Abstract
Purpose
Ibrutinib, a Bruton’s tyrosine kinase (BTK) inhibitor, is approved for the treatment of relapsed CLL and CLL with del17p. Mechanistically, ibrutinib interferes with BCR signaling as well as multiple CLL cell to microenvironment interactions. Given the importance of ibrutinib in the management of CLL, a deeper understanding of factors governing sensitivity and resistance is warranted.
Experimental Design
We studied 48 longitudinally sampled paired CLL samples, 42 of which were procured before and after standard CLL chemotherapies, and characterized them for well-studied CLL molecular traits as well as by whole exome sequencing and SNP 6.0 array profiling. We exposed these samples to 0.25 μM – 5 μM of ibrutinib ex vivo and measured apoptosis fractions as well as BCR signaling by immunoblotting. We disrupted TP53 in HG3, PGA1 and PG-EBV cell lines and measured BCR signaling and ibrutinib responses.
Results
CLL samples demonstrated a surprisingly wide range of ex vivo sensitivities to ibrutinib with IC50 values ranging from 0.4 μM – 9.7 μM. Unmutated IGVH status, elevated ZAP70 expression and trisomy 12 were associated with heightened sensitivity to ibrutinib treatment. Five CLL samples were substantially more resistant to ibrutinib following relapse from chemotherapy; of these, three had acquired a del17p/TP53 mutated status. A validation sample of 15 CLL carrying TP53 mutations, of which 13 carried both del17p and a TP53 mutation confirmed substantially less sensitivity to ibrutinib-induced apoptosis.
Conclusions
This study identifies that CLL harboring del17p/TP53 mutated cells are substantially less sensitive to ibrutinib-induced apoptosis than del17p/TP53 wild type cells.
Keywords: Ibrutinib, CLL, del17p/TP53 mutations, apoptosis
INTRODUCTION
The therapy of chronic lymphocytic leukemia is evolving. Inhibitors of B-cell receptor signaling are demonstrating substantial activity in the absence of traditional chemotherapy(1–5). Ibrutinib, an inhibitor of the BTK tyrosine kinase is approved for the treatment of relapsed CLL and CLL with del17p(6). Ibrutinib irreversibly inhibits the BTK kinase through covalent binding(7). The vast majority of CLL patients treated with ibrutinib derive prolonged clinical benefit and most patients respond to the drug(8–10).
Following prolonged therapy with ibrutinib, some CLL patients stop responding to the drug and eventually develop progressive disease. Such acquired or secondary resistance to ibrutinib has been associated in some cases with acquired mutations in BTK or its downstream target PLCγ, almost always in patients with structural genomic lesions including del17p and/or complex karyotype at study entry(11–13). The currently available data suggest that patients with most high-risk genomic lesions associated with traditional chemotherapy respond well to ibrutinib, with only patients with del17p or complex karyotypes and possibly patients with del11q showing shortened remission durations compared to the others(14–16). The mechanism for this phenomenon is largely unknown but it is of interest to note that the same CLL subgroups have shorter remissions following standard therapies(17, 18).
In the setting of CLL treated with conventional therapy, del17p/TP53 mutations confer direct cellular resistance to chemotherapeutics and also are characterized by a high degree of genomic complexity; the later allows for outgrowth of genomically highly complex cases at relapse that somehow are more resistant to therapy in vivo or regrow faster at relapse(18–21). However, del17p/TP53 mutations have not been associated with increased nucleotide mutation rates in genes and thus are not expected to directly facilitate or enable the generation of BTK or PLCγ mutations; therefore there is no immediate explanation why patients with del17p/TP53 mutations have a shorter duration of response to ibrutinib than those without. It is also unclear if such clinical resistance is solely caused by acquired point mutations in genes, as documented for BTK and PLCγ mutations, or if additional mechanisms affecting ibrutinib response in CLL are operational. In other B-cell neoplasms, like DLBCL or Waldenstrom macroglobulinemia, specific gene mutations have been associated with lower or higher response rates to ibrutinib(22, 23). In CLL, such information is currently unavailable as the number of patients progressing after ibrutinib therapy remains low to date, but is of potential interest.
In this study we have exposed 42 highly characterized paired CLL samples procured before and after traditional chemotherapy to ibrutinib ex vivo and have correlated ibrutinib-induced cell death with genomic and other CLL characteristics. Through these efforts we have identified a heightened sensitivity of CLL cases carrying unmutated IGVH genes or elevated ZAP70 expression or trisomy 12 to ibrutinib-mediated apoptotic cell death. Importantly, we identify del17p/TP53 mutated status as a cause of partial CLL cell-intrinsic resistance to ibrutinib offering a credible mechanism for shorter remission durations of ibrutinib-treated del17p/TP53 mutated CLL patients and providing further impetus for development of novel drugs or combinations to treat this high-risk disease group(24, 25).
METHODS
Patients
Between January 2005 and June 2011, 300 patients evaluated at the University of Michigan Comprehensive Cancer Center were enrolled onto this study. As specified in the protocol, patients were resampled, where applicable, at multiple time points following initial enrollment. The trial was approved by the University of Michigan Institutional Review Board (IRBMED #2004-0962) and written informed consent was obtained from all patients prior to enrollment. DNA from 48 paired pre-treatment and relapsed CLL patients that were subjected to WES and SNP 6.0 profiling constituted the discovery cohort for ibrutinib ex vivo apoptosis assays(19). Of these 48 patients, 42 patients had available paired samples procured before therapy and at subsequent relapse from prior chemotherapy and 6 patients had longitudinal samples analyzed without receiving intercurrent therapy (Supplementary Tables 1 and 2).
CLL treatment was defined as cytotoxic chemotherapy (usually fludarabine, pentostatin, bendamustine or cyclophosphamide) with or without monoclonal antibody therapy for CLL. Clinical information, including Rai stage and all treatments given, was collected on all patients. Patient samples were characterized for selected CLL-associated chromosomal aberrations on the day of trial enrollment as a routine clinical test at the Mayo Clinic (Rochester, MN) using FISH (CLL-FISH). Measurements of CLL-associated molecular characteristics were as described(18). Further details of prior therapy and standard CLL prognostic factors including CLL-FISH results are shown in Supplementary Table 1.
CLL cell purification and ibrutinib apoptosis assays
CLL cell column purification through negative selection and ibrutinib treatment: Cryopreserved CLL PBMC samples were thawed, washed and re-suspended in degassed BSA/EDTA (1×PBS, 0.5% BSA, 1mM EDTA) buffer at an approximate concentration of 107 cells per 85μl. Cells were treated with 10μl/107 cells of Milteny CD3 magnetic microbeads (human cat#130050101) and 5μl/107 cells of Milteny CD14 magnetic microbeads (human cat#130050201) and incubated at 4°C for 25′. Cells were washed with BSA/EDTA and re-suspended in 500μl of BSA/EDTA and passed through Milteny LS columns (cat#130042401) loaded onto a QuadriMax magnet as per manufacturer’s recommendations. The flow through fraction containing CD19+ enriched CLL cells was centrifuged and re-suspended in RPMI1640 medium supplemented with 10% heat inactivated FBS and cultured in 24 well tissue culture plates (Fisher, Nunc low-cell-binding, cat#145387). Ibrutinib (Selleckchem cat#S2680) was diluted serially in medium and was added to corresponding wells at final concentrations ranging from 0.25 μM – 5 μM or 0μM (DMSO only). Plates were incubated at 37°C at 5% CO2 for 72h.
After 72h of incubation cells were washed twice with HBSS. The annexin V-FITC reagent was combined with annexin V binding buffer (10mM HEPES, 140mM NaCl, 2.5mM CaCl2, 0.1% BSA, pH 7.4) in a ratio of 5μl to 100μl and added to each sample and incubated on ice in the dark for 15′. After 15′ 400μl of annexin V binding buffer and 20μl of PI solution were added to each sample and samples immediately analyzed by flow cytometry. IC50 values to ibrutinib treatment were calculated using the curve fitting program XLfit.
Ex vivo BCR signaling studies in purified CLL cells
CLL cells purified through negative column selection as detailed above, were plated at 5×106 cells/500μl RPMI1640 medium without FCS and rested for 1 hour at 37°C and subsequently pre-incubated for 1 hour at 37°C as indicated with ibrutinib at 0 μM (DMSO only) or 0.25 μM, 0.5 μM or 1 μM. Cells were stimulated with goat anti-human IgM (Southern Biotech, cat #2020-08) at 10μg/ml for 15′ and subsequently pelleted through centrifugation at 4°C. Cell pellets were lyzed in lysis buffer containing 1% NP-40 detergent (#DSC41010; Dot Scientific), 150mM NACL, 25mM Tris pH 8.0 (#T6066; Sigma Aldrich), 20mM NAF, 2mM EGTA, 2mM EDTA (#ED2SS; Sigma Aldrich), supplemented with protease inhibitors (#P8340) and phosphatase inhibitors (#P0044), and sodium orthovanadate (#450243; Sigma-Aldrich, St. Louis, MO), and PMSF (#36978; Thermo Scientific). The detergent-soluble fraction of the cell lysates was cleared by centrifugation at 14,000 rpm for 10 min. Protein was fractionated through SDS-PAGE and prepared for immunoblotting using standard procedures. For immunoblotting the following antibodies were used (all rabbit anti-human antibodies from Cell Signaling Technologies): PLCy2: #3872; p1217-PLCy2: #3871; BTK: #8547; p223-BTK: #5082s; AKT: #9272; p473-AKT: #9271s; ERK: #4695; p202/204-ERK: #4370.
Crispr-Cas9-mediated disruption of TP53 in cell lines and ex vivo BCR signaling studies
TP53 targeting
Oligonucleotides encoding guide RNAs targeting exon 4 of TP53 were cloned into the pLentiCRISPRv2 (Addgene #52961) plasmid(26). HEK293T cells were transfected with recombinant pLentiCRISPRv2 together with viral packaging plasmids using the polyethylenimine (PEI; Polyscience Inc., #23966) transfection protocol.
Viral supernatant fractions were collected 48–72 h after transfection by low speed centrifugation to remove cells and debris. HG3, PGA1 and PG-EBV cell lines (all procured directly and authenticated from the Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures GmbH, Germany) were infected by spin-inoculation at 30°C at 2600 rpm using 8μg/ml of Polybrene for 2 h before seeding into fresh medium and subsequent selection in puromycin (Sigma #P9620) at a concentration of 2μg/ml for 3 days. Cells were subsequently plated in 96 well plates at an average density of 3 cells per well. After cell expansion, recombinant cell clones were identified using diagnostic PCR performed directly on aliquots of 20,000 cells per well using the TP53 forward primer: 5′ tctgtctccttcctcttcctaca 3′ and reverse primer: 5′ GGGCCAGACctaagagcaat 3′. PCR products were directly sequenced and analyzed using Mutation surveyor and visual inspection of sequence traces. Positive clones were subcloned using TA cloning (Invitrogen) and 10 clones each analyzed by sequence analysis.
Nutlin3a treatment and ibrutinib treatment of recombinant cell lines: Cells were cultured in RMPI-1640 with 10% heat-inactivated FCS with or without 10μM Nutlin3a (Cayman #10004372) for 16 hours and cells harvested. Cell lysates were prepared in lysis buffer containing 1% NP-40 detergent (#DSC41010; Dot Scientific), 150mM NACL, 25mM Tris pH 8.0 (#T6066; Sigma Aldrich), 20mM NAF, 2mM EGTA, 2mM EDTA (#ED2SS; Sigma Aldrich), supplemented with protease inhibitors (#P8340) and phosphatase inhibitors (#P0044), and sodium orthovanadate (#450243; Sigma-Aldrich, St. Louis, MO), and PMSF (#36978; Thermo Scientific), the lysates cleared by centrifugation and protein fractioned by SDS-PAGE and prepared for immunoblotting using standard methods. Immunoblotting for p53 and actin was performed as described(27).
Ibrutinib treatment of recombinant cell lines and analysis of cell apoptosis and cell death was performed as detailed above. Anti-Ig crosslinking employed anti-IgM or anti-IgG (SouthernBiotech #2020 and #2040) at 10μg/ml for 15′. Cell harvest, immunoblotting and antibodies are as described above.
Solution-based exome capture, HiSeq2000-based massively parallel sequencing and Bioinformatic pipeline analysis of WES data
Solution-based exome capture and HiSeq2000-based massively parallel sequencing and bioinformatic pipeline analysis of WES data was performed as described(28).
Statistical analyses
We used multiple linear regression analysis to better understand the relationship between Ibrutinib IC50 and four potential predictive factors: del17p/TP53 mutated, ZAP70 expresion, IGVH status, and trisomy 12 status. We modeled IC50 as the dependent variable on its original quantitative scale. The four predictive factors were dichotomized (IGVH-UM ≥98% homology to germline; ZAP70 positive ≥20% and trisomy 12 or del17p present in >25% of nuclei).
RESULTS
Ibrutinib-induced apoptosis in purified CLL cells
This study deals with CLL patients samples procured longitudinally before and after standard chemotherapy. The mean elapsed time between procurement dates for the 42 paired pre-treatment and post-treatment (relapsed) CLL cases was 53 months and the relapsed CLL samples were procured within 3–12 months from clinical recognition of relapse status and always in the setting of rising absolute lymphocyte counts (ALCs); see Supplementary Table 1. The CLL cases selected for this study were highly characterized for known clinically relevant traits. In addition, the relapsed CLL samples were analyzed for gene mutations through whole exome sequencing (WES) and subsequent Sanger sequencing of all recurrently mutated genes as detected through WES, SNP 6.0 profiling of genomic copy number changes and deep panel-based gene re-sequencing of TP53, NOTCH1 and other genes as described(28).
To identify CLL cell-intrinsic molecular characteristics that govern response to the BTK inhibitor ibrutinib in CLL, we exposed purified CLL B-cells cultured in RPMI1640 medium supplemented with 10% heat-inactivated fetal bovine serum to escalating concentrations of ibrutinib (0 μM and 0.25μM – 5μM); after 72h, we measured the fraction of cells alive (double negative population) and the apoptotic or dead cell fraction by FACS using annexin-V/PI staining and normalized the surviving fraction of ibrutinib-treated cells to the value for cells exposed to solvent only (e.g. intra-patient normalization for spontaneous apoptosis).
CLL cells demonstrated a surprisingly wide range of ex vivo sensitivities to ibrutinib with IC50 values ranging from 0.37 μM – 9.69 μM in pre-treatment samples and 0.56 μM – >10 μM in post treatment samples. Overall, relapsed samples (after intercurrent chemotherapy) were modestly less sensitive to ibrutinib than the corresponding paired pre-treatment samples (mean IC50 values for pre-treatment and relapsed samples were 2.7 μM and 3.7 μM, respectively). The ibrutinib dose response curves for pre- and post-treatment samples are displayed in Figures 1A and 1B. Complete data including biomarker associations and gene mutations findings are detailed in Supplementary Table 2.
Figure 1. Ibrutinib-induced cell death in purified paired CLL cells (ibrutinib dose-response curves).
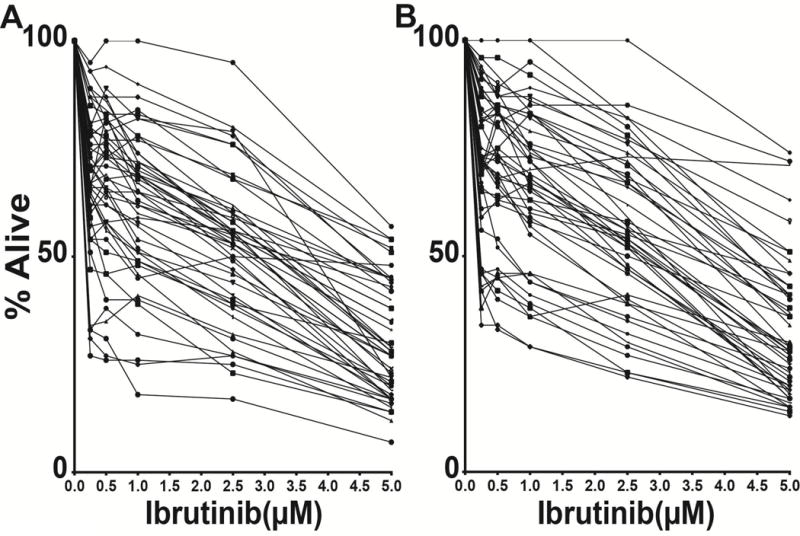
CD19+ CLL cells purified through negative selection were incubated with escalating doses of ibrutinib for 72 hours and the apoptotic and necrotic cell population measured using annexinV/PI staining and FACS. Displayed is the viable double-negative (DN) cell fraction normalized to the individual DN fraction measured in the solvent only controls. The mean of duplicate drug incubations and measurements is displayed. A: pre-treatment CLL samples; B: post-treatment (relapsed) CLL samples.
An unmutated IGVH status, ZAP70 positivity and trisomy 12 predict for heightened apoptotic CLL cell sensitivity to ibrutinib ex vivo
We grouped CLL cases by molecular traits and plotted IC50 values accordingly. Through these efforts, we uncovered that an unmutated IGVH status, ZAP70 positivity and trisomy 12 predict for heightened sensitivity to ibrutinib ex vivo (see Figure 2A–D and Supplementary Figure 1A–D; mean IC50 values for pre-treatment CLL samples were: IGVH-UM (2.1 μM) and IGVH-M (3.5 μM; p=0.02), and for relapsed samples of IGVH-UM (2.8 μM) and IGVH-M (4.0 μM; p=0.04), respectively. Similar findings were derived for ZAP70 status (measured pre-treatment only), with CLL samples with ≤20% ZAP70 positivity by FACS being less sensitive to ibrutinib than ZAP70 samples with >20% positivity (mean IC50 values for pre-treatment CLL samples were 3.5 μM and 2.2 μM, respectively; p=0.04). Trisomy 12 present in >25% of cells was also associated with increased sensitivity to ibrutinib (mean IC50 values for trisomy 12 positive and negative pre-treatment CLL samples were 1.9 μM and 3 μM, respectively; p=0.04). Given the known associations of an unmutated IGVH status or elevated ZAP70 with heightened BCR signaling in CLL these results suggest that strength of BCR signaling underlies some of the differential responses to ibrutinib in CLL and that such differences are CLL cell-intrinsic(3, 29).
Figure 2. Ibrutinib-induced cell death is influenced by CLL cell-intrinsic traits.
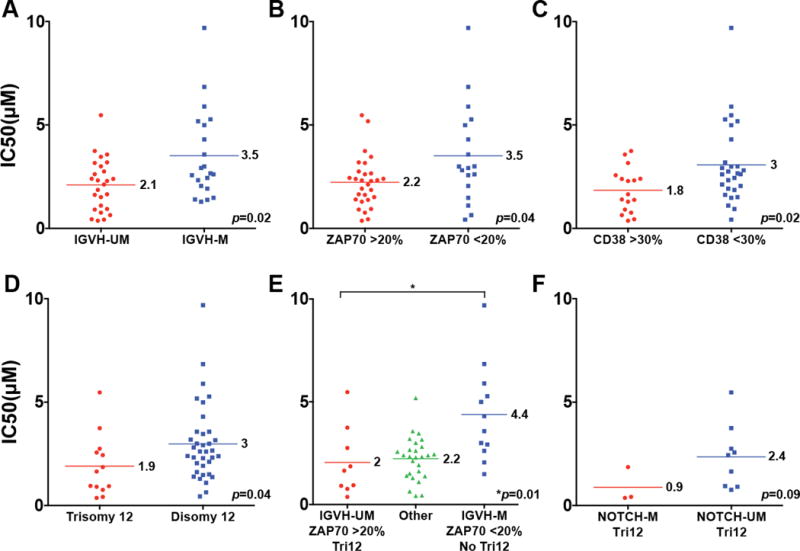
Groupings of ibrutinib CLL IC50 values by biomarker. The mean IC50 values are indicated by horizontal bars and a numerical value. A–D: single dichotomized biomarkers as indicated. E: Left: IGVH-UM AND ZAP70 >20% AND trisomy 12 versus right: IGVH-M AND ZAP70 <20% AND disomy 12; middle: all other cases. F: trisomy 12 cases only with or without NOTCH1 mutations.
We also analyzed the Ibrutinib IC50 values for CLL cases that were IGVH-UM and ZAP70 positive and carried trisomy 12 compared with the cases that carried none of these traits and also compared with the remaining CLL that carried 1 or 2 such traits: As can be seen in Figure 2E, CLL cases that were IGVH-UM/ZAP70+/trisomy12 were significantly more sensitive to ibrutinib than cases that were IGVH-M/ZAP70-/disomy12 (IC50 of 2 μM and 4.4 μM; p=0.01).
Recurrent gene mutations in relation to Ibrutinib-induced apoptosis in CLL
Gene mutations (MYD88, CARD11 and others) have been identified that modulate the efficaciousness of ibrutinib in diffuse large B-cell lymphoma (DLBCL) or Waldenstrom’s macroglobulinemia (WM)(22, 23). To identify potential modifier mutations to ibrutinib in CLL, we grouped CLL cases by gene mutation status for genes commonly mutated in CLL and plotted IC50 values and means (see Supplementary Figure 2). Overall, mutations in MYD88, NOTCH1, SF3B1, XPO1, POT1 and NXF1 did not seem to influence ibrutinib-mediated ex vivo CLL cell apoptosis significantly. Mutations in EGR2, CHD2 and FBXW7 were associated with higher mean IC50 values but case numbers were too small for definitive conclusions. However, EGR2 may be deserving of further study, given downstream involvement in BCR signaling(30).
Given the known enrichment of NOTCH1 mutations in CLL with trisomy 12, we also grouped trisomy 12 CLL cases by NOTCH1 mutation status(31, 32). While all trisomy 12/NOTCH1 mutated cases were very sensitive to ibrutinib, ultimately, the small group of cases fulfilling these criteria (N=3) prevented definitive conclusions to be drawn (Figure 2F).
Analysis of paired CLL samples uncovers a subset of cases with acquired ibrutinib resistance at relapse
The analysis of paired tumor samples procured over extended time periods or after therapy can be informative with regards to identification of acquired changes affecting drug sensitivities. We compared ibrutinib’s IC50 values in 48 paired CLL samples treated with conventional therapies and overall measured a high degree of concordance (see Figure 3 and Supplementary Figure 3). This high degree of concordance provided further confidence in the reliability and validity of the assay and confirmed that the ex vivo ibrutinib CLL kill threshold is largely determined by CLL cell-intrinsic factors that were maintained for years within individual cases.
Figure 3. Ibrutinib CLL IC50 values in paired pre-treatment and post-treatment (relapsed from chemotherapy) paired CLL samples.
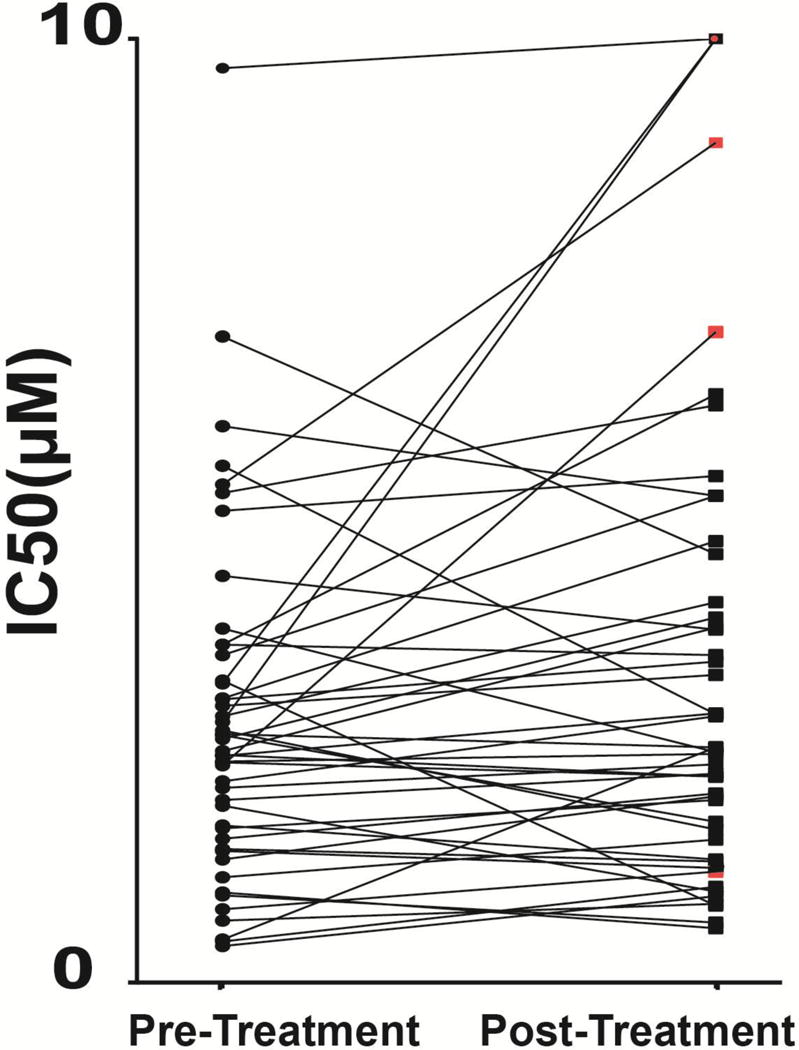
CLL cases with acquired del17p/TP53 mutations are marked with a red square.
A few CLL cases however were markedly more resistant to ibrutinib at the time of relapse from prior conventional CLL therapy (see Figure 3 and Supplementary Table 2). Interestingly, three of the five cases showing a substantial increase in IC50 had acquired a del17p/TP53 mutated status or increased the del17p/TP53 mutant fraction. Specifically, CLL #90 (acquired a del17p and a clonal TP53 mutation (variant allele frequency [VAF] of 0.569) at relapse; CLL #104 positive for del17p before therapy and at relapse increased the mutant TP53 VAF from 0.718 pre-treatment to 0.969 at relapse; and, CLL #117 acquired a del17p at relapse and three distinct TP53 mutations with VAFs of 0.166, 0.243 and 0.419, respectively. The two other cases (CLL #78 and #94) demonstrating a substantial increase in IC50 did not have recurrent genomic changes that could be readily implicated in relative ibrutinib resistance. Specifically, CLL #78 harbored clonal mutations in FBXW7, NOTCH1, POT1 and ZMYM3 and a short del13q-I before and after chemotherapy overall demonstrating no genomic clonal evolution; CLL #94 harbored clonal mutations in FAT3 and NOTCH1 and two stable aCNAs before and after chemotherapy also without evidence for genomic clonal evolution.
Of potential interest, three CLL cases with acquired TP53 mutations at relapse did not markedly change their ibrutinib sensitivities. Two of these cases did not acquire a del17p. Specifically, CLL #125 acquired two distinct TP53 frameshift mutations at clonal VAFs (0.539 and 0.448, respectively) indicative of complete p53 protein inactivation, while CLL # 219 acquired a mono allelic TP53 splice site mutation with a clonal VAF of 0.491. There were no CLL cases carrying del17p and wildtype TP53 available for analysis.
Overall, these findings allowed for the hypothesis to be formulated that the combined del17p/TP53 mutated status is a major determinant of ibrutinib sensitivity in CLL.
We also identified a few CLL cases that had markedly increased sensitivity to ibrutinib at relapse. We reviewed their genomic evolution or lack thereof over time for signals that could potentially explain findings: CLL #170 carried a clonal FBXW7 mutation and 4 acquired copy number changes (aCNAs) all of which were stable over time; CLL #218 demonstrated evidence for a rising clone post chemotherapy identified through gains in the mutant allele frequencies of the genes FAT3, MGA and SAMHD1 together with the marked decline of a clone carrying a SF3B1 mutation and carried no aCNA in either disease phase; CLL #72 was marked by at least two rising clones post chemotherapy identified through gains in the mutant allele frequencies of the genes ATP10A and SF3B1, respectively, and also acquired a chromosomal microdeletion on chr.7 (delineated by the SNPs rs1962039 and rs2727500); and, CLL #2 was marked by at least one rising clone post chemotherapy identified through gains in the mutant allele frequencies of the genes FAT3 and BATF1, respectively, and also acquired a chromosomal microdeletion on chr.16 (delineated by the SNPs rs130002 and rs9936111). Therefore, three out of four cases with marked shifts to ibrutinib sensitivity post chemotherapy displayed clonal evolution; however no shared genomic event was identified.
Del17p/TP53 mutated status is associated with substantially reduced sensitivity to ibrutinib-induced apoptosis in CLL
Based on the findings detailed above, we identified an additional 15 cases of CLL that carried TP53 mutations, 13 of which also carried del17p and subjected these cases to the ibrutinib CLL cell kill assay (Supplementary Table 2). Overall, CLL cells carrying del17p/TP53 mutated status were substantially less sensitive to ibrutinib than CLL without del17p (the mean IC50 values for del17p/TP53 mutated status or no del17p cases were 6.4 μM and 2.6 μM, respectively; p<0.001; of note only two cases carried clonal TP53 mutations in the absence of del17p; Figure 4A).
Figure 4. Del17p/TP53 mutations confer partial resistance to Ibrutinib-induced cell death in CLL.
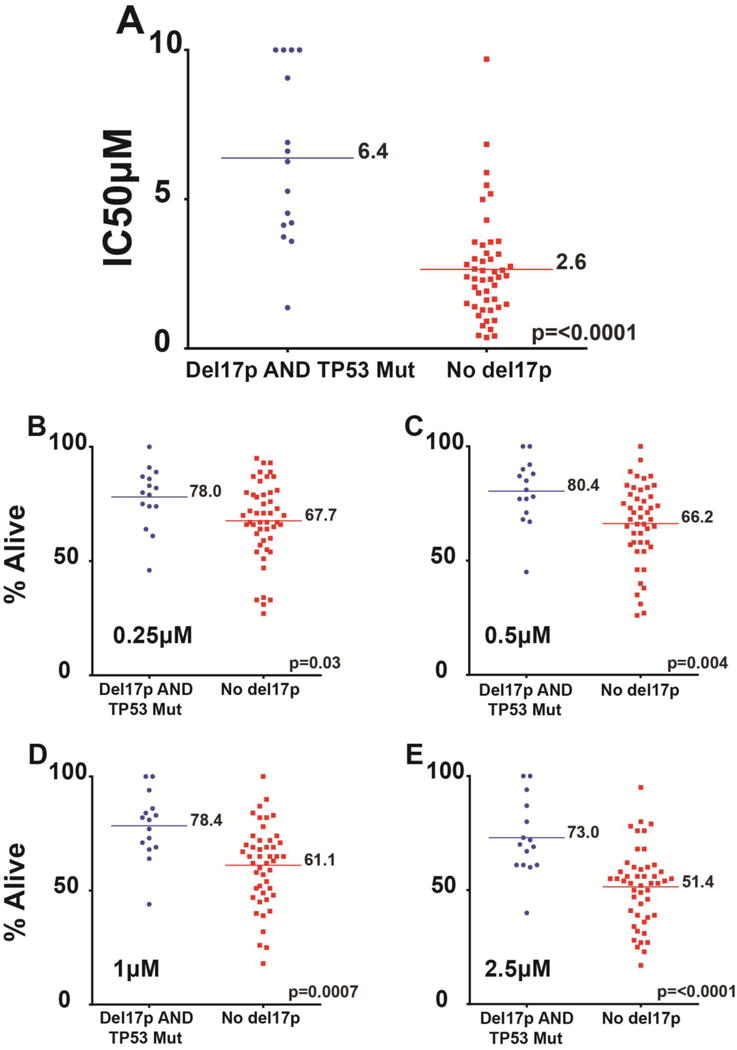
A: Groupings of ibrutinib CLL IC50 values by del17/TP53 status. The mean IC50 values are indicated by horizontal bars and a numerical value. B–E: Cell fraction alive following ibrutinib treatment grouped by del17/TP53 status and by ibrutinib concentrations used.
We also grouped CLL cases by del17p/TP53 mutated status and analyzed the fraction of CLL cells alive after ibrutinib treatment (the annexinV-PI double negative fractions) for varying ibrutinib concentrations (0.25, 0.5, 1 and 2.5 μM). As can be seen in Figure 4B–E, for all ibrutinib concentrations a substantial and significantly higher fraction of CLL cells resisted ibrutinib-induced apoptosis.
Next, we grouped CLL cases that did not carry del17p/TP53 mutated status by the degree of genomic complexity as measured through SNP 6.0 profiling at varying thresholds and analyzed ibrutinib IC50 values for apoptosis. No significant associations were detected (see Supplementary Figure 4A–B).
Finally, we used multiple linear regression analysis to better understand the relationship between ibrutinib IC50 and four potential predictive factors: del17p/TP53 mutated, ZAP70 expression, IGVH status, and trisomy 12. We modeled IC50 as the dependent variable on its original quantitative scale. The four predictive factors were dichotomized (IGVH-UM ≥98% homology to germline; ZAP70 positive ≥20% and trisomy 12 or del17p present in >25% of nuclei). In the fitted model, only del17p/TP53 mutated and trisomy 12 showed statistically significant association with ibrutinib IC50. IC50 is approximately 4 units (μM) higher (95% CI 2.73 to 5.28; p<0.001) for patients with del17p/TP53 mutated compared to those without and IC50 is approximately 1.6 units (μM) lower (95% CI −2.88 to −0.32; p=0.02) for patients with trisomy 12 compared to those without. These associations are estimated using multiple regressions so the del17p/TP53 mutated association should be interpreted as being controlled for trisomy 12, and vice versa.
BCR signal transduction studies in CLL cells ex vivo
We proceeded with ex vivo BCR signaling studies in a subset of the CLL cases analyzed for ibrutinib sensitivity. CLL were grouped by presence (N=6) or absence (N=11) of del17p/TP53 mutated status. Purified CLL cells were left untreated, or pre-treated for 1h with 0,25 μM, 0.5 μM or 1 μM of ibrutinib followed by treatment with anti-IgM for 15′. Cells were pelleted, lysates made, protein fractionated and prepared for immunoblotting with antibodies to p1217-PLCy2; PLCy2; p223-BTK; BTK; p473-AKT; AKT; p202/204-ERK and ERK. Data are displayed in Figure 5A and B (please note that the results for individual blots and epitopes cannot be directly quantitatively compared across different patients as exposure times for various blots are different).
Figure 5. A–B: Results of ex vivo B-cell receptor signaling studies in CLL.
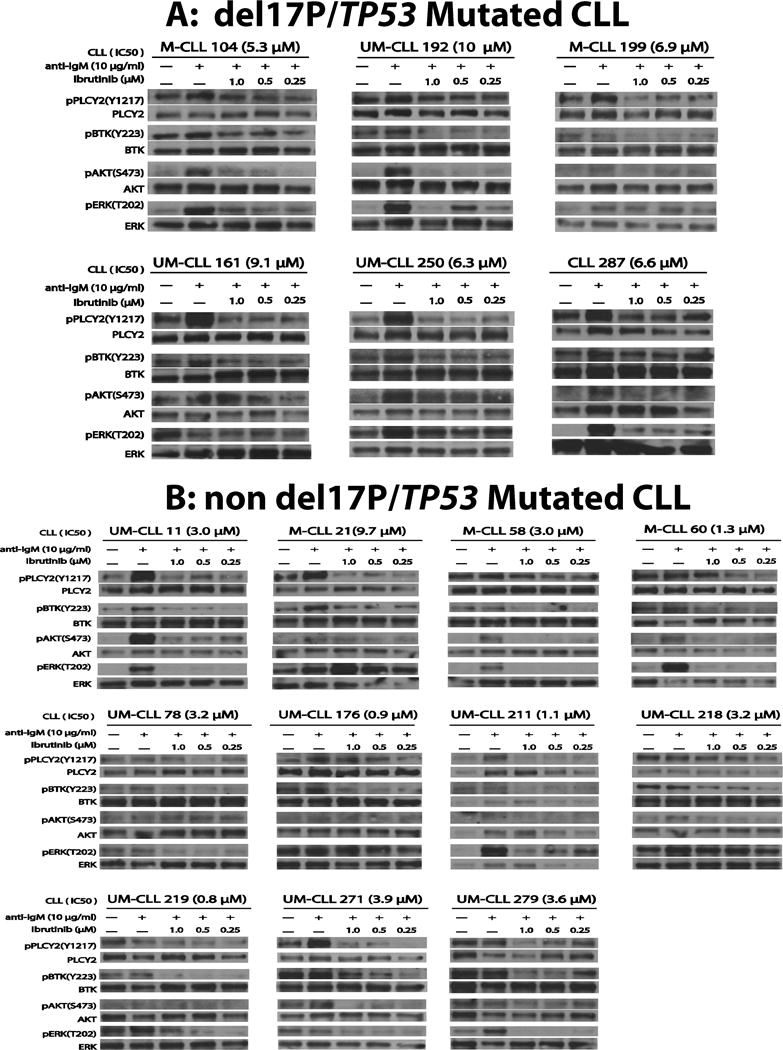
Purified CLL cells were cultured in serum-free medium for 2 hours and left untreated, or pre-treated with ibrutinib at 0 μM (DMSO only) or 0.25 μM, 0.5 μM or 1 μM for 60′ and subsequently treated with anti-IgM at 10μg/ml for 15′. Cells were pelleted, lysates made, protein fractionated and prepared for immunoblotting with antibodies to p1217-PLCy2; PLCy2; p223-BTK; BTK; p473-AKT; AKT; p202/204-ERK and ERK. Figure 5; Panel A (del17p/TP53 mutated CLL) and Figure 5; Panel B (non-del17p/TP53mutated CLL). The results for individual blots and epitopes cannot be directly quantitatively compared across different patients as exposure times for various blots are different. The IGVH mutation status for each CLL case is indicated (UM: unmutated; M: mutated). The ex vivo IC50 values to ibrutinib are indicated in brackets.
Overall data support multiple relevant conclusions including: i) that BCR signaling is active ex vivo in most CLL samples even prior to anti-IgM BCR crosslinking, as evidenced by substantial baseline phosphorylation of p1217-PLCy2 and p223-BTK(3, 33); ii) that CLL samples display substantial variability in baseline and anti-IgM crosslinking-induced substrate phosphorylation and that only a subset of cases display phosphorylation of all 4 interrogated substrates and phosphorylation sites; iii) that PLCy2 is frequently strongly phosphorylated at aminoacid residue 1217 after IgM crosslinking; iv) that ibrutinib treatment at clinically achievable concentrations in most CLL cases prevented anti-IgM crosslinking-induced phosphorylation of the 4 tested substrates; however, it is noteworthy and of potential relevance that for a subset of cases target phosphorylation was not reduced below baseline levels while in other cases much more significant inhibition was achieved. Specifically, in most of the del17/TP53mut CLL cases, ibrutinib did not fully abolish the phosphorylation on p1217-PLCy2 or p223-BTK while a subset of the more sensitive non-del17/TP53wt cases demonstrated inhibition below baseline (for instance see p223-BTK in Figure 5A in CLL # 104, 161, 250 and 287 and in Figure 5B in CLL # 58, 60, 176, 219, 271 and 279). These data allow for the hypothesis to be formulated that incomplete inhibition of BCR signaling may underlie some of the observed differences in CLL cell sensitivity to ibrutinib treatment. Given overall preserved BCR signaling in CLL ex vivo, post BCR signaling defects in the apoptotic machinery and altered thresholds are likely to also contribute to the overall phenomena observed.
Studies of CLL- and B-cell lymphoblastoid cell lines carrying crispr-Cas9-mediated disruption of TP53
To study if TP53 disruption directly affects BCR signaling, we employed three cell lines: HG3 and PGA1, which are CD5+/CD19+ CLL lymphoblastoid cell lines and PG-EBV, which is an EBV transformed B-cell lymphoblastoid cell line, all of which are TP53 unmutated as verified by us through sequencing(34). Over a dose range of 0.25 μM to 5 μM of ibrutinib the HG3 and PGA1 lines were not sensitive to ibrutinib while the PG-EBV line displayed an IC50 value of ~2.5 μM (Figure 6A; Supplementary Figure 5A; Supplementary Figure 6A). Next, we disrupted or functionally inactivated TP53 using crispr-Cas9-mediated targeting of TP53 exon 4. Resulting cell lines (see methods) carried TP53 frameshift mutations or in-frame deletions and displayed p53 protein responses to the MDM2 inhibitor Nutlin3a that is typical for inactive p53 (either no p53 induction and high baseline expression or no p53 expression as in the case of the frameshift mutants as compared with the typical patterns of strong p53 induction detected in control cells) (Figure 6B–C; Supplementary Figure 5B–C; Supplementary Figure 6B–C).
Figure 6. A–E: Effects of TP53 disruption on BCR signaling in the HG3 CLL lymphoblastoid cell line.
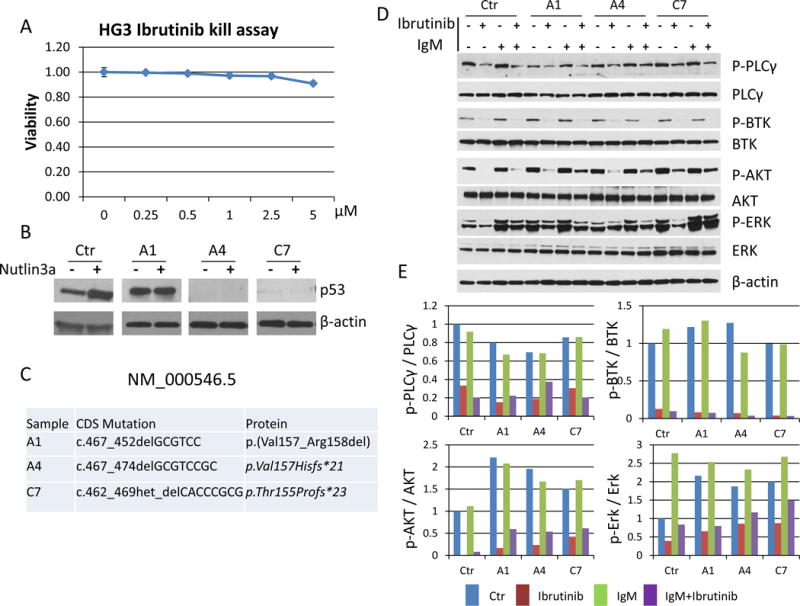
A: Ibrutinib treatment at indicated dosing for 72 hours and annexin V/PI quantitation of cell apoptosis and death. B: p53 immunoblotting before and after 16 hours of Nutlin3a treatment; C: details of TP53 mutations induced using crispr-Cas9 targeting; D: Results of immunoblotting for control and TP53 disrupted cell clones following anti-Ig treatment; E: quantification of immunoblot results using Image J software. Displayed are ratios of p1217-PLCy2/PLCy2; p223-BTK/BTK; p473-AKT/AKT; p202/204-ERK/ERK. Ctr: un-manipulated cells; A1, A4, C7; individual TP53 disrupted cell clones.
In HG3 and PGA1 cell lines which are insensitive to ibrutinib at baseline, we detected robust anti-Ig-induced BCR signaling as measured through immunoblotting for p1217-PLCy2; PLCy2; p223-BTK; BTK; p473-AKT; AKT; p202/204-ERK and ERK, performed in two separate experiments per cell line (Figure 6D–E; Supplementary Figure 5D–E). We find that despite >90% inhibition of p223-BTK (and also p473-AKT) these cells were not induced to undergo apoptosis but we note that the p202/204-ERK signal was only modestly reduced by ibrutinib. Despite some clone to clone variation, the disruption of TP53 did not markedly affect BCR signal transduction. Data overall support that BCR signaling, ibrutinib-mediated inhibition of key signaling intermediates (especially p223-BTK) and cell death are dissociate in these lines.
Finally, an interesting finding emerged from parallel studies of the ibrutinib sensitive cell line PG-EBV. Following crispr-Cas9-mediated TP53 disruption, resulting clones dramatically altered their phenotype, loosing expression of PLCy2 and BTK and substantially upregulating expression of p473-AKT (Supplementary Figure 6D–E; plz note that ß-actin is a p53 response gene, explaining substantially lower expression in TP53 mutant clones). These changes were associated with acquired resistance to ibrutinib. These unexpected findings should motivated follow-up studies in resistant CLL, transformed CLL or MCL to determine if such changes occur in vivo.
DISCUSSION
In this study, we have performed ibrutinib CLL cell kill assays ex vivo in serum-supplemented medium with the goal of identifying CLL factors that govern ibrutinib sensitivities and apoptosis thresholds. The experimental design used highly characterized paired CLL samples procured before and after conventional non-targeted CLL therapies (mostly chemo-immunotherapies).
Summarizing the major findings from this study, we identify multiple CLL cell-intrinsic factors including i) an unmutated IGVH status and elevated ZAP70 expression as contibutors to ibrutinib sensitivity, ii) presence of trisomy 12 as an independent determinant of ibrutinib sensitivity, and importantly, iii) identify del17p/TP53 mutated status as a major cell-intrinsic independent resistance factor to ibrutinib treatment, independent of acquired BTK or PLCγ mutations(12).
BTK is of major importance to B-cell development and B-cell receptor signaling and it was therefore of interest that CLL with unmutated IGVH or elevated ZAP70 demonstrated heightened sensitivity to ibrutinib even in the absence of antigen or micro environmental stimuli. Data suggest a greater dependence on tonic BCR signaling for survival for these CLL cases that translates into greater ibrutinib sensitivities(33). Similarly, in patients treated with ibrutinib an unmutated IGVH status has been correlated with less pronounced ibrutinib-induced lymphocytosis possibly due to more effective killing of CLL cells in the bloodstream(35, 36). Similar findings have been reported for trisomy 12 status(37).
Overall, the most impactful finding from this study is the surprising identification of del17p/TP53 mutated status as a strong independent cell-intrinsic resistance factor to ibrutinib. The del17p/TP53 mutated status is enriched upon prior chemotherapy(19, 38, 39). Clinically, although CLL patients with del17p generally fare better when treated with ibrutinib than with other standard chemotherapies, they appear to relapse faster than other genomically defined CLL subgroups in ibrutinib trials; a finding that hitherto lacked an explanation. While CLL with del17p/TP53 mutations is characterized by a high degree of genomic instability, it has not been demonstrated that such instability also applies to acquisition of gene mutations that could directly confer resistance to ibrutinib(18). It was therefore unclear, what mechanism underlies the relatively reduced efficaciousness to ibrutinib in CLL patients with del17p/TP53. Our data suggest that this reduced ibrutinib efficaciousness is CLL cell- intrinsic and due to reduced sensitivity to ibrutinib-induced apoptosis in CLL cells carrying del17p/TP53. In two CLL-like lymphoblastoid cell lines, in which we engineered TP53 disruptions, we detected increased p-AKT and p-ERK at baseline and after IgM crosslinking. While the molecular mechanisms for this observation remain unidentified, possibilities center on miR deregulations, PTEN expression changes or less specific effects due to increased cell growth and proliferation. These data thereby identify a clinically relevant CLL cell-intrinsic ibrutinib resistance mechanism that is independent of BTK itself or its immediate downstream target PLCγ, which have been linked with cases of acquired resistance to ibrutinib(12). Whether this reduced ibrutinib sensitivity that is observed in vivo and ex vivo is due to greater resistance of TP53-mutated CLL cells to apoptosis in general or if more specific mechanisms are operational remains unresolved. In this context it is noteworthy, that our ex vivo BCR signal transduction studies identified multiple CLL cases in which BTK autophosphorylation as measured through p223-BTK was not suppressed below the baseline levels that was measured in unstimulated CLL. It is therefore possible that such incomplete suppression of BTK and also of PLCy2 may translate into differential effects of ibrutinib in CLL patients and ultimately may effect clinical remission durations. A quantitative analysis of BCR signaling intermediates in CLL cells isolated from patients responding to ibrutinib and at relapse may further inform on ibrutinib mechanisms of action and acquired resistance.
In this study, we used simple assay conditions to measure ibrutinib-induced CLL cell apoptosis ex vivo. By avoiding experimental substitutes for the CLL microenvironment that would have masked the CLL cell-intrinsic variability in ibrutinib-mediated apoptosis we were able to identify unexpected modifiers of this response. In contrast, in vivo, CLL cells in secondary lymphoid organs are in contact with other cells and secreted molecules that together constitute the CLL microenvironment(40). Upon exposure to ibrutinib, CLL cell to microenviromental interactions are weakened and a fraction of CLL cells egresses the lymphoid organs and relocates to the blood stream(41–44). How relevant are our ex vivo findings to CLL patients treated with ibrutinib in vivo? To answer this question, it is worth noting that fluctuations in micro-environmental inputs or the strength of CLL cell to micro-environmental interactions have not been linked to differential ibrutinib effects in patients. Instead, traditional genomic markers including del17p/TP53 mutations or complex karyotypes (which are highly associated with del17p/TP53) are associated with shorter remission durations(16). Therefore, our findings of del17p/TP53 mutation-mediated CLL cell-intrinsic partial resistance to ibrutinib-induced apoptosis together with the reported inhibitory effects of ibrutinib on CLL cell proliferation may well be clinically relevant mechanisms especially for CLL cells that have egressed into the bloodstream(45).
Finally, as combination therapies with ibrutinib are advancing into the clinic it would be important to test combinations of drugs that are able to bypass del17p/TP53 mutated status-mediated apoptosis blocks. Of the currently tested or available therapeutics, the BH3 mimetics or therapeutic antibodies may well offer benefits for del17p/TP53 mutated CLL in combinations with ibrutinib(25, 46, 47).
Supplementary Material
Translational Relevance.
Ibrutinib, a Bruton’s tyrosine kinase (BTK) inhibitor, is approved for the treatment of relapsed CLL and CLL with del17p and is substantially changing the approach to clinical CLL management. Mechanistically, ibrutinib interferes with BCR signaling as well as multiple CLL cell to microenvironment interactions. Given the importance of ibrutinib in the therapeutic management of CLL, a deeper understanding of factors governing sensitivity and resistance is warranted. In this study we have exposed highly characterized paired CLL samples longitudinally procured from patients before and after traditional chemotherapy to ibrutinib ex vivo. We demonstrate that multiple CLL cell-intrinsic traits including an unmutated IGVH status, ZAP70 positivity and trisomy 12 confer sensitivity to ibrutinib. In contrast, a combined del17p/TP53 mutated status in CLL cells confers substantial resistance to ibrutinib-induced apoptosis ex vivo. Therefore, in addition to acquired mutations in BTK, other CLL cell-intrinsic mechanisms influence ibrutinib sensitivity or resistance in CLL.
Acknowledgments
We are grateful for services provided by the DNA sequencing and bioinformatics cores of the University of Michigan Comprehensive Cancer Center.
Funding Support
Supported by a research grant by Janssen R&D, LLC (SM) and a Clinical Scholars Award of the Leukemia and Lymphoma Society of America (SM). This research is supported (in part) by the National Institutes of Health through the University of Michigan’s Cancer Center Support Grant (5 P30 CA46592).
Footnotes
Conflict of Interest
Sriram Balasubramanian is an employee of Janssen R&D, LLC.
Sami Malek owns stock in Abbvie.
Individual contributions:
Nisar Amin, Kamlai Saiya-Cork, Nan Hu and Sami Malek performed the laboratory research.
Sriram Balasubramanian assisted with data analysis and writing of the paper.
Nisar Amin and Sami Malek wrote the paper.
Sami Malek conceived the study and supervised the work.
References
- 1.Spaargaren M, de Rooij MF, Kater AP, Eldering E. BTK inhibitors in chronic lymphocytic leukemia: a glimpse to the future. Oncogene. 2015;34:2426–36. doi: 10.1038/onc.2014.181. [DOI] [PubMed] [Google Scholar]
- 2.Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood. 2014;123:3390–7. doi: 10.1182/blood-2013-11-535047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118:4313–20. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 4.Advani RH, Buggy JJ, Sharman JP, Smith SM, Boyd TE, Grant B, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–84. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Claro RA, McGinn KM, Verdun N, Lee SL, Chiu HJ, Saber H, et al. FDA Approval: Ibrutinib for Patients with Previously Treated Mantle Cell Lymphoma and Previously Treated Chronic Lymphocytic Leukemia. Clin Cancer Res. 2015;21:3586–90. doi: 10.1158/1078-0432.CCR-14-2225. [DOI] [PubMed] [Google Scholar]
- 7.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–80. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd JC, Furman RR, Coutre SE, Burger JA, Blum KA, Coleman M, et al. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125:2497–506. doi: 10.1182/blood-2014-10-606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien S, Furman RR, Coutre SE, Sharman JP, Burger JA, Blum KA, et al. Ibrutinib as initial therapy for elderly patients with chronic lymphocytic leukaemia or small lymphocytic lymphoma: an open-label, multicentre, phase 1b/2 trial. Lancet Oncol. 2014;15:48–58. doi: 10.1016/S1470-2045(13)70513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddocks KJ, Ruppert AS, Lozanski G, Heerema NA, Zhao W, Abruzzo L, et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients With Chronic Lymphocytic Leukemia. JAMA Oncol. 2015;1:80–7. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–94. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furman RR, Cheng S, Lu P, Setty M, Perez AR, Guo A, et al. Ibrutinib resistance in chronic lymphocytic leukemia. N Engl J Med. 2014;370:2352–4. doi: 10.1056/NEJMc1402716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson PA, O’Brien SM, Wierda WG, Ferrajoli A, Stingo F, Smith SC, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–21. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farooqui MZ, Valdez J, Martyr S, Aue G, Saba N, Niemann CU, et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: a phase 2, single-arm trial. Lancet Oncol. 2015;16:169–76. doi: 10.1016/S1470-2045(14)71182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd JC, O’Brien S, James DF. Ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:1278–9. doi: 10.1056/NEJMc1309710. [DOI] [PubMed] [Google Scholar]
- 17.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 18.Ouillette P, Collins R, Shakhan S, Li J, Peres E, Kujawski L, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–61. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19:2893–904. doi: 10.1158/1078-0432.CCR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malek SN. The biology and clinical significance of acquired genomic copy number aberrations and recurrent gene mutations in chronic lymphocytic leukemia. Oncogene. 2013;32:2805–17. doi: 10.1038/onc.2012.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–9. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 22.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21:922–6. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treon SP, Xu L, Hunter Z. MYD88 Mutations and Response to Ibrutinib in Waldenstrom’s Macroglobulinemia. N Engl J Med. 2015;373:584–6. doi: 10.1056/NEJMc1506192. [DOI] [PubMed] [Google Scholar]
- 24.Jain P, Keating M, Wierda W, Estrov Z, Ferrajoli A, Jain N, et al. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood. 2015;125:2062–7. doi: 10.1182/blood-2014-09-603670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaglowski SM, Jones JA, Nagar V, Flynn JM, Andritsos LA, Maddocks KJ, et al. Safety and activity of BTK inhibitor ibrutinib combined with ofatumumab in chronic lymphocytic leukemia: a phase 1b/2 study. Blood. 2015;126:842–50. doi: 10.1182/blood-2014-12-617522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanjana N, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116:71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amin N, Seymour EK, Saiya-Cork K, Parkin B, Shedden K, Malek S. A Quantitative Analysis of Subclonal and Clonal Gene Mutations Pre- and Post-therapy in Chronic Lymphocytic Leukemia. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Widhopf G, Huynh L, Rassenti L, Rai KR, Weiss A, et al. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100:4609–14. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–96. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2012;119:329–31. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shedden K, Li Y, Ouillette P, Malek SN. Characteristics of chronic lymphocytic leukemia with somatically acquired mutations in NOTCH1 exon 34. Leukemia. 2012;26:1108–10. doi: 10.1038/leu.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489:309–12. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 34.Lanemo Myhrinder A, Hellqvist E, Bergh AC, Jansson M, Nilsson K, Hultman P, et al. Molecular characterization of neoplastic and normal “sister” lymphoblastoid B-cell lines from chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1769–79. doi: 10.3109/10428194.2013.764418. [DOI] [PubMed] [Google Scholar]
- 35.Herman SE, Niemann CU, Farooqui M, Jones J, Mustafa RZ, Lipsky A, et al. Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: correlative analyses from a phase II study. Leukemia. 2014;28:2188–96. doi: 10.1038/leu.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woyach JA, Smucker K, Smith LL, Lozanski A, Zhong Y, Ruppert AS, et al. Prolonged lymphocytosis during ibrutinib therapy is associated with distinct molecular characteristics and does not indicate a suboptimal response to therapy. Blood. 2014;123:1810–7. doi: 10.1182/blood-2013-09-527853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson PA, Ferrajoli A, O’Brien S, Wierda WG, Keating MJ, Burger JA. Trisomy 12 is associated with an abbreviated redistribution lymphocytosis during treatment with the BTK inhibitor ibrutinib in patients with chronic lymphocytic leukaemia. Br J Haematol. 2015;170:125–8. doi: 10.1111/bjh.13269. [DOI] [PubMed] [Google Scholar]
- 38.Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29:877–85. doi: 10.1038/leu.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Fama R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–47. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herman SE, Mustafa RZ, Jones J, Wong DH, Farooqui M, Wiestner A. Treatment with Ibrutinib Inhibits BTK- and VLA-4-Dependent Adhesion of Chronic Lymphocytic Leukemia Cells In Vivo. Clin Cancer Res. 2015;21:4642–51. doi: 10.1158/1078-0432.CCR-15-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–9. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Rooij MF, Kuil A, Geest CR, Eldering E, Chang BY, Buggy JJ, et al. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor- and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood. 2012;119:2590–4. doi: 10.1182/blood-2011-11-390989. [DOI] [PubMed] [Google Scholar]
- 43.Herman SE, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–96. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wodarz D, Garg N, Komarova NL, Benjamini O, Keating MJ, Wierda WG, et al. Kinetics of CLL cells in tissues and blood during therapy with the BTK inhibitor ibrutinib. Blood. 2014;123:4132–5. doi: 10.1182/blood-2014-02-554220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo A, Lu P, Galanina N, Nabhan C, Smith SM, Coleman M, et al. Heightened BTK-dependent cell proliferation in unmutated chronic lymphocytic leukemia confers increased sensitivity to ibrutinib. Oncotarget. 2016;7:4598–610. doi: 10.18632/oncotarget.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cervantes-Gomez F, Lamothe B, Woyach JA, Wierda WG, Keating MJ, Balakrishnan K, et al. Pharmacological and Protein Profiling Suggests Venetoclax (ABT-199) as Optimal Partner with Ibrutinib in Chronic Lymphocytic Leukemia. Clin Cancer Res. 2015;21:3705–15. doi: 10.1158/1078-0432.CCR-14-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger JA, Keating MJ, Wierda WG, Hartmann E, Hoellenriegel J, Rosin NY, et al. Safety and activity of ibrutinib plus rituximab for patients with high-risk chronic lymphocytic leukaemia: a single-arm, phase 2 study. Lancet Oncol. 2014;15:1090–9. doi: 10.1016/S1470-2045(14)70335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


