Abstract
AUF1 is a family of four RNA-binding proteins generated by alternative pre-mRNA splicing, with canonical roles in controlling the stability and/or translation of mRNA targets based on recognition of AU-rich sequences within mRNA 3′ untranslated regions. However, recent studies identifying AUF1 target sites across the transcriptome have revealed that these canonical functions are but a subset of its roles in posttranscriptional regulation of gene expression. In this review, we describe recent developments in our understanding of the RNA-binding properties of AUF1 together with their biochemical implications and roles in directing mRNA decay and translation. This is then followed by a survey of newly discovered activities for AUF1 proteins in control of miRNA synthesis and function, including miRNA assembly into miRISC complexes, miRISC targeting to mRNA substrates, interplay with an expanding network of other cellular RNA-binding proteins, and reciprocal regulatory relationships between miRNA and AUF1 synthesis. Finally, we discuss recently reported relationships between AUF1 and long noncoding RNAs and regulatory roles on viral RNA substrates. Cumulatively, these findings have significantly expanded our appreciation of the scope and diversity of AUF1 functions in the cell, and are prompting an exciting array of new questions moving forward.
Graphical Abstract
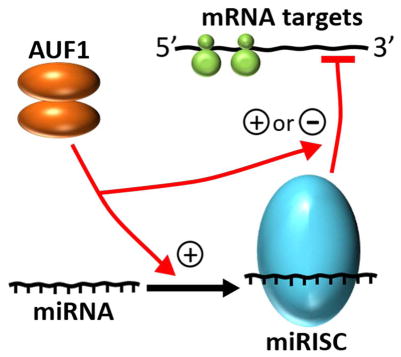
AUF1 enhances loading of select miRNAs into RISC complexes but can also regulate miRISC access to mRNA substrates.
Introduction
In eukaryotes, gene expression occurs via a highly orchestrated series of compartmentalized steps. For protein-coding genes, transcription by RNA polymerase II generates a pre-mRNA that is subsequently processed and exported to the cytoplasm, allowing it to then be translated by ribosomes to yield its encoded protein product. However, each step in gene expression is also subject to regulatory control. Following transcription, most gene regulatory processes involve selective recognition and manipulation of RNA targets. In the nucleus, functional consequences of these interactions may include altering the position or location of pre-mRNA splicing or 3′-cleavage and polyadenylation events, targeted nuclear RNA decay, or modulating the efficiency with which an mRNA is exported to the cytoplasm. RNA-targeted interactions in the cytoplasm can regulate mRNA decay kinetics and subcellular localization, while also influencing the rates of ribosome engagement and translation.
Traditionally, mechanisms regulating gene expression at post-transcriptional levels were considered principally in terms of selective transcript recognition by RNA-binding proteins (RBPs). In eukaryotes a notable exception is nuclear splicing, where core spliceosome ribonucleoprotein complexes are recruited to 5′-splice and branch point sites via base pair complementarity to small nuclear RNA subunits.1 Nevertheless, other nuclear and cytoplasmic RNA-targeted mechanisms controlling mRNA transport, localization, degradation, and recruitment to ribosomes have canonically been defined in terms of site-specific RBP recognition.2,3 However, the discovery of microRNA (miRNA) and other endogenous non-coding RNA species revealed critical deficiencies in that paradigm, and it is now well-established that post-transcriptional control of gene expression involves targeted recognition of substrate mRNAs by both protein and RNA molecules acting in trans. Recently, advances in transcriptome-wide surveys of RBP and miRNA targeting events have demonstrated that many mRNAs are recognized by a plurality of trans-acting proteins and/or RNAs,4,5 raising the prospect that combinatorial and/or competitive relationships may exist between diverse RNA-binding partners. Furthermore, newly discovered substrate targeting and metabolic relationships between RBPs and trans-acting RNAs also indicate that extensive cross-talk may exist between these post-transcriptional gene regulatory modalities.6,7
In recent years, a paradigm for interrelationships between RBP and miRNA-directed control of gene expression has emerged about a family of RBPs collectively termed ARE/poly(U)-binding/degradation factor 1 (AUF1). In this review, we discuss the canonical functions of AUF1 in control of mRNA decay and translation via recognition of specific sequence elements in mRNA 3′-untranslated regions (3′UTRs), then extend these models of post-transcriptional gene regulation by AUF1 to include: (i) reciprocal regulatory relationships between AUF1 and miRNA synthesis, (ii) roles for AUF1 in several facets of miRNA metabolism and targeting, and (iii) a new frontier involving AUF1 interactions with long non-coding RNA (lncRNA) and viral RNA substrates. Cumulatively, these diverse functions of AUF1 suggest that its role extends far beyond directly targeting specific mRNA substrates for degradation, but instead may serve as a key integrator of post-transcriptional control, impacting expression of genes both directly and indirectly by coordinating multiple RNA-targeted regulatory pathways.
THE AUF1 PROTEIN FAMILY
The AUF1 (also known as hnRNP D) family of proteins encompasses four members generated by alternative splicing of a common pre-mRNA.8 Differences in protein size and amino acid composition result from exclusion or inclusion of exon 2 and/or 7 in the mature mRNA, encoding a 19-amino acid insert near the N-terminus and a 49-amino acid sequence near the C-terminus, respectively. The resulting four protein isoforms are named by their apparent molecular weights and designated p37AUF1 (contains neither exon 2 nor 7-encoded domains), p40AUF1 (contains exon 2-encoded domain), p42AUF1 (contains exon 7-encoded domain) and p45AUF1 (contains both exon 2 and exon 7-encoded domains) (Fig. 1). Despite the selective inclusion of distinct protein domains resulting from differential splicing events, all four isoforms of AUF1 share some common structural elements, including two tandem, non-identical RNA Recognition Motif (RRM) domains containing canonical RNP-1 and RNP-2 sequence motifs, as well as an 8-amino acid glutamine-rich sequence located C-terminal to RRM2.8,9 Also, all isoforms form stable dimers in solution.10
Fig. 1.

Domain organization of AUF1 isoforms. All contain tandem RRM domains, each with characteristic RNP-2 and RNP-1 sequence motifs (red boxes), followed by a short glutamine-rich domain. Isoform-specific sequences encoded by alternatively spliced exons 2 (yellow) and 7 (blue) are indicated.
The various AUF1 isoforms can localize to both nuclear and cytoplasmic compartments, although not equally as p42AUF1 and p45AUF1 are typically enriched in nuclei while the smaller isoforms are generally more uniformly distributed throughout the cell.9,11–13 While there remains little consensus on the mechanisms controlling the subcellular localization of AUF1 isoforms, several possibilities have been proposed. One factor that may direct AUF1 protein localization is a 19-amino acid sequence located near the C-terminus of all four isoforms that binds the nuclear transport factor transportin 1.14 However, another model proposes that sequences encoded by exon 7 interrupt a nuclear import activity found in the smaller isoforms (p37AUF1 and p40AUF1); although such an arrangement would require a distinct nuclear targeting mechanism to minimize cytoplasmic accumulation of p42AUF1 and p45AUF1.15 An alternative strategy that may contribute to the subcellular distribution of AUF1 isoforms is selective interaction with specific nuclear (scaffold attachment factor-β) or cytoplasmic (14-3-3σ) factors.12,16 The potential for AUF1 proteins to form heterodimeric structures in solution 15 further amplifies the complexity of possible localization mechanisms. Finally, activation of select cellular signaling pathways17,18 and cellular stresses including those resulting from viral infection can re-localize AUF1 proteins.19–21 In the case of coxsackievirus B3, cytoplasmic enrichment of AUF1 appears to be linked to deposition in nascent stress granules.22
RNA recognition by AUF1
Early biochemical analyses of AUF1, generally using the p37AUF1 isoform as a model, demonstrated direct, robust, and specific binding to U-rich RNA targets derived from the AU-rich elements (AREs) responsible for rapid decay of many mRNAs.23,24 AREs are potent, cis-acting determinants of mRNA stability and translational efficiency found in the 3′UTRs of many labile mRNAs, including those encoding cytokines, oncoproteins, inflammatory receptors and G protein-coupled receptors.25–27 The canonical ARE sequence is distinguished by the presence of AUUUA pentamers, frequently in multiple and/or overlapping arrangements, within a U-rich region that may span in size from 40 to 150 nucleotides.25 Besides AUF1, these sequences provide binding sites for a diverse array of factors collectively termed “ARE-binding proteins”. Depending on the specific factor recruited, the functional consequences of protein binding may stabilize or destabilize the mRNA,18,28–33 or alternatively alter its translational potential.34–36
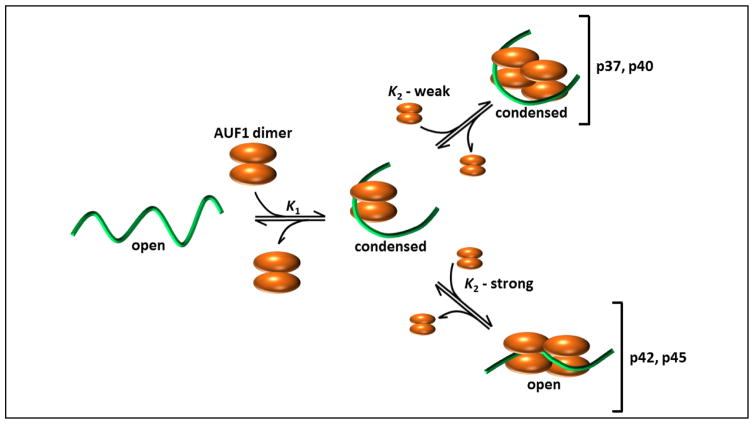
AUF1 binding to ARE-like substrates has been rigorously characterized at the biochemical level, from the perspectives of both protein and RNA sequence requirements. Studies using protein truncation mutants revealed that the RRM domains are necessary but not sufficient for high affinity RNA binding, requiring contributions from N- and C-terminal protein domains to maximize the stability of resulting ribonucleoprotein (RNP) complexes.37 Furthermore, AUF1 isoforms show different affinities for RNA targets. Specifically, p37AUF1 and p42AUF1, which lack the exon 2-encoded domain, show higher affinity for model ARE substrates relative to isoforms that contain the exon 2-encoded domain (p40AUF1 and p45AUF1).8,10 On extended RNA substrates, initial AUF1 dimer binding events can be followed by subsequent rounds of protein recruitment to yield oligomeric AUF1 complexes (Fig. 2).23,37 The proclivity of AUF1 dimers to form larger multi-subunit structures on RNA targets is also isoform-dependent, with those containing exon 7-encoded sequences (p42AUF1 and p45AUF1) forming the most stable oligomeric complexes.10
Fig. 2.
Assembly of AUF1 RNPs. Initial binding between an AUF1 dimer (orange) and an RNA substrate (green) generates an RNP complex with a P2R stoichiometry concomitant with adoption of a locally condensed RNA structure (center). Recruitment of a subsequent p37AUF1 or p40AUF1 dimer (top right) occurs with low affinity and maintains the condensed RNA fold. However, subsequent dimer binding events on p42AUF1 or p45AUF1 RNPs (bottom right) occur with high affinity and induce extended RNA conformations. Figure adapted from Ref. 55
The precise RNA requirements for AUF1 binding have been more elusive, and accumulated data suggest a degree of promiscuity among RNA target sites. Using ARE-based RNA substrates, biochemical approaches have shown that the RNA footprint necessary for maximal p37AUF1 or p42AUF1 binding affinity is approximately 30 nucleotides,10 surprisingly large for proteins that exist as dimers of 70 to 80 kDa in size. However, binding is not exclusive for canonical ARE domains, since AUF1 RNPs will form on comparably sized polyuridylate substrates with no energetic penalty,23,38 in contrast to more stringent ARE-binding factors like tristetraprolin.39 AUF1 recruitment is also weakened by localized folding of RNA targets, consistent with prominent roles for direct protein contacts with unpaired bases.40 Not all RNA contacts are base specific, however. Analyses of p37AUF1 binding to modified RNA substrates revealed that a contiguous AU-rich domain of as few as 15 nucleotides was sufficient to nucleate assembly of a high affinity (Kd < 10 nM) RNP, provided that it included a 3′-purine residue and at least 19 nucleotides 5′ of the U-rich sequence (Fig. 3). While interactions with the upstream domain only contribute a net enhancement of 0.5 – 1 kcal/mol to RNP complex stability and are largely independent of RNA sequence, these contacts are required for AUF1 to remodel local RNA structure.33 AUF1 substrate requirements resolved biochemically have also been supported by recent screens for cellular RNA targets. A predicted AUF1-binding motif elucidated by microarray analysis of RNP-immunoprecipitation reactions contained 79% A/U content across 29–39 nucleotides, although one caveat was that the motif did not explicitly register to sequences of known AUF1 target mRNAs.41 More recent photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analyses of AUF1 binding sites indicated that RNA targets were not limited to the canonical AU-rich sequences, but also showed an abundance of U- and GU-rich motifs36 consistent with the relaxed stringency suggested by biochemical studies. However, a paradoxical result from the PAR-CLIP screens was the identification of several miRNA partners for AUF1 which, measuring as small as 20 nt in length, are far shorter than the minimal ARE-based substrates required for high affinity RNP formation. Subsequent experiments revealed that one GU-rich miRNA, let-7b, bound p37AUF1 directly in vitro with a Kd of 6 nM,42 far superior to the Kd = 26 nM observed for p37AUF1 binding to a comparably sized AU-rich RNA substrate.10 How AUF1 is able to form such stable RNP complexes with let-7b and possibly other miRNA targets is unknown, but it is likely that distinct sequence and/or structural determinants within these substrates present optimal contacts for protein recognition. Unfortunately, the current dearth of crystallographic or NMR-based structural information on AUF1 RNP complexes remains a major impediment to resolving the biochemical basis for substrate selectivity by these proteins.
Fig. 3.
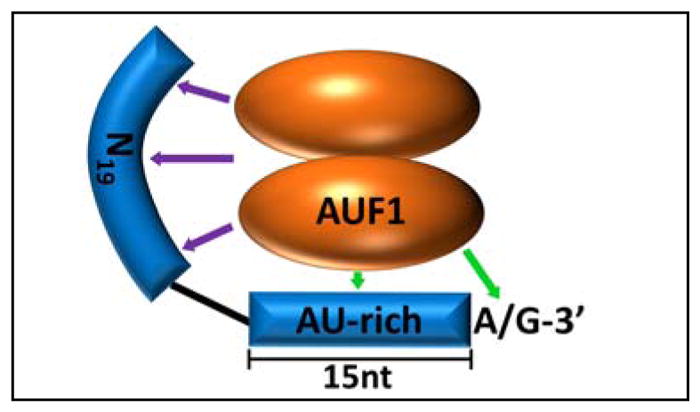
RNA domains contributing to high affinity binding of p37AUF1 to RNA substrates. Green arrows denote base-specific contacts to AU-rich RNA sequences, while purple arrows show nonspecific contacts upstream of the nucleating U-rich domain. Protein contacts with the 5′-domain are also required for AUF1-induced remodeling of local RNA structure. Figure adapted from Ref. 33
Formation of AUF1 RNPs can also have dramatic but very specific consequences on the local structure of RNA targets.10,43 Association of an initial AUF1 dimer induces a condensed conformation on RNA substrates, detected by shortening of the distance between their 5′- and 3′-termini in solution (Fig. 2). While this behavior is observed for all AUF1 isoforms, the structural consequences of subsequent binding events vary depending on the presence of the protein domain encoded by exon 7. As such, oligomeric structures formed by the smaller isoforms maintain RNA substrates in condensed states, while p42AUF1 and p45AUF1 induce extended RNA conformations as successive protein dimers are recruited.10 Current understanding of relationships between AUF1 binding and local RNA conformations is further complicated by observations that they are also influenced by protein phosphorylation events.44 While the global significance of RNA conformational changes induced by AUF1 remains unclear, an appealing hypothesis is that they may obstruct or enhance access for other trans-acting proteins or RNAs that target nearby sites on mRNA substrates. This possibility will be discussed further in the context of miRNA recruitment (below).
Modulation of AUF1-directed RNA folding by protein phosphorylation is likely only one example of an ever growing list of AUF1 functions that might be regulated by posttranslational modifications, which conceivably could have major impacts on AUF1 subcellular localization, RNA-binding activity, protein turnover, and engagement with other protein factors. Three distinct phosphorylation sites have been identified within the exon 2-encoded domain, at Ser83, Ser87, and Thr91.44,45 This domain is limited to the p40AUF1 and p45AUF1 isoforms, yet other forms of AUF1 have also been identified as phosphoproteins in cells,46,47 indicating that the catalog of phosphorylation sites remains incomplete. Furthermore, AUF1 modifications are not limited to phosphorylation, as AUF1 is also subject to proline isomerization by Pin1, which inhibits AUF1 binding to and destabilization of GM-CSF mRNA,48 arginine methylation within its C-terminal RGG motifs,49 and ubiquitination.50 This range of posttranslational modifications is not unique to AUF1, as other ARE-binding factors including HuR can exist in similarly diverse populations of covalently modified forms.51
AUF1 functions in control of mRNA turnover and translation
The major mRNA decay pathway in eukaryotes is initiated by 3′→5′ excision of the poly(A) tail, principally through the Ccr4/Not and Caf1/Not deadenylase complexes.52 The decaying mRNA is subsequently shuttled to P-bodies which contain 5′-decapping factors and 5′→3′ and 3′→5′ exonucleases.53 Deadenylation is normally rate-limiting and AREs generally promote mRNA decay by accelerating deadenylation.54 Numerous studies have associated AUF1 binding to specific mRNA substrates with accelerating their degradation; these are reviewed extensively elsewhere.53–55 This canonical function was recently supported by observations that expression of a sizeable fraction of AUF1-targeted mRNAs identified by PAR-CLIP were inversely related to AUF1 levels.36 Similarly, in AUF1 knockout mice TNFα and IL-1β mRNAs were dramatically induced in macrophages following even modest endotoxin challenge owing to defects in the degradation of these transcripts.56
The mechanism(s) by which AUF1 binding to an ARE enhances mRNA decay remain obscure, but some details have emerged. AUF1 proteins appear to possess no intrinsic nucleolytic function but rather are believed to recruit various components of the mRNA decay machinery to ARE-containing transcripts. Ancillary factors identified in AUF1 complexes include eukaryotic translation initiation factor 4G (eIF4G), poly(A)-binding protein (PABP), and heat shock proteins Hsp70, Hsc70 and Hsp27.57–59 Although specific roles for these co-purifying factors in AUF1-enhanced mRNA degradation are largely unknown, Hsp27 recruitment appears to exert a destabilizing function since ARE-containing transcripts are stabilized when Hsp27 expression is silenced.59 By contrast, Hsp70 can form direct high affinity complexes with AREs that stabilizes mRNA targets,60 suggesting that it might co-purify with AUF1 by virtue of binding at adjacent sites on RNA substrates rather than as part of an AUF1-initiated trans-factor assembly. Finally, some evidence has been presented that AUF1 might accelerate mRNA degradation by recruiting the exosome 3′-exonuclease complex61 or even targeting mRNAs to the proteasome.57 However, as neither of these activities has been shown to enhance the rate-limiting step of mRNA decay (i.e., deadenylation), it appears likely that a distinct mechanism linking AUF1 to the recruitment or activation of specific deadenylase complexes remains to be discovered, possibly analogous to the Ccr4/Not-recruiting function of tristetraprolin.62 Another intriguing possibility is that assembly of AUF1-initiated trans-acting complexes might enhance accessibility of deadenylases to the poly(A) tail by altering the stability or dynamics of PABP:poly(A) complexes.
While most examples to date support an mRNA-destabilizing role for AUF1, isolated cases have emerged contradicting that paradigm. For example, AUF1 binding is associated with stabilization of some transcripts. In a breast fibroblast cell model, SDF-1, α-SMA, TGF-β1, and IL-6 mRNAs were collectively stabilized by AUF1 as part of an IL-6/STAT3/NF-κB-driven pro-inflammatory positive feedback loop.63,64 Similarly, AUF1 silencing suppressed expression of a cohort of AUF1-targeted mRNAs identified by PAR-CLIP. These included mRNAs encoding several proteins associated with chromosome maintenance such as HP1α, centromere protein D, and DNAJ,36 although possible AUF1-dependent effects on transcription65,66 cannot yet be discounted. Other examples have indicated a role for AUF1 in regulating translation. AUF1 promotes translation of MYC mRNA by competing with the translational suppressor TIAR for a common binding site.34 AUF1 also enhances translation of the transforming growth factor-β-activated kinase TAK1, which maintains the NF-κB activation circuit in lipopolysaccharide-activated monocytes.67 Analyzing ribosome profiles of AUF1-binding mRNAs identified over 100 transcripts where ribosome occupancy was substantially increased or decreased when AUF1 was depleted.36 In 70% of these cases, no accompanying change in mRNA steady-state level was observed, indicating that modulating translation efficiency is not a rare consequence of AUF1 binding, nor need it be coupled to alterations in mRNA decay kinetics.
The examples listed above highlight a major question currently dogging the field: how can AUF1 exert such distinct functional roles on different RNA targets? An appealing model is that the consequences of AUF1 binding may be highly context-dependent. Conceivably, this could be influenced by: (i) RNA sequence, (ii) local RNA structural environment, (iii) protein post-translational modifications, (iv) proximal or cooperative binding with other RBPs or miRNAs, or (v) competition with other trans-factors. These features might also be impacted by the specific AUF1 isoform involved. For example, the PAR-CLIP datasets indicate significant overlap among RNA targets associated with each AUF1 isoform, but also substantial isoform-specific contacts.36 Other reports also highlight isoform-dependent differences in AUF1 function. Early studies indicated that a reporter mRNA containing the ARE from GM-CSF mRNA was only destabilized robustly by overexpressed p37AUF1,31 while either p37AUF1 or p42AUF1 could destabilize a reporter containing the 3′UTR of IL-6 mRNA.68 Furthermore, more recent work indicates AUF1 isoform-dependent roles in inhibiting mRNA decay. For example, IL-10 mRNA was only stabilized in lipopolysaccharide-stimulated monocytes if p40AUF1 was present,69 while stabilization of estrogen receptor α mRNA in uterine tissue was specific for the p45AUF1 isoform.70 Isoform-selective functions of AUF1 may reflect differences in RNA-binding specificity, a hypothesis supported by PAR-CLIP data showing that the various isoforms bind overlapping but distinct subpopulations of cellular RNA targets, frequently varying by over 50% in pairwise comparisons.36 However, isoform-specific variations in RNA-binding affinity, oligomerization potential, and consequences on local RNA structure may also contribute to differential regulatory outcomes (described above). Finally, current data do not assess potential roles for AUF1 heterodimers or RNA-mediated hetero-oligomeric complexes on the functional consequences of AUF1:RNA interactions. While the isoform composition of AUF1 dimers and higher-order assemblies can conceivably be regulated by relative expression levels and co-localization in specific cellular compartments, another intriguing possibility is that RNA-induced hetero-oligomers might be targeted to specific mRNA substrates by tandemly arranged RNA binding sites for distinct protein isoforms.
MODULATION OF miRNA FUNCTION BY AUF1
miRNAs are small (generally 21–24 nt) RNAs with diverse roles as trans-regulators of gene expression. They are produced from hairpin-shaped precursors71 that may synthesized within a dedicated primary transcript (pri-miRNA) or as a byproduct of other RNA processing events (reviewed in Refs. 7,72,73). Pri-miRNAs are transcribed in the nucleus, where double-stranded hairpin precursor-miRNAs (pre-miRNA) are then excised by the microprocessor complex which includes the RNase III enzyme Drosha and DGCR8, an essential co-factor. Once transported to the cytoplasm via exportin 5 and Ran GTPase, the pre-miRNA is further processed by Dicer, another RNase III protein, to yield its mature duplex miRNA form. The guide strand of the miRNA is then assembled with a member of the Argonaute (AGO) family of proteins to generate the RNA-induced silencing complex (RISC).74 This miRNA-loaded RISC complex (miRISC) is then targeted to specific mRNAs by base pair complementarity with the miRNA, frequently emphasizing the miRNA seed region, spanning nucleotides 2–7 from the 5′-end.75 Canonical consequences of miRISC recruitment are mRNA degradation and/or translational suppression, although a growing body of evidence also suggests nuclear roles for miRISC complexes in regulating transcription.76
AUF1 regulation of miRISC recruitment
Since the post-transcriptional gene regulatory effects of miRNAs require association with cognate mRNA targets, cellular mechanisms that alter miRNA:mRNA recognition could presumably manipulate this process. In recent years, collaboration and/or competition amongst RBPs and miRNAs has emerged as a critical factor in the regulated expression of many genes, and general principles are thoroughly reviewed elsewhere.5,6,77 In 2013, AUF1 was added to the growing number of RBPs that could influence miRISC binding to mRNA targets. These observations were prompted initially by in vitro screens for AUF1-binding mRNAs, when a subpopulation of candidates validated to bind AUF1 in cells was noted to be enriched in transcripts known to be regulated by miRNAs.78 The hypothesis that functional relationships might exist between AUF1 and miRISC recruitment (Fig. 4, arrow A) were supported by similar observations involving the mRNA-stabilizing, ARE-binding protein HuR. For example, HuR binding to MYC mRNA is required for let-7-directed repression of MYC translation,79 while binding to lincRNA-p21 promotes destabilization of this transcript, again involving let-7 recruitment.80 Conversely, HuR can inhibit miRNA-mediated suppression of gene expression in other contexts.81,82 Additional support for crosstalk between AUF1 and miRISC recruitment was given by observations that ARE-like sequences are enriched near functional miRNA binding sites in mRNA 3′UTRs.83 To determine whether AUF1 impacted miRISC recruitment or function on select AUF1-binding mRNAs, interactions between these transcripts and AUF1 or AGO2, the sole AGO family member with endonuclease activity,84 were measured in cells where one or both trans-factors were depleted. Intriguingly, AUF1 and AGO2 recruitment to MYC mRNA were mutually inhibitory, supporting a competitive relationship, while binding of each protein enhanced recruitment of the other on HOXB8 mRNA.78 Furthermore, for several mRNA targets rapid decay was only observed when both AUF1 and AGO2 were present, while in other cases functional outcomes were independent of each other. Subsequently, collaboration between AUF1 and miRISC-directed control of gene expression has been identified in other systems, including suppression of DNA methyltransferase 1 (DNMT1) expression in endometrial stromal cells under hypoxic stress. Here, destabilization of DNMT1 mRNA required coordinated recruitment of miR-148b and AUF1 to proximal sites within the mRNA 3′UTR.85
Fig. 4.
Functional and regulatory interrelationships between AUF1 and miRNAs. Major pathways described in the text are indicated by red arrows: [A] positive and negative consequences of AUF1 on miRISC recruitment to specific target sites on mRNA substrates, [B] AUF1-enhanced miRNA loading into RISC complexes, [C] suppression of miRNA synthesis by AUF1-targeted degradation of DICER mRNA, and [D] miRNA-directed control of AUF1 expression.
While these examples highlight positive and negative functional relationships between AUF1 and miRISC recruitment and function, the biochemical mechanisms by which these events are coupled remain unknown although several possibilities may be proposed. First, miRISC recruitment could be enhanced (or potentially even inhibited) by direct contact between AUF1 and AGO2 or other RISC components. Some support for this model may be interpreted from observations that AUF1 and AGO2 can be co-immunoprecipitated, and that co-purification is enriched under hypoxic stress concomitant with DNMT1 mRNA destabilization,85 although the possibility that these proteins are tethered by common RNA molecules cannot yet be excluded. Alternatively, reciprocal inhibition of AUF1 and AGO2 recruitment, as observed for MYC mRNA,78 could be mediated by competition between AUF1 and miRISC for cognate binding sites. Finally, the ability of AUF1 proteins to modify the local RNA structural landscape (described above) may contribute to allosteric relationships with miRISC recruitment, by exposing or occluding nearby target sites.
AUF1 as part of a global network of RBPs that can regulate miRISC function
The examples above demonstrate potential competition or collaboration between AUF1 and miRISC function, but it must also be remembered that AUF1 is only one of 25 or more proteins that bind AREs and related sequences.86 Furthermore, substantial evidence for coincident or competitive RNA-binding events involving multiple proteins has been accumulating over the past decade. Considering just AUF1 and HuR, for example, array analyses of co-purifying transcripts identified many common targets, including a subset that could bind AUF1 and HuR simultaneously87 although spatial relationships between protein binding events on individual RNA molecules could not be rigorously resolved. However, a subsequent in-cell FRET study detected both proteins in homo- and hetero-oligomeric complexes in both nuclei and cytoplasmic compartments, supporting a model whereby they may bind in close proximity on common mRNA targets.88 Other examples have identified competitive relationships between AUF1 and HuR in mRNA recognition. In vitro experiments showed identical RNA footprints for both AUF1 and HuR binding to androgen receptor mRNA.89 Both proteins also compete for a binding site within the 3′UTR of JUND mRNA, although this equilibrium is shifted in favor of HuR by the presence of polyamines.90 However, forthcoming biochemical and functional analyses of HuR and AUF1 binding events will be greatly aided by the availability of PAR-CLIP datasets for both proteins,36,91 which have provided a wealth of new model transcripts upon which their interrelationships may be interrogated. Comparisons of RNA targets recognized by each factor have revealed 6550 overlapping sites to date (Fig. 5) mapping predominantly to introns and 3′UTRs, and representing 7% of all HuR targets but 23% of all AUF1 hits.36 Notably, HuR targeted 82% of all AUF1-binding domains within mRNA 3′UTRs, particularly at U/AU-rich motifs, suggesting that it is in these regions where combinatorial or competitive roles of AUF1 and HuR are most likely to be manifested.
Fig. 5.

Distribution and overlap of RNA binding sites recognized by AUF1 and/or HuR resolved by PAR-CLIP studies. The pie chart at right summarizes the locations of RNA targets recognized by both trans-factors. Figure adapted from Ref. 36
To this point we have considered regulatory relationships between AUF1 and miRISC complexes, but also widespread competition (and possible collaboration) for AUF1 and HuR binding to common RNA sequences. Combining these themes together with known relationships between HuR and miRISC (described above) augur that miRISC function can be intimately entwined with a spectrum of RBP:mRNA interactions. A recent example of such multifactorial control was observed for bone morphogenetic protein 2 (BMP2) mRNA, which contains a 3′UTR ARE-like sequence targeted by HuR, AUF1, and miR-633. In a pre-osteoblast cell model, HuR inhibited the suppressive role of miR-633 at this element, likely by competing for binding to the target site.92 However, an elegant series of mRNA point mutations showed that AUF1 recruitment was required for miR-directed gene suppression at this site, since abrogating AUF1 binding or miR-633 base pair potential increased reporter expression, but combining these mutations had no further effect. This example shows how competition between AUF1 and HuR can direct the activity of a specific miRNA.
The regulatory potential of multifactorial RBP:miRNA networks could conceptually be profound, and manipulable by a variety of conditions. For example, alterations in the levels of individual RBPs based on tissue specificity, developmental stage, or acute changes resulting from signaling events would be expected to alter this equilibrium, potentially enhancing or restricting miRISC access. Returning to the DNMT1 mRNA example, enhanced decay owing to AUF1-coupled miR-148b recruitment under hypoxic stress was accompanied by a modest decrease in HuR expression, which likely contributed to diminution of HuR binding to mRNA target site and concomitant enhancement of AUF1 access.85 Beyond RBP availability, RNA-centric features could also conceivably influence the equilibrium between RBPs and miRISC at specific sites, including the relative affinity and positioning of diverse trans-factors on common mRNA targets (influenced by both primary and higher-order RNA structural considerations), as well as the potential effects of RNA structural remodeling by associated trans-factors. In the case of AUF1 at least, the complexity of this last regulatory mode is already amplified by its protein concentration-, isoform-, and phosphorylation-dependence (Fig. 2). Cumulatively, these concepts may provide a means by which gene-specific consequences of RBP and miRNA functions may be manifested. We anticipate that forthcoming studies will determine which, if any, of these models is correct, and will doubtless include novel mechanisms by which RBPs and miRNA can reciprocally influence their activities.
AUF1 binding to miRNAs
Beyond modulating miRISC recruitment to mRNA target sites, new evidence that AUF1 can bind some miRNAs directly indicates additional roles for this protein in miRNA biology. PAR-CLIP analyses from human embryonic kidney cells identified 14 distinct miRNA targets of AUF1 in this cell model, including 4 members of the let-7 family (let-7a-2, -7b, -7f-2, and -7i).36 Subsequent biochemical assays verified that all AUF1 isoforms directly interacted with let-7b and miR-21, both of which include UU- and UG-rich sequences,42 and in the case of let-7b demonstrated surprisingly strong affinity given the short length (22 nt) of the miRNA substrate (described above). Several functional consequences could be envisioned for RBP binding to specific miRNAs, including sequestering them from assembly into miRISC complexes or altering miRNA turnover kinetics. However, in the case of AUF1, focusing principally on binding to let-7b, a different role was identified. Silencing AUF1 expression in cells had no effect on levels of let-7b or other tested miRNAs,42 indicating that AUF1 binding was unlikely to impact their turnover. However, in the absence of AUF1, let-7b loading onto AGO2 was dramatically inhibited (Fig. 4, arrow B), which in turn abrogated its ability to accelerate decay of let-7b-targeted mRNAs. The mechanism whereby AUF1 enhances let-7b loading onto AGO2 is unknown, but in vitro experiments indicate that no additional co-factors are required. Furthermore, it is unlikely to exclusively involve AUF1:AGO2 contacts, since AGO2 loading of miR-10 (not an AUF1 target) was unaffected by AUF1. Rather, the loading mechanism appears to be coupled to AUF1:miRNA contacts, since in addition to let-7b, AGO2 association with the weak AUF1-binding miRNAs miR-21 and miR-130b was also reduced when AUF1 was silenced.42 Regardless of mechanism, these data also implicate AUF1 as a major regulator of the let-7-driven gene silencing program, which, given the diverse roles of let-7 family members in development and suppression of oncogenic phenotypes,93,94 suggest a novel mechanism for some of AUF1’s anti-proliferative properties (reviewed in Ref. 95). Finally, direct interactions with miRNAs might also be subject to competitive and/or combinatorial control by RBPs. For example, HuR also binds select miRNA targets91, and like AUF1, can form very stable complexes (Kd = 1.4 nM) with let-7b.96
Reciprocal relationships between AUF1 and miRNA synthesis
In addition to regulating the assembly and substrate targeting of select miRISC complexes, AUF1 is also a general regulator of miRNA synthesis. This role is mediated via AUF1’s canonical mRNA-destabilizing function, by targeting and accelerating decay of DICER mRNA (Fig. 4, arrow C) through interactions at multiple sites within its coding region and 3′UTR.97 In AUF1-overexpressing cells, the resulting decrease in DICER protein levels reduced accumulation of several miRNA species, including miR-24, miR-130a, miR-203 and let-7. A physiological application of this mechanism was demonstrated across an array of healthy and tumor tissues, where increased expression of AUF1 in many tumors was accompanied by diminution of DICER levels.97 A subsequent study demonstrated that AUF1-directed suppression of DICER could also be manipulated pharmacologically.98 The hypoglycemic agent metformin induced nuclear retention of AUF1, particularly the larger isoforms, via an AMP kinase-dependent mechanism. This relocalization inhibited AUF1 binding to cytoplasmic DICER mRNA, resulting in increased production of DICER protein and concomitant enhancement in the synthesis of several microRNAs, including a subset associated with senescence and aging (miR-20a, miR-34a, miR-130a, miR-125b and let-7c).98
Reciprocally, AUF1 itself is suppressed by miRNA-directed events (Fig. 4, arrow D), consistent with a model of interrelated regulation of RBP and miRNA production recently reviewed by Ciafre and Galardi.99 The tumor suppressor microRNAs miR-141 and miR-146b-5p interact with distinct regions within the 3′UTR of AUF1 mRNA and target the transcript for degradation.100 Curiously, the impact of AUF1 suppression by these miRNAs on inhibiting osteosarcoma cell proliferation and migration is based on accelerated decay of mRNA substrates that AUF1 normally stabilizes in this cell model: PDK1, which encodes an AKT-activating kinase,101 and ZEB1, a transcription factor that drives the epithelial-mesenchymal transition.102 This experimentally defined example of miRNA regulation of AUF1 was previously predicted in a computational study that highlighted an abnormally high concentration of potential miRNA target sites among mRNAs encoding ARE-binding proteins.103 In the case of AUF1, this model would appear to present a potential feed-forward cycle, where elevated miRNA levels suppress AUF1 production, which in turn enhances DICER synthesis leading to production of more miRNAs. As new studies emerge, it will be very interesting to see how this circuit is applied and controlled within the global networks regulating miRNA synthesis and function.
AUF1 INTERACTIONS WITH OTHER NON-CODING AND VIRAL RNAS
Beyond mRNA and miRNA targets, PAR-CLIP analyses identified over 1700 binding sites for AUF1 proteins within endogenous lncRNAs, including contacts at multiple distinct locations within the NEAT1, MALAT1, and XIST transcripts.36 LncRNAs are defined as RNA molecules longer than 200 nucleotides that lack protein coding roles. Like mRNAs, most but not all lncRNAs are transcribed by RNA polymerase II and subject to subsequent processing steps including splicing, 5′-capping, and 3′-polyadenylation.104 The lncRNA field is currently expanding at an exponential pace as novel examples and functions continue to be defined. However, in general lncRNAs typically play regulatory roles involving one or more of three mechanisms: as molecular decoys, guides, and/or scaffolds, which are mediated by their abilities to interact variously with DNA, RNA, and/or protein targets. In the nucleus, these functions allow lncRNAs to selectively promote or prevent assembly and/or targeting of diverse multi-subunit complexes including transcriptional regulators and chromatin-modifying activities.105,106 Additional roles influencing pre-mRNA splicing have also been detected.107 In the cytoplasm, lncRNAs perform a host of different tasks, including modulating mRNA turnover and translation kinetics as well as facilitating protein-targeted functions such as post-translational modification and degradation.108 Regulatory control of mRNA targets may involve sequestering miRNAs and/or RBPs from the transcript (termed “sponging”) or recruiting specific trans-factors. Recent data suggest that many cytoplasmic lncRNAs also associate with ribosomes, interactions that may contribute to their translational regulatory roles109 or control their decay kinetics.110
One AUF1-targeted lncRNA identified by PAR-CLIP is NEAT1, a principally nuclear RNA that plays a structural role in the formation of nuclear paraspeckles and a regulatory role in the export of specific mRNA substrates.111,112 Binding by AUF1 suppresses NEAT1 levels by accelerating its degradation, leading to enhanced nuclear retention of NEAT1-regulated transcripts.36 However, accelerated decay is not a uniform consequence for lncRNAs associated with nuclear AUF1, since levels of MALAT1 and XIST were not sensitive to AUF1 concentration, despite robust association with this protein. Future studies will have to define whether AUF1 binding imposes distinct functional consequences on these alternative lncRNA targets. A second example of an AUF1-regulated lncRNA is linc-RoR, best known for maintaining the dedifferentiated state of stem cells by sequestering specific miRNAs to prevent suppression of key pluripotency factors Oct4, Nanog, and Sox2.113 The miRNA sponge activity of linc-RoR is also responsible for its promotion of tumor development and metastatic pathways in breast cancer.114,115 By contrast to its roles as a miRNA sponge, linc-RoR binding to AUF1 appears to function as a protein sponge, sequestering AUF1 from access to canonical mRNA substrates such as MYC mRNA.116 Accordingly, suppressing linc-RoR levels concomitantly decreases MYC expression by liberating AUF1, which in turn binds and accelerates degradation of the MYC transcript. Although specific roles for AUF1:lncRNA interactions are currently limited to these few examples, the diversity of lncRNA targets recently revealed by AUF1 PAR-CLIP presages a significant expansion in our understanding of the interrelationships between AUF1 and lncRNA expression and function in the coming years.
A final class of AUF1 targets that have recently been described are viral RNAs. A growing number of cases have been reported where AUF1 binding to non-coding viral RNA sequences is associated with positive or negative consequences for the virus. For example, AUF1 inhibits the replication cycle of picornaviruses such as poliovirus and human rhinovirus, involving recruitment to a distinct structured element in the 5′UTR of the viral RNA.117,118 While AUF1 binding appears to function by preventing downstream translation of this RNA, viruses overcome this obstacle by proteolytically cleaving AUF1 using virally-encoded proteinases. AUF1 also inhibits translation and replication of the picornavirus enterovirus 71 (EV71) by a similar mechanism, involving direct binding to a specific structural feature (stem-loop 2) within its internal ribosome entry site (IRES).20 Curiously, AUF1 binding to stem-loop 2 is in competition with the RBPs HuR and Ago2, whose association promotes translation from the IRES and subsequent viral replication.119 This model is complicated further by observations that the EV71 IRES is also targeted by DICER, which generates at least four small RNAs, termed viral small RNAs (vsRNAs), from this substrate. One such product (vsRNA1) is derived from stem-loop 2 sequence, and paradoxically enhances recruitment of AUF1, HuR, and Ago2 to the stem-loop 2 binding site.120 While the precise mechanisms by which AUF1 inhibits picornaviral translation remain unknown, it is conceivable that a major contributor is AUF1’s ability to dynamically compete with antagonistic factors for common binding sites, analogous to an emerging paradigm in ARE-directed control of cellular mRNA decay and translation. However, a newly discovered role for AUF1 in replication of West Nile virus (WNV) appears to function through a distinct mechanism. Like other flaviviruses, WNV RNA requires a large-scale conformational transition in order to initiate the replication process.121 A recent report demonstrates that p45AUF1 can enhance this structural transition and activate viral RNA synthesis.122 This AUF1 isoform appears to bind at multiple locations within the WNV RNA, but with highest affinity at an AU-rich region in the 3′UTR, a domain that is required for viral RNA replication in cells. The presentation of this RNA chaperone-like activity initiated by contact with an AU-rich sequence would appear to be consistent with the intrinsic local RNA remodeling activity of AUF1 described above, which is readily observable on AU-rich RNA substrates and also presents isoform-specific features.10
Conclusion
The AUF1 family of RNA-binding proteins constitute four isoforms that canonically function to positively or negatively regulate mRNA decay and/or translation by binding AREs and related sequences within target transcripts. The biochemical mechanisms that mediate and discriminate each function remain largely unknown, but are likely coupled to mRNA and protein context-specific features which may include mRNA target sequence or structure, selectivity for ancillary factors, and competition or collaboration with other trans-factors including RBPs and miRISC complexes that bind proximal sites on the mRNA substrate. However, PAR-CLIP analyses of RNA-binding sites for AUF1 across the transcriptome have added exciting new functions to this protein family, particularly with respect to interactions with miRNAs and lncRNAs. Notably, recent studies identified several mechanisms linking AUF1 to control of gene expression directed by these trans-acting RNAs. First, AUF1 binding to select miRNAs enhances their assembly into AGO2 complexes. Second, AUF1 binding to mRNA targets can enhance or inhibit miRISC recruitment and function at nearby or overlapping target sites, roles that may be coupled to AUF1’s RNA structural remodeling activity but are also influenced by the actions of other cellular RBPs including HuR. Third, AUF1 can limit global miRNA production by suppressing DICER expression, while reciprocally being regulated itself by select miRNAs. Finally, AUF1 can also form functional complexes with some lncRNAs and viral RNAs. These interactions have already led to a variety of observed outcomes including control of lncRNA degradation, AUF1 sequestration, and viral RNA translation, although this list is likely to grow as new noncoding RNA targets of AUF1 are investigated.
Moving forward, work in the field will continue to elucidate and refine the mechanisms by which AUF1 impacts posttranscriptional gene regulatory events. Key questions that remain include: (i) what signals, cellular conditions, RNA determinants, and/or binding partners direct the various functional outcomes resulting from AUF1 binding to mRNA (and now noncoding RNA) targets? (ii) how does AUF1 enhance miRNA loading onto AGO2? and (iii) what biochemical mechanisms mediate competitive or cooperative binding and/or functional relationships between AUF1, other RBPs, and miRISC complexes on mRNA substrates? Addressing these emerging questions will significantly enhance our understanding of the dynamic and complex network of trans-factors responsible for controlling gene expression at posttranscriptional levels, and the multi-faceted roles of AUF1 in these processes.
Acknowledgments
This study was supported by NIH grant GM102428 and a pilot grant from the Marlene and Stewart Greenebaum Comprehensive Cancer Center (GMW).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest for this article.
Contributor Information
Elizabeth J.F. White, Department of Biochemistry and Molecular Biology and Marlene and Stewart, Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore MD 21201
Aerielle E. Matsangos, Department of Biochemistry and Molecular Biology and Marlene and Stewart, Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore MD 21201
Gerald M. Wilson, Department of Biochemistry and Molecular Biology and Marlene and Stewart, Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore MD 21201
References
- 1.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishore S, Luber S, Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 6.Connerty P, Ahadi A, Hutvagner G. RNA Binding Proteins in the miRNA Pathway. Int J Mol Sci. 2016;17:E31. doi: 10.3390/ijms17010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem. 2010;285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J Biol Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 12.Arao Y, Kuriyama R, Kayama F, Kato S. A nuclear matrix-associated factor, SAF-B, interacts with specific isoforms of AUF1/hnRNP D. Arch Biochem Biophys. 2000;380:228–236. doi: 10.1006/abbi.2000.1938. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, Arao Y, Omori A, Ichinose S, Nishio K, Yamamoto N, Kinoshita Y, Mita S. Identification of S1 proteins B2, C1 and D1 as AUF1 isoforms and their major role as heterogeneous nuclear ribonucleoprotein proteins. Biochem J. 2003;372:775–785. doi: 10.1042/BJ20021719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 16.He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Sun Y, Haycraft C, Palanisamy V, Kirkwood KL. MKP-1 regulates cytokine mRNA stability through selectively modulation subcellular translocation of AUF1. Cytokine. 2011;56:245–255. doi: 10.1016/j.cyto.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arao Y, Kikuchi A, Kishida M, Yonekura M, Inoue A, Yasuda S, Wada S, Ikeda K, Kayama F. Stability of A+U-rich element binding factor 1 (AUF1)-binding messenger ribonucleic acid correlates with the subcellular relocalization of AUF1 in the rat uterus upon estrogen treatment. Mol Endocrinol. 2004;18:2255–2267. doi: 10.1210/me.2004-0103. [DOI] [PubMed] [Google Scholar]
- 19.Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G, Jagdeo J, Wang T, Zhong Z, Jan E, et al. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J. 2013;27:2777–2787. doi: 10.1096/fj.12-226498. [DOI] [PubMed] [Google Scholar]
- 20.Lin JY, Li ML, Brewer G. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One. 2014;9:e103827. doi: 10.1371/journal.pone.0103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cathcart AL, Semler BL. Differential restriction patterns of mRNA decay factor AUF1 during picornavirus infections. J Gen Virol. 2014;95:1488–1492. doi: 10.1099/vir.0.064501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Lin L, Zhao W, Li X, Wang Y, Si X, Wang T, Wu H, Zhai X, Zhong X, et al. AUF1 is recruited to the stress granules induced by coxsackievirus B3. Virus Res. 2014;192:52–61. doi: 10.1016/j.virusres.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 24.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 25.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 26.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 28.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen TM, Hsu CH, Tsai SJ, Sun HS. AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay. Nucleic Acids Res. 2010;38:8061–8071. doi: 10.1093/nar/gkq717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X, Dong H, Lin C, Sheng J, Zhang F, Su J, Xu Z. Reduction of AUF1-mediated follistatin mRNA decay during glucose starvation protects cells from apoptosis. Nucleic Acids Res. 2014;42:10720–10730. doi: 10.1093/nar/gku778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zucconi BE, Wilson GM. Assembly of functional ribonucleoprotein complexes by AU-rich element RNA-binding protein 1 (AUF1) requires base-dependent and -independent RNA contacts. J Biol Chem. 2013;288:28034–28048. doi: 10.1074/jbc.M113.489559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, Kim SH, Kim HJ, Kim W, Lee HR, Jung Y, Choi JH, Hong KY, Jang SK, Kim KT. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 2014;42:3590–3606. doi: 10.1093/nar/gkt1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 38.Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 39.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 40.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon JH, Jo MH, White EJ, De S, Hafner M, Zucconi BE, Abdelmohsen K, Martindale JL, Yang X, Wood WH, III, et al. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015;29:1599–1604. doi: 10.1101/gad.263749.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson GM, Sutphen K, Moutafis M, Sinha S, Brewer G. Structural remodeling of an A + U-rich RNA element by cation or AUF1 binding. J Biol Chem. 2001;276:38400–38409. doi: 10.1074/jbc.M106509200. [DOI] [PubMed] [Google Scholar]
- 44.Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- 45.Shen ZJ, Malter JS. Regulation of AU-rich element RNA binding proteins by phosphorylation and the prolyl isomerase Pin1. Biomolecules. 2015;5:412–434. doi: 10.3390/biom5020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 47.Blum JL, Samarel AM, Mestril R. Phosphorylation and binding of AUF1 to the 3′-untranslated region of cardiomyocyte SERCA2a mRNA. Am J Physiol Heart Circ Physiol. 2005;289:H2543–H2550. doi: 10.1152/ajpheart.00545.2005. [DOI] [PubMed] [Google Scholar]
- 48.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 49.Fellows A, Deng B, Mierke DF, Robey RB, Nichols RC. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3′ UTR-dependent gene expression. Int Immunopharmacol. 2013;17:132–141. doi: 10.1016/j.intimp.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li ML, Defren J, Brewer G. Hsp27 and F-box protein beta-TrCP promote degradation of mRNA decay factor AUF1. Mol Cell Biol. 2013;33:2315–2326. doi: 10.1128/MCB.00931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1372. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2. 0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2013;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 58.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kishor A, Tandukar B, Ly YV, Toth EA, Suarez Y, Brewer G, Wilson GM. Hsp70 is a novel posttranscriptional regulator of gene expression that binds and stabilizes selected mRNAs containing AU-rich elements. Mol Cell Biol. 2013;33:71–84. doi: 10.1128/MCB.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 62.Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendrayani SF, Al-Harbi B, Al-Ansari MM, Silva G, Aboussekhra A. The inflammatory/cancer-related IL-6/STAT3/NF-kappaB positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget. 2016 doi: 10.18632/oncotarget.9633. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendrayani SF, Al-Khalaf HH, Aboussekhra A. The cytokine IL-6 reactivates breast stromal fibroblasts through transcription factor STAT3-dependent up-regulation of the RNA-binding protein AUF1. J Biol Chem. 2014;289:30962–30976. doi: 10.1074/jbc.M114.594044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pont AR, Sadri N, Hsiao SJ, Smith S, Schneider RJ. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol Cell. 2012;47:5–15. doi: 10.1016/j.molcel.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panda AC, Abdelmohsen K, Yoon JH, Martindale JL, Yang X, Curtis J, Mercken EM, Chenette DM, Zhang Y, Schneider RJ, et al. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol Cell Biol. 2014;34:3106–3119. doi: 10.1128/MCB.00423-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, Pestka S. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol Cell Biol. 2011;31:602–615. doi: 10.1128/MCB.00835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paschoud S, Dogar AM, Kuntz C, Grisoni-Neupert B, Richman L, Kuhn LC. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J Interferon Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor alpha mRNA. J Biol Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. [DOI] [PubMed] [Google Scholar]
- 71.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 74.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 75.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269–273. doi: 10.4161/rna.19354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iadevaia V, Gerber AP. Combinatorial control of mRNA fates by RNA-binding proteins and non-coding RNAs. Biomolecules. 2015;5:2207–2222. doi: 10.3390/biom5042207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Chesoni S, Rondeau G, Tempesta C, Patel R, Charles S, Daginawala N, Zucconi BE, Kishor A, Xu G, et al. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 82.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–1019. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 85.Hsiao KY, Wu MH, Chang N, Yang SH, Wu CW, Sun HS, Tsai SJ. Coordination of AUF1 and miR-148a destabilizes DNA methyltransferase 1 mRNA under hypoxia in endometriosis. Mol Hum Reprod. 2015;21:894–904. doi: 10.1093/molehr/gav054. [DOI] [PubMed] [Google Scholar]
- 86.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J Biochem. 2012;151:423–437. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- 90.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fotinos A, Fritz DT, Lisica S, Liu Y, Rogers MB. Competing repressive factors control bone morphogenetic protein 2 (BMP2) in mesenchymal cells. J Cell Biochem. 2016;117:439–447. doi: 10.1002/jcb.25290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Mondol V, Pasquinelli AE. Let’s make it happen: the role of let-7 microRNA in development. Curr Top Dev Biol. 2012;99:1–30. doi: 10.1016/B978-0-12-387038-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 95.Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Front Biosci. 2011;16:2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abdelmohsen K, Tominaga-Yamanaka K, Srikantan S, Yoon JH, Kang MJ, Gorospe M. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Res. 2012;40:11531–11544. doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noren HN, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, Becker KG, Gorospe M, de CR, Evans MK. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell. 2016;15:572–581. doi: 10.1111/acel.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ciafre SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 2013;10:935–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Al-Khalaf HH, Aboussekhra A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. J Biol Chem. 2014;289:31433–31447. doi: 10.1074/jbc.M114.593004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gagliardi PA, di BL, Primo L. PDK1: A signaling hub for cell migration and tumor invasion. Biochim Biophys Acta. 2015;1856:178–188. doi: 10.1016/j.bbcan.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 102.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Rashid F, Shah A, Shan G. Long Non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de BE, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carlevaro-Fita J, Rahim A, Guigo R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 114.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, Zhang X, Xu M, Mo YY. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cathcart AL, Rozovics JM, Semler BL. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol. 2013;87:10423–10434. doi: 10.1128/JVI.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rozovics JM, Chase AJ, Cathcart AL, Chou W, Gershon PD, Palusa S, Wilusz J, Semler BL. Picornavirus modification of a host mRNA decay protein. MBio. 2012;3:e00431–12. doi: 10.1128/mBio.00431-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin JY, Brewer G, Li ML. HuR and Ago2 bind the internal ribosome entry site of enterovirus 71 and promote virus translation and replication. PLoS One. 2015;10:e0140291. doi: 10.1371/journal.pone.0140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weng KF, Hung CT, Hsieh PT, Li ML, Chen GW, Kung YA, Huang PN, Kuo RL, Chen LL, Lin JY, et al. A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells. Nucleic Acids Res. 2014;42:12789–12805. doi: 10.1093/nar/gku952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, Liebert UG, Golbik RP, Behrens SE. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol. 2014;88:11586–11599. doi: 10.1128/JVI.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]