Abstract
An eleven amino acid ribosomal peptide was shown to completely neutralize Western Diamondback Rattlesnake (Crotalus atrox) venom in mice when a lethal dose of the venom was pre-incubated with the peptide prior to intravenous injection. We have expressed the peptide as a concatenated chain of peptides and cleaved them apart from an immobilized metal affinity column using a protease. After ultrafiltration steps, the mixture was shown to partially neutralize rattlesnake venom in mice. Preliminary experiments are described here that suggest a potential life-saving therapy could be developed. To date, no recombinant therapies targeting cytotoxic envenomation have been reported.
Introduction
Pit viper envenomations occur in the United states, resulting in approximately 2,700 hospitalizations annually1 as well as over 100,000 bites of cats and dogs2. Pit vipers can be found in warm climates, including on the Asian and American continents. Their bites can be fatal if not treated and cause severe morbidity such as swelling, edema, necrosis, blood coagulation and kidney failure3. The FDA-approved treatment for pit viper envenomation in the US is a Fab cocktail produced in sheep4. The sheep are injected with immunogens from the snake venom and produce antibodies. The sheep serum containing the antibodies is processed, including incubation with pepsin, to produce the Fab antivenom. The wholesale cost of this treatment is more than 1000 USD per vial and immunogenic effects are reported.
The North American Opossum (Didelphis virginiana) is able to survive the bite of the C. atrox (CA) snake. Several laboratories purified proteins from the serum of opossum species and showed these proteins could neutralize various toxic components of cytotoxic snake venoms5–8. Interestingly, the N-terminus of several of these proteins was shown to be highly conserved. Work was published by B. Lipps demonstrating the ability of a short peptide composed of first 10–15 amino acids of one of these proteins (named Lethal Toxin Neutralizing Factor, LTNF) from the opossum serum to maintain the activity of the complete protein9.
Our work confirmed the ability of a chemically synthesized 11-mer (LTNF11-CS), the first 11 amino acids (LKAMDPTPPLW) of the peptide published by Lipps, to neutralize CA venom in mice when a lethal dose of the venom was pre-incubated with the peptide followed by intravenous injection. Additionally, we have expressed the 11-mer (LTNF-11) as a concatenated chain of eight peptides with a C-terminal 6xHIS tag to facilitate downstream purification of the peptide chain. The 11-mer was chosen because the eleventh amino acid is a unique tryptophan on the peptide and serves as a protease cleavage site to sever apart the peptides. On-column cleavage of the peptides was carried out and the final product was tested in mice and shown to have some neutralizing capacity against CA venom. LC/MS analysis of the product showed partial purification of the peptide. This preliminary work can potentially lead to the development of the first recombinant antivenom therapy10.
Materials and Methods
Strain and genetic construct
A gene construct was purchased from Eurofins MWG Operon (Huntsville, AL) to express the sequence shown in Figure 1. The sequence was optimized for expression in E. coli. The genes were cut and inserted into a pET29A plasmid (Novagen) between NdeI and XhoI (Promega) sites for transformation of BL21(DE3) E. coli (Novagen singles) by heat shock. The pET29a plasmid contains a termination sequence downstream of a 6xHIS tag that is immediately following the XhoI cutting site.
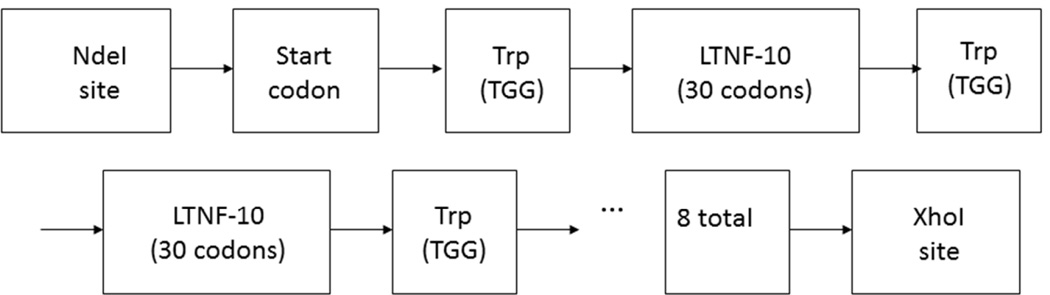
Figure 1.
Synthetic construct for expression of tandem repeat of eight LTNF-11 peptides (snk8) in E. coli. LTNF-10 is the original 10-mer published by Lipps that has the sequence LKAMDPTPPL. The natural 11th amino acid is W.
Chemicals and media
Unless otherwise stated, chemicals were purchased from Fisher. Chemically synthesized LTNF-11 peptide (20 mg, LTNF11-CS) was purchased from Thermo Scientific (Pierce Biological Products; Illinois, US) with a specification of 95% purity. Shake flask cultures were done with Luria-Bertani (LB) broth containing 30 Ig/mL of kanamycin. Frozen stocks of transformed E. coli were prepared from LB overnight cultures and maintained at −80°C with 15% glycerol.
Media for fermentation was based on the media reported in Reisenberg11. The trace metal and salt composition was identical to that described in Reisenberg. Solutions of KH2PO4, (NH4)2HPO4, citric acid, MgSO4·7H2O, EDTA and yeast extract (there is no yeast extract in the media reported by Reisenberg) all in deionized water were prepared separately. The salt solutions were added to a bottle through a 0.45Im filter in the order of KH2PO4, (NH4)2HPO4, citric acid, then EDTA. The pH of the mixture was adjusted to 6.2 using 10N NaOH then poured into the 2 liter bioreactor. The level of the liquid was adjusted to about 1200 mL with deionized water and then the filtered trace metal solution was added. The headplate of the bioreactor (Applikon round-bottom) was fastened loosely and all exit ports from the headplate were clamped. Solutions of MgSO4·7H2O, yeast extract, glucose feed solution in DI H2O (400 g/L) and a 50 mL shake flask of LB for the inoculum were autoclaved (Hirayama HA-300MW) separately. The solution of Thiamine-HCL was sterile filtered. After the solutions cooled, the MgSO4·7H2O, yeast extract (to a final concentration of 2 g/L), thiamine-HCL, and glucose (to a final concentration of 15 g/L) were added followed by enough water to achieve a final volume of 1450 mL.
Production of LTNF-11 peptide chain in E. coli
The BL21(DE3) pET29A-Snk8 strain (named snk8) was grown at 35°C overnight in a shake flask (New Brunswick Scientific Innova 4080) shaken at 185 rpm. The pH in the fermentor was adjusted to 6.9 with NH4OH and the media was warmed with a heating jacket to 36°C. The temperature in the bioreactor was controlled through the Applikon ADI 1030 controller connected with both the heating jacket and a circulating water bath (Neslab CFT-33 Refrigerated Recirculator). The cooling water was also pumped through a condenser on the headplate of the bioreactor. The overnight culture was added (50 mL) to complete the 1.5 L volume in the bioreactor once the media reached the desired temperature. The initial stir speed in the bioreactor was kept at 500 RPM until the dissolved oxygen dropped below 50% of saturation. The stir speed was increased manually to keep the DO above 50%. Once the OD600 reached about 20, IPTG (Gold Bio) was added for induction to a final concentration of 400 Ig/mL. One additional bolus of 10 grams of glucose (in solution) was added until the OD 600 reached about 40, at which point the glucose was allowed to drop to approximately zero g/l and the prepared glucose feed solution was fed at a rate of approximately 130 mg glucose per minute. The run was shut down at about 24 hours and the final OD600 was measured at approximately 60. Samples (~5 mL each) were collected from the fermentor and analyzed. One milliliter of the sample was centrifuged for one minute at 15,000 g in a refrigerated microcentrifuge (Thermo IEC Micromax RF) to pellet the cells and the supernatant was analyzed for glucose concentration with a YSI glucose analyzer calibrated at 20 g/L. Some of the fermentation sample was used to measure the optical density. The sample was appropriately diluted to measure OD600 with a spectrophotometer (Genesys 5). From the samples, smaller volumes were taken for SDS-PAGE analysis such that the amount of cells in each sample would be comparable, according to the relation
Vsample (mL) = 1.2/OD600
These small volume samples were centrifuged at 15,000 g, supernatants were aspirated off, and the cell pellets stored frozen at −30°C for later analysis.
SDS-PAGE analysis of fermentation samples
Thawed samples were incubated with 200 IL of BugBuster HT solution (EMDMillipore) with added lysozyme (Fisher Bioreagents, 0.2 Ig) and after spinning, the pellet was incubated with 8 M urea to solubilize the inclusion bodies. These were added to SDS-PAGE sample buffer and boiled for 10 min prior to loading on a 12% gel.
Cell separation and lysis
At the end of the fermentation run, the broth was divided equally into eight 50 mL centrifuge tubes and spun down for 10 min in a Beckman GS centrifuge at 1225g. Supernatants were discarded and more broth was added to the tubes and spun. This was repeated until all of the fermentation broth was processed. The cell pellets were stored at −30°C. On the day of purification, the cell pellets (approximately 5 mL of packed cell volume in each tube) were thawed at room temperature and incubated with end-over-end mixing for 20 min with 5 mL Bugbuster HT plus 20 Ig lysozyme, followed by centrifugation at 30,000 g for 20 min in a centrifuge (DuPont Sorvall RC5B). The pellet was incubated with 8 M urea for 20 min with end-over-end mixing and centrifuged again at 30,000 g for 20 min. The resulting supernatant underwent two pH modification steps (to pH 5 by addition of 4M urea dissolved in 1M acetic acid followed by centrifugation at 3,200 g (Beckman GS-6 centrifuge), then the pellet was discarded and the pH of the supernatant was readjusted to pH 8 with 4M urea in 0.4M disodium hydrogen phosphate).
Purification and on-column cleavage of LTNF-11
Immobilized metal affinity chromatography (Pharmacia 501 Plus FPLC with Direktor software) followed by ultrafiltration was carried out to purify LTNF peptide as follows: A 0.5 cm3 volume of Ni-NTA (GE Ni sepharose fast flow 6) gel in a 1 cm diameter column was equilibrated with binding buffer composed of saline solution (9 g/L NaCl in deionized water) with 10 mM sodium phosphate. 15 mL of the processed cell pellet was applied to the column then washed with binding buffer until no further absorbance was measured. After washing, the column was incubated with 5 mL of a solution of 5.6×10−6M chymotrypsin (Creative Enzymes trypsin-free chymotrypsin) in saline with 10 mM phosphate and 20 mM CaCl2. After 15 min, the column was flushed with the bind buffer until the absorbance began to drop. The eluate was centrifuged immediately with a 10,000 MWCO Centricon (Amicon) unit to separate the chymotrypsin from the eluted peptides. The solution was concentrated using a 1000 MWCO membrane (Amicon concentrator, 1kD EMD-Millipore membrane) followed by 3kD membrane (Millipore Amicon Ultra 2mL cartridge).
Analysis of the product by LC/MS
An Agilent 1200 Series HPLC with diode array detection (DAD) containing a 150mm×2.1 mm Agilent Poroshell 120 EC-C18 column was coupled to an Agilent QTOF6520 for analysis of peptides. The column was kept at 55°C temperature during the run. A binary mobile phase system of solvent A (0.1% formic acid) and solvent B (100% acetonitrile) was used to elute material from the column. A flow rate of 0.3 mL/min was used. The column was first equilibrated into 5% B before 10–20 Il of sample was injected onto the column. A gradient was used to separate compounds from the column. The gradient began with 5% B for 3 min and increased linearly to 75% B at 23 min, at which time the gradient increased linearly to 95% B at 27 min and finally returned to originally starting conditions at 30 min. A diode array MS data were collected in full scan positive mode over the mass range of 100–3000 m/z. Ion voltages and gas settings were as follows: fragmentor, 150 V; skimmer, 65 V; drying gas, 8L/min; gas temperature, 350°C. The DAD window was established between 200 and 320 nm. Agilent MassHunter Acquistion software version B.06.00 was used for data acquisition and analysis.
In vivo activity assays
All in vivo assays were carried out at the National Natural Toxins Research Center (Texas A&M University Kingsville, Kingsville, TX). All necessary IACUC protocols were approved by animal welfare committees at both the NNTRC and SJSU sites. C. atrox venom was extracted from the snake, filtered and lyophilized. Mice, BALB/c, M&F, 18–20 g were used for the experiments. For these experiments the injections were done intravenously through the tail vein. Control mice (n=8) were injected with 100 Ig of venom each following pre-incubation (30 min at 37°C) with saline, while a separate test group (n=8) were injected with 100 Ig of venom each following pre-incubation (30 min at 37°C) with 200 Ig of LTNF11-CS. Finally, another set of control mice (n=8) were injected with 200 Ig of LTNF11-CS peptide dissolved in saline.
The partially purified E. coli-produced LTNF-11 peptide was sent as the frozen filtrate from the 3000 MWCO ultrafiltration step of the purification to the NNTRC. The solutions were shipped frozen on dry ice to the NNTRC where they were lyophilized (Labconco) and reconstituted with saline prior to use. One control solution was sent: a solution prepared by repeating the FPLC and all downstream purification steps using cell-free 8 M urea pH 8. This solution was also lyophilized and reconstituted with saline. The volumes of the solutions were recorded and the lyophilized material was weighed to determine appropriate amounts to be injected. Control solution (27 Ig delivered to each of 2 mice) was reconstituted and pre-incubated with the CA venom (n=2), while 247 Ig of the purified E. coli extracts lyophilized then resuspended in saline were pre-incubated with the CA venom (n=2).
Results and Discussion
The goal of this work was to introduce a recombinant product that can serve as an antivenom therapy. Current antivenoms are produced in livestock, and the resulting antivenom products can cause allergenic reactions in the patients. Although E. coli is known to produce endotoxin and potentially allergenic host cell proteins, strategies have been developed for safely producing protein therapies with E. coli12. In contrast, the currently available snake antivenom against US venomous snakes that is produced in sheep has been reported to result in allergenic reactions in 14.3% of patients13. Thus, the use of a recombinant therapeutic for snake bite treatment may result in reduced patient discomfort.
There are no literature reports of bioactive peptides that are able to neutralize whole cytotoxic snake venom with the exception of the LTNF peptide9. The initial publications on this peptide claimed that the peptide is able to neutralize the venom of several snakes, including those with neurotoxic venom. Our experiments with the chemically synthesized LTNF-11 showed that the peptide is not able to neutralize neurotoxic snake venoms such as cobra or Russell Viper (data not shown). The experiments reported here are only with the Western Diamondback Rattlesnake (C. atrox) that contains primarily cytotoxic and hemotoxic components14. The major component of C. atrox venom is snake venom metalloproteinases, although the venom also contains disintegrins, L-amino acid oxidases, phospholipase A2, serine proteases and other proteins and peptides toxic to humans. While Lipps proposed that the mechanism of the peptide is to serve as a metalloproteinase inhibitor, this does not explain how the peptide could adequately neutralize the cocktail of toxins found in CA venom.
This work is directed to a US snake where the market may or may not be large enough to merit a recombinant process, but the global toll of snakebite, according to the World Health Organization may be as high as five million15. Thus the production of a recombinant antivenom could assist developing countries where serum-based antivenoms are often difficult to acquire.
Activity experiments with chemically-synthesized LTNF11-CS peptide
Although the 10-mer was shown by Lipps to be active against snake venom, we included the natural 11th amino acid of the chain that is a tryptophan. Tryptophan serves as a cleavage site for chymotrypsin16. To be able to compare the activity experiments between the chemically synthesized and recombinant peptide we selected the 11-mer for all our experiments.
Our initial experiment with the LTNF11-CS peptide demonstrated the ability of LTNF to neutralize snake venom. The lethal dose of C. atrox venom was determined to be 71 Ig per 18–20 g mouse. The lethal dose is higher than the LD50 and is determined to be a dose that kills all the mice. Eight mice were injected with 100 Ig of venom that had been incubated with saline solution at 37°C for 30 min prior to injection in the mouse tail veins. All eight of these control mice died overnight. A second set of eight mice were injected with 100Ig of the same lot of venom that had been incubated with LTNF11-CS peptide at 37°C for 30 min prior to injection. None of these eight mice died or showed any symptoms of envenomation. The pre-incubation step is normally performed to determine the effective doses of standard (animal-derived) antivenoms. An additional experiment tested mice (n=8) receiving only 200 Ig of LTNF11-CS peptide by intravenous injection, and all of these remained healthy. This experiment demonstrates the ability of the peptide to neutralize the venom. In addition, the peptide can be readily dissolved in its active form in saline solution and in our experiments did not show any deleterious effects on the mice.
Expression of tandom-multimer of LTNF-11
We have chosen to express the peptide as a tandem repeat chain of peptides. The use of the tandem-multimer expression strategy can reduce their proteolytic cleavage during microbial expression17. The pET expression vectors utilize a T7 promoter and the T7 RNA polymerase is expressed from the chromosome of the BL21 E. coli under control of a lac promoter. Thus, IPTG or lactose can be used for the expression of the target protein. BL21(DE3) E. coli is a protease knock-out strain (Ion and OmpT proteases) for increased yield of protein as compared with the parent strain. We chose eight peptides on the tandem-multimer as that number was the maximum number that could be optimized for production as an Express Gene Construct from Eurofins MWG Operon. Additionally, the molecular weight of the expressed peptide chain with eight peptides is 11.7 kD, which is an appropriate size for forming inclusion bodies in E. coli. A map of the gene construct is shown in Figure 1. The 6xHis tag is expressed downstream of the XhoI restriction site and is followed by a termination sequence. The N terminus has a methionine start codon followed by a tryptophan codon so the start codon can be cleaved together with the consequent eight peptides but leave a short, two-amino acid piece that can be removed during the ultrafiltration step.
Production and purification of recombinant LTNF-11
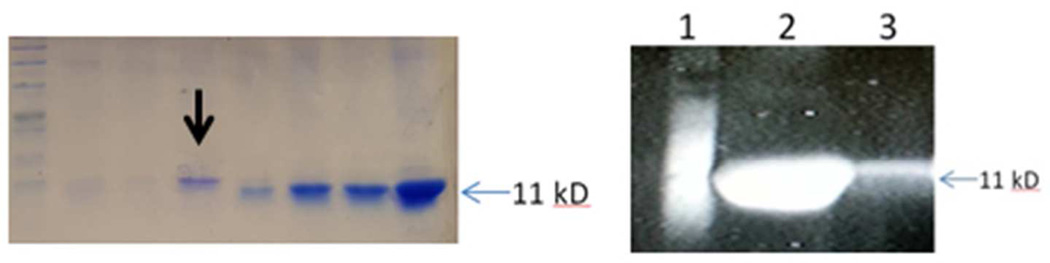
The media recipe was chosen based on the original work of Reisenberg11. The media recipe reported does not contain yeast extract and we have found that protein expression with this media is facilitated by the addition of yeast extract (2 g/L) to the media. The media is prepared according to the original paper with the exception that yeast extract, MgSO4 solution, glucose and thiamine solutions are sterilized separately. The remaining media components are autoclaved for 15 min at 121°C in the fermentor vessel. We found that 400Ig/mL final concentration of IPTG induction at an OD600 of 20 resulted in the highest final titer of the peptide chain (data not shown). Samples taken based on the OD600 were taken to unify the cell concentration in the SDS-PAGE samples. The gel in Figure 2A shows solubilized pellet fractions of the samples taken during the fermentation run prior to and after induction. The gel suggests that the expression of the 11.7 kD peptide chain increases after induction. Gels prepared from the supernatant post lysis did not show any significant band that increased after induction, suggesting that the peptide chain forms inclusion bodies when expressed in E. coli. The gel in Figure 2B shows that the fermentation product cells processed at the end of the run have a large amount of a protein slightly larger than 11 kD that has a 6xHis tag on it. This corroborates that we produced the peptide chain with a 6xHis tag.
Figure 2.
SDS-PAGE gels showing expression of 11.7 kD product in E. coli (pellet fraction in 8 M urea) from a 2 liter fermentation. 2A (left gel) Commassie Blue stain with black arrow indicating the induction time. 2B Pierce 6xHis tag stain gel (lane 1 marker, lane 2 solubilized and processed pellet fraction post cell lysis, lane 3 1/50 dilution of material shown in lane 2.
The solubilized inclusion bodies were processed to reduce the viscosity by performing the two pH modifications. Reduction of the pH to 5.0 caused a white precipitate to form that could be easily separated by centrifugation. The supernatant was returned to pH 8.0 and no additional precipitate was formed. This solution (15 mL) was added directly to a Ni-NTA column equilibrated with saline containing 10 mM sodium phosphate. This bind buffer was selected to obviate the need for a buffer exchange after elution. The goal was to prepare a purified peptide devoid of large amounts of salt that could affect the mouse. A large amount of protein did not bind to the column but flushed immediately as could be seen by the trace on the absorbance detector at 280 nm (data not shown). After flushing the column until the absorbance at 280 nm reduced to zero, a mixture of bind buffer with added CaCl2 and chymotrypsin was added to the column and incubated at room temperature. The concentration of chymotrypsin was chosen as 10-fold higher than the KM reported in a literature study using a colorimetric substrate with chymotrypsin16. After incubation with chymotrypsin, the column was eluted with bind buffer until the absorbance reduced to 0.5 mAu. The solution was immediately spun through a 10 kD ultrafiltration membrane to separate the chymotrypsin from the peptide. The filtrate was then concentrated with a 1kD ultrafiltration membrane to a total volume of 5 mL. The final step in the process was to pass the concentrated peptide through a 3 kD ultrafiltration membrane to separate any remaining large protein molecules and any large particles that could be pyrogenic in the mice. The final solution was frozen and stored at −30°C until shipped to the NNTRC for activity testing.
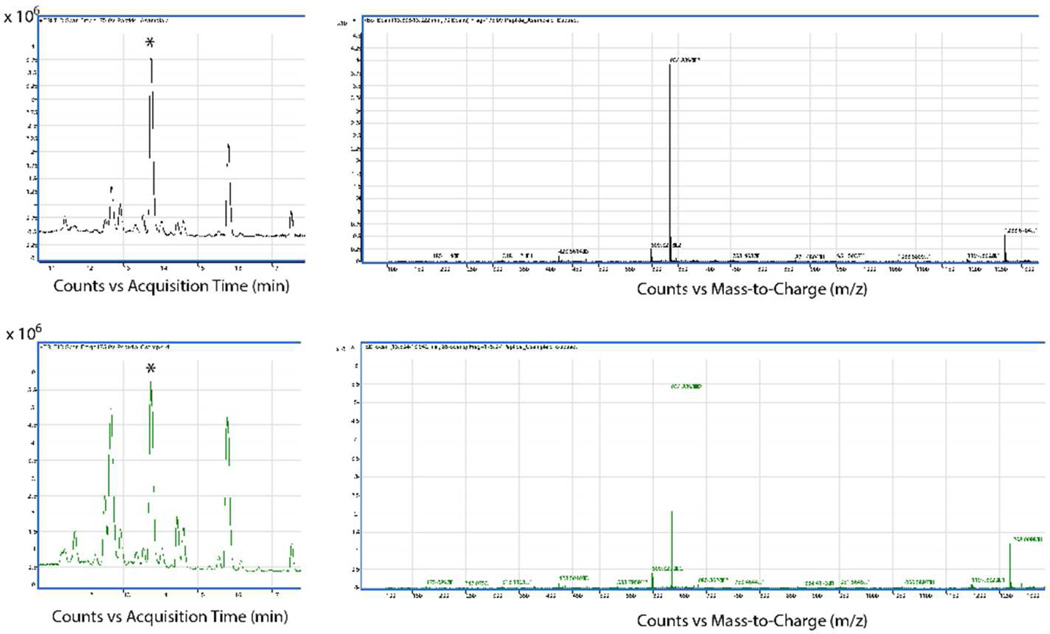
We tested different chymotrypsin incubation times to determine the optimal time that did not result in cleaved peptide. LC/MS analysis of the chemically synthesized LTNF11-CS peptide showed a single peak at a retention time at about 13.7 min with the MS trace giving two peaks, one at the single charge mass of 1268 and one at a double charge mass of 634, corresponding with the molecular weight of the LTNF11-CS peptide. Figure 3 shows the LC/MS analysis of the E. coli product from two different chymotrypsin incubation times, 15 and 30 min. The 15 minute incubation time (top figures) shows some impurities present with peaks at around 12.5 and also at 15.8 min. The 30 minute incubation time shows larger peaks at 12.5 and 15.8 min relative to the desired peak at 13.7 min. Additionally, 2 hours of incubation with chymotrypsin resulted in no peak present at 13.7 min (data not shown). Further work to optimize the chymotrypsin incubation step is warranted.
Figure 3.
LC/MS traces of E. coli-produced LTNF-11 peptide from our lab. Asterisks identify peak from HPLC column eluting at the proper retention time based on determination using a chemically synthesized LTNF-11 standard. Top chromatogram/trace taken from 15 minute chymotrypsin incubation batch, Bottom chromatogram/trace taken from 30 minute chymotrypsin incubation batch. Peptide molecular weight is 1268.7 d.
Activity of LTNF-11 produced in E. coli
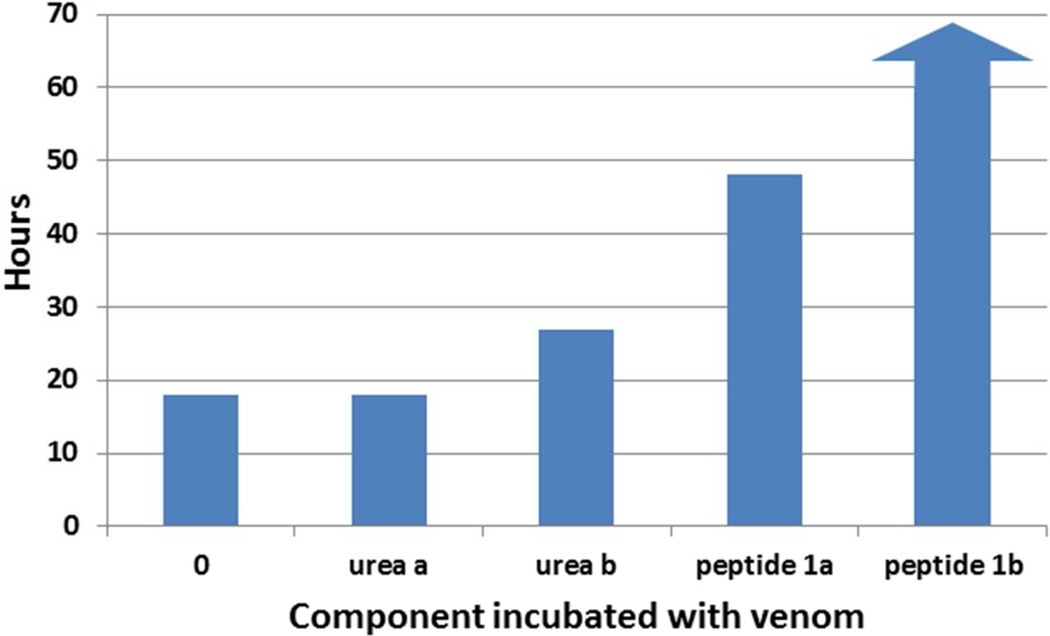
A control solution was prepared by running cell-free 8 M urea through all the purification steps in the process. The masses of lyophilized control and peptide solutions were chosen based on the mass of salt in the control solution. The control solution was resuspended to the same volume using water and the actual volume injected in the mice contained 27 Ig of salt matter. It was assumed that the concentration of salt in the peptide solution was the same as in the control, but in order to reach an equivalent amount of peptide as in the activity assays with the chemically synthesized peptide, 247 Ig was chosen. This weight was determined assuming that the peptide in the E. coli-produced batch is perfectly pure. The result of the mouse assay is shown in Figure 4. The two control mice receiving no peptide with the venom both died sooner than the mice receiving the peptide. Indeed, one of the test mice survived the experiment. Mice receiving only the control solution or only the peptide solution without any venom survived without showing any deleterious effects. Although this result did not demonstrate complete neutralization of the venom, as with the chemically synthesized peptide, there is partial neutralization with the E. coli produced peptide. As the dose was calculated assuming the E. coli-produced peptide was pure, it is likely that there was less than 200Ig of peptide injected which would reasonably explain the loss of the mouse. Further work is needed to establish a purification protocol that results in complete neutralization of the venom.
Figure 4.
Survival Time post injection of venom in five mice incubated with saline (0), processed 8 M urea (urea), processed snk8 E. coli fermentation cell pellets. Arrow indicates survival. Two mice (a and b) were used for the urea control and peptide test experiments.
Conclusion
This work demonstrates the first recombinant biomolecule that is able to neutralize cytotoxic snake venom. The expression of a tandem repeat chain of peptide molecules forms inclusion bodies in E. coli and can be purified by a series of chromatography and ultrafiltration steps. The use of an enzyme for the cleavage of peptides from the tandem multimer is also shown to result in partially purified, active peptide.
Acknowledgments
The author gratefully acknowledges a Fulbright-Nehru award to carry out research at the IIT-Delhi. Fermentation and other equipment was provided through an NSF CCLI grant (0088653, C. Komives PI). The LC/MS instrumentation was provided through an NSF MRI grant (0923573, JBW, PI). Additional funding from the Charles W. Davidson College of Engineering at SJSU was helpful. Special thanks to graduate students S. Singh, V. Thakur and V. Kumar in the Rathore lab and to graduate students Z. Ahmad and Y. Khilco in the Komives lab.
References
- 1.Lavonas EJ, Schaeffer TH, Kokko J, Mlynarchek SL, Bogdan GM. Crotaline Fab antivenom appears to be effective in cases of severe North American pit viper envenomation: An integrative review. BMC Emergency Medicine. 2009;9 doi: 10.1186/1471-227X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson M. Snake bite: pit vipers. Clinical Techniques in Small Animal Practice. 2006;21:174–182. doi: 10.1053/j.ctsap.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez JM, Theakston RDG, Warrell DA. Confronting the Neglected Problem of Snake Bite Envenoming: The Need for a Global Partnership. PLoS Medicine. 2006;3:1–5. doi: 10.1371/journal.pmed.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavonas EJ, Ruha A-M, Banner W, et al. Unified treatment algorithm for the management of crotaline snakebite in the United States: results of an evidence-informed consensus workshop. BMC Emergency Medicine. 2011;11 doi: 10.1186/1471-227X-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catanese JJKL. Isolation from opossum serum of a metalloproteinase inhibitor homologous to human alpha 1 B-glycoprototein. Biochemistry. 1992;31:410–418. doi: 10.1021/bi00117a015. [DOI] [PubMed] [Google Scholar]
- 6.Neves-Ferreira AGC, Cardinale N, Rocha SLG, Perales J, Domont GB. Isolation and characterization of DM40 and DM43, two snake venom metalloproteinase inhibitors from Didelphis marsupialis serum. Biochimica et Biophysica Acta. 2000;1474:309–320. doi: 10.1016/s0304-4165(00)00022-2. [DOI] [PubMed] [Google Scholar]
- 7.Lipps BV. Anti-lethal factor from opossum serum is a potent antidote for animal, plant and bacterial toxins. Journal of Venomous Animals and Toxins. 1999;5:56–66. [Google Scholar]
- 8.Jurgilas PB, Neves-Ferreira AGC, Domont GB, Perales J. PO41, a snake venom metalloproteinase inhibitor isolated from Philander opossum serum. Toxicon. 2003;42:621–628. doi: 10.1016/j.toxicon.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Lipps BV. Small synthetic peptides inhibit the lethality in mice for toxins derived from animal, plant and bacteria. Journal of Venomous Animals and Toxins. 2000;6:77–86. [Google Scholar]
- 10.Komives C, Rathore AS, inventors. A low cost process for production of a snake antivenom peptide. Indian patent application 778/DEL/2015
- 11.Reisenberg D, Schulz V, Knorre WA, et al. High cell density cultivation of Escherichia coli at controlled specific growth rate. Journal of Bacteriology. 1991;20:17–27. doi: 10.1016/0168-1656(91)90032-q. [DOI] [PubMed] [Google Scholar]
- 12.Huang CJ, Lin H, Yang X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. Journal of Industrial Microbiology & Biotechnology. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- 13.Juckett G, Hancox J. Venomous Snakebites in the United States: Management Review and Update. American Family Physician. 2002;65:1367–1375. [PubMed] [Google Scholar]
- 14.Calvete JJ, Fasoli E, Sanz L, Boschetti E, Righetti PG. Exploring the Venom Proteome of the Western Diamondback Rattlesnake Crotalus atrox, via Snake Venomics and Combinatorial Peptide Ligand Library Approaches. Journal of Proteome Research. 2009;8:3055–3067. doi: 10.1021/pr900249q. [DOI] [PubMed] [Google Scholar]
- 15.Arnold C. The Snakebite Fight. Nature. 2016;537:26–28. doi: 10.1038/537026a. [DOI] [PubMed] [Google Scholar]
- 16.Bender M, Kezdhy F, Wedler F. Alpha-chymotrypsin: Enzyme concentration and kinetics. Journal of Chemical Education. 44:84–88. doi: 10.1021/ed044p84. [DOI] [PubMed] [Google Scholar]
- 17.Wang F-J, Song H-L, Wang X-M, et al. Tandem Multimer Expression and Preparation of Hypoglycemic Peptide MC6 from Momordica charantia in Escherichia coli. Appl Biochem Biotechnol. 2012;166:612–619. doi: 10.1007/s12010-011-9452-3. [DOI] [PubMed] [Google Scholar]