Abstract
DNA damage present a barrier to DNA-templated biochemical processes, including gene expression and faithful DNA replication. Compromised DNA repair leads to mutations, enhancing the risk for genetic diseases and cancer development. Conventional experimental approaches to study DNA damage required a researcher to choose between measuring bulk damage over the entire genome, with little or no resolution regarding a specific location, and obtaining data specific to a locus of interest, without a global perspective. Recent advances in high-throughput genomic tools overcame these limitations and provide high-resolution measurements simultaneously across the genome. In this review, we discuss the available methods for measuring DNA damage and their repair, focusing on genome-wide assays for pyrimidine photodimers, the major types of damage induced by ultraviolet irradiation. These new genomic assays will be a powerful tool in identifying key components of genome stability and carcinogenesis.
Graphical abstract
Traditional experimental approaches to study DNA damages required a researcher to choose between measuring bulk damage over the entire genome, with little or no resolution regarding a specific location, and obtaining data specific to a locus of interest, without a global perspective. Recent advances in high-throughput genomic methods overcome these limitations and provide high-resolution measurements simultaneously across the genome. These new genomic assays will be a powerful tool in identifying key components of genome stability and carcinogenesis.
INTRODUCTION
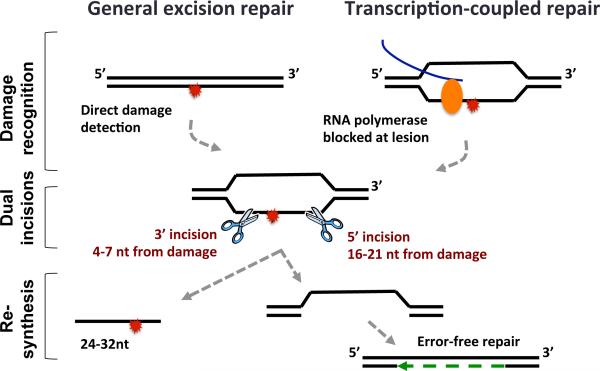
Ultraviolet radiation (wavelength 100-400 nm) consists of UVA (315-400nm), UVB (280-315nm) and UVC (100-280nm). More than 95% of UV radiation to reach the earth's surface is the less-energetic UVA, and the rest is UVB. The more-energetic UVC is absorbed or blocked by the atmosphere. Pyrimidine photodimers including cyclobutane pyrimidine dimers (CPDs), pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs] and their related Dewar valence isomers are the most abundant types of damage induced by UV (1, 2). UVB and UVC can induce CPDs, (6-4)PPs and Dewar valence isomers. In contrast, UVA can only induce CPDs but not (6-4)PPs. However, UVA can convert (6-4)PPs to Dewar valence isomers and indirectly induce oxidative DNA damage in a much lower yield than CPDs. Pyrimidine dimers block transcription and replication, and their removal is necessary to preserve genomic integrity and function. Photodamages are efficiently and faithfully repaired by both photoreactivation and Nucleotide Excision Repair (NER, excision repair) (3). In photoreactivation a single photolyases enzyme harnesses the energy in light photons to reverse the damage. Photolyases specific for either CPD or (6-4)PP have been identified in bacteria, yeast, insects, plants and marsupial mammals. In other mammals, including humans, in the absence of a photolyase homologue, nucleotide excision repair is the sole mechanism for removing photodimers. Mammalian excision repair is orchestrated by at least 11 enzymes (comprised by 30 polypeptides) in a multistep process (Figure 1) (4). Repair is divided into three major steps: 1) Recognition of the damage, which can occur either directly (in general repair), or in a transcription-coupled manner (in transcription-coupled repair), 2) Incision 3’ and 5’ of the damage, removing a nucleotide stretch (24-32 nt) and leaving a single stranded gap, and 3) Gap-filling DNA synthesis and ligation to restore intact double-stranded DNA. Mutations that inactivate excision repair genes cause the severe genetic diseases xeroderma pigmentosum (XP), Cockayne syndrome (CS) and trichothiodystrophy (TTD) (5). Even attenuated repair resulting from polymorphism and not inactivation of excision repair genes can lead to enhanced cancer risk (6, 7).
Figure 1.
Schematic of mammalian nucleotide excision repair.
In over three decades of biochemical research, Prof. Aziz Sancar's seminal contributions have led to detailed mechanistic understanding of both the photoreactivation and excision repair mechanisms, which are both reviewed in detail in this issue (3,8-10).
While the mechanism of damage formation and repair is well understood, there are still many open questions regarding the effect of chromatin context, transcription and replication on DNA damage sensitivity. A comprehensive understanding of damage formation and DNA repair in an active nucleus requires high-resolution genome-wide measurements. In this review we discuss the available methods for measuring UV-induced DNA damage and DNA repair, and the current genome-wide methodologies available for their analysis.
METHODS FOR MEASURING UV INDUCED DNA DAMAGE AND DNA REPAIR
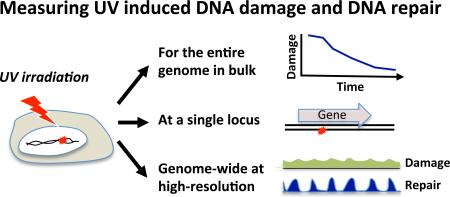
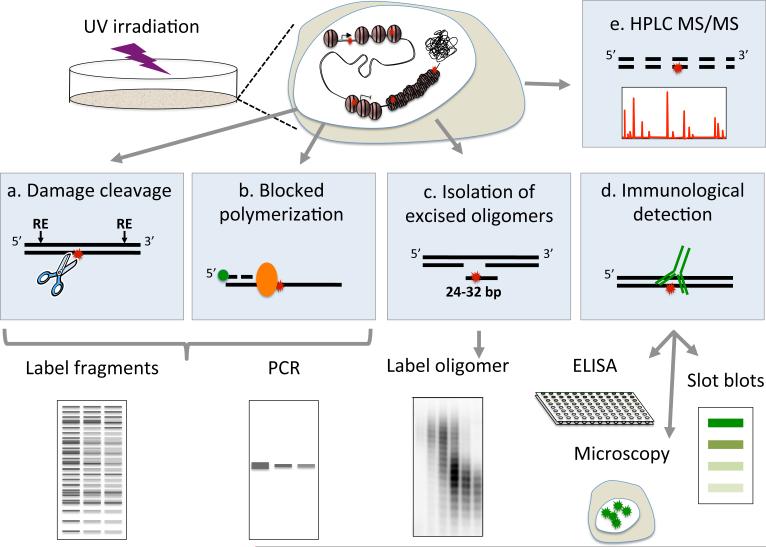
The first evidence for DNA repair came from observation of “unscheduled DNA synthesis” which was then identified as repair replication. However, measuring repair by unscheduled DNA synthesis is indirect and limited to cells outside of S-phase (11). To date, the majority of methods used to quantify DNA damage and repair either study damage levels in the entire genome in “bulk”, or study a specific locus at high resolution but with limited global perspective. There are three general approaches for photodimer detection: 1) Enzymatic approaches that identify damage sites 2) Damage-specific antibodies and 3) Chromatographic techniques incorporating mass spectrometry (Figure 2).
Figure 2.
Illustration of the different approaches for measurement of UV induced DNA damage and their repair.
The most common enzyme for photodimer detection is the T4 endonuclease V, which specifically nicks CPD sites. Similarly, the Saccharomyces Pombe Uve1p (UVDE) nicks DNA at damage sites but cannot discriminate between CPDs and (6-4)PP. In methods relying on these enzymes (Figure 2a), the genome is first fragmented with restriction nucleases, and then digested with damage specific endonucleases. After digestion, single stranded DNA is either labeled with site-specific radioactive or fluorescent probes (southern blot) (12, 13), PCR amplified with primers (ligation-mediated PCR, or LM-PCR) (14), or end labeled by incorporation of radioactive deoxynucleotides (15), and separated by gel electrophoresis under denaturing conditions to sensitively detect damages at specific genomic loci. These methods provide single-nucleotide resolution (with the exception of southern blot) and strand-specific information under physiological-relevant conditions. The damage level can be quantified by comparing digested and full-length products. Alternatively, quantitative assessment of global damage levels can be achieved by subjecting the digested DNA to single cell alkali gel electrophoresis (16). An alternative enzymatic strategy with a broader damage-spectrum is based on DNA polymerization, which is blocked by the damage (17). This method is simple and straightforward, and does not require nuclease treatment or labeling. However, it only has gene-level resolution, lower accuracy and cannot separate two strands.
Using these sensitive and specific enzymatic approaches Hanawalt and colleagues were able to first identify transcription coupled repair in the DHFR gene locus (12), and the effect of nucleosome binding on DNA damage formation was first assayed by Smerdon and colleagues (18, 19).
The development and commercialization of antibodies that specifically recognized photodimers opened the door to a variety of commonly used immunological methods (Figure 2d). These include damage quantification by Enzyme-Linked ImmunoSorbent Assay (ELISA), radioimmunoassay (RIA), flow cytometry, immunoblots, immunoprecipitation and visualization of damages in single cells using immunofluorescent microscopy (20, 21)
While the enzymatic and immunological assays sensitively measure the overall damage levels, they cannot easily distinguish between the different di-pyrimidines composing each type of UV photodamage. High performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS, Figure 2e) of hydrolyzed DNA has the benefit of sensitively measuring the levels of all three photodimer types (CPD, (6-4)PP and Dewar isomers) in a single experiment, as well as being able to distinguish between damages in each of the 4 possible di-pyrimidine combinations. Experiments in UVB and UVC irradiated DNA showed that (6-4)PP occur at an 8 fold lower frequency than CPDs, and that the majority of photoproducts occur in T-T and T-C pairs (22, 23). However, since these measurements are performed on hydrolyzed DNA, information on their genomic positions is lost.
With all the technologies mentioned above, repair is measured based on the calculated disappearance of damage. They are therefore limited in their sensitivity as they may not be able to accurately measure low levels of repair. A more direct assay measures excision repair based on the abundance of the excised oligomer released in-vivo (Figure 2c). While information on the overall levels of damage is lost, this assay can sensitively detect relatively low repair levels (24-26).
GENOME-WIDE MAPPING OF UV-INDUCED DNA DAMAGE
The development of DNA microarrays and high-throughput sequencing technologies has given rise to a plethora of genomic methodologies that probe the entire genome in a single experiment. Methods for mapping protein binding (e.g. ChIP-chip/seq, (27)), nucleosomes (Mnase-seq (28, 29)), nucleosome free regions (DNase-,FAIRE-,and ATAC-seq (30-33)), and transcription (RNA-seq (34)) have been constantly evolving and improving. However, methods for mapping DNA damage and repair have lagged behind. Mapping damages has several additional and unique challenges. First, as opposed to protein bound or transcribed regions that span dozens to thousands of nucleotides, damages are generally in one or two nucleotides. Second, damages can occur essentially everywhere in the genome (CPD dimers can occur in any pyrimidine pair, covering a total of 28% of the human genome) and third, in each cell in the population, damage will likely occur in a different position. As a result, accurate genome wide characterization of DNA damage levels and their repair requires high specificity, high resolution and high genomic coverage.
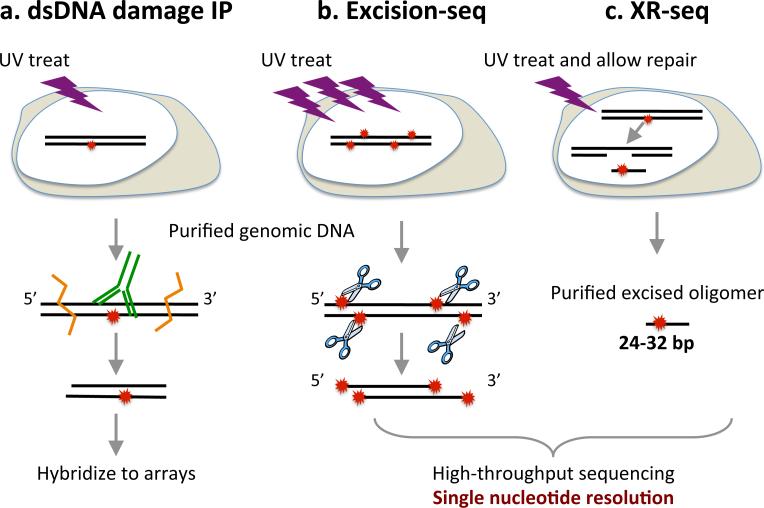
The first method to map UV-induced lesions genome-wide was based on immunoprecipitation with damage-specific antibodies, followed by microarray hybridization (Figure 3a) (35). Briefly, genomic DNA is sheared by sonication and damaged DNA is immunoprecipitated. Damages are repaired in vitro, the dsDNA fragments are ligated to adapters, amplified by PCR and hybridized to microarray chips. This method has now been used to map UV induced CPDs in yeast and human cells (36, 35, 37). In yeast and humans, CPD formation appears to be dictated mostly by the di-pyrimidine frequency in the underlying sequence. In human cells, Zavala et al. reported hot-spots for CPDs at sites adjacent to repeat sequences. The immunoprecipitation-based method has certain limitations in terms of sensitivity, specificity and resolution. The sensitivity and specificity depend on, and are limited by, the quality and availability of an antibody. Since the resolution of the data is low, and the damage may be anywhere within the isolated double stranded DNA, it is difficult to evaluate the specificity of the result by the sequence composition of the data itself. The resolution, currently in the range of several hundred base pairs, is determined by the fragments’ size and the density of the microarray probes. Thus, this method cannot provide the precise position or strand of the lesion. However, these limitations can be improved by moving to a high-throughput sequencing platform that incorporates strand information, and perhaps by reducing fragments size after antibody binding, similarly to what was achieved in high-resolution ChIP-exo (38).
Figure 3.
Methods for genome-wide mapping of UV-induced photoproducts and their repair.
More advanced methods for mapping UV induced dimers at single nucleotide resolution utilize enzymes that cleave DNA at damage sites. These methods are based to the previous damage-detection assays, except that high-throughput sequencing allows mapping of all the damage sites in the genome instead of detecting a single locus, by a specific probe or PCR primer. Excision-seq (Figure 3b) makes use of the damage-specific endonuclease UVDE to map photodimers at single-nucleotide resolution. To date this method has only been applied to the yeast genome (39). UVDE nicks the DNA 3’ to photodimers. The photodimers at enzyme-generated ends were repaired by (6-4)PP or CPD photolyases to allow adapter-ligation. After PCR-amplification, the exact position of the damage is identified by high-throughput sequencing, based on the position of the fragment end. CPD and (6-4)PP were discriminated by the use of respective photolyase. The specificity and accuracy of this method are determined by UVDE. Because photodimers are only formed at dipyrimidine positions, this specificity can be evaluated by the enrichment of dipyrimidine sequences at the fragment ends. This single-nucleotide resolution mapping of damage uncovered a sequence preference for an “A” downstream of (6-4)-TC dimers. The major drawback of this method is that it requires a very high and very genotoxic dose of UV (the study used 10,000J/m2 254nm UVC). In contrast to other damage-specific endonuclease-based methods, this high dose is necessary in order to generate the small fragment sizes for sequencing. The high resolution of this method therefore comes at a cost of not being able to study physiological conditions, or repair. “Post-digestion Excision-seq” was therefore developed to map physiological levels of damage (39). In this modification of excision-seq, the DNA is fragmented and ligated to adapters, and only incubated with excision enzymes prior to PCR. The damage-containing fragments are thus excluded from the final sequencing libraries. This allows identification of damage sites based on signal depletion. However, the ability to identify physiological damage levels comes with a resolution cost, which is dictated once more by the fragment sizes.
Recently, Mao et al developed CPD-seq (Figure 3c), a method that provides single nucleotide resolution mapping of CPDs in yeast (40). This method is a modification of a previous method that mapped ribonucleotide incorporations (41, 42). In CPD-seq, after UVC treatment, genomic DNA was sheared and ligated to adapters, followed by digestion with T4 endonuclease V and APE1 to produce a nick 5’ to the CPD. The generated ends were ligated to another adaptor to determine the exact position of CPDs by next-generation sequencing. Due to relatively high background, however, CPD-seq has currently only be performed in yeast irradiated with 100J/m2 UVC, which is much lower than the dose used in Excision-seq, but still higher than the common dose used in human cells (normally no more than 20 J/m2 UVC).
Both damage IP and Excision-seq have been applied to damages beyond UV dimers. Immunoprecipitation followed by microarray hybridization has been used to map cisplatin- and oxaliplatin- induced damages, and can be modified for any damage that meets two basic conditions: 1) That specific antibodies are available for the damage, and 2) That the damage can either be reversed or does not block DNA polymerase activity, allowing amplification of damage-enriched DNA. Excision-seq has also been used to measure incorporation of uracils in the genome using a combination of the uracil DNA glycosylase (UDG) and the T4 endonuclease IV, which then cleaves the abasic site that is formed. In the future, Excision-seq and CPD-seq could be applied to other damage types for which specific glycosylases are available.
We have also recently reported the development of Damage-seq (43), a method to map DNA-polymerase blocking damages at single nucleotide resolution in the human genome (Figure 3d). In Damage-seq, damaged genomic DNA is incubated with a high-fidelity polymerase in vitro to precisely map the position of the damaged nucleotide. Damage-seq has been used to map damages induced by cisplatin and oxaliplatin, but it can be used to map UV induced photodimers.
It is important to note that due to the nature of the genome-wide measurements, none of the current methods inform on absolute damage or repair levels. However, incorporating an internal standard such as UV-irradiated plasmid controls, could be used to quantify relative damage levels.
GENOME-WIDE MAPPING OF ADDITIONAL TYPES OF DAMAGE
Several methods have been developed for mapping other type of DNA damages in a genome-wide manner. These methods could be modified to map UV-induced dimers and we therefore briefly review them.
Genome wide mapping of double-strand breaks
Double strand breaks (DSBs) are one of the most studied types of damage, and were the first to be targeted by genome-wide methods. DSBs can be measured by detecting indirect markers (e.g. γH2AX, RPA, or ssDNA) or consequence of incorrect repair (e.g. translocation or end-joining with insertion). Here we focus on methods which directly capture break ends. The challenge for direct mapping of DSBs is to distinguish breaks induced in vivo from breaks formed during and after DNA extraction. To resolve this issue, break ends were labeled with biotin in situ. In BLESS (direct in situ Breaks Labeling, Enrichment on Streptavidin and next-generation Sequencing) (44), DSBs are ligated to a biotinylated linker in formaldehyde-fixed nuclei. In Break-seq (45), yeast cells are embedded in agarose to protect DNA integrity. After cell lysis and protein removal, breaks are labeled with biotinylated nucleotides by T4 DNA polymerase. For both methods, genomic DNA is extracted and sheared only after labeling, and the biotin-labeled fragments are isolated and sequenced. By avoiding DSBs formed in vitro these methods have been found to sensitively and accurately map breaks sites and found breaks induced by replication stress were associated with cancer rearrangements and cancer-linked genes. These strategies could be modified and used for capturing enzyme-generated breaks at UV-induced damages.
High-resolution detection of oxidatively damaged bases
Similar to mapping DSBs, the major challenge for mapping oxidatively damaged bases is preventing high level of artifactual oxidation during DNA extraction. This is confounded by the fact that for a given biological effect, oxidatively damaged bases are formed on average at a two order of magnitude lower level than pyrimidine dimers (46). There are several enzymatic approaches to mapping these damages with high sensitivity and resolution, though to our knowledge, none to-date have been performed in a genome wide manner.
An oxidatively generated base modification that has been mapped on a genome-wide scale is the oxidation of the epigenetic DNA mark 5-methylcytosine (5mC) at CpG sites to 5-hydroxymethylcytosine (5hmC). This oxidation is created by DNA deoxygenase enzymes of the TET family, and is perhaps therefore not a damage per se. The genome-wide mapping of 5hmC is generally based on modifications of mapping of 5mC by bisulfite sequencing. For review see ref (47).
Mapping DNA damage by third-generation sequencing
The new generation of single-molecule sequencing technologies, or third-generation sequencing, differ from the current sequencing by synthesis (SBS) technologies in that they identify the nucleotides based on the kinetics of single incorporation events. Since polymerization kinetics differ for modified bases, these methods, in theory, can detect them directly without complicated biochemical treatment. One of these technologies, single-molecule sequencing in real time (SMRT) by Pacific Biosciences has been shown to directly detect various DNA damage including 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxo-dG) and CPD (48-50). The nanopore sequencing technology also has the potential to detect DNA damage such as interstrand crosslink or 8-oxo-dG, directly or after labeling (51, 52). To date, these methods have only been used with simple sequence contexts such as synthesized templates, plasmid DNA, and mitochondrial DNA. Though it is still far from practical, genome-wide use, third-generation sequencing has the potential to be the future of DNA damage mapping, including of UV-induced lesions.
HIGH RESOLUTION MAPPING OF PHOTOPRODUCT REPAIR
Genome wide mapping of nucleotide excision repair by XR-seq
Current assays that measure repair do so by quantifying the disappearance of damages from the genome. Similarly, genome-wide mapping of repair could be achieved by subtraction of the genome-wide damage maps. However, with existing damage-mapping methods, these repair maps would have limited resolution, and may not be sensitive to small differences in damage levels.
A breakthrough in the ability to map repair came with the development of the method to isolate the excised oligomers as they are released from the genome. In eXcision-Repair-seq (XR-seq, Figure 3e, (53, 54)), the excised oligomers are captured by immunoprecipitation of TFIIH, a transcription and repair factor that remains bound to the excised oligomer after its release. Following this, adapters are ligated to the single stranded DNA, the DNA is repaired in vitro to allow PCR amplification, and sequencing libraries are subjected to next generation sequencing. To date, XR-seq has been applied to CPD and (6-4)PP repair in human skin fibroblasts. The two UV-induced damages were distinguished by specific immunoprecipitation with anti-damage antibodies. The XR-seq method produces single-nucleotide resolution, strand-specific maps of excision repair across the genome.
Excised oligomers captured by XR-seq were on average 26nt long, in agreement with the previous biochemical analyses (53, 54, 25, 26). These previous works also showed that the incision events were not symmetrical, and therefore damages were predicted to be ~5-7nt from the 3’ end. Indeed, for both CPD- and (6-4)PP- XR-seq, strong enrichment of pyrimidines was observed in the 3’ region of the reads. However, the identity of the enriched pyrimidine dimers was different for the two damages, with predominantly TT enriched in XR-seq for CPD, and TC and TT for (6-4)PP (54). Sequence context analysis of the enriched TC and TT (6-4)PP revealed a preference for C upstream and A downstream of the (6-4)-TC and (6-4)-TT, in agreement with the high-resolution mapping of damages by “excision-seq” in yeast (39).
New insights gained with genomics of DNA repair
XR-seq in normal human skin fibroblasts corroborated previous reports that the repair of CPDs is primarily transcription-coupled, requiring at least 12h, whereas (6-4)PPs were recognized efficiently by general excision repair and repaired within 4h of irradiation. Conducting the assay in cells proficient only in transcription coupled repair (54) showed that this repair was exclusive to the template strand of actively transcribed genes. Furthermore, enhanced repair was not limited to genes, but occurred in other regions transcribed by RNA pol II, including bi-directional promoters and enhancers. It is not clear if the RNAs transcribed in these regulator regions have a functional role, but the results show that as a consequence of this transcription, they are preferentially repaired.
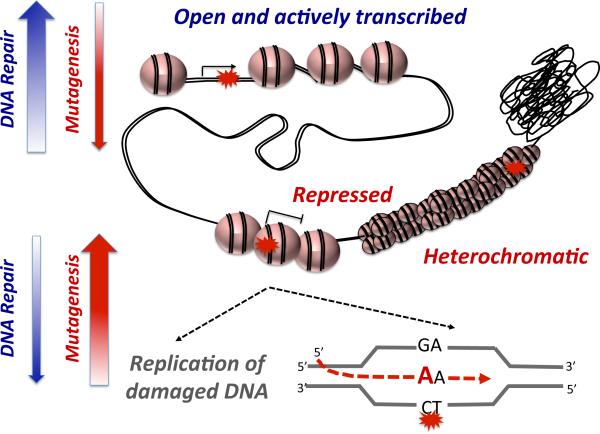
Time-course XR-seq experiments for both CPD and (6-4)PP (53) showed open and actively transcribed regions of the genome, characterized by DNase hypersensitivity and active histone marks, are preferentially repaired at earlier time points (Figure 4). In contrast, repair in the heterochromatic and repressed regions is low and can persist even 48h after irradiation.
Figure 4.
Current model based on genome-wide observations for the relationship between chromatin state, repair efficiency and mutagenesis.
RELEVANCE FOR HUMAN HEALTH
Sun exposure increases skin aging and cancer risk. During DNA replication, UV-induced damages block the replicative polymerases. If unresolved, specialized bypass polymerases can be recruited to synthesize past the lesion. However, due the miscoding nature of damages, translesion synthesis has a higher risk of forming a mutation (55). The prediction was therefore that inefficient repair would lead to higher mutations rates. Indeed, using the XR-seq genome-wide data, three groups, including us, have been able to show that inefficient excision repair of UV induced CPDs and (6-4)PP at hard-to-access regions is associated with higher levels of cancer mutagenesis (53, 56, 57). While these works showed a positive association, the development of genome-wide mutagenesis assays will allow direct experiments connecting damage and repair to mutagenesis (58, 59).
In cancer research, studying DNA damage and its repair is important not only for understanding carcinogenesis, but also for therapy. The major anti-cancer drugs kill cancer cells by damaging their DNA. These include agents that cause double strand breaks (doxorubicin) as well as bulky adducts (cisplatin, mitomycinC). Methods that map DNA damage and their repair in a comprehensive and sensitive manner will be advantageous in studying the effect of chemotherapy drugs, and identifying novel approaches to sensitize cells to these chemotherapies.
The majority of studies on UV induced damage assay damage and repair after acute UVC or UVB exposure. The more accurate treatment would be constant exposure of sunlight-mimicking radiation. Such studies will be technically challenging, as a low dose exposure requires very high specificity, however, as the technologies improve these experiments become feasible and will lead to better models for UV damage formation DNA repair, and their roles in human disease.
CONCLUSIONS
These are exciting times for the DNA repair field as it finally enters the genomic era. A comprehensive understanding of DNA repair requires understanding how it occurs despite the packaging of DNA in nucleosomes and chromatin, and how it is coordinated with transcription and replication. Comparing the genome-wide patterns of DNA damage, DNA repair, chromatin components, transcription and replication, will allow us to deduce how these processes influence each other. Ideally, as technologies advance, we will be able to study DNA damage and DNA repair not just in human cell lines, but also in disease relevant human tissues and in single cells.
ACKNOWLEDGEMENTS
We would like to thank Prof. Aziz Sancar for his mentorship; for sharing the breadth of his knowledge, his scientific curiosity, his dedication to science and his love and enthusiasm for discovery. J.H and S.A are supported by NIH grant GM118102.
Footnotes
This article is part of the Special Issue highlighting Dr. Aziz Sancar's outstanding contributions to various aspects of the repair of DNA photodamage in honor of his recent Nobel Prize in Chemistry.
REFERENCES
- 1.Cadet J, Grand A, Douki T. Solar UV radiation-induced DNA Bipyrimidine photoproducts: formation and mechanistic insights. Top Curr Chem. 2015;356:249–275. doi: 10.1007/128_2014_553. [DOI] [PubMed] [Google Scholar]
- 2.Schreier WJ, Gilch P, Zinth W. Early events of DNA photodamage. Annu Rev Phys Chem. 2015;66:497–519. doi: 10.1146/annurev-physchem-040214-121821. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture). Angew Chem Int Ed Engl. 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 4.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Re. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 5.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 6.Kiyohara C, Takayama K, Nakanishi Y. Lung cancer risk and genetic polymorphisms in DNA repair pathways: a meta-analysis. J Nucleic Acids. 2010;2010:701760. doi: 10.4061/2010/701760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mocellin S, Verdi D, Nitti D. DNA repair gene polymorphisms and risk of cutaneous melanoma: a systematic review and meta-analysis. Carcinogenesis. 2009;30:1735–1743. doi: 10.1093/carcin/bgp207. [DOI] [PubMed] [Google Scholar]
- 8.Kemp M, Hu J. PostExcision Events in Human Nucleotide Excision Repair. Photochem. Photobio. 2016;(This issue) doi: 10.1111/php.12641. DOI: 10.1111/php.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozturk N. Phylogenetic and Functional Classification of the Photolyase/Cryptochrome Family. Photochem. Photobio. 2016;(This issue) doi: 10.1111/php.12676. DOI:10.111/php.12676. [DOI] [PubMed] [Google Scholar]
- 10.Zhong D. Electron transfer mechanisms of DNA repair by photolyase. Annu Rev Phys Chem. 2015;66:691–715. doi: 10.1146/annurev-physchem-040513-103631. [DOI] [PubMed] [Google Scholar]
- 11.Hanawalt PC. Historical perspective on the DNA damage response. DNA Repair. 2015;36:2–7. doi: 10.1016/j.dnarep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard H, Wellinger RE, Aguilera A. Methods to study transcription-coupled repair in chromatin. Methods Mol Biol. 2015;1288:273–288. doi: 10.1007/978-1-4939-2474-5_15. [DOI] [PubMed] [Google Scholar]
- 14.Besaratinia A, Pfeifer GP. Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR. Methods in molecular biology. 2012;920:189–202. doi: 10.1007/978-1-61779-998-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SS, Waters R, Smerdon MJ. Low- and high-resolution mapping of DNA damage at specific sites. Methods-a Companion to Methods in Enzymology. 2000;22:170–179. doi: 10.1006/meth.2000.1058. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol Biol. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale JM, Nissen KA, Smerdon MJ. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc Natl Acad Sci U S A. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale JM, Smerdon MJ. Photofootprint of nucleosome core DNA in intact chromatin having different structural states. J Mol Biol. 1988;204:949–958. doi: 10.1016/0022-2836(88)90054-x. [DOI] [PubMed] [Google Scholar]
- 20.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell DL, Nguyen TD, Cleaver JE. Nonrandom induction of pyrimidine-pyrimidone (6-4) photoproducts in ultraviolet-irradiated human chromatin. J Biol Chem. 1990;265:5353–5356. [PubMed] [Google Scholar]
- 22.Douki T, Cadet J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry. 2001;40:2495–2501. doi: 10.1021/bi0022543. [DOI] [PubMed] [Google Scholar]
- 23.Douki T, Court M, Sauvaigo S, Odin F, Cadet J. Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J Biol Chem. 2000;275:11678–11685. doi: 10.1074/jbc.275.16.11678. [DOI] [PubMed] [Google Scholar]
- 24.Choi JH, Gaddameedhi S, Kim SY, Hu J, Kemp MG, Sancar A. Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 2014;42:e29. doi: 10.1093/nar/gkt1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu JC, Choi JH, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide Excision Repair in Human Cells FATE OF THE EXCISED OLIGONUCLEOTIDE CARRYING DNA DAMAGE IN VIVO. J Biol Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem. 2012;287:22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John S, Sabo PJ, Canfield TK, Lee K, Vong S, Weaver M, Wang H, Vierstra J, Reynolds AP, Thurman RE, Stamatoyannopoulos JA. Genome-scale mapping of DNase I hypersensitivity. Curr Protoc Mol Biol. 2013 doi: 10.1002/0471142727.mb2127s103. Chapter 27, Unit 21 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 35.Teng Y, Bennett M, Evans KE, Zhuang-Jackson H, Higgs A, Reed SH, Waters R. A novel method for the genome-wide high resolution analysis of DNA damage. Nucleic Acids Res. 2011;39:e10. doi: 10.1093/nar/gkq1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell JR, Bennett MR, Evans KE, Yu S, Webster RM, Waters R, Skinner N, Reed SH. 3D-DIP-Chip: a microarray-based method to measure genomic DNA damage. Sci Rep. 2015;5:7975. doi: 10.1038/srep07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavala AG, Morris RT, Wyrick JJ, Smerdon MJ. High-resolution characterization of CPD hotspot formation in human fibroblasts. Nucleic Acids Res. 2014;42:893–905. doi: 10.1093/nar/gkt912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan DS, Ransom M, Adane B, York K, Hesselberth JR. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 2014;24:1534–1542. doi: 10.1101/gr.174052.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. P Natl Acad Sci USA. 2016;113:9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding J, Taylor MS, Jackson AP, Reijns MAM. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat Protoc. 2015;10:1433–1444. doi: 10.1038/nprot.2015.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reijns MA, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. P Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman EA, McCulley A, Haarer B, Arnak R, Feng WY. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 2015;25:402–412. doi: 10.1101/gr.180497.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H, Zhang Y. Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol. 2015;22:656–661. doi: 10.1038/nsmb.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schadt EE, Banerjee O, Fang G, Feng Z, Wong WH, Zhang X, Kislyuk A, Clark TA, Luong K, Keren-Paz A, Chess A, Kumar V, Chen-Plotkin A, Sondheimer N, Korlach J, Kasarskis A. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 2013;23:129–141. doi: 10.1101/gr.136739.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark TA, Spittle KE, Turner SW, Korlach J. Direct detection and sequencing of damaged DNA bases. Genome Integr. 2011;2:10. doi: 10.1186/2041-9414-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Z, Fang G, Korlach J, Clark T, Luong K, Zhang X, Wong W, Schadt E. Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLoS Comput Biol. 2013;9:e1002935. doi: 10.1371/journal.pcbi.1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Price NE, Fang X, Yang Z, Gu LQ, Gates KS. Characterization of Interstrand DNA-DNA Cross-Links Using the alpha-Hemolysin Protein Nanopore. ACS Nano. 2015;9:11812–11819. doi: 10.1021/acsnano.5b03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An N, Fleming AM, White HS, Burrows CJ. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano. 2015;9:4296–4307. doi: 10.1021/acsnano.5b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci U S A. 2016;113:E2124–2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Gene Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JW. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 57.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 58.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]