Abstract
In 1989, transcription-repair coupling (TRC) was first described in Escherichia coli, as the transcription-dependent, preferential nucleotide excision repair (NER) of UV photoproducts located in the template DNA strand. This finding led to pioneering biochemical studies of TRC in the laboratory of Professor Aziz Sancar, where, at the time, major contributions were being made toward understanding the roles of the UvrA, UvrB and UvrC proteins in NER. When the repair studies were extended to TRC, template but not coding strand lesions were found to block RNA polymerase (RNAP) in vitro, and unexpectedly, the blocked RNAP inhibited NER. A transcription-repair coupling factor, also called Mfd protein, was found to remove the blocked RNAP, deliver the repair enzyme to the lesion, and thereby mediate more rapid repair of the transcription-blocking lesion compared to lesions elsewhere. Structural and functional analyses of Mfd protein revealed helicase motifs responsible for ATP hydrolysis and DNA binding, and regions that interact with RNAP and UvrA. These and additional studies provided a basis upon which other investigators, in following decades, have characterized fascinating and unexpected structural and mechanistic features of Mfd, revealed the possible existence of additional pathways of TRC, and discovered additional roles of Mfd in the cell.
Graphical Abstract
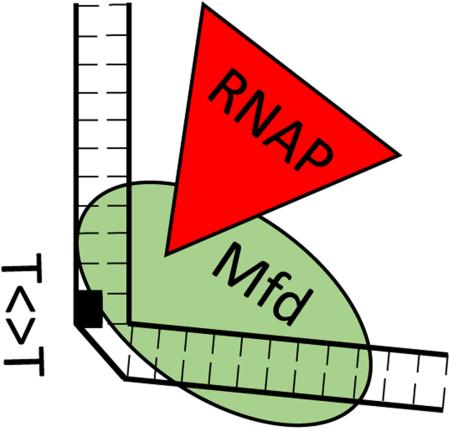
Pivotal Intermediate in Transcription-Repair Coupling. RNA polymerase (RNAP) is tethered to DNA via Mfd protein, with DNA partially wrapped around Mfd. This structure forms after RNAP becomes blocked by a template strand lesion (here, a cyclobutane thymine dimer, T<>T). Mfd removes blocked RNAP and RNA from DNA. Mfd in this resulting tethered intermediate assumes a conformation that binds strongly to the UvrA-UvrB nucleotide excision repair proteins. The next step, binding to and delivering the repair proteins to the damage, destabilizes Mfd, which dissociates from DNA. The net result is preferential, relatively rapid repair of transcription-blocking damage.
INTRODUCTION
Bulky DNA adducts such as UV photoproducts constitute potentially lethal and mutagenic impediments to replication and transcription. The NER pathway removes these lesions and restores the original DNA sequence. Some adducts, such as cyclobutane pyrimidine dimers (CPDs), are poorly recognized by NER enzymes and are repaired slowly. Others, such as 6-4 pryimidine-pyrimidone photoproducts (6-4PP), are more rapidly repaired (1).
The global NER pathway functions throughout the genome. A supplemental transcription-repair coupling pathway acts upon template strand lesions that block transcription by RNAP. In TRC, RNAP essentially aids in scanning for damage, and enables faster repair of transcription-blocking lesions, especially lesions that are poorly recognized via global NER, such as the CPD (2).
TRC in E. coli was discovered based upon the enhanced repair rate of UV damage located in the template strand during transcription (3). The transcription-dependent template strand repair was faster than repair of the non-transcribed strand which was repaired at the same slow rate as both strands under non-transcribing conditions. A transcription-repair coupling factor (TRCF) was found to be necessary for the transcription-stimulated repair. TRCF is the product of the mfd gene and thus is also called Mfd protein (4-6). Mfd protein controls a phenotype - mutation frequency decline - in which there is a decrease in the frequency of certain suppressor mutations when UV-irradiated cells are incubated, before plating, in an environment in which protein synthesis is inhibited (7,8). The phenomenon was inferred to arise from the specific repair of template strand lesions (9, 10). Knockout of mfd greatly reduces mutation frequency decline, increases the frequency of mutations induced in growing cells, and influences the location of mutations, but has only a modest effect on UV sensitivity.
NUCLEOTIDE EXCISION REPAIR IN E. COLI
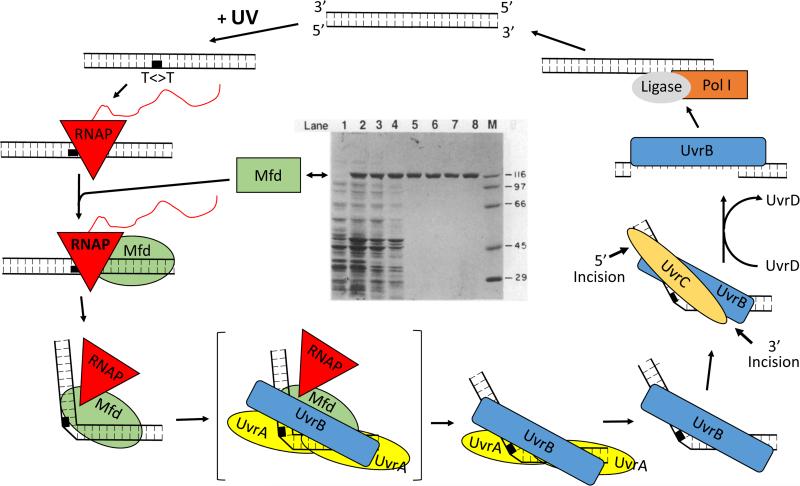
In E. coli, global NER (Fig. 1) is initiated by the action of the UvrA, UvrB and UvrC proteins. Sancar and Rupp (11) made the fundamental discovery that these three subunits recognize bulky adducts in dsDNA and remove them in the form of 12- to 13-base oligonucleotides, by incising the phosphodiester backbone of the damaged strand approximately 5 bases 3’ to the damage and 8 bases 5’ to the damage. Following the incisions, NER is completed with synthesis and ligation of a repair patch by DNA helicase II (uvrD gene product), DNA polymerase I and DNA ligase (12-14).
Figure 1.
Model for nucleotide excision repair (NER) in E. coli. Figure courtesy of Aziz Sancar ©The Nobel Foundation.
The Uvr proteins function in a stepwise manner, as shown in Fig. 1. UvrA exists as a dimer that binds with higher affinity to damaged versus undamaged DNA. UvrB has no affinity for DNA. Together they form a complex that locates damage in DNA and binds to the damaged site in a specific orientation. Footprinting and hydrodynamic studies of UvrA-UvrB preincision complexes have shown that following damage recognition, UvrA dissociates, leaving a stable UvrB-damaged DNA preincision complex. The presence of a DNaseI hypersensitive site on the damaged strand several bases 5’ to the lesion is diagnostic of the presence of UvrB in the preincision complex, and indicative of an altered DNA structure at the site of the damage. Interaction with UvrC then produces the coupled, dual incisions. The enzyme is referred to as (A)BC excinuclease or simply (A)BC with A in parentheses since UvrA is absent during the incisions. Following incision, removal of the UvrB and UvrC subunits, as well as the damaged oligonucleotide, is brought about by the action of UvrD and DNA Pol I, and repair is completed by synthesis and ligation of a repair patch (12-19).
The overall process of ATP-dependent, stepwise substrate recognition, binding and excision by (A)BC has been referred to as ‘molecular matchmaking’ (20). An interesting aspect of this multistep mechanism is that together with the controlled expression of UvrA and UvrB during the SOS response, it provides a path for specific damage-dependent excision of practically all bulky adducts without extensive nonspecific nuclease activity directed towards undamaged DNA (21).
FACTORS MODULATING REPAIR EFFICIENCY
Variations are seen in the repair rate for different DNA damages, for example, (A)BC repairs the 6-4PP about 9-fold faster than the CPD (1). Other circumstances influencing NER may be helpful to consider when modelling TRC.
In one case, (A)BC exhibits an enhanced rate of repair of CPDs when E. coli photolyase is bound to the dimer (22). E. coli photolyase binds to CPDs and photoenzymatically repairs the CPDs. In the absence of photoreactivating light, the CPD with photolyase bound appears to be much more readily recognized by (A)BC than the CPD without photolyase. Interestingly, E. coli photolyase also binds weakly to a cisplatin intrastrand crosslink, another relatively poor substrate for (A)BC, and enhances repair of the cisplatin by (A)BC (23).
Another variation in repair rate was observed when optimizing (A)BC reaction conditions in vitro. Incision activity was higher when using relatively low UvrA concentrations, and addition of undamaged plasmid to reactions greatly increased the extent of incision. These relationships likely involve the conversion of the 2UvrA-1UvrB-damaged DNA complex to the UvrB preincision complex. The lower UvrA concentration and the added plasmid may promote dissociation of UvrA from the preincision complex, perhaps by preventing UvrA from rebinding. Dissociation of UvrA from the preincision complex appears to be necessary for binding of UvrC and excision (16-19).
The action of DNA Pol I and UvrD bring about turnover of Uvr subunits (Fig. 1). This explains why, in (A)BC incision reactions conducted with excess substrate (UV-irradiated plasmid), addition of UvrD and Pol I did not enhance the rate of repair, but delayed a plateau in the number of photoproducts repaired with time (24). The moderate UV sensitivity of uvrD cells (25) has been assumed to arise from reduced Uvr subunit turnover, although it may also reflect involvement of UvrD in Mfd-independent TRC (26; discussed below).
Inhibition of NER is produced by caffeine, which sensitizes cells towards killing by UV. Caffeine, a weak intercalator, and other noncovalent DNA-binding chemicals, when bound to DNA, constitute substrates for nonproductive binding by UvrA. UvrA bound to DNA via caffeine becomes essentially ‘trapped’, and unavailable to participate in NER of covalent damage present in the same cell or in vitro reaction (17, 27). An analogous UvrA trapping mechanism has been invoked (below) to explain inhibition of NER by mutant Mfd proteins and by overexpressed Mfd.
DISCOVERY OF MFD PROTEIN
Studies of TRC in vitro began with experiments using linear, dsDNA templates that included a promoter, and a uniquely located DNA damage downstream. In reactions with purified transcription and repair proteins, RNAP was found to form a stable, stalled elongation complex at the site of a lesion in the template. Unexpectedly, RNAP stalled at the lesion in the template strand inhibited excision of the lesion by (A)BC. When the lesion was in the coding strand, it did not block RNAP and its repair was unaffected by transcription (28). It appeared that a factor required to couple transcription and repair was absent from the reconstituted reactions and that such a factor might be present in cells and cell extracts.
A method (29) for preparing cell-free extracts capable of both transcription and NER was then identified. A UV-irradiated plasmid bearing a gene (uvrC) under the control of the tac promoter was used as substrate for transcription-repair reactions. Following transcription-repair synthesis reactions with radiolabeled dNTPs, a region of the uvrC gene was digested with restriction enzymes in a manner that allowed separation of complementary strands using a low percentage sequencing gel. Results showed that the ratio of radiolabel in template/coding strands was over 4/1 in transcribing conditions, while it was near unity in non-transcribing conditions (4). Thus it appeared that in fact there was a transcription-repair coupling factor in wild-type cell extract. With this assay it was found that mfd mutant cell extract lacked TRC (5), and this was confirmed by other methods (30). The TRCF or factors absent in mfd extracts was unknown. Several candidate factors including photolyase, Rho protein, UvrD, topoisomerase I, MutL and MutS did not confer TRC in vitro (4, 28, 31).
TRC activity was purified from TRC-competent extracts by adding fractions of chromatographically separated extract to semi-reconstituted transcription/repair-synthesis reactions containing a UV-irradiated plasmid template/substrate, RNAP, (A)BC, UvrD, PolI and DNA ligase. Following successive purification steps, a large protein with apparent molecular mass of approximately 121 kDa was identified as TRCF based upon its ability to confer TRC to the reconstituted system. In addition, TRC activity was restored by adding the partially purified TRCF to extract from mfd− cells. It was concluded that TRCF was encoded by the mfd gene (5).
Mapping information was used to select a phage, presumed to possess the mfd gene, from the Kohara phage library (32). The gene was subcloned from the selected phage based upon ability to confer UV-resistance to an mfd− recA- strain of E. coli. The purified protein product of the cloned mfd gene was found to be necessary and sufficient to reconstitute TRC with purified transcription and repair proteins (6), and the sequence obtained from the cloned gene matched that of the partial N-terminal sequence of the TRCF/Mfd protein purified from extracts.
In vitro, Mfd protein was found to dissociate RNAP stalled at a template strand lesion. This is a central feature of the coupling reaction. Also, the mfd gene sequence exhibited a region of homology with UvrB, and, like UvrB, Mfd specifically interacted with UvrA. These two features of the enzyme, RNAP removal and UvrA binding, constitute the basis for the TRC model that was developed (Fig. 2): Mfd locates RNAP stalled at a lesion in the template, it removes the polymerase, and it delivers the repair enzyme to the site of the lesion (6).
Figure 2.
Model for transcription-repair coupling (TRC) in E. coli. Mfd in its “closed”, repressed state (rectangle) exhibits an altered, “open”, active state (oval) after binding to RNAP blocked by a template strand lesion. The active state Mfd exhibits higher ATPase activity, binds with higher affinity to DNA, and exhibits translocase activity. Upon binding to RNAP, a DNA structure with a 90 degree bend forms, in which the DNA is partially wrapped around Mfd. Also, RNAP and RNA are dissociated from the template by Mfd, and RNAP remains tethered to Mfd. In this conformation, Mfd binds strongly to the 2UvrA-1UvrB damage recognition complex. Binding of UvrA-UvrB to Mfd at this stage results in dissociation of Mfd and RNAP, and UvrA-UvrB are delivered to the damage site. The figure models the arriving Uvr proteins and the departing Mfd and RNAP proteins in a bracketed structure to represent a transient intermediate. The following steps of repair are the same as in transcription-independent repair. Adapted from figure courtesy of Aziz Sancar ©The Nobel Foundation.
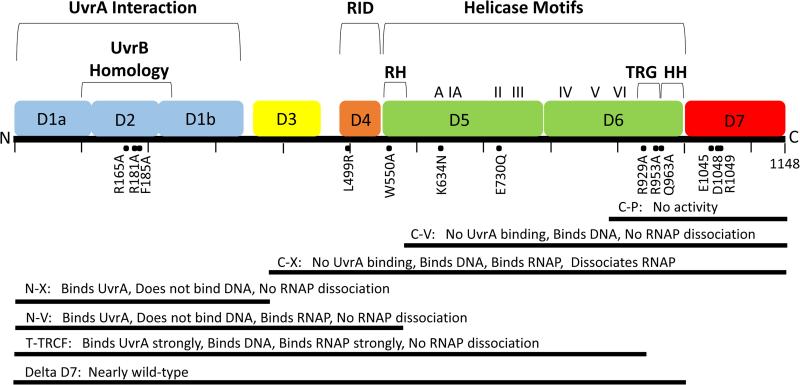
The sequence of mfd predicts a large, 130 kDa protein of 1148 residues (6). Homologies and motifs predicted from the sequence are indicated in Fig. 3. In addition to the UvrB homology region, there is a helicase motifs region (33) that has sequence similarity to the RecG protein of E. coli, which is involved in branch migration of Holliday structures (34). Functional analyses using truncated mutant proteins (Fig. 3, bottom) identified other mechanistically important areas, including UvrA and RNAP interacting regions, a DNA binding region, and a region associated with removal of stalled RNAP (discussed below). Structural analysis of the protein revealed an architecture consisting of seven discreet structural domains (D1-D7, Fig. 3), with most domains associated with specific functions (35).
Figure 3.
Structural and Functional Domains of Mfd. Upper: locations of domains D1-D7 (35) are indicated on the 1148 residue Mfd protein together with a region of sequence homology with UvrB, the RNA polymerase interaction domain (RID), the helicase motifs region where homology exists with RecG, the translocase in RecG (TRG) motif, the relay helix (RH) and the hook helices (HH). D3 is a species-specific domain of unknown function. Locations of mutations discussed in the text are indicated. Lower: Seven deletion mutants represented by bars are shown with the names of the proteins and summary information. See text for details.
FUNCTIONAL/STRUCTURAL FEATURES OF MFD
Perhaps owing to its modular structure, Mfd is amenable to experimental manipulations. Investigations of the structural modules of Mfd and their functions, described below, have had a common theme. Many of the deletion and point mutants studied have expressed greater activity than full-length, wild-type Mfd, suggesting that the wild-type protein exists in a repressed state that becomes de-repressed during TRC (35).
UvrA Interaction Module
An interaction of Mfd with UvrA was predicted based partly upon a region of sequence homology with UvrB, which was known to bind to UvrA. An Mfd-UvrA interaction was in fact observed using affinity chromatography and pull-down assays but was not detected by gel exclusion chromatography (6). In contrast, the UvrB-UvrA interaction was detected with gel exclusion chromatography (18), and therefore the UvrB-UvrA interaction was presumed to be stronger than the Mfd-UvrA interaction (36).
Sequence homology with UvrB is present among residues 82-219 of Mfd (6). In pull-down assays, UvrA interacted with full-length Mfd or with constructs containing the first 378 amino acids of Mfd (N-X, N-V, and t-TRCF, Fig. 3). A construct containing residues from 378 to the C-terminus (C-X) did not bind UvrA, but was active; it did bind DNA and RNAP, and dissociated stalled RNAP (37, 38). Similarly, a Mfd triple mutant (R165A, R181A, F185A) with mutations at important Mfd-UvrA interaction sites (see below) was found to remove RNAP stalled at a template strand lesion, but it did not support coupled repair of the lesion in vitro or in vivo (38). These data clearly show that it is the amino terminal region of Mfd that interacts with UvrA.
UvrB also interacts with UvrA via its amino terminus, and the crystal structures of the amino termini of both Mfd and UvrB exhibit three structurally similar, identically named subdomains D1A, D2, D1B (Fig. 3; 35, 38-41). The region of highest sequence and structural similarity with UvrB resides mainly in domain 2, residues 127-212 of Mfd.
The regions of Mfd and UvrA that interact with one another have been co-crystallized, and the structure of the interaction is similar to the structure seen in the UvrB-UvrA co-crystal. Two regions of Mfd were found to contribute to the interaction with UvrA, one surrounding residue F185, which is solvent exposed, and one surrounding residue R165, which is occluded by interaction with domain 7 of Mfd in the full-length Mfd structure. Occlusion of the binding site likely contributes to the weaker binding of Mfd to UvrA compared to UvrB (40).
Evidence supports an occluded UvrA binding site in solution. UvrA has been shown to have a GTPase activity that is inhibited by the addition of UvrB. Inhibition of the UvrA GTPase was also observed upon addition of either of two Mfd mutant proteins; the Mfd delta D7 mutant, which lacks the D7 domain, or an Mfd construct with three mutations (E1045, D1048 and R1049) in D7 that reduce the interaction with D2. Full-length Mfd did not inhibit GTPase, suggesting that in solution, the D2-D7 interaction represses binding to UvrA (38).
Clearly, Mfd and UvrB bind to the same site of UvrA. Initially it was reported that UvrA functions as a 2UvrA:1UvrB complex (18) and, in this scenario, both Mfd and UvrB could theoretically bind simultaneously to different UvrAs in a 1Mfd:2UvrA:1UvrB complex. It has also been reported that a 2UvrA:2UvrB complex functions as the damage sensor (42). In this case a more complex scenario must be envisaged for the transfer of the repair enzyme to the damage site by Mfd.
UvrB possesses the two RecA-like domains (domains 1A and 3 of UvrB) that together comprise a region of helicase motifs. As noted above, domains D1A, D2, and D1B of UvrB and Mfd are structurally similar. This region of helicase motifs in UvrB is involved in both ATP hydrolysis, and in binding of UvrB to damaged DNA in a preincision complex. Domain 1A of UvrB possesses a loop that has been associated with damage-specific DNA binding. The corresponding loop in domain 1A of Mfd is truncated, the corresponding Walker A and B sequences are absent from Mfd, and amino-terminal constructs of Mfd did not hydrolyse ATP. Mfd does not possess a structure homologous to UvrB's RecA-like domain 3. Thus, the amino terminus of Mfd cannot perform the damaged-DNA and ATP-hydrolysis related functions of UvrB (39).
Whether the UvrA interaction region of Mfd and especially domain 1A retains any DNA binding capacity is of interest. Amino-terminal constructs of Mfd did not bind to DNA in a gel shift assay, although weak binding may not be detected by this assay (37). Mfd is known to bind to DNA via a separate helicase motifs region (domains 5 and 6, see below). It is possible that binding of Mfd to DNA by its helicase motifs region effectively increases the concentration of DNA in the vicinity of motif 1A, leading to DNA binding at this second site. A hypothetical structure with DNA bound to two sites on Mfd and thus partially wrapped around Mfd is consistent with binding results described below.
RNAP Interacting Domain (RID)
A structure-function analysis of Mfd, utilizing a series of N- and C-terminal truncation mutants, identified a region (residues 379 – 571, Fig. 3) responsible for interaction with RNAP (37). A point mutant (L499R) in this region prevented RNAP interaction (35). This location for an RNAP interaction domain was confirmed by yeast two hybrid analysis, which also indicated that Mfd interacts with the β subunit of RNAP (35, 43). Further investigation of RNAP identified mutations in the amino terminal region of the β subunit that conferred resistance of the stalled polymerases to removal by Mfd (44).
The crystal structure of E. coli Mfd shows that residues ca. 479-545, or D4, constitute a discreet structure, described as an all Tudor-like domain (35). A Tudor-like domain such as seen in Mfd was also identified in the CarD (45) and CdnL (46) proteins of M. xanthus. Both CarD and CdnL interact with the β subunit of RNAP (45-47), and both appear to be transcriptional regulators in Myxococcus xanthus. CarD has a DNA binding domain adjacent to the Tudor-like domain. Thus D4 of Mfd, the RID, constitutes a module that has been conserved among bacteria. The Pfam database identifies the CarD_CdnL_TRCF family (PF 02559) based upon a pattern recognition/multiple sequence comparison algorithm designed to identify common domains. The algorithm is able to assemble family members having modest sequence similarity, as is the case with Mfd compared to CarD and CdnL (47). This family includes over two thousand members from 1540 species.
A co-crystal structure of the interacting domains of Mfd and RNAP β from Thermus thermophiles has been described. Interestingly, the structure of the RNAP portion of the co-crystal revealed a localized change in conformation of nine consecutive residues described as a register shift, as compared to the free RNAP structure. The register shifted residues are located in a β-sheet and are oriented differently and make contact with different internal residues. The authors suggested that the two observed conformations exist in equilibrium, and that Mfd binds to and stabilizes the novel conformation (48). It is of interest whether the novel conformation is associated with RNAP pausing and thus serves as a signal for Mfd to bind.
An analysis of available structural information placed the Mfd-RNAP interaction site at the upstream face of a stalled RNAP (35). Consistent with this location, it was reported that TRC failed to occur near the transcription start site, where sigma factor remains bound to the upstream face of RNAP and presumably blocks Mfd (6, 37, 38). Similarly, a protein bound immediately (within approximately 25-30 bp) upstream of a paused RNAP blocked Mfd from acting upon the polymerase (49), and DNaseI footprinting showed that Mfd protects a region 25 nt upstream of stalled RNAP from digestion (40).
Helicase Motifs/RecG Homology Region
Immediately C-terminal to the RID of Mfd is the helicase motifs region (residues 548-968, D5 and D6 in Fig. 3), which is presumed to be responsible for its DNA binding and ATPase (and dATPase; 49) activities (6, 35). Mfd was originally described as a weak, DNA-independent ATPase (6). Studies with other Mfd preparations reported moderately higher ATPase activity, and modest stimulation of ATPase by DNA. An Mfd with a mutated Walker B DEEH consensus sequence (E730Q) failed to hydrolyse ATP (40). A Mfd construct with the C-terminus deleted demonstrated relatively high ATPase activity that was stimulated by DNA (41, 50), thus ATPase activity may be latent, and repressed in the full-length protein.
Proteins with the helicase motifs commonly exhibit binding to nucleic acids that varies as a function of NTP binding/hydrolysis (51). Mfd binding to DNA was observed only in the presence of the slowly hydrolysed ATP-Ɣ-S, although a mutant with an altered Walker A GKT consensus sequence (K634N) that hydrolyses ATP slowly did bind to DNA in the presence of ATP. Mfd thus binds to DNA when bound to ATP and may dissociate following ATP hydrolysis. In a comparative analysis of binding to different nucleic acid structures, Mfd bound most strongly to double-stranded nucleic acid structures, bound with less affinity to structures (including bubbles) with single-stranded regions, bound relatively weakly to ssDNA, and notably, did not bind to ssRNA (37).
The interaction of Mfd with dsDNA oligonucleotides hundreds of bp in length was examined using DNase I footprinting. Binding in the presence of ATP-Ɣ-S revealed a pattern of alternating protected and DNase I hypersensitive sites, which suggested wrapping of the DNA around Mfd, and phased binding of Mfd along DNA (37). Interestingly, confirmatory results were obtained in studies using defined DNA templates immobilized at one end to a glass slide, and linked at the other end to a magnetic bead. Visualization of the magnetic bead in a magnetic field (“magnetic trapping”) allowed measurements to be made of the extension/length change of the DNA produced by the binding/action of added RNAP and Mfd (52). In these studies, ATP-Ɣ-S dependent binding of Mfd to a dsDNA produced a distortion described as “most likely due to partial, left-handed wrapping of DNA about Mfd.” The distortion in DNA increased with Mfd concentration, suggesting cooperative binding, for which there is precedent among helicase motif proteins (51). An interesting possibility is that the wrapping of DNA around Mfd could be a consequence of DNA binding to both the helicase motifs region and Motif 1A. It is also possible that the wrapped DNA structure facilitates delivery of repair proteins. There is evidence for wrapping of DNA around UvrB in a preincision complex (13, 14). However, during TRC, Mfd does not simply substitute for UvrB in a preincision complex (38)
Removal of stalled RNAP by Mfd requires the RID and the helicase motifs. Structure-function analysis has shown that neither the N-terminal UvrA interaction module (37, 38, 41), nor the C-terminal domain 7 downstream of the helicase motifs (35) are required for removing stalled RNAP. Furthermore, it has been shown that RNAP removal requires ATP (37, 40, 49).
The ability of Mfd to dissociate RNAP has been demonstrated with RNAP stalled at a DNA lesion, stalled by nucleotide starvation, or blocked by a template-bound protein roadblock to transcription (35, 37, 38, 40, 41, 44, 49, 50, 53, 54). Polymerase transcribing slowly due to nucleotide deficiency was also dissociated by Mfd, likely becoming a substrate as it transiently paused (49). In cases where Mfd dissociates RNAP stalled by a DNA-bound protein roadblock, it seems unlikely that Mfd interacts with downstream DNA, due to the presence of the protein roadblock. In a more direct test of template requirements, Mfd was found to remove stalled RNAP from a template having only 3 to 4 bp of DNA downstream from the forward edge of the RNAP (49).
It has been noted that under certain circumstances, a protein roadblock to transcription may be removed by successive RNAPs upstream ‘pushing’ the leading RNAP forward (55). RNAP stalled in this circumstance, and in the presence of Mfd, may either be removed by Mfd, or transcribe through the roadblock, depending largely upon promoter strength and roadblock protein binding affinity (56).
An important discovery was made when Mfd was incubated with an arrested RNAP. The arrested state arises at certain template sites when RNAP, paused due to nucleotide starvation, is incubated for several minutes, during which time the RNAP loses the ability to continue elongation when the missing NTP is added. In this experimental system, both RNAP and its active site “backtrack” with respect to the template and the 3’ end of the RNA. Apparently the 3’ end becomes dislocated from the active site of RNAP (57). Surprisingly, addition of Mfd to this system restored the ability of arrested RNAP to elongate, and elongation proceeded efficiently without dissociation (49). Transcription elongation factor GreB performs the same function, by stimulating RNAP to trim the 3’end of the nascent, dislocated RNA (58). Mfd restored transcription without RNA hydrolysis, and it was proposed that Mfd ‘pushed’ the arrested RNAP forward to re-establish a functional active site using a translocase activity (49). Such an activity seems reasonable since helicase-motif containing proteins, most of which unwind nucleic acids, do so by a combination of strand separation and translocation (51). Furthermore, many helicases, including possibly RecG (59), are able to translocate along duplex DNA without strand separation (51).
Mfd was then tested for translocation activity by measuring the dissociation of a triplex forming oligonucleotide (TFO), a 22-nt ssDNA specifically bound to a much longer dsDNA via Hoogsteen base pairing. Experiments with a C-terminal deletion mutant of Mfd, delta D7 (Fig. 3), showed that the mutant was quite active at translocation, while full-length Mfd was not. Interestingly, full-length Mfd did remove the TFO when it was located downstream from a stalled RNAP, suggesting that translocation is a latent activity revealed upon interaction with the stalled RNAP. Also of interest, for the Mfd mutant to remove the TFO from DNA required the presence of free DNA on one side or the other of the TFO. Conceptually, then, it appears that TFO dissociation requires that Mfd get a “head start” and progress through the TFO, starting from either stalled RNAP (full-length Mfd) or from free DNA (truncated Mfd; 38, 50).
Relationships between Mfd and the structurally related RecG protein are consistent with a translocase activity of Mfd. The co-crystal structure of RecG with a forked DNA substrate suggested that the helicase motifs region (where there exists homology with Mfd) binds to a double stranded section of the substrate, while a separate domain of RecG (with no homology to Mfd) binds to a melted area of the substrate (60). This general organization models a possible TRC intermediate in which the helicase motifs region of Mfd is bound to the dsDNA upstream of stalled RNAP, and the RNAP to which it is bound constitutes a ‘module’ of the complex that binds to the melted area of the substrate.
A more specific relationship links Mfd to RecG and translocation. A sequence of 37 amino acids at the C-terminus of the helicase motifs region of RecG from different species has been found to be so highly homologous that it is essentially diagnostic for putative RecG proteins in bacteria. This sequence, called the TRG motif (translocation in RecG) is also present in Mfd (59). Residues in the TRG motif form a hairpin structure comprised of two antiparallel alpha helices that are connected by a loop of four (RecG) or seven (Mfd) residues. The hairpin projects into the cleft between the two helicase subdomains (D5 and D6), where dsDNA is presumed to be located, and across motif VI (35). In UvrD, motif VI was shown to be involved in ATP hydrolysis and DNA binding (61). In RecG, mutation of three different residues within the TRG motif that are highly homologous among bacterial RecGs each inhibited translocation and DNA-dependent ATPase activity, and, if anything, enhanced dsDNA binding (59). The corresponding Mfd mutants (R929A, R953A and Q963A) were able to bind to stalled RNAP, but inefficiently dissociated the RNAP. Furthermore, the Mfd mutants were capable of ATP hydrolysis, and unlike wild-type protein, bound to DNA in the presence of ATP (53). Thus, in both RecG and Mfd, ATP hydrolysis appears to be coupled to translocation via the TRG hairpin structure (35).
Interestingly, early in vitro studies were conducted with a C-terminal deletion mutant of Mfd (missing residues 940-1148) that lacked the C-terminal 26 residues of the TRG motif. This mutant, termed trunc, or t-TRCF (Fig. 3), demonstrated normal DNA binding, and bound very stably to stalled elongation and initiation complexes. However, similar to the TRG mutants described above, t-TRCF did not dissociate the stalled elongation complex in vitro (37) or in vivo (53). Another C-terminal deletion mutant, delta D7 (Fig. 3), possessing the complete TRG motif but lacking C-terminal residues 998-1148, was able to dissociate stalled RNAP (35). Together, these studies also attribute RNAP dissociation activity to residues 940-998, which contain most of the TRG motif.
Thus the evidence supports a model in which the TRG structure of Mfd is responsible for a latent translocase activity that acts, after Mfd has bound to both stalled RNAP and the DNA upstream of RNAP, to pull DNA through the RNAP, which ultimately dissociates RNA and RNAP stalled at DNA damage (62).
The t-TRCF mutant exhibited an interesting property that suggests a possible additional role of the TRG motif/region in TRC. The t-TRCF mutant conferred UV-sensitivity when expressed in cells. In vitro, t-TRCF bound UvrA very tightly, prevented UvrA from binding to damaged DNA, and prevented global NER. It was concluded that “t-TRCF appears to bind to UvrA in a manner different from wild-type TRCF, and t-TRCF effectively traps this Uvr subunit, making it unavailable to catalyze repair” (37). Delta D7, in contrast, supported TRC and did not interfere with global NER and thus appeared to bind to UvrA productively (38). The exceptionally strong UvrA binding capacity revealed by t-TRCF suggests that the strong binding to UvrA is a latent activity of the full-length protein, and the TRG region may be involved in both regulating binding to UvrA, and delivering UvrA to the damage.
Two additional motifs have been described as part of the helicase motifs region, a “relay helix” (RH) and “hook helices” (HH), at the N-terminus and the C-terminus, respectively, of the helicase motifs region. These motifs may also be considered linkers between D4-D5 and D6-D7, and are shown in Fig. 3 as part of the helicase motifs region, as they were originally described (35). The RH and HH are described in more detail below. The HH, located adjacent to the TRG motif, is absent in the t-TRCF mutant and present in the delta D7 mutant and therefore could be involved with the TRG motif in regulating strong binding to UvrA, and delivering UvrA to the damage.
D7
D7 in the C-terminus of Mfd comprises a discreet structural domain located adjacent to the helicase motifs (Fig. 3; 35). The structure of full-length Mfd shows that domain D7 covers much of the UvrA interaction site in D2, and regulatory functions for D7 in modulating the latent activities noted above have been discussed (35). The delta D7 Mfd construct is capable of TRC in vitro and in vivo, indicating that D7 is dispensable for activity. Residues E1045, D1048 and R1049 in D7 are located near D2 in the crystal structure and have been proposed to be important in stabilizing a D2-D7 interaction and maintaining a “repressed” state of Mfd (38).
CONFORMATIONAL REGULATION OF TRC
The modular structure of Mfd, its latent activities, and the multiple enzymes and substrates involved in TRC strongly suggest that TRC results from a coordinated interplay among modules and substrates. Investigations have focused on the role of conformational changes of Mfd in a coordinated TRC mechanism.
As noted above, point and deletion mutants designed to stabilize the open conformation by disrupting or removing the D2-D7 interaction revealed enhanced ATPase activity. The ATPase activity revealed in the mutants was comparable to that exhibited by helicase proteins, and was stimulated by DNA, suggesting that DNA binding is also enhanced in the open conformation. More direct manipulation of the D2-D7 interaction showed enhanced binding of the open Mfd conformation to naked DNA (40). Several mutants revealed translocase activity absent in wild-type Mfd, and in some cases demonstrated enhanced rates of stalled RNAP removal compared to wild-type Mfd (38, 40). Enhanced ATPase and translocase activities were also demonstrated by an M. tuberculosis Mfd mutant lacking its C-terminus (63).
Regarding UvrA binding, the D1a-D2-D1b region was found to maintain a relatively rigid structure both in isolation and as a component of the full-length protein (41). Indirect support for structural integrity was obtained by expressing the UvrA binding module from M. tuberculosis Mfd in vivo. Expression of this domain conferred UV sensitivity; apparently it assumed native conformation, and bound to, sequestered, and trapped the cellular pool of UvrA (63). It was suggested that conformational opening of the Mfd structure, by the UvrA binding region moving away from the body of the enzyme, could simultaneously activate UvrA binding and the helicase domain (41).
More direct observations of repositioning of domains within MFD were made using small-angle x-ray scattering to examine the Mfd structure in solution. These experiments employed Mfd with a mutation E730Q in the Walker B motif which allows Mfd to bind but not hydrolyse ATP. In the presence of ADP, or in apo-Mfd, the structure resembled the “closed” conformation demonstrated by the crystal structure, whereas an “open” conformation was observed in the presence of ATP. This open conformation was characterized by D7 having moved away from D2 and the body of Mfd, as well as a more subtle repositioning of domains 1-3 outward, away from the body of the enzyme. One consequence was that the UvrA binding site was at least partly revealed in the open conformation (40).
A complementary set of experiments utilized a mutant Mfd engineered with cysteine residues to form a disulfide bond between D2 and D7 under oxidizing conditions. Formation of the crosslink was inhibited by the presence of ATP-Ɣ-S compared to ATP or no nucleotide, consistent with stabilization of the open conformation with ATP-Ɣ-S bound. D2-D7 crosslink formation was also inhibited by addition of a peptide containing the UvrA sequence of amino acids that interacts with Mfd. A parallel set of crosslinking experiments detected greater interaction between D2 and the RID upon addition of ATP-Ɣ-S or the UvrA peptide (40).
Variation of Mfd conformation arising as a function of NTP binding status seems reasonable. Helicases in general display motion along DNA as a consequence of stepwise changes in NTP binding followed by hydrolysis and nucleotide release, which produce associated changes in conformation and DNA binding properties (51). As noted earlier, binding of Mfd to DNA was dependent upon binding to nucleotide. It is interesting that with Mfd, a conformation in which D2-D7 crosslinking was inhibited was stabilized by either nucleotide or UvrA peptide binding.
While the opening of the Mfd structure may facilitate interaction with UvrA, opening was unnecessary to initiate removal of stalled RNAP, since the D2-D7 crosslinked Mfd was able to dissociate stalled RNAP. D2-D7 crosslinked Mfd did exhibit reduced ATPase and DNA binding activities, as would be expected with Mfd locked in the repressed conformation. Importantly, ATPase and DNA binding activities were restored to the D2-D7 crosslinked Mfd following the addition of a stalled RNAP. Thus, interaction with stalled RNAP appears to have created sufficient opening of Mfd to enable ATPase and DNA binding needed for RNAP dissociation. Interaction with stalled RNAP thus appears to precede, and likely contributes to formation of the open conformation (40).
Evidence for conformational flexibility was also obtained by analyzing tryptic and chymotryptic digestion patterns. Mutations in D2 expected to destabilize the D2-D7 interaction conferred protease sensitivity to Mfd, and this sensitivity was reversed upon addition of either ADP or ATP-Ɣ-S (64). The effect of ATP-Ɣ-S was relatively minor and was seen principally in the helicase motifs region, though it is curious since binding of this nucleotide would be expected to stabilize the open conformation and maintain protease sensitivity. It was concluded that Mfd exists in equilibrium between open (active) and closed (repressed) conformations, and the equilibrium is influenced by mutations and ligand binding.
The initial structural characterization of full-length Mfd described an intraprotein interaction between the N- and C-terminal ends of the helicase motifs, via the “relay helix” (RH), a linker that connects the RID with the helicase motifs, and a pair of “hook helices” (HH), adjacent to the TRG motif (Fig. 3). This interaction could provide a conduit for communicating or coordinating ligand interaction and conformational states between the RID and the helicase motifs (35). An interesting possibility is that this connection could assist in RNAP dissociation by “prying” RNAP from the template, by transmitting motion originating from the helicase motifs TRG region to the RID contact site with RNAP. It is also possible that this conduit regulates opening of the protein to facilitate UvrA binding. As noted above, the t-TRCF mutant lacking the HH and TRG motifs bound very strongly to UvrA, and the t-TRCF mutant may represent an intermediate in the TRC reaction that is regulated by these motifs and the RH-HH connection.
A further test of the importance of the RH-HH interaction was conducted by mutating a contact point in the relay helix, W550A (65). This mutation produced a protein structure with novel protease-sensitive sites, altered DNA binding properties, and exceptional DNA-dependent ATPase, RNAP-independent translocase, and stalled RNAP removal activities that were dependent upon a functional TRG motif. The novel DNA binding properties included more than one bandshifted species in DNA mobility shift assays. This multiplicity of bandshifted species is consistent with either multiple W550A proteins per DNA, or different DNA structures, each producing a different electrophoretic mobility shift. Interestingly, these results may reflect phased binding of multiple Mfds and/or DNA wrapping around Mfd such as was seen with wild-type Mfd in the presence of ATPƔS. The W550A translocation activity was so strong it displaced an initiation complex formed by an RNAP mutant that lacked an Mfd binding site. The term “molecular bulldozer” was used to refer to this apparently indiscriminant protein-DNA dissociation activity. Overall, the findings with W550A support the importance of the RH-HH intraprotein interaction in regulating structure and function. One plausible interpretation is that binding of Mfd to stalled RNAP induces a conformational change transmitted through the relay and hook helices so as to initiate opening and activation of the Mfd structure, and the W550A mutant mimics the conformation of the activated state.
Structural changes initiated upon Mfd interaction with stalled RNAP were directly examined using magnetic trapping experiments (52). These experiments measured the length/extension change of an anchored, defined DNA template upon which RNAP was stalled by nucleotide starvation. Mfd added to stalled RNAP was found to create a long-lived intermediate structure in which the transcription bubble was largely collapsed and the DNA was sharply bent (by ca. 90 degrees), and was also apparently wrapped around protein. The wrapping of DNA around Mfd in the absence of RNAP and in the presence of ATP-Ɣ-S (discussed above) suggests that the wrapping in the intermediate is also around Mfd. Both formation and resolution of the DNA-bound intermediate required ATP binding and hydrolysis by Mfd (52).
Additional magnetic trapping experiments utilized fluorescently labeled proteins and DNA probes to enable localization and movement of reactants to be measured together with DNA extension. In these experiments, RNAP and RNA were dissociated from DNA by Mfd. However, RNAP remained bound to DNA via Mfd, as an RNAP-Mfd-DNA intermediate, and Mfd was capable of translocating thousands of bp with RNAP tethered, at a rate of around 3 to 4 bp/sec (66). The RNAP tethered to Mfd apparently serves to maintain the conformational state of Mfd in the active, open conformation, that is, long-lived, DNA-bound, and translocation-competent, “possibly acting as a cofactor for the translocase” (66). This opened conformational state is also expected to possess a relatively accessible UvrA binding site.
Subsequent steps of TRC were studied by analyzing the effect of Uvr proteins on the Mfd-tethered RNAP intermediate (67). The template designed for these experiments allowed Mfd tethered to RNAP to translocate about 7,000 bp before arriving at the end of the template and dissociating. In this system, about half of the translocating complexes were “released” prematurely. Interestingly, 75% and 93% were released prematurely in the presence of UvrA and UvrA plus UvrB, respectively. These data strongly support a model in which a 2UvrA-1UvrB complex interacts with Mfd, with UvrB interacting with one UvrA, and Mfd interacting with the other UvrA. Of great interest is the ability of UvrA with UvrB to dissociate Mfd-RNAP from the template.
To examine steps more directly involved in TCR, a template with a uniquely located CPD was utilized (67). When Mfd was added to RNAP stalled at the CPD, RNAP was dissociated, and the Mfd-tethered RNAP intermediate persisted for about 548 sec. This lifetime was reduced to about 141 sec after adding UvrA and UvrB. The UvrA-UvrB preincision complex that formed in 141 sec was devoid of RNAP and Mfd. As noted above, adding UvrC to a preincision complex results in dual incisions, the central reaction of NER. Adding Mfd to a reaction with UvrA, UvrB, UvrC, and RNAP stalled at a CPD resulted in incision after about 389 to 390 sec. Uvr proteins added to naked, damaged DNA without stalled RNAP or Mfd produced incisions after about 1200 sec. The 3+ fold enhanced rate of transcription-coupled repair, compared to global repair, measured in this system, is very close to the 4-fold stimulation of repair seen when TRC was first detected in vitro and measured by incorporation of radiolabeled dNTPs during repair synthesis (4). Measurements of dissociation constants of UvrA-UvrB from the DNA-Mfd-tethered RNAP intermediate (67) indicated that the Uvr proteins bind 20 to 200 times more strongly to the Mfd-tethered RNAP intermediate than to naked CPD damage (with the naked DNA damage value obtained from the literature). These measurements support the original proposal made when Mfd was found to be the transcription-repair coupling factor (5), that Mfd binds to RNAP stalled at a template strand lesion and “constitutes a high affinity binding site for the damage recognition subunit of (A)BC excinuclease, resulting in rapid excision repair on the template strand”.
While the conformational changes leading to activation of Mfd provide a coordinated path to carry out TRC, reversion of Mfd to a repressed state following release from the DNA substrate is probably also valuable towards restraining potentially deleterious actions such as molecular bulldozing and sequestration of UvrA.
TRC MODEL
An overall scenario for TRC consistent with the findings discussed above and shown in Fig. 2 is as follows. RNAP becomes blocked at lesions located in the template but not coding strand. Stalling of the polymerase may cause the Mfd binding site of RNAP to assume a distinct “register shifted” structure that Mfd binds with high affinity. Whether or not this is the case, Mfd binds to the β subunit of stalled RNAP, at a site on the upstream face of the stalled RNAP. Interaction of Mfd with RNAP triggers opening of the Mfd structure altogether or initiates incremental opening that increases allosterically with subsequent interactions. Opening is possibly mediated via the relay and hook helices. Opening of Mfd facilitates binding to ATP. Cycles of ATP binding, hydrolysis and release produce another, possibly repetitive conformational change which may stabilize and even enhance the open conformation during the ATP-bound state. After binding to RNAP and ATP, Mfd binds to DNA upstream of RNAP. Then, hydrolysis of ATP within the helicase motifs transmits a conformational change involving the TRG motif that pulls a length of DNA through the RNAP. This translocase action leads to collapse of the transcription bubble, dissociation of RNA and RNAP from DNA, and bending of DNA so that it is partially wrapped around Mfd. RNAP remains bound to the template-bound Mfd through the RID of Mfd, and the nascent RNA is released. At this stage, Mfd is capable of binding Uvr proteins with very high affinity. The 2UvrA-1UvrB complex binds to Mfd, and is transferred to the damage site. Transfer of Uvr proteins to the damage is associated with dissociation of Mfd from DNA, which then equilibrates with the closed, repressed conformation. The dissociated RNAP equilibrates with a conformation that binds Mfd with lower affinity. The ensuing steps of incision, polymerization, and Uvr subunit and damage-containing oligonucleotide removal are presumably the same as in global repair.
A somewhat speculative aspect of the above TRC model is the manner in which the Uvr proteins are delivered to the damage. The notion of Mfd facilitating damage recognition by Uvr proteins is supported by the observation that several UvrA mutants that were deficient in their damage recognition function in global repair were capable of participating normally in TCR (38).
The TRC mechanism described above may be extended to the case in which TRC was observed at a site of damage located up to hundreds of bp downstream of a stalled RNAP (68). Repair of the downstream lesion was dependent upon the presence of RNAP stalled upstream, and was template strand specific. As discussed above, after Mfd dissociates RNAP stalled by nucleotide starvation, it translocates with RNAP tethered, albeit at a slow rate of around 4 bp/sec (66, 67). Translocation by the RNAP-tethered Mfd was found to be in the downstream direction. Presumably, the TRC reaction ensues upon encountering a lesion in the template strand downstream.
The finding of TRC of damage downstream of stalled RNAP raises the issue of DNA template/substrate requirements for Mfd activities. In fact, Mfd requires essentially no DNA template downstream of stalled RNAP to remove RNAP. Interestingly, a different template/substrate requirement was seen for the repair component of TRC. Early studies demonstrated that TRC occurred in vitro with a UV-irradiated plasmid whether it was supercoiled, relaxed or linear (31). On the other hand, TRC failed to occur with a 138-bp linear duplex with a unique, centrally-located psoralen monoadduct downstream from a promoter. Repair of the psoralen adduct was inhibited by transcription, and Mfd overcame the inhibition, but did not stimulate repair (6). This lack of stimulation seemed possibly related to the small size of the substrate, which led to an experiment using as substrates UV-irradiated, linearized plasmids restricted to give downstream ends 102, 401, or more from the transcriptional start site. Interestingly, it was found that TRC occurred with these substrates - except for regions within about 87 to 90 bp from the downstream end. That is, 87 to 90 bp of DNA between the damage and the downstream end of the duplex was required for TRC (31).
A related phenomenon was observed in experiments using the magnetic bead-tethered DNA template assay, in which the lifetime of Mfd bound to DNA after dissociating RNAP was measured (67). Experiments were done with two templates, 1kbp and 2 kbp in length. The intermediate lifetime was shorter when the smaller template was used and the Mfd-RNAP intermediate was closer to the end of the template. Thus a nearby downsteam DNA terminus may destabilize the (damaged DNA-Mfd-tethered RNAP) intermediate and cause the intermediate to dissociate before Uvr protein binding and repair.
From the above observations, the question arises how does Mfd ‘sense’ the presence or absence of downstream DNA? Helicase proteins in general are known to function in both monomeric and oligomeric states, function cooperatively in the oligomeric state, and coat DNA (51). Phased and cooperative binding of Mfd to DNA has been observed in defined experimental settings, and oligomerization of Mfd is a possible mechanism by which it could sense downstream DNA. However, there is no direct evidence for Mfd sensing downstream DNA in the context of TRC. As an aside, it would be interesting to find out if protein binding and DNA metabolism downstream of a template strand lesion also destabilize the (damaged DNA-Mfd-tethered RNAP) intermediate, and whether any such effects are relevant in vivo.
ALTERNATIVE TRC PATHWAYS
Interestingly, additional pathways for TRC have been proposed. A motivating factor for seeking alternative pathways has been the modest UV sensitivity of mfd− cells. Additional inadequacies regarding the idea of Mfd-dependent TRC as the sole TRC pathway have been reported. In one such study (69), the rate of repair of individual CPDs in wild-type and mfd− cells was measured in two regions of the transcribed stand of lacZ: an initially transcribed region “A” corresponding to approximately +1 to +34, and a region”B” of similar length immediately downstream. Region “A” contained the area where the lac repressor binds and protects the template from DNaseI in vitro (70), and repression of repair in this Region “A” was relieved by addition of IPTG, as would be expected if binding by the lac repressor were to inhibit repair. Stimulation of repair in region “B” was seen in wild-type cells following IPTG addition, as expected (3), although stimulation was modest. Surprisingly, IPTG also stimulated repair in region “B” in mfd− cells, and this stimulation was specific to the transcribed strand (69).
The original discovery of TRC in E. coli cells was made in the lac operon by measuring repair not at the single nucleotide level, but the cumulative repair across several kbp of each of the two DNA strands (3). The same method was used more recently using a lac operon in which the lac promoter was replaced with a T7 promoter, and T7 RNAP expression was controlled by IPTG. Surprisingly, TRC was seen in this system (71). Furthermore, TRC was absent in similarly constructed mfd− cells. The TRC, and especially the Mfd-dependence were unexpected, since earlier in vitro experiments had shown that T7 RNAP stalled at a lesion inhibited repair by (A)BC, and Mfd did not remove the stalled T7 RNAP or reverse the inhibition of repair (6). The authors concluded that a cellular factor in addition to Mfd, absent from the in vitro experiments, was required for T7 RNAP-dependent TRC (71).
The existence of an Mfd-independent TRC pathway may be invoked to explain another observation. An RNAP with a point mutation R529C in the β subunit was found to confer a TRC-deficient phenotype. However, this mutation is not close to the site that is known to interact with Mfd. The mutant RNAP was found to transcribe normally in vivo, and was able to form a stable, stalled elongation complex that was removed by Mfd in vitro (72). The existence of the mutant suggests that the wild-type RNAP interacts with another factor besides Mfd for TRC, although it is also possible that the mutant RNAP impedes the delivery of Mfd or the Uvr proteins to the damage site, or it may not form a stable Mfd-tethered RNAP intermediate.
NusA-Dependent TRC
A specific, Mfd-independent TRC pathway has been proposed, and it involves the transcription termination/antitermination factor NusA, which is known to bind to RNA and RNAP, and is associated with RNAP during transcription. Evidence exists for roles of NusA in responding to damage. One role involves translesion DNA synthesis, which is upregulated as part of the SOS response. An interaction between NusA and DinB, one of the translesion polymerases in E. coli, has been observed and may be important in this role. NusA also interacts with UvrA, and epistasis group analysis and other supporting information suggest that NusA participates in (A)BC-dependent repair, and in RNAP-dependent, Mfd-independent repair (73).
UvrD-Dependent TRC
Another study proposed that not only NusA, but UvrD is involved in an Mfd-independent pathway of TRC (26). This study confirmed the observation that UvrD interacts with RNAP (74), and showed unexpected effects of UvrD on transcription. UvrD promoted transcriptional pausing, and more importantly, it promoted backtracking of the transcription bubble within a stalled RNAP. In vivo footprinting showed that the presence of UvrD shifted the transcription bubble in a stalled RNAP about 2 nt in the upstream direction. In vitro experiments showed that incubation of stalled RNAP with UvrD caused the 3’ end of the nascent transcript to become susceptible to cleavage up to about 16 nt or more from the 3’end following addition of GreB (26). NusA also promotes backtracking of RNAP. In one in vitro experimental setting, it was shown that NusA caused the leading edge of stalled RNAP to recede by about 2 nt in about 25% of the population of stalled polymerases (75). It was claimed that NusA and UvrD function synergistically in backtracking RNAP. As evidence, NusA and UvrD were shown in vitro to produce more transcriptional pausing together than individually (26).
Regarding TRC and UvrD, in vitro DNA repair reactions were conducted with a defined, damaged DNA template and purified proteins. The results confirmed that RNAP stalled at a lesion (CPD) inhibits (A)BC excinuclease (6, 38), and most importantly, addition of UvrD reversed the inhibition (26). It was concluded that UvrD caused backtracking of RNAP along with the transcription bubble, exposing the damage to NER enzymes. It was suggested (but not shown) that UvrD not only reverses RNAP inhibition of repair, but also enhances repair, as is seen with Mfd-dependent TRC (26). This suggestion was based upon reports of an UvrD-UvrB interaction (76, 77; see Fig. 1), and the NusA-UvrA interaction noted above.
The distance of backtracking produced by UvrD and NusA, described above, seems limited; however, different transcription-blocking lesions advance different distances within the leading edge of RNAP towards the active site, and in some cases, limited backtracking could significantly uncover the lesion (78).
Pushing and Pulling Stalled RNAP
The backtracking functions of UvrD and NusA are, in a sense, opposed by transcription factors GreA and GreB, which re-activate transcription by inducing stalled RNAP to cleave the 3’ end of the nascent transcript. It was predicted that GreA and GreB would inhibit repair by opposing the role of NusA and UvrD in backtracking and revealing lesions hidden by RNAP. To investigate this possibility, NER of a template strand undergoing transcription in E. coli cells was measured. Elevated repair was observed in greA greB double mutants compared to wild-type cells, thus demonstrating the expected anti-repair effect (26).
Studies of the anti-backtracking/anti-repair activity of GreA and GreB were extended using survival assays (26). The greA greB double mutation did not confer enhanced UV survival compared to wild-type cells, as predicted from the repair experiment, but the greA greB double mutation did confer enhanced UV survival in the uvrD, the nusA, and the mfd mutant backgrounds. Thus, at face value, considering survival to be a simple function of RNAP positioning at a lesion, a reasonable interpretation would be that in vivo, GreA-GreB can overcome backtracking by NusA in uvrD- cells, overcome UvrD in nusA- cells, overcome both NusA and UvrD in mfd− cells, but curiously, cannot overcome backtracking in wild-type cells with Mfd present. In a related survival test, a sublethal concentration of chloramphenicol was added to DNA-damaged cells to effectively decrease the propensity of successive ribosomes to ‘push forward’ the leading RNAP stalled at a lesion. Chloramphenicol conferred enhanced survival in an uvrD- background and conferred minimal or no enhanced survival in a wild-type background. It was argued that these results together showed that “UvrD competes with anti-backtracking factors during genotoxic stress to promote NER” (26).
The Mfd-induced forward-translocation of stalled RNAP is similar to the effect of GreA and GreB in the sense that it theoretically opposes the action of UvrD, and consequently it was proposed that Mfd is an anti-repair factor, similar to GreA and GreB. This reasoning was used to explain why, in an uvrD background, the absence of Mfd conferred enhanced survival in cells treated with UV or mitomycin C (26). (In UvrD- cells, by the above reasoning, Mfd would enhance killing by opposing the backtracking effect of NusA.) This backtracking - opposing effect of Mfd was also invoked to explain why overexpression of Mfd sensitized otherwise wild-type cells to DNA-damaging treatments (55), although an alternative explanation is that overexpressed Mfd inhibits NER by an UvrA sequestration and trapping mechanism. Although Mfd expressed in its repressed conformation would not efficiently bind UvrA, overexpression to a sufficient concentration could promote UvrA binding.
The Stringent Response in TRC
Another proposed Mfd-independent pathway for TRC involves the stringent response (55). During the stringent response, ppGpp accumulates as a result of the action of the enzymes RelA and SpoT. Elevated levels of ppGpp destabilize RNAP, inhibit initiation, and may also inhibit elongation. The end result is that ppGpp reduces the number of RNAPs stalled behind an RNAP blocked by nucleotide starvation or DNA damage (79, 80). DksA is also important in the stringent response, it binds to RNAP and potentiates the effect of ppGpp (81). DksA, ppGpp, RelA and SpoT have been reported to contribute to TRC either directly or indirectly by promoting backtracking of RNAP blocked by DNA damage (55). In vitro, addition of ppGpp alone was shown to directly induce backtracking of stalled RNAP, and it was suggested that ppGpp and UvrD function together in backtracking RNAP in vivo, since induction of ppGpp in vivo did not induce backtracking unless UvrD was expressed. The reduced number of RNAPs backed up at lesions produced during the stringent response would indirectly facilitate backtracking in a manner analogous to chloramphenicol reducing the number of ribosomes, as described above.
An important experiment measured repair in the two strands of the induced lac operon in wild-type and relA-spoT- double knockout cells. Increased repair of the template strand of the lac operon, compared to the coding strand, was seen in control wild-type cells, but increased repair of the template was absent from relA-spoT- cells (55). The repair data is consistent with the involvement of RelA and SpoT in enhanced repair where RNAP becomes stalled. In general, mechanistically, backtracking could provide a means to overcome inhibition of NER produced by stalled RNAP, and may be responsible for the apparent “push-pull” effects of stalled RNAP positioning on repair and survival discussed above. Overcoming inhibition of repair by stalled RNAP and enhancement of repair where RNAP is stalled are two different things, and the repair data with relA-spoT- cells are notable as evidence for Mfd-independent enhancement of repair, although a mechanism for this effect is unclear.
In survival assays, sensitivity to UV and other DNA damaging agents was observed in the relA- spoT- double mutant, and in the dksA mutant (55). uvrD (25) and nusA (73) mutations, in wild-type backgrounds, also conferred moderate UV sensitivity, in contrast to mfd mutants. However, the survival data are not completely consistent with the scenario in which pushing or pulling of RNAP and consequent hiding or revealing DNA damage corresponds with survival outcome. Notably, it was concluded from epistasis analysis and in vivo backtracking results that UvrD, and RelA-SpoT act in the same pathway (55), and functional data suggested that UvrD and NusA acted together (26). Although all four of these are backtracking factors, the mfd mutation conferred enhanced survival in UV-irradiated uvrD- cells (26), but sensitized nusA- cells (73), and relA- spoT- cells (55) to killing by UV. Inconsistent survival data obtained when the mfd mutation was introduced into different backtracking mutants suggests that the observed phenotypes reflect more complex processes. Clearly, as discussed above, NusA and UvrD have non-TRC roles in response to DNA damage. DksA, RelA, SpoT and ppGpp may also influence survival through alternative pathways (80). Thus, additional characterization will be valuable to establish the existence and role of alternative TRC pathways and make it possible to predict the outcome of these numerous factors targeting stalled RNAP in wild-type cells. It will be interesting to discover if alternative TRC pathways exist for not simply overcoming the inhibition of repair by stalled RNAP, but for RNAP-stimulated repair, such as is seen in the TRC pathway catalyzed by Mfd (4, 38).
The SOS Response and UvrD-Dependent TRC
It was suggested that under SOS conditions, UvrD provides the predominant pathway for TRC (26). Under non-SOS, basal conditions, the amounts of UvrD and RNAP were calculated to be approximately equimolar. However, one UvrD per stalled RNAP was deemed insufficient for backtracking, and a sufficient amount was expected to be produced during the SOS response (26). Consistent with this role of the SOS response is the finding that while the mfd− mutation conferred only modest UV sensitivity in a wild-type background, mfd− conferred substantial UV sensitivity in the recA- background (6) in which induction of UvrD is expected to be blunted. An effect of UvrD on TRC was not observed in early biochemical assays of TRC using wild type or mfd− cell extracts (6, 37) perhaps because extracts were made from cells that were not undergoing the SOS response and thus had only basal levels of UvrD (6, 37).
Uvr proteins are also induced during the SOS response, which likely influences rates and pathways of repair. In an effort to examine which Uvr subunit was limiting in TRC, in vitro reactions were performed with a plasmid template/substrate, wild-type cell extract, and supplemental, purified Uvr proteins (31). Adding a low amount of purified UvrA to such a reaction stimulated both transcription-coupled and global repair, but higher amounts of UvrA specifically inhibited TRC while having a modest additional stimulatory effect on global repair. The anti-coupling effect of UvrA was overcome by adding a high concentration of Mfd. UvrA is complexed with UvrB in vivo (18) and it appeared that a large amount of added UvrA, in the absence of UvrB, was in dimer form, and the UvrA dimer inhibited coupling by interacting with Mfd. On the other hand, adding exogenous UvrB to extract-catalysed TRC reactions stimulated both TRC and global repair. Combinations of exogenous UvrA and UvrB were not studied, but would be important since both UvrA and UvrB are induced in the SOS response. It would also be important to directly assay the repair capacity of wild-type and mutant extracts made from unirradiated and UV-induced cells. The experiments involving supplementing extract-catalysed TRC reactions with purified proteins were completed by testing addition of purified Mfd or UvrC. Both were without effect. It was concluded that UvrA and UvrB, and probably also the turnover of the UvrA subunit, were limiting factors in the coupling reaction in vitro (31).
ROLE OF TRANSCRIPTION RATE IN TRC
Mutagenesis studies have suggested a role of transcription levels on TRC. Ito et al. (82) observed two patterns of strand bias for mutation induction following treatment of cells with UV and alkylating agents. Among a set of target genes called class I, mutations induced by damage in the nontranscribed strand were more prevalent, and this is the expected consequence of TRC. Class II genes showed the opposite prevalence. Interestingly, a region from the strongly transcribed rpsL (class II) gene exhibited reduced strand bias for mutations induced by MNNG when the gene was expressed from the relatively weak lac promoter. The authors stated that “an effect of preferential repair might be concealed under conditions where the target sequence is transcribed very frequently.”
An in vivo study of repair of the tyrT tRNA gene suggested an inverse relation between transcription rate and TRC (83). This study showed Mfd-dependent repair of the tyrT gene in stationary phase cells in minimal media, and the absence of this preferential repair in log phase cells in rich medium. Stationary phase cells in minimal media approximates the culture conditions in which cells undergo mutation frequency decline. Thus, the in vivo repair data indicate, as previously suggested (36), that there is sufficient transcription of tRNA to support TRC under the Mfd condition, and furthermore show that the high level of transcription associated with log phase growth conditions is associated with inhibition of TRC.
In our own in vitro studies, we observed a negligible level of TRC in a tRNA gene, and this diminished TRC was attributed to inhibition by a high level of transcription (Selby and Sancar, not shown). Under the same conditions, robust Mfd-dependent TRC was observed downstream of the tac promoter, and modest TRC was observed in the tet gene. The result with the tet gene was assumed to reflect a very low level of transcription (4). The inhibition of repair associated with strong transcription has been suggested to result from successive RNAPs interrupting Uvr protein function (36), and successive RNAPs may also interfere with the Mfd-tethered RNAP intermediate formed in TRC. Overall, from the available data, it appears that relatively high and low transcription levels may not fully support TRC, and further study of this parameter seems warranted.
TRC- AND NON-TRC ROLES OF MFD
Mfd has no essential function, as cell lines with a disrupted mfd gene are viable (37, 84). mfd− cells demonstrate an elevated frequency of induced mutations, and modest sensitization to UV killing (7, 8, 26, 37, 85, 86); however, mfd mutant cells have been reported to be substantially more sensitive than wild type cells to killing by nitrofurazone (73), although this was not confirmed (55). A role for Mfd in stationary phase mutagenesis in B. subtilis has been reported (87). In E. coli, although TRC in stationary phase has been observed (88), stationary phase mutagenesis was not dependent upon Mfd (89). To date, Mfd is the only factor known to contribute to the mutation frequency decline response in E. coli. This response is NER-dependent (8) and is likely a result of strand bias in NER (9, 10, 90).
Oller et al. (86) identified a causal role of Mfd in strand bias in UV-induced mutagenesis among dividing cells. In a study of a region of the lacI gene, mutants were isolated and sequenced, and the presumptive dipyrimidine damage sites responsible for each mutation was identified. Among these, on average, 4.5-fold more of the premutagenic sites were observed in the template versus the nontranscribed strand in mfd− cells, and 3.2-fold more premutagenic sites were observed in the nontranscribed versus the template strand in wild-type cells. A comparable strand bias in UV-mutagenesis and its association with Mfd was also observed in irradiated B. subtilis (91). The findings are consistent with mutations being more prevalent in the template strand in mfd− cells because the stalled RNAP blocks repair, and more prevalent in the nontranscribed strand of wild-type cells because the template is preferentially repaired.
Mfd participates in catabolite repression of the hut and gnt genes in B. subtilis. In these genes, the catabolite repressor binding element is located downstream of each promoter, and Mfd presumably facilitates removal of RNAP stalled at these elements due to the repressor CapA binding, since repression is relieved in the mfd mutant (92). Mfd was found to have an analogous role in repression of the ybgE gene by the B. subtilis transcription factor CodY, which also binds to an element downstream from the promoter (93). CodY is involved in regulation of over 100 genes, and a more recent study identified over 300 CodY binding sites in the genome. Interestingly, nearly half of the CodY binding sites are within coding regions, where a roadblock mechanism of repression susceptible to Mfd action is possible (94). Several of the binding sites within coding regions were tested directly by reporter assay and did in fact repress transcription in vivo.
RNAP removal by Mfd is utilized in the mechanism whereby phage HK022 excludes superinfecting phage lambda. The Nun protein of HK022 binds to specific sites in nascent lambda RNA, which causes transcription to stall, and leads to RNAP removal by Mfd (95).
In all organisms, transcription presents a potential block to replication, and factors that regulate stalled RNAP such as Mfd, GreA, and DksA may participate in resolving such conflicts (79, 80). Mfd was reported to displace RNAP in the case of a head-on collision with replication, thereby facilitating replication in E. coli (96). Another study found that when replication overtakes an arrested RNAP from the upstream direction, double stranded breaks are produced, and factors including Mfd, which overcome or prevent RNAP arrest, also prevent the strand breaks (97). It was suggested that the role of Mfd in reducing the number of induced mutations in cells is to some degree a consequence of eliminating stalled RNAPs that are a potential source of double strand breaks and ensuing mutations (26). However, in the study described above (86), the UV-induced mutations in the lacI gene of wild-type and mfd− mutant cells varied principally in the quantities and locations of dipyrimidine-targeted mutations. A unique spectra associated with double-strand break repair was not identified in wild-type mfd+ cells.
A number of the roles of Mfd described above rely solely upon its RNAP displacement function. In this light it is worth noting that organisms with Mfd but no UvrA, B, or C have been identified. Further, the Mfds in these organisms lack domains D1 and D2 of the UvrA interacting region. Thus the primary if not sole function of Mfd in these organisms appears to be dissociation of stalled RNAP (65).
The induction of UvrA during the SOS response is followed by its degradation by a protease, ClpXP. The half-life of UvrA degradation was found to decrease from about 70 min to about 150 min in mfd− cells, suggesting that Mfd facilitates the degradation of UvrA. UvrB was found to have the opposite effect, stabilizing UvrA (98).
CONCLUSIONS
The initial characterization of Mfd protein and TRC in vitro in the Sancar lab provided a valuable framework for further investigation, and the initial, overall model of TRC has been largely confirmed in the ensuing decades. The protein has been amenable to experimental manipulation, and further investigation has revealed amazing and unexpected findings. Notably, Mfd posseses discrete structural domains, including a domain that interacts with UvrA, a domain that interacts with RNAP, a helicase domain with ATP-dependent DNA binding and translocation activities, and an autoinhibitory domain. Binding of Mfd to stalled RNAP induces conformational changes that reveal otherwise repressed activities. Mfd activated by binding to blocked RNAP then binds ATP, then DNA, and then dissociates RNAP and RNA from the template by means of a latent translocase activity. The dissociated RNAP remains bound to Mfd and functions to maintain the active conformation of Mfd, which, unlike Mfd in the repressed conformation, binds Uvr proteins with high affinity. Interaction of the Uvr damage recognition proteins with RNAP- and template-bound Mfd destabilizes the interaction of Mfd with the template, and delivery of Uvr proteins to the damage is associated with release of RNAP-bound Mfd from the template. In a situation in which RNAP stalls upstream of a template strand lesion, Mfd dissociates the polymerase, and with RNAP tethered, translocates to the template strand lesion, and repair ensues.
The two main steps of TRC are, first, overcoming inhibition of repair by RNAP stalled at DNA damage, and second, stimulation of repair of the transcription-blocking damage. The stalled RNAP displacement activity of Mfd is relatively well characterized, and Mfd appears to be the only cellular factor that efficiently removes RNAP blocked by DNA damage. The stimulation of repair where RNAP is blocked is clearly produced by the action of Mfd, and may be a process uniquely performed by Mfd.
The biochemical evidence characterizing the involvement of Mfd in transcription-stimulated repair is unambiguous. Evidence at the cellular level also indicates that Mfd plays a role in TRC in vivo. This evidence includes direct measurements of repair, the Mfd phenotype, and effects of Mfd on the strand bias of mutagenesis. However, observations inconsistent with Mfd as the sole mediator of TRC, and the effects of NusA, UvrD, and the stringent response regulators on stalled RNAP suggest the possibility of alternative routes for TRC, depending upon cellular conditions following DNA damage. The evidence linking these alternative mediators to TRC is largely indirect, and it is not clear whether they may participate in Mfd-dependent TRC, or bring about TRC by Mfd-independent mechanisms. Study of these factors will be important to clarify their role not only in the stringent response, but also under circumstances that efficiently induce the SOS response (high dose, lesions that effectively block replication), and with high levels of transcription. The assortment of potential consequences of RNAP stalling at DNA damage, the growth conditions in which bacteria may encounter various DNA damages, and the varied levels of expression of each gene make it challenging at this time to confidently predict in detail how E. coli will repair its genome under any given circumstance.
ACKNOWLEDGEMENTS
Dr. Yi-Ying Chiou generously provided constructive criticism, and Dr. Wentao Li stimulated discussion on the topic. Supported by NIEHS grant RO1ES-027255 to A. Sancar, who also provided valuable insights and reference material.
Abbreviations
- Mfd
mutation frequency decline protein
- TRC
transcription-repair coupling
- TRCF
transcription-repair coupling factor
- NER
nucleotide excision repair
- RNAP
RNA polymerase
- CPD
cyclobutane pyrimidine dimer
- 6-4PP
6-4 pyrimidine-pyrimidone photoproduct
- RID
RNA polymerase interacting domain
- TFO
triplex forming oligonucleotide
- TRG
translocation in RecG
- HH
hook helices
- RH
relay helix
Footnotes
This article is part of the Special Issue highlighting Dr. Aziz Sancar's outstanding contributions to various aspects of the repair of DNA photodamage in honor of his recent Nobel Prize in Chemistry.
REFERENCES
- 1.Svoboda DL, Smith CA, Taylor J-SA, Sancar A. Effect of sequence, adduct type, and opposing lesions on the binding and repair of ultraviolet photodamage by DNA photolyase and (A)BC excinuclease. J. Biol Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 2.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 4.Selby CP, Sancar A. Gene-and strand-specific repair in vitro: Partial purification of a transcription-repair coupling factor. Proc. Natl. Acad. Sci. USA. 1991b;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: In vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 7.Witkin EM. Time, temperature and protein synthesis: a study of ultraviolet-induced mutations in bacteria. Cold Spring Harbor Symp. Quant. Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Witkin EM. Radiation induced mutations and their repair. Science. 1966;152:1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 9.Bockrath RC, Palmer JE. Differential repair of premutational UV-lesion of tRNA genes in E. coli. Mol. Gen. Genet. 1977;156:133–140. doi: 10.1007/BF00283485. [DOI] [PubMed] [Google Scholar]
- 10.Bockrath R, Barlow A, Engstrom J. Mutation frequency decline in Escherichia coli B/r after mutagenesis with ethyl methanesulfonate. Mutat. Res. 1987;183:241–247. doi: 10.1016/0167-8817(87)90006-x. [DOI] [PubMed] [Google Scholar]
- 11.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 12.Lin JJ, Sancar A. (A)BC excinuclease: the Escherichia coli nucleotide excision repair enzyme. Mol. Microbiol. 1992;6:19–24. doi: 10.1111/j.1365-2958.1992.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 13.Sancar A. DNA Excision Repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 14.Kisker C, Kuper J, Van Houten B. Cold Spring Harb Perspect Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Houten B, Gamper H, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 16.Bertrand-Burggraf E, Selby CP, Hearst JE, Sancar A. Identification of the different intermediates in the interaction of (A)BC excinuclease with its substrates by DNaseI footprinting on two uniquely modified oligonucleotides. J. Mol. Biol. 1991;219:27–36. doi: 10.1016/0022-2836(91)90854-y. [DOI] [PubMed] [Google Scholar]
- 17.Selby CP, Sancar A. Noncovalent drug-DNA binding interactions that inhibit and stimulate (A)BC excinuclease. Biochemistry. 1991;30:3841–3849. doi: 10.1021/bi00230a006. [DOI] [PubMed] [Google Scholar]
- 18.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc. Natl. Acad. Sci. USA. 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orren DK, Selby CP, Hearst JE, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J. Biol. Chem. 1992;267:780–788. [PubMed] [Google Scholar]
- 20.Hearst JE, Sancar A. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 21.Branum ME, Reardon JT, Sancar A. DNA repair excision nuclease attacks undamaged DNA. A potential source of spontaneous mutations. J. Biol. Chem. 2001;276:25421–25426. doi: 10.1074/jbc.M101032200. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A, Franklin KA, Sancar GB. Escherichia coli DNA photolyase stimulates UvrABC excision nuclease in vitro. Proc. Natl. Acad. Sci. USA. 1984;81:7397–7401. doi: 10.1073/pnas.81.23.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozer Z, Reardon JT, Hsu DS, Malhotra K, Sancar A. The other function of DNA photolyase: stimulation of excision repair of chemical damage to DNA. Biochemistry. 1995;34:15886–15889. doi: 10.1021/bi00049a002. [DOI] [PubMed] [Google Scholar]
- 24.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. USA. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuemmerle NB, Masker WE. Effect of the uvrD mutation on excision repair. J. Bact. 1980;142:535–546. doi: 10.1128/jb.142.2.535-546.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby CP, Sancar A. Molecular mechanism of DNA repair inhibition by caffeine. Proc. Natl. Acad. Sci. USA. 1990;87:3522–3525. doi: 10.1073/pnas.87.9.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 29.Lu A-L, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl. Acad. Sci. USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunala S, Brash DE. Excision repair at individual bases of the Escherichia coli lacI gene: relation to mutation hot spots and transcription coupling activity Proc. Natl. Acad. Sci. USA. 1992;89:11031–11035. doi: 10.1073/pnas.89.22.11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor II. Catalytic properties. J. Biol. Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 32.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: Application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 33.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opinion Struct. Biol. 1993;3:419–429. [Google Scholar]
- 34.Lloyd RG, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 36.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 38.Manelyte L, Kim Y-IT, Smith AJ, Smith RM, Savery N. Regulation and rate enhancement during transcription-coupled DNA repair. Molec. Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assenmacher N, Wenig K, Lammens A, Hopfner K-P. Structural basis for transcription-coupled repair: the N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J. Mol. Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 40.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc. Natl. Acad. Sci. USA. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy MN, Gong P, Ralto K, Manelyte L, Savery NJ, Theis K. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat. Struct. Mol. Biol. 2012;19:291–299. doi: 10.1038/nsmb.2240. [DOI] [PubMed] [Google Scholar]
- 43.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, Chemama Y, Labigne A, Legtrain P. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 44.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernal-Bernal D, Gallego-Garcia A, Garcia-Martinez G, Garcia-Heras F, Jimenez MA, Padmanabhan S, Elias-Arnanz M. Structure-function dissection of myxococcus xanthus CarD N-terminal domain, a defining member of the CarD_CdnL_TRCF family of RNA polymerase interacting proteins. PLOS ONE. 2015;10(3):e0121322. doi: 10.1371/journal.pone.0121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallego-Garcia A, Mirasou Y, Garcia-Moreno D, Elias-Arnanz M, Jiminez MA, Padmanabhan S. Structural insights into RNA polymerase recognition and essential function of Myxococcus xanthus Cdnl. PLOS ONE. 2014;9(10):e1047. doi: 10.1371/journal.pone.0108946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Moreno D, Abellon-Ruiz J, Garcia-Heras F, Murillo FJ, Padmanabhan S, Elias-Arnanz M. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 2010;38:4586–4598. doi: 10.1093/nar/gkq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, Darst S. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park J-S, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 50.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel SS, Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 52.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, Darst SA, Savery NJ, Strick TR. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription-repair coupling factor. Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Proshkin SA, Mironov AS. Stalled RNA polymerase is a target of the Mfd factor. Molec. Biol. 2016;50:381–384. doi: 10.7868/S0026898416020191. [DOI] [PubMed] [Google Scholar]
- 55.Kamarthapu V, Epshtein V, Benjamin B, Proshkin S, Mironov A, Cashel M, Nudler E. ppGpp couples transcription to DNA repair in E. coli. Science. 2016;352:993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao N, Krishna S, Ahlgren-Berg A, Cutts EE, Shearwin KE, Dodd IB. Road rules for traffic on DNA – systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res. 2014;42:8861–8872. doi: 10.1093/nar/gku627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 58.Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 59.Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 61.Hall MC, Ozsoy AZ, Matson SW. Site-directed mutations in motif VI of Escherichia coli DNA helicase II result in multiple biochemical defects: evidence for the involvement of motif VI in the coupling of ATPase and DNA binding activities via conformational changes. J. Mol. Biol. 1998;277:257–271. doi: 10.1006/jmbi.1997.1614. [DOI] [PubMed] [Google Scholar]
- 62.Park J-S, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prabha S, Rao DN, Nagaraja M. Distinct properties of hexameric but functionally conserved Mycobacterium tuberculosis transcription-repair coupling factor. PLoS ONE. 2011;6(4):e191931. doi: 10.1371/journal.pone.0019131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava DB, Darst SA. Derepression of bacterial transcription-repair coupling factor is associated with a profound conformational change. J. Molec. Biol. 2011;406:275–284. doi: 10.1016/j.jmb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith AJ, Pernstich C, Savery NJ. Multipartite control of the DNA translocase, Mfd. Nucleic Acids Res. 2012;40:10408–20416. doi: 10.1093/nar/gks775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graves ET, Duboc C, Fan J, Stransky F, Leroux-Coyau M, Strick TR. A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence. Nat. Struct. Mol. Biol. 2015;22:452–457. doi: 10.1038/nsmb.3019. [DOI] [PubMed] [Google Scholar]
- 67.Fan J, Leroux-Coyau M, Savery N, Strick TR. Reconstruction of bacterial transcription-coupled repair at single molecule resolution. Nature. 2016;536:234–237. doi: 10.1038/nature19080. [DOI] [PubMed] [Google Scholar]
- 68.Haines NM, Kim Y-IT, Smith AJ, Savery NJ. Stalled transcription complexes promote DNA repair at a distance. Proc. Natl. Acad. Sci. USA. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kunala S, Brash DE. Intragenic domains of strand-specific repair in Escherichia coli. J. Mol. Biol. 1995;246:264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 70.Straney SB, Crothers DM. Lac repressor is a transient gene-activating protein. Cell. 1987;51:699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- 71.Ganesan AK, Hanawalt PC. Transcription-coupled nucleotide excision repair of a gene transcribed by bacteriophage T7 RNA polymerase in Escherichia coli. DNA Repair. 2010;9:958–963. doi: 10.1016/j.dnarep.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ganesan AK, Smith AJ, Savery NJ, Zamos P, Hanawalt PC. Transcription coupled nucleotide excision repair in Escherichia coli can be affectd by changing the arginine at position 529 of the β subunit of RNA polymerase. DNA Repair. 2007;6:1434–1440. doi: 10.1016/j.dnarep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gwynn EJ, Smith AJ, Guy CP, Savery NJ, McGlynn P, Dillingham MS. The conserved C-terminus of the PcrA/UvrD helicase interacts directly with RNA polymerase. PLoS ONE. 2013;8(10):e78141. doi: 10.1371/journal.pone.0078141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 76.Ahn B. A physical interaction of UvrD with nucleotide excision repair protein UvrB, Mol. Cells. 2000;10:592–597. doi: 10.1007/s10059-000-0592-5. [DOI] [PubMed] [Google Scholar]
- 77.Manelyte L, Guy CP, Smith RM, Dillingham MS, McGlynn P, Savery NJ. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair. 2009;8:1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen SE, Walker GC. New discoveries linking transcription to DNA repair and damage tolerance pathways. Transcription. 2011;2:37–40. doi: 10.4161/trns.2.1.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 80.Trautinger BW, Jaktaji RP, Rusakova ER, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between replication and transcription. Molec. Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 82.Ito T, Nakamura T, Maki H, Sekiguchi M. Roles of transcription and repair in alkylation mutagenesis. Mutation Res. 1994;314:273–285. doi: 10.1016/0921-8777(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 83.Li S, Waters R. Induction and repair of cyclobutane pyrimidine dimers in the Escherichia coli tRNA gene tyrT: Fis protein affects dimer induction in the control region and suppresses preferential repair in the coding region of the transcribed strand, except in a short region near the transcription start site. J. Mol. Biol. 1997;271:31–46. doi: 10.1006/jmbi.1997.1154. [DOI] [PubMed] [Google Scholar]
- 84.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schalow BJ, Courcelle CT, Courcelle J. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J. Bact. 2012;194:2637–2645. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oller AR, Fijalkowska IJ, Dunn RL, Schaaper RM. Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1992;89:11036–11040. doi: 10.1073/pnas.89.22.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J. Bact. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch P. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Molec. Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 89.Bridges BA. Starvation-associated mutation in Escherichia coli strains defective in transcription repair coupling factor. Mutation Res. 1995;329:49–56. doi: 10.1016/0027-5107(95)00016-c. [DOI] [PubMed] [Google Scholar]
- 90.Fabisiewicz A, Janion C. DNA mutagenesis and repair in UV-irradiated E. coli K-12 under condition of mutation frequency decline. Mutat. Res. 1998;402:59–66. doi: 10.1016/s0027-5107(97)00282-0. [DOI] [PubMed] [Google Scholar]
- 91.Ayora S, Rojo F, Ogasawara N, Nakai S, Alonso JC. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 1996;256:301–318. doi: 10.1006/jmbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- 92.Zalieckas JM, Wray LV, Jr., Ferson AE, Fisher SJ. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Molec. Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]
- 93.Belitsky BR, Sonenshein AL. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA. 2013;110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belitsky BR, Soneneshein AL. Roadblock repression of transcription by Bacillus subtilis CodY. J. Mol. Biol. 2011;411:729–743. doi: 10.1016/j.jmb.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Washburn RS, Wang Y, Gottesman ME. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 96.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pruteanu M, Baker TA. Controlled degradation by ClpXP protease tunes the levels of the excision repair protein to the extent of DNA damage. Mol. Microbiol. 2009;71:912–924. doi: 10.1111/j.1365-2958.2008.06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]