a specific mutation manifested by a specific disease) status, as well as drugs interacting with mutations from multiple genes. For example, ABL1, from the 8th ranked kinase cluster, interacts with four tyrosine kinase inhibitors (TKIs): bosutinib, dasatinib, imatinib, and nilotinib; each has been used for treating chronic myelogenous leukemia (CML) patients with the BCR-ABL fusion29, 30. Although there are only three unique mutations (Val390Leu, Asp400Tyr, and Phe401Leu) observed in the ABL1 drug cluster, the cluster closeness measure is significantly increased due to the four drugs involved. Each of the Asp400 and Phe401 residues, from the DFG motif, controls blocking of the binding pocket by conformational changes and therefore modulates the binding of imatinib and nilotinib. The gatekeeper in ABL1, Thr315, which controls ATP access to the binding pocket, was not found to be mutated in the TCGA dataset studied, but the gatekeeper in EGFR, Thr790, is found in its own TKI drug-mutation cluster with erlotinib, gefitinib, and lapatinib. Both Thr315 in ABL1 and Thr790 in EGFR are shown to confer drug resistance to TKI therapy, indicating similarly positioned mutations in drug families have the same effects within a drug class31. Further, we found that the DFG motif is also mutated in BTK (PheGly540LeuCys), another tyrosine kinase. Notably, mutations in three genes, ABL1, BTK (including Leu528Phe), and BMX (Gly424Glu), are within the spatial interaction range of dasatinib (Supplementary Fig. 3). Overall, HotSpot3D provides the means to identify complex, multi-dimensional interactions among drugs and mutations and consequently to find alternative therapeutics that may provide greater flexibility in treating a wide range of genetic diseases.
Discussion
The enormous numbers of available variants and protein structures offer an unprecedented resource for investigating the direct impact these variants have upon protein structures, which is fundamentally important to the design of targeted cancer drugs. Here, we developed HotSpot3D to provide novel capabilities not found in existing tools: 1) It handles any mutation and variation data, has no limitation on the number of clusters per protein, and considers all available structures, thus maximizing the potential for novel cluster/interaction discovery for studies not limited to cancer. 2) It unifies discovery of many different entities under a single algorithm: significant clusters within a single protein, at the interface of protein-protein complexes, and near drugs. It is the first tool to effectively handle drug-mutation clusters. 3) It provides comprehensive downstream analyses in prioritizing clusters that are significantly enriched in mutations from multiple patient samples and supports rare/medium recurrent functional mutation discovery.
We used HotSpot3D to analyze TCGA Pan-Cancer data, discovering a large set of mutations and revealed their relationships with known drivers. This is a rich resource for future functional explorations (Supplementary Table 22). Our HotSpot3D drug analysis also indicated that only 14 unique mutations in the significant mutation-drug clusters have been reported in the four standard databases we searched, implying discovery of over 800 novel drug interacting candidate mutations. The larger implications of this work are threefold: 1) using non-cancer drugs for treating cancers, 2) applying cancer-type specific drugs for treating patients with other types of cancers, and 3) employing targeted drugs for treating patients with non-canonical cancer mutations that cluster with known druggable mutations.
Although we have experimentally validated a small subset of predictions using high throughput phosphorylation data and in vitro cell-based assay, additional experimental testing of all putative novel drivers and drug interacting mutations discovered in our study is required to confirm their biological functions. We envision that structure-based analyses using HotSpot3D will lead to discoveries of many types of relationships among variants undetectable by conventional approaches, for example, in human variations identified from population-based studies, as well as germline variations and de novo mutations that play roles in many common diseases.
Online Methods
HotSpot3D and code comparison
HotSpot3D (see URLs) has three parts: data preprocessing, structural analyses, and visualization (Figure 1a). For SpacePAC comparison, we used the “SimMax” option, cluster radii 2-10 angstroms, up to 3 clusters, and 1000 simulated configurations. We restricted HotSpot3D to the single molecule information available to SpacePAC and configured its parameters for an unbiased comparison: no linear separation, links formed with distance p-values, and 10 angstrom maximum cluster radius. We retained only the most significant clusters for SpacePAC and used the average inner cluster distance between constituent residues as a test statistic. Permutation testing was performed for each cluster residue mass (number of residues in a cluster) for each structure. For cluster k of mass m, there are n = m(m-1)/2 residue pairs among all residues, which have an average of d̄k. For each m, we sampled 106 sets of n random pairs, and for the lth set we obtained the average inner cluster distance, d̄l. The p-value for the kth cluster of mass m is the proportion of sets with average inner distance less than d̄k.
Data preprocessing
Genes and their transcripts and proteins are procured from public sources, including the Human Genome Organization (HUGO). Preprocessing extracts four features from the HUGO Gene Nomenclature Committee (HGNC) (see URLs): HGNC gene name, Universal Protein Resource (UniProt32) ID, gene synonyms, and description.
UniProt is a comprehensive database for protein sequence and annotation data. For each HUGO gene, UniProt ID was used to retrieve PDB IDs from the Protein Data Bank (PDB) (see URLs), transcript and protein IDs from Ensembl, sequence from UniProt, and region of interest (ROI) information. For each ROI, corresponding information contains initial and destination coordinates of UniProt sequence and specific function description. By comparing each UniProt sequence with all known and novel peptide sequences of human build GRCh37 (Ensembl release 74), we identified and kept only those transcripts having the same translated length and sequence identity ≥98%. We only allowed one top Ensembl transcript match based on alignments with UniProt sequences.
This process culminates in an association table containing each HUGO gene, its UniProt, PDB, and transcript IDs, and sequence identity with UniProt sequence (Supplementary Table 23). This table was used for PDB-related 3D distance calculations and conversion between PDB and UniProt coordinates. This information is stored in a MySQL database and a flat file.
3D proximal pairs analysis
3D distance calculation
UniProt ID enables protein structure data to be extracted from PDB33. For each of the 25,627 PDB structures, one or more chains could correspond to the UniProt sequence. Here, we used the longest chain containing the amino acid of interest to calculate 3D distances between amino acids. In case of multiple identical MODELs, one is picked randomly. We take intra-molecular interactions as any pair from the same UniProt ID, regardless of chain in homomer complexes. Inter-molecular pairs are between amino acid pairs from different UniProt ID’s within the same PDB structure.
Distance is calculated as follows. Given a pair, AA0 and AA1, and their respective sets of atomic coordinates in space, AA0 and AA1, the distance between them, D(AA0,AA1), is the minimum 3D distance between all atoms of AA0 and of AA1:
| (1) |
where d is the distance between atoms i and j from AA0 and AA1, respectively, and the amino acids range either over a single chain or over two chains, depending on context.
Significance determination and prioritization
To calculate significance of distance between mutations, we statistically analyzed all possible 3D distances within each PDB structure. Permutation-based P-value for each pair of amino acids is the proportion of all pairwise 3D distances less than or equal to D(AA0,AA1). To reduce false-positives due to proximal residues in primary sequence, amino acid pairs must be separated by at least ΔN residues along the protein sequence. Here, we use the following empirically derived criteria: P < 0.05, D ≤ 10Å, and ΔN > 20 for intra-molecular clusters, while D ≤ 20Å was allowed for inter-molecular and drug-mutation clusters. This procedure generates a data set consisting of the residue pairs and their 3D distance, linear distance, and p-value for each PDB structure.
Variant List Input
For a given MAF or VCF input, transcript ID and amino acid change information from Ensembl annotation must be provided for each variant. Based on the association table, variants map to specific UniProt IDs. From the 3D proximity results, the amino acid change information was then used to map the variant to a specific location within the UniProt sequence. Using 3D proximity results, COSMIC annotation information, and ROI information, we conducted 3D proximal pairs analysis for a given variant list. Ultimately, our method reports 5 kinds of proximity information: mutations in ROI, close to ROI, close to each other, at COSMIC locations, and close to COSMIC mutations. Users can extract pairs of mutations that are in close proximity to each other within a single protein, as well as on protein-protein complexes.
Drug interaction module
HotSpot3D includes a drug-protein interaction module based on data from DrugPort (see URLs), which contains structures of drugs and their target proteins in PDB, the latter derived from DrugBank34. The version of DrugPort used here contains 1,492 approved drugs and 1,664 unique protein targets, in which there are 480 molecules in all (425 drugs and 55 nutraceuticals) contained within 21,603 PDB structures. Each drug, has four attributes: number of different targets, number of targets with known structure in PDB, number of drug-bound target structures, and total number of drug-bound structures. There is an important preprocessing step to establish the relationship between mutations and PDB structures containing each pharmaceutical. Using the DrugPort API, we parsed the raw DrugPort data file, obtaining DrugPort ID, PDB Het Group, drug molecule position in the PDB structure, and flag information. Het records describe non-standard residues, such as prosthetic groups, inhibitors, solvent molecules, and ions for which coordinates are supplied. Flag information identifies whether the structure is a target protein or not a target protein but which nevertheless contains this drug molecule. Using these pre-processing results as input for each drug, the HotSpot3D drug-protein interaction module can search mutations to determine whether any are within the three-dimensional distance cutoff of each drug.
Cancer mutation data set and cancer types
We analyzed somatic mutations (Supplementary Table 24) from 4,405 TCGA tumor samples from 19 cancer types: bladder urothelial carcinoma (BLCA), breast adenocarcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), colon and rectal carcinoma (COAD, READ), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukaemia (LAML; conventionally called AML), low-grade glioma (LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous carcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC).
Identifying mutation and drug-mutation clusters
Mutations in proximal pairs are assigned to different clusters. To seed initial clusters, we start from significant proximal pairs, iteratively adding new mutations if they are significantly paired with a mutation already in that cluster. Because this procedure can form large clusters by the “chaining effect”, as each addition lacks knowledge of the overall cluster size, we require a “stopping rule” to limit growth. Specifically, we identify the centroid of the cluster as the mutation having the highest closeness centrality and discard mutations outside its threshold radius (see below).
Formally, a cluster is an undirected graph G = (V,E), where V is a subset of the nonsynonymous mutations from the input and E is the set of proximal pairs from V identified by HotSpot3D. Two options are available for selecting V: 1) the set of all non-truncational mutations, V = V1 2) the set of unique mutations affected by the mutation cohort without recurrence, V = V2 (a proximity only approach). Let νi, νj ∈ V for i, j ∈ {1,2,…,N} where N is the number of vertices in V. Edges ei,j ∈ E are distances, where |ei,j| = di,j for paired elements vi and vj, and |ei,j| = ∞ for vertices that are not paired. For V = V1, |ei,j| = di,j = 0 if vi and vj are recurrent mutations as well as different amino acid changes at the same residue, and for V = V2, |ei,j| = di,j = 0 if vi and vj are different amino acid changes at the same residue. Clusters are built-up by the Floyd-Warshall shortest paths algorithm, initialized by the distance matrix of the edges, to obtain the geodesics, gi,j between each vi and vj. Unique clusters emerge as disjoint subsets in V having infinite geodesics between any two elements from different clusters. For each νi ∈ V, we then calculate the closeness centrality35, c(vi),
| (2) |
where N is the number of vertices in the cluster. For each cluster, the centroid is the vertex whose closeness centrality is the maximum. Finally, clusters can be focused according to user input for the cluster radius limit. The cluster radius limit is the maximum geodesic measured from the cluster centroid; any vertices outside this bound are pruned. For intra-molecular clusters, we used a radius limit of 10Å to keep clusters small and dense spatially. For inter-molecular, we used a larger limit of 20Å, since we are spanning across multiple proteins.
Clustering for drug-mutation pairs follows the same approach. Multiple instances of the same drug in a single protein are considered a single entity, despite the possibility of binding in several places. All mutations significantly paired with the drug, regardless of binding location, are included in the initial cluster, even if the mutations themselves are not close to one another. Conversely, one drug binding within a protein is treated separately from the same drug bound to other proteins, forming disjoint clusters; each cluster only includes mutations from a single protein. The cluster radius is again 20Å.
Prioritizing clusters with high cluster closeness
We focused on top clusters for downstream analyses using cluster closeness (Cc) as a measure to establish thresholds. Cc is simply the sum of the closeness centralities over each mutation in a cluster. High Cc indicates spatially dense clusters enriched in mutations from multiple patient samples. Here, we distinguished between clusters with cancer genes and non-cancer genes. We generated Cc distributions for both groups, using Wilcoxon testing to verify that they were significantly different and that Cc was in fact a good metric to determine functionality of clusters. We observed that clusters with cancer genes had significantly higher Cc than clusters without (P ≈ 5.3e-13). We could use the Cc threshold to identify novel cancer genes that exhibit similar tightness and enrichment of mutations in clusters as cancer genes. We wanted a stringent Cc threshold focusing on a small, conservative subset of intra-molecular clusters, so we defined the threshold as the top 5% cutoff of the cancer gene group (Cc = 10.283) (Figure 2a). To get an idea of the spatial “tightness” this threshold implies, an idealized equilateral tetrahedron having all equal geodesic distances, g, would indicate threshold of N2/2g ≥ 10.283 from Eq. (2), whereby g ≤ ln(N2/10.283)/ln(2). Substituting N=4 for the tetrahedron, each vertex would be a distance of 0.64Å at most from all the others. For inter-molecular analysis, we distinguished clusters with all cancer genes, at least one cancer gene, and no cancer genes. We created Cc distributions for all three groups. Here, clusters with cancer genes also had significantly higher Cc than clusters having none. Due to significantly fewer inter-molecular clusters, we defined the threshold as the top 20% cutoff for the all cancer gene group (Cc = 4.118) (Figure 2b), which equates to a maximum geodesic distance of 1.96 Å in the idealized tetrahedron model.
Cluster conservation score
The phastCons score36 quantifies conservation of mutated and deleted bases. Each cluster is scored by the weighted average of its variants’ phastCons scores, with variants weighted by recurrence. For each intra-molecular cluster, we compared Cc to cluster conservation score to evaluate whether clusters occur in functionally important regions: 70% (4,083 out of 5,822 intra-molecular clusters) have a high score (above 0.95). T-testing on mutations within clusters versus mutations not in clusters showed clustered mutations’ preference for conserved regions (P < 2.2e-16). Clusters with high Cc tend to have a high conservation score, and we found 547 clusters from 542 cancer genes, including all 38 of the top intra-molecular clusters, among the high cluster conservation score group. Clusters of cancer genes segregate as oncogene, TSG, or unclassified (general) cancer genes, and cluster conservation between groups is compared for clusters exhibiting high Cc. T-tests on clusters with top Cc failed to show significant difference between oncogenes and TSGs in terms of cluster conservation, both for the top 38 intra-molecular clusters and the top 100 clusters (p-values of 0.1036 and 0.7733, respectively).
Cluster validation
Reverse Phase Protein Array (RPPA) data
Using the subset of the TCGA cohort having available RPPA data, we examined EGFR protein expression and site-specific phosphorylation at major autophosphorylation sites pTyr1173 and pTyr1068. Here, we discarded the linear limit on clustering because proximal mutations in the linear sequence may be functionally significant. We examined GBM samples, dividing them into 3 categories: having mutations from the EGFR Ala289 cluster, having mutations outside of any cluster, and having no EGFR mutation. The same method was applied to LUAD samples, the cluster of interest being Leu858Arg. Protein and phosphoprotein levels were retrieved for the 3 categories. Welch’s t-test was used to determine if the mean protein and phosphoprotein levels were significantly higher in samples from the first category, as compared to samples from the other two categories. Similar methodology was used for ERBB2.
Phosphorylation functional experiments
NIH3T3 (clone2.2) cells were kindly provided by Dr. Robert Friesel (Maine Medical Center Research Institute). These cells have typical fibroblast morphology, undetectable levels of endogenous EGF receptor, characteristic of this subclone37, and were negative for mycoplasma, based on the absence of extranuclear signals by DAPI (4',6-diamidino-2-phenylindole) staining. Cells were cultured in DMEM (Corning) supplemented with 10% calf serum (ThermoFisher) and penicillin/streptomycin (Life Technologies). All plasmids for the expression of EGFR variants were generated from the wild-type EGFR plasmid (Sino Biological) using Q5 site–directed mutagenesis (New England Biolabs). All constructs were confirmed by sequencing. Cells were transiently transfected with wild-type or mutant EGFR constructs using Lipofectamine 2000 reagent (Life Technologies) in 6-well plates. 24 hours after transfection, cells were switched to medium containing 0.5% calf serum for 24h before stimulation with 50ng/ml recombinant human EGF (R&D Systems) for 10 minutes. Cells were lysed in buffer containing 20mM Tris-HCl (pH7.5), 150mM NaCl, 1mM Na2EDTA, 1mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1mM c-glycerophosphate, 1%, 1mM Na3VO4, 1ug/ml leupeptin (Cell Signaling). Protease and phosphatase inhibitors (Roche) were added immediately before use. Samples were boiled in buffer and subjected to SDS-PAGE on 10% polyarcrylamide gels and Western blotting was done on Immobilon-P PVDF membranes (Millipore). The following antibodies were used for immunoblotting: anti-phospho-EGFR Tyr1068 (Abcam, Tyr1092 in the unprocessed EGFR), anti-EGFR (Abcam) and anti-β-Tubulin (DSHB). Appropriate secondary antibodies with infrared dyes (LI-COR) were used. Protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR).
Mutation and drug annotations
ClinVar contains clinical variant annotation for 19,801 genes and 129,758 variants (see URLs). The Pancan19 MAF was annotated with available ClinVar clinical variant information. Of the 549,295 unique mutations observed in the TCGA dataset, 805 had pathogenic information from ClinVar.
We curated mutations from 4 databases: MyCancerGenome, PCT, GDKD, and CancerDR. MyCancerGenome catalogs cancer mutations, therapeutic options, available clinical trials, and druggability information for 43 genes (including receptor tyrosine kinases like EGFR, KIT, and PDGFRA) and 289 relevant variants. PCT, or the Personalized Cancer Therapy, contains druggability information for variants of 24 cancer-related genes and over 140 gene variant-drug interactions supported by clinical evidence. GDKD, or the Gene-Drug Knowledge Database, provides information on predictive genomic markers for over 40 malignancies and tumor-type sensitivity/resistance for specific gene variants to approved or experimental drugs. More than 700 variant-specific gene–drug interactions with therapeutic relevance were curated for this effort. CancerDR lists 148 anticancer drugs and their effectiveness against 1000 cancer cell lines. Pharmacological profiles of these drugs were collected from the CCLE and COSMIC databases as IC50 values. CancerDR contains information for 116 drug targets, including their corresponding gene sequences in cancer cell lines. Drug/sequence interactions that resulted in an IC50 value ±2 S.D. of the mean were used.
Prioritized variant list for functional validation
We prioritized putative drivers that would be good candidates for experimental validation (Supplementary Table 22), based on rare and medium recurrent variants appearing in clusters above the intra-molecular and inter-molecular Cc thresholds. The variants were ranked according to closeness centralities and only the top 10 variants were included per gene.
Software engineering aspects
We developed an interactive browser-based visualization portal (see URLs) to help assess whether a mutation interaction is likely to have functional importance. It maps individual mutations onto a PDB structure, displays potentially interacting mutation pairs or clusters, and provides for graphic annotation. Users can load individual mutations, multiple mutations, or HotSpot3D results and review all protein structures that contain the residues of the mutations. As an example, Supplementary Figure 4 shows two mutations from TCGA kidney cancer data, one from TCEB1 and the other from VHL. The client side of the portal runs within any native browser implementation, depending only on the Java plug-in to run the open-source Jmol Java applet for displaying protein structures. The webserver is Apache Tomcat 7 running JSP programs and a Java servlet as an interface to access the underlying MySQL database of pre-processed biological information. The entire server runs on a Dell PowerEdge M620 blade server, with one 8-core Intel Xeon E-2603 1.8 GHz CPUs, and 128 GB of RAM.
We analyzed clustering algorithm performance using robustness trials (Supplementary Note), where random mutations were chosen and run through the HotSpot3D clustering module. We observed O(n3) time where n represents the number of input mutations, which is consistent with the characteristic time complexity of the Floyd-Warshall algorithm. Other algorithms that might provide performance gains would do so only under special constraints on the graph that are not guaranteed to exist for problems of this type.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute grants R01CA180006 and R01CA178383 and the National Human Genome Research Institute grant U01HG006517 to L.D. F.C. is supported in part by the Department of Defense grant PC130118 and the National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK087960. M.H.B. is in part supported by the Genetics and Genomics of Disease Pathway at Washington University School of Medicine. We thank Dr. Robert Friesel for the NIH3T3 clone 2.2 cells. We thank Kimberly Johnson, Cyriac Kandoth, Maheetha Bharadwaj, and Kuan-lin Huang for suggestions on data analysis. We also thank Mingchao Xie, Reyka Jayasinghe, and Heidi Greulich for suggestions on experiments.
Footnotes
URLs
HotSpot3D code, https://github.com/ding-lab/hotspot3d; HUGO, http://www.genenames.org; PDB, http://www.rcsb.org; DrugPort, http://www.ebi.ac.uk/thornton-srv/databases/drugport#/; ClinVar, http://www.clinvar.com; HotSpot3D Portal code, https://github.com/ding-lab/hotspot3d_portal
AUTHOR CONTRIBUTIONS
L.D. and F.C. designed and supervised research. B.N., A.D.S., S.S., J.N., M.H.B., P.B., J.W., M.D.M., P.T., C.L., K.Y., S.Q.S., W.L., and F.C., L.D. analyzed the data. M.C.W. B.N., A.D.S., and Q.Z. performed statistical analysis. M.A.W., B.N., A.D.S., S.S., and M.H.B. prepared figures and tables. B.N., A.D.S., S.S., J.W., and R.J.M. contributed to HotSpot3D code. L.D., F.C., B.N., A.D.S., S.S., and M.H.B. wrote the manuscript. F.C., M.C.W., A.D.S., S.S. and L.D. revised the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Dees ND, et al. MuSiC: Identifying mutational significance in cancer genomes. Genome research. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter H, Samayoa J, Hruban RH, Karchin R. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM) Cancer biology & therapy. 2010;10:582–587. doi: 10.4161/cbt.10.6.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Perez A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nature methods. 2013 doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic acids research. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 7.Niknafs N, et al. MuPIT interactive: webserver for mapping variant positions to annotated, interactive 3D structures. Human genetics. 2013;132:1235–1243. doi: 10.1007/s00439-013-1325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teyra J, Kim PM. Interpreting protein networks with three-dimensional structures. Nature methods. 2013;10:43–44. doi: 10.1038/nmeth.2300. [DOI] [PubMed] [Google Scholar]
- 10.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A, et al. MutDB: update on development of tools for the biochemical analysis of genetic variation. Nucleic acids research. 2008;36:D815–D819. doi: 10.1093/nar/gkm659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.H, R.G.a.Z. SpacePAC: Identification of Mutational Clusters in 3D Protein Space via Simulation. R package version 1.6.0. 2013 [Google Scholar]
- 14.Kamburov A, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci U S A. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betts MJ, et al. Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic Acids Res. 2015;43:e10. doi: 10.1093/nar/gku1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature genetics. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 17.Choi YL, et al. Oncogenic MAP2K1 mutations in human epithelial tumors. Carcinogenesis. 2012;33:956–961. doi: 10.1093/carcin/bgs099. [DOI] [PubMed] [Google Scholar]
- 18.Fleming NI, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer research. 2013;73:725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 19.Berman HM, et al. The Protein Data Bank. Nucleic acids research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen M, Potapov V, Schreiber G. Four distances between pairs of amino acids provide a precise description of their interaction. PLoS computational biology. 2009;5:e1000470. doi: 10.1371/journal.pcbi.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO journal. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerner GS, et al. Common and rare EGFR and KRAS mutations in a Dutch non-small-cell lung cancer population and their clinical outcome. PLoS One. 2013;8:e70346. doi: 10.1371/journal.pone.0070346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009;15:460–467. doi: 10.1158/1078-0432.CCR-08-1757. [DOI] [PubMed] [Google Scholar]
- 25.de Biase D, et al. Next-generation sequencing of lung cancer EGFR exons 18-21 allows effective molecular diagnosis of small routine samples (cytology and biopsy) PLoS One. 2013;8:e83607. doi: 10.1371/journal.pone.0083607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel VG, et al. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA : the journal of the American Medical Association. 2006;295:2727–2741. doi: 10.1001/jama.295.23.joc60074. [DOI] [PubMed] [Google Scholar]
- 28.Hardman WE. (n-3) fatty acids and cancer therapy. J Nutr. 2004;134:3427S–3430S. doi: 10.1093/jn/134.12.3427S. [DOI] [PubMed] [Google Scholar]
- 29.Redaelli S, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:469–471. doi: 10.1200/JCO.2008.19.8853. [DOI] [PubMed] [Google Scholar]
- 30.Ohanian M, Cortes J, Kantarjian H, Jabbour E. Tyrosine kinase inhibitors in acute and chronic leukemias. Expert Opin Pharmacother. 2012;13:927–938. doi: 10.1517/14656566.2012.672974. [DOI] [PubMed] [Google Scholar]
- 31.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nature structural & molecular biology. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 32.Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic acids research. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman HM. The Protein Data Bank: a historical perspective. Acta crystallographica. Section A, Foundations of crystallography. 2008;64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 34.Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dangalchev C. Residual closeness in networks. Physica A: Statistical Mechanics and its Applications. 2006;365:556–564. [Google Scholar]
- 36.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friesel R, Burgess WH, Maciag T. Heparin-binding growth factor 1 stimulates tyrosine phosphorylation in NIH 3T3 cells. Mol Cell Biol. 1989;9:1857–1865. doi: 10.1128/mcb.9.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.