SUMMARY
The xyloglucan endo-transglycosylase/hydrolase (XTH) gene family encodes enzymes of central importance to plant cell wall remodelling. The evolutionary history of plant XTH gene products is incompletely understood vis-à-vis the larger body of bacterial endo-glycanases in Glycoside Hydrolase Family 16 (GH16). To provide molecular insight into this issue, high-resolution X-ray crystal structures and detailed enzyme kinetics of an extant transitional plant endo-glucanase (EG) were determined. Functionally intermediate between plant XTH gene products and bacterial licheninases of GH16, Vitis vinifera EG16 (VvEG16) effectively catalyzes the hydrolysis of the backbones of two dominant plant cell wall matrix glycans, xyloglucan (XyG) and β(1,3)/β(1,4)-mixed-linkage glucan (MLG). Crystallographic complexes with extended oligosaccharide substrates reveal the structural basis for the accommodation of both unbranched, mixed-linked (MLG) and highly decorated, linear (XyG) polysaccharide chains in a broad, extended active-site cleft. Structural comparison with representative bacterial licheninases, a xyloglucan endo-tranglycosylase (XET), and a xyloglucan endo-hydrolase (XEH) outline the functional ramifications of key sequence deletions and insertions across the phylogenetic landscape of GH16. Although the biological role(s) of EG16 orthologs remains to be fully resolved, the present biochemical and tertiary structural characterization provides key insight into plant cell wall enzyme evolution, which will continue to inform genomic analyses and functional studies across species.
Keywords: Vitis vinifera, Accession numbers: NCBI refseq: XP_002273975.1, PDB identifiers 5DZE, 5DZF, 5DZG, and 5SV8, GH16, XET, XEH, XTH, mixed-linkage endo-glucanase, licheninase, xyloglucan, X-ray crystallography
INTRODUCTION
Plant cell walls are complex barriers constructed from a wide variety of carbohydrate and non-carbohydrate polymers (Albersheim et al. 2010, Carpita and McCann 2000). The wall serves as a rigid structural support, yet has a dynamic morphology that enables plant cells to assume a vast array of structures. The strength and flexibility of plant cell walls comes from the carefully programmed assembly of a composite matrix of cellulose microfibrils, matrix glycans (“hemicelluloses”), pectins, structural proteins, and polyphenolics. This complexity necessitates a large complement of proteins and enzymes for assembling, remodelling, and recycling the diverse cell wall components. Despite significant advances, many of these enzymes remain uncharacterized or unknown (Mewalal et al. 2014).
The matrix glycans, including mannans, xylans, mixed-linkage glucans (MLGs), and xyloglucans (XyGs), are biosynthesized in the Golgi apparatus by a diversity of glycosyltransferases (GTs). These glycans are then shuttled to the apoplast for assembly into the cell wall (Albersheim, et al. 2010, Carpita and McCann 2000, Pauly et al. 2013, Scheible and Pauly 2004). Wall remodelling may occur subsequently through hydrolysis (leading to glycan degradation) and transglycosylation (leading to glycan rearrangement), catalyzed by enzymes in diverse glycoside hydrolase (GH) families (Buchanan et al. 2012, Eklöf and Brumer 2010, Fincher 2009, Frankova and Fry 2013, Kaewthai et al. 2013, Lopez-Casado et al. 2008, Minic and Jouanin 2006, Sampedro et al. 2012, Simmons et al. 2015, Tyler et al. 2010).
In this context, the xyloglucan endo-transglycosylases (XETs, EC 2.4.1.207), which catalyze the non-hydrolytic cleavage and re-ligation of XyG chains, are the archetype of apoplastic wall remodelling enzymes (Eklöf and Brumer 2010, Rose et al. 2002). XET activity has been implicated in the incorporation of nascent XyG into primary walls, the expansion of primary walls, the morphogenesis of secondary cell walls, and the function of reaction wood (Bourquin et al. 2002, Gerttula et al. 2015, Scheller and Ulvskov 2010). XETs are encoded by the large and diverse xyloglucan endo-transglycosylase/hydrolase (XTH) gene subfamily of Glycoside Hydrolase Family 16 (GH16), members of which number 20–60 in all vascular plant genomes sequenced thus far (Eklöf and Brumer 2010, Goodstein et al. 2012). As implied by their name, a select number of XTH genes encode predominantly hydrolytic xyloglucan endo-hydrolases (XEH, EC 3.2.1.151) (Baumann et al. 2007, Kaewthai, et al. 2013).
Given their widespread distribution across monocots, dicots, and early vascular plants (Eklöf and Brumer 2010), revealing the functional diversity of XTH gene products is fundamental to understanding plant cell wall development, and thus remains an extremely active research area (for examples, see (Becnel et al. 2006, Hara et al. 2014, Lee et al. 2010, Ye et al. 2015, Ye et al. 2012) and references therein). In this context, the evolution of catalytic specificity in these enzymes is particularly intriguing. The majority of XTH gene products characterized thus far appear to encode strict transglycosylases (XETs, EC 2.4.1.207) with little to no hydrolytic (XEH, EC 3.2.1.151) activity (reviewed in (Eklöf and Brumer 2010); for recent examples, see (Hara, et al. 2014, Maris et al. 2011, Ye, et al. 2015)). On the other hand, previous work has indicated that XEH activity is likely to have arisen from ancestral XETs through a specific amino acid loop insertion in a small clade of XTH gene products (Baumann, et al. 2007). This fundamental structure-function analysis has enabled the further refinement of XTH phylogeny, which significantly facilitates functional prediction from sequence data (Eklöf and Brumer 2010, Hara, et al. 2014, Kaewthai, et al. 2013). Continuing efforts to characterize XTH gene products are uncovering new catalytic function that can be readily mapped onto, and thus further enhance, the predictive power of this molecular phylogeny (Simmons, et al. 2015).
A number of fundamental questions remain regarding the evolutionary origin of the XTH gene products in the broader context of GH16. GH16 is a large family of β-jelly-roll proteins whose individual activities include the hydrolysis and/or transglycosylation of a range of β-glucans and β-galactans (Lombard et al. 2014). In early phylogenetic analyses of GH16, it was noted that the closest homologs of plant XTH gene products are bacterial licheninases (β(1,3)/β(1,4)-β-D-glucan 4-glucanohydrolases, EC 3.2.1.73) (Barbeyron et al. 1998, Michel et al. 2001) (Figure 1A). This is particularly remarkable, in light of the significant structural and catalytic differences between both groups of enzymes; beyond the conserved active site residues and a common overall fold, primary and tertiary structural similarity is generally low due to major loop differences and the presence of the canonical XTH C-terminal extension (InterPro Domain IPR010713, “XET_C”) (Johansson et al. 2004). Both groups also exhibit widely different substrate specificities: the licheninases hydrolyze linear, undecorated MLG chains, whereas XETs and XEHs act upon the heavily decorated xyloglucan backbone. These large differences obfuscate the evolutionary pathway in the diversification of extant licheninases and XTH gene products.
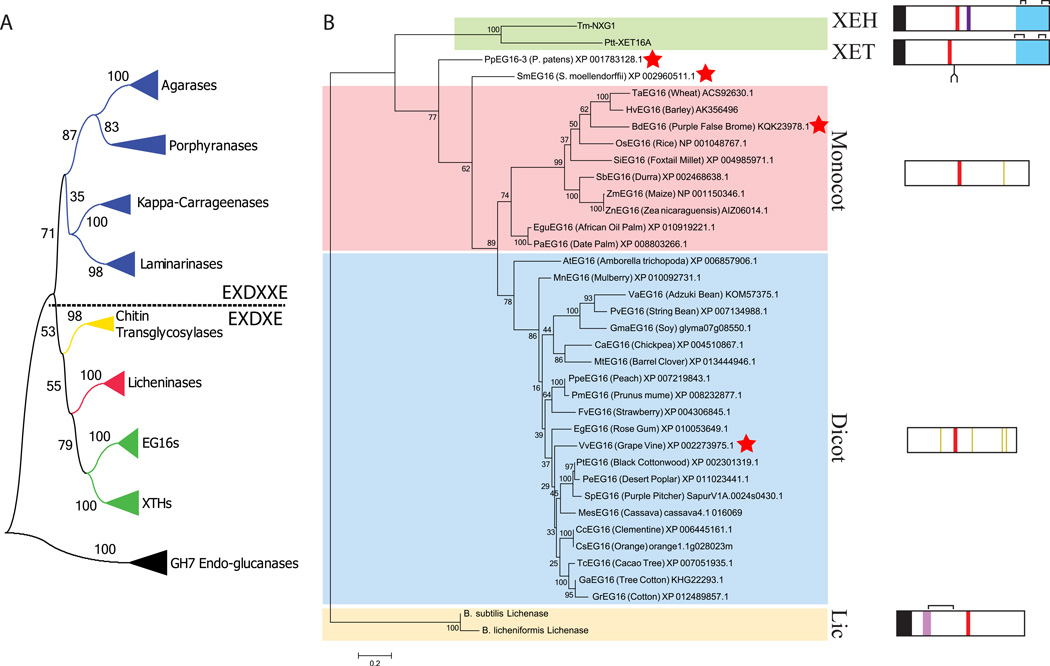
Figure 1.
Protein sequence-based phylogeny EG16 homologs within Glycoside Hydrolase Family 16 (GH16). A) Overall phylogeny of GH16 encompassing all specificities identified to date. Each collapsed branch represents 5 sequences (Table S5). The tree is rooted with 5 GH7 cellulases; GH7 and GH16 together form Clan GH-B. A dashed line separates enzymes containing the EXDXXE “beta-bulge” active-site motif from those with the regular beta-strand EXDXE active-site motif. B) Phylogenetic tree of EG16 homologs identified in Genbank and Phytozome, based on the protein sequence alignment shown in Figure S1. Abbreviated protein names are derived from the genus and species of origin (see corresponding GenBank entries); the common name (where available) is given along with the accession code. The tree is rooted using two Bacillus licheninases and additionally includes a xyloglucan endo-transglycosylase (PttXET16A, (Johansson, et al. 2004)) and a xyloglucan endo-hydrolase (TmNXG1, (Baumann, et al. 2007)) as outliers with known tertiary structures. Sequences indicated with a red star were selected for recombinant expression in E. coli (Table S1). Box diagrams, to scale, indicate key protein sequence features: Black, signal peptides; red, active site EXDXE motif; light purple, licheninase loop extension; dark purple, XEH loop extension; blue, C-terminal XET/XEH extension (XET_C); yellow, conserved cysteine residues in the monocot or dicot EG16 clades; brackets, crystallographically observed disulfide bonds; fork, conserved XET N-glycosylation site. Bootstrap values from 100 maximum likelihood resamplings are shown next to each branch of both trees.
Aided by the significant recent expansion in the number of sequenced plant genomes (Goodstein, et al. 2012), we recently identified a small clade of homologs that represent a key phylogenetic link between the plant XTH gene products and the bacterial licheninases in GH16 (Eklöf et al. 2013). These homologs lack several highly conserved features of XETs and XEHs, including disulfide-bridged C-terminal extensions, N-linked glycosylation sites, and cell-wall-targeting N-terminal signal peptides. Also in contrast with XTH gene products, this emergent clade is uniquely represented by a single member in those genomes in which they are found; various exemplar dicots (e.g. Arabidopsis thaliana) have none. Enzymological characterization of a heterologously expressed representative from Populus trichocarpa (black cottonwood), PtEG16, demonstrated that this enzyme is a broad specificity endo-β(1,4)-glucanase, strikingly capable of hydrolyzing both of the major distinguishing cell wall glycans of dicots and Commelinoid monocots, XyG and MLG, respectively (Eklöf, et al. 2013). However, the commonality of this catalytic ability across the clade, and especially its structural basis, remained experimentally unresolved.
We now report the recombinant production, enzymological characterization, and X-ray crystallography of a new EG16 homolog from a Vitis vinifera (wine grape) expressed-sequence tag (EST) library (Peng et al. 2007). Hydrolytic kinetics on a variety of polysaccharide, oligosaccharide, and synthetic substrates, together with MLG and XyG oligosaccharide crystal complexes, reveal the molecular basis for the substrate plasticity of EG16 active-sites. In turn, a refined phylogenetic analysis enabled by these new data provides unprecedented molecular insight into the evolution of GH16 endo-glucanases and XTH gene products in plants.
RESULTS
Molecular Phylogeny of the EG16 Clade
Thirty-three EG16 sequences were identified by BLAST queries of both the NCBI GenBank and JGI Phytozome databases using PtEG16 (Eklöf, et al. 2013) as a query sequence. Protein sequence alignment revealed high similarity among EG16 homologs, and clear differences with bacterial licheninases and plant XETs and XEHs of GH16 (Figure S1). GH16 enzymes hydrolyze glycosidic bonds using the canonical double-displacement, anomeric configuration-retaining mechanism, which employs both a catalytic nucleophile and a general acid/base residue (Eklöf and Brumer 2010, Koshland 1953, Planas 2000). The consensus active-site motif, DEID(F/I)EFLG, contains the key catalytic glutamate/glutamic acid residues, as well as a catalytic “helper” aspartate residue that is thought to electrostatically modulate catalysis (underlined). This motif is strictly conserved among bacterial licheninases and all plant GH16 members. A phylogenetic tree constructed from the alignment shown in Figure S1 and rooted with two licheninase sequences reveals a division between monocot and dicot EG16s with moderate support (bootstrap value 82%), in addition to outlying Physcomitrella patens and Selaginella moellendorffii sequences (Figure 1B). Inspection of the alignment indicates that the monocot/dicot division likely arises from sequence differences in β-strand 6 and loops 6 and 7, as well as in the Glu/Asp/Ala-rich C-terminal tails (Figure S1). Notably, these variable C-termini bear no homology to the distinctive C-terminal extensions of XETs and XEHs (InterPro Domain IPR010713, “XET_C”) (Baumann, et al. 2007, Johansson, et al. 2004). Unlike the licheninases and the XTH gene products in GH16, EG16 members are rich in cysteine residues throughout the main β-jelly-roll domain, containing between 3 (e.g., BdEG16, HvEG16) and 12 (e.g., PeEG16) per protein. Positional conservation of these cysteine residues is generally poor, although there are four homologous cysteine residues in dicot EG16s, and one in monocot EG16s (Figure 1B). In contrast, the four cysteine residues in the C-terminal domain of XTH gene products (InterPro Domain IPR010713, “XET_C”; which is absent in EG16 homologs) are widely conserved and participate in the formation of 2 structural disulfide bonds (Baumann, et al. 2007, Johansson, et al. 2004).
Recombinant production of VvEG16 in E. coli
From this phylogeny, four genes encoding EG16 targets, representing bryophyte (Physcomitrella patens EG16), lycophyte (Selaginella moellendorffii EG16), monocot (Brachypodium distachyon EG16), and dicot sequences (Vitis vinifera EG16), were selected for heterologous expression in E.coli in native form and in tandem with solubilizing fusion polypeptides (Table S1). Of these, only native VvEG16 and an N-terminal SUMO conjugate could be produced in a stable, soluble form in an initial screen. The expression and purification of native VvEG16, a fortuitous mutant VvEG16(ΔV152) (having a single amino acid deletion of Val152)., as well as the corresponding site-directed catalytic-nucleophile mutants (VvEG16(E89A) and VvEG16(ΔV152/E89A)) and surface-cysteine deletion mutants (VvEG16(C22S/C188S) and VvEG16(ΔV152/C22S/C188S)) under optimized conditions gave exceptional yields (~100 mg/L of E. coli culture) of electrophoretically pure protein following TEV protease cleavage of the His6 affinity tag (Figure S2). All recombinant VvEG16 variants were significantly more stable than PtEG16, which was prone to cysteine oxidation and aggregation upon storage (Eklöf, et al. 2013). VvEG16(ΔV152), in particular, was amenable to crystallization (vide infra). In light of this observation, the majority of the subsequent biochemical analyses were performed on this variant.
VvEG16 is a bi-functional MLG/XyG endo-glucanase
Recombinant VvEG16(ΔV152) hydrolyzed barley MLG (bMLG) with the highest apparent kcat value and lowest apparent KM value of any polysaccharide tested, at the pH optimum of 6.0 and 30°C (Table 1, Figure S3A, Figure S4A,B). Under these conditions, tamarind seed XyG (tXyG) was hydrolyzed less efficiently, evident from a >5-fold higher apparent KM value and a 2-fold lower kcat value. The recombinant wild-type enzyme had nearly identical kinetics on both substrates, thus providing further justification for our focus on the crystallisable VvEG16(ΔV152) variant (Table 1, Figure S3B). Very poor activity of VvEG16(ΔV152) was observed on konjac glucomannan (kGM), carboxymethylcellulose (CMC), and hydroxyethylcellulose (HEC), all of which contain β(1,4)-glycosyl backbone residues, while no activity was detected with wheat arabinoxylan, crystalline cellulose, laminarin, or guar galactomannan. Under reducing conditions (dithiothreitol-containing buffer), VvEG16(ΔV152) is thermally unstable above 50°C (Figure S4C), which is near the thermal stress limit of V. vinifera (Greer and Weston 2010).
Table 1.
Apparent Michaelis-Menten kinetic constants for hydrolysis and transglycosylation reactions catalyzed by wild-type VvEG16 and VvEG16(ΔV152).
| Substrate | Enzyme |
kcat,
app (min−1) |
Km,
app (mM) |
kcat,
app/Km,
app (M−1s−1) |
Assay |
|---|---|---|---|---|---|
| bMLG | VvEG16(ΔV152) | 143±7 | 0.34±0.03* | 7.00* | BCA |
| VvEG16 | 163±5 | 0.36±0.02* | 7.55* | BCA | |
| tXyG | VvEG16(ΔV152) | 68±8 | 1.8±0.4* | 0.63* | BCA |
| VvEG16 | 70±6 | 2.1±0.4 | 0.56* | BCA | |
| HEC | VvEG16(ΔV152) | 10.4±0.8 | 1.1±0.2* | 0.16* | BCA |
| CMC | VvEG16(ΔV152) | 17±2.2 | 2.8±0.5* | 0.10* | BCA |
| kGM | VvEG16(ΔV152) | ND | ND | 0.040±5* | BCA |
| Guar Galactomannan | VvEG16(ΔV152) | ND | ND | ND | BCA |
| Wheat Arabinoxylan | VvEG16(ΔV152) | ND | ND | ND | BCA |
| Laminarin | VvEG16(ΔV152) | ND | ND | ND | BCA |
| GG-PNP | VvEG16(ΔV152) | 0.023±0.002 | 5.9±0.6 | 0.065 | PNP |
| GG-CNP | VvEG16(ΔV152) | 0.45±0.02 | 3.5±0.4 | 2.1 | CNP |
| GG-DNP | VvEG16(ΔV152) | 20±1 | 2.0±0.2 | 170 | DNP |
| GGG-PNP | VvEG16(ΔV152) | 0.15±0.006 | 2.8±0.2 | 0.9 | PNP |
| GGG-CNP | VvEG16(ΔV152) | 1.03±0.04 | 1.1±0.1 | 16 | CNP |
| VvEG16 | 0.98±0.03 | 0.8±0.1 | 20 | CNP | |
| GGG-DNP | VvEG16(ΔV152) | 95±2 | 0.80±0.06 | 2000 | DNP |
| XXXG-CNP | VvEG16 | 12.0±0.5 | 1.1±0.3 | 180 | CNP |
| VvEG16(ΔV152) | 13.9±0.6 | 1.2±0.1 | 190 | CNP | |
| GG→2×G | VvEG16(ΔV152) | ND | ND | ND | HPAEC-PAD |
| GGG→GG+G | VvEG16(ΔV152) | 1.44±0.02 | 0.43±0.02 | 56 | HPAEC-PAD |
| GGGG→2×GG | VvEG16(ΔV152) | 66±1 | 0.094±0.005 | 1.2×104 | HPAEC-PAD |
| GGGG→GGG+G | VvEG16(ΔV152) | 70±10 | 0.5±0.2 | 2300 | HPAEC-PAD |
| 2×GGGG→GG+GGGGGG | VvEG16(ΔV152) | 98±6 | 1.3±0.2 | 1250 | HPAEC-PAD |
| GGGGG→GGG+GG | VvEG16(ΔV152) | 62±2 | 0.26±0.04 | 3500 | HPAEC-PAD |
| G3GGG | VvEG16(ΔV152) | ND | ND | ND | HPAEC-PAD |
| GG3GG→2×GG | VvEG16(ΔV152) | ND | ND | 0.3±0.05 | HPAEC-PAD |
| GGG3G→GGG+G | VvEG16(ΔV152) | ND | ND | 28±5 | HPAEC-PAD |
| GGG3G→GG+G3G | VvEG16(ΔV152) | 36±3 | 1.0±0.1 | 600±20 | HPAEC-PAD |
| G3GGG3GGG→ G3GGG+GGG |
VvEG16(ΔV152) | ND | ND | 25±1 | HPAEC-PAD |
| XXXGXXXG→2×XXXG | VvEG16 | 14±1 | 0.055±0.007 | 4200 | HPAEC-PAD |
| XXXGXXXG→2×XXXG | TmNXG1 (Baumann, et al. 2007) |
9.32±0.16 | 0.076±0.007 | 2000 | HPAEC-PAD |
ND = not determinable;
Concentrations expressed as (mg/mL).
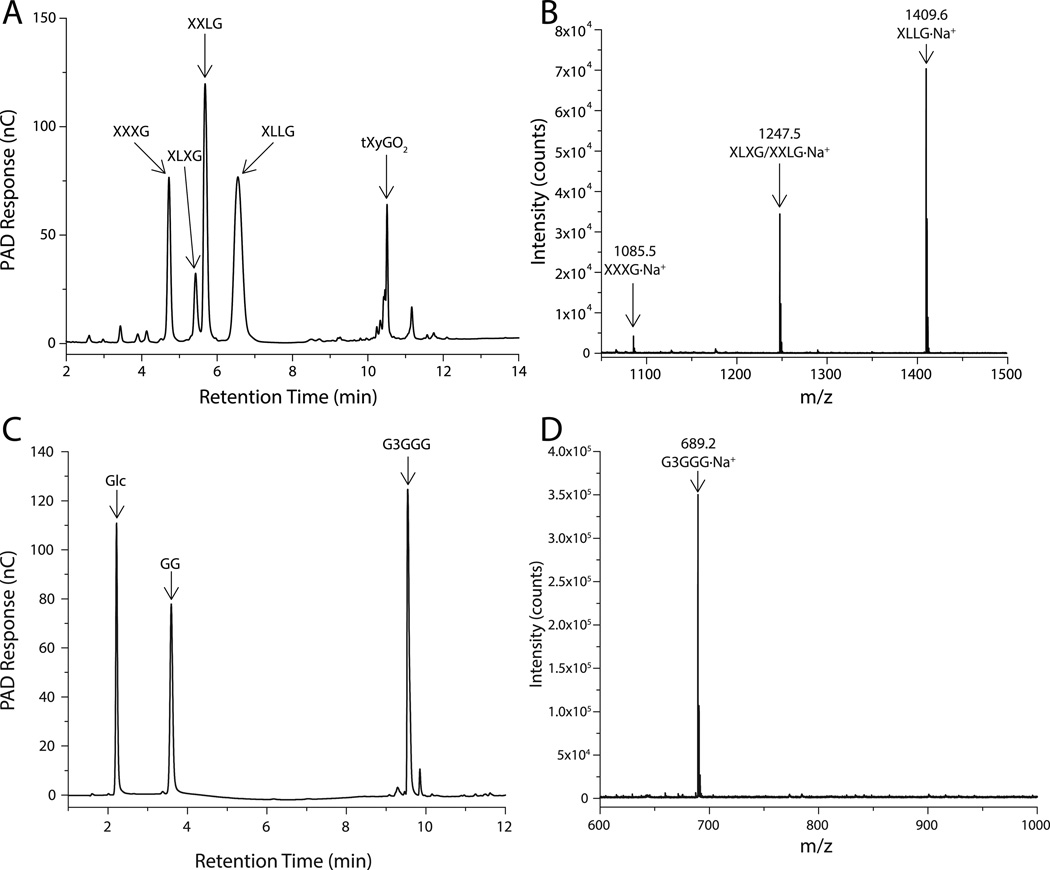
HPLC analysis of the limit digest of tXyG by VvEG16(ΔV152) revealed the production of the canonical mixture of tXyGOs arising from hydrolysis at the anomeric position (C-1) of the unbranched backbone glucosyl residue (Figure 2A,B; oligosaccharide nomenclature is according to (Tuomivaara et al. 2015)). However, prior to the completion of xyloglucan hydrolysis, a notable increase in the proportion of the Xyl3Glc4 heptasaccharide XXXG was observed relative to the galactosylated congeners XLXG, XXLG and XLLG (XXXG:XLXG:XXLG:XLLG observed: 2.2:1:3.9:5.6, versus 1.4:1:3:5.4 (York et al. 1990)). This result corroborates the kinetic analyses, and collectively suggests that extended polysaccharide sidechains are disfavored in the active-site.
Figure 2.
VvEG16 limit-digest products. A) HPAEC-PAD chromatogram of XyGOs produced by the action of VvEG16(ΔV152) on tXyG. B) MALDI-TOF analysis of the mixture shown in A). C) HPAEC-PAD chromatogram of oligosaccharides produced by the action of VvEG16(ΔV152) on bMLG. D) MALDI-TOF analysis of the oligosaccharide mix shown in C).
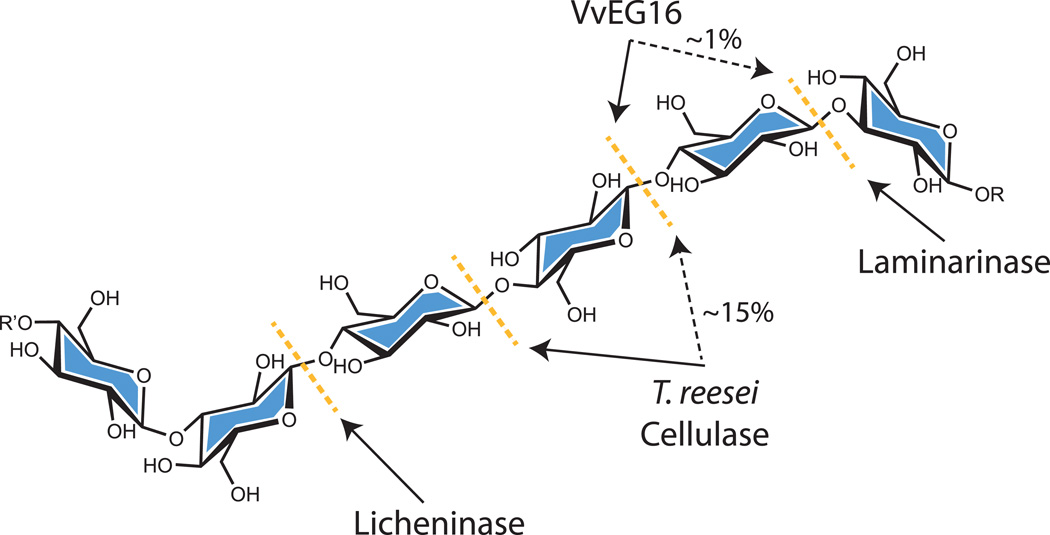
Digestion of bMLG using a similar amount of VvEG16(ΔV152) resulted in the production of an oligosaccharide series corresponding to 3n+1 glucose residues (where n is an integer ≥1), along with less abundant 3n+2 and 3n+3 series (Figure S5). Exhaustive digestion of bMLG using a higher concentration of the enzyme resulted in three products: glucose, cellobiose and the mixed-linkage tetrasaccharide β-D-Glcp-(1,3)-β-D-Glcp-(1,4)-β-D-Glcp-(1,4)-β-D-Glcp (G3GGG), based on comparison with elution times of standard samples in HPLC (Figure 2C,D, Figure S6A). Due to impurities in commercial G3GGG, the identity of this product was further confirmed by HPLC analysis following partial digestion of the purified oligosaccharide with Agrobacterium sp. β-glucosidase (Wakarchuk et al. 1986) (Figure S6B) and by independent LC-MS/MS analysis (Figure S7). Notably, the production of G3GGG indicates that VvEG16 selectively hydrolyzes the β(1,4) glycosidic bond to a β(1,3)-Glc residue. This further implies that the dominant oligosaccharide series observed in partial digests (Figure S5) is comprised of repeats of β(1,3)-linked cellotriose with a β(1,3)-Glc residue at the non-reducing end. To the best of our knowledge, this strict linkage specificity is unique not only among GH16 members, but also among all known endo-glucanases with activity toward MLG (Henriksson et al. 1995, Malet et al. 1993, Zverlov and Velikodvorskaya 1990) (Figure 3).
Figure 3.
Site of MLG hydrolysis by VvEG16 compared to other known MLG-active endo-glucanases. The primary bond targeted by each enzyme is indicated with a solid black arrow and any known secondary activity is indicated with a dashed black arrow and an approximate relative activity.
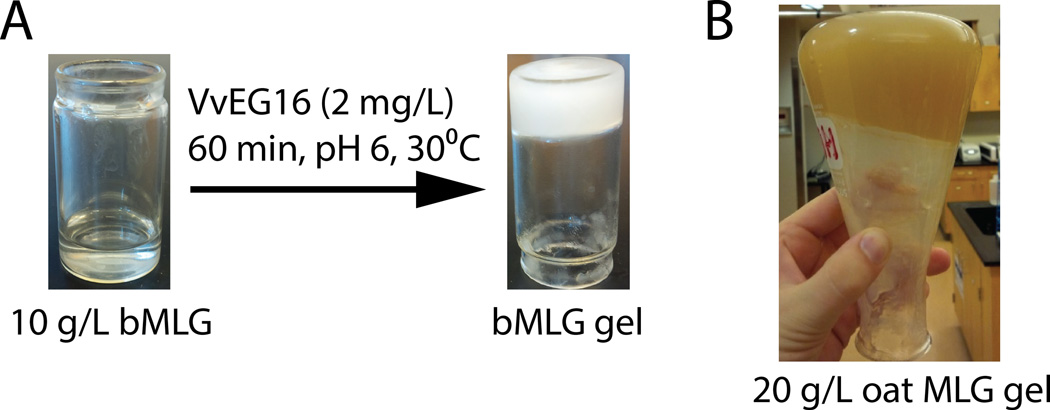
Remarkably, we observed that hydrolysis of bMLG and oat MLG at elevated concentrations (>10 g/L) by VvEG16 resulted in the formation of a gel (Figure 4). The structural changes in the polysaccharide that gave rise to this gelation were initially investigated by following the hydrolysis of bMLG by VvEG16 using HPAEC-PAD analysis and licheninase digestion (Figure S8). Consistent with the product analysis above, a complex mixture of barley MLG oligosaccharides (bMLGOs) is formed after 5 minutes of digestion and the solution remains fluid (Figure S8A). After heat-inactivation of VvEG16, complete digestion with a bacterial licheninase revealed a depletion of stretches of β(1,4)-linked glucosyl residues longer than Glc4 (Figure S8B) in this sample. Extending the VvEG16 incubation time to 60 minutes resulted in gel formation. This gel could be dissolved by heating to 65°C, and subsequent HPLC analysis revealed that the oligosaccharide mixture had become simpler (Figure S8C). Licheninase digestion of this material indicated a selective depletion of β(1,4)-linked glucan motifs longer than Glc3 (Figure S8D).
Figure 4.
Gel formation by VvEG16-catalyzed hydrolysis of MLG. A) Gel formation with purified barley MLG. B) Gel formation with partially purified oat MLG.
Kinetic Subsite Mapping of the VvEG16 Active Site
The active-sites of GHs can be delineated into positive subsites, which extend toward the reducing-end of oligosaccharide and polysaccharide substrates from the point of cleavage, and negative subsites extending toward the non-reducing-end (Davies et al. 1997). To better understand the substrate and product specificity of VvEG16, enzyme kinetics on a series of pure, native oligosaccharides and synthetic chromogenic oligosaccharides were quantified to map the contributions of individual active-site subsites to catalysis (Table 1).
In the first instance, the contributions of the negative subsites to catalysis were elucidated using substituted phenyl β-glycosides as chromogenic substrates (Figure S9). 4-Nitrophenyl β-glucoside (G-PNP) was not hydrolyzed by VvEG16(ΔV152), indicating that the presence of single Glc residue capable of binding in subsite −1 is not sufficient for catalysis. In contrast, 4-nitrophenyl and 2-chloro-4-nitrophenyl β-cellobiosides were competent substrates, with kcat/KM values inversely dependent on leaving group pKa (Table 1), as expected (Planas 2000). The corresponding cellotriosyl congeners were consistently hydrolyzed with ca. 10-fold greater kcat/KM values, thereby indicating that a third negative subsite contributes approximately −6 kJ/mol (ΔΔG‡) to catalysis. Analysis of the contribution of a potential −4 subsite using GGGG-CNP was not possible due to internal cleavage of the oligosaccharide to give primarily GG and GG-CNP. A ca. 10-fold greater kcat/KM value was observed for XXXG-CNP hydrolysis over GGG-CNP that likely reflect specific interactions with internal xylosyl branches observed crystallographically (vide infra).
To extend this analysis to the positive subsites, initial-rate kinetics of the hydrolysis of a series of cello-oligosaccharides (Glc2-Glc6) were quantified by HPLC (Figure S10A–D). Hydrolysis of cellobiose could not be detected under any conditions, including at high enzyme and substrate concentrations. This recapitulates the observation with G-PNP; binding in subsite −1, with or without a contribution of Glc binding in +1, is insufficient for catalysis. Hydrolysis of cellotriose (GGG) to cellobiose and glucose (GG + G) by VvEG16(ΔV152) was poor (kcat/Km = 56 M−1s−1). In contrast, the hydrolysis of cellotetraose (GGGG) via either of two modes, yielding two molecules of cellobiose (2 × GG), or yielding cellotriose plus glucose (GGG + G), was considerably more efficient, with 200- and 40-fold greater kcat/KM values, respectively. Cellopentaose was hydrolyzed through a single mode, producing only cellotriose and cellobiose, with a kcat/KM value comparable to those of cellotetraose as a substrate.
Additional product analysis by mass spectrometry from assays performed in 18O-enriched water indicated that for asymmetrical cleavage, cellotriose yielded GG(18O)+G though a −2→+1 subsite binding mode, cellotetraose yielded GGG(18O)+G though a −3→+1 binding mode, and cellopentaose yielded GGG(18O)+GG though primarily a −3→+2 binding mode (Figure S11, Table S2). The kinetically favored hydrolysis of cellotetraose via the −2→+2 subsite binding mode over the −3→+1 indicates a slightly stronger contribution of the +2 versus the −3 subsite, which is not fully realized in the hydrolysis of cellopentaose in either the −3→+2 or −2→+3 binding modes.
In light of the clear specificity of VvEG16 for bMLG, we determined the hydrolysis kinetics of VvEG16(ΔV152) acting on various model mixed-linkage glucan oligosaccharides, to understand the effects of β(1,3) linkages on catalysis (Figure S10E–G, Table 1). Notably, VvEG16(ΔV152) did not hydrolyze G3GGG to any detectable degree, indicating that the enzyme discriminates against Glcβ(1,3) residues in the −2 and −3 subsites (cf. GGGG, Table 1). This observation also explains the accumulation of G3GGG as a major product in the limit-digest of the bMLG polysaccharide (vide supra). GG3GG was hydrolyzed only to two molecules of GG, with a low specificity constant (kcat/Km = 0.3 M−1s−1) that is 40000- and 7600-fold lower than the −2→+2 and −3→+1 hydrolysis modes of GGGG, respectively. This indicates a strong preference for the hydrolysis of β(1,4)- versus β(1,3)-glucan linkages by VvEG16. Correspondingly, GGG3G was preferentially hydrolyzed to GG and G3G via a −2→+2 binding mode. Interestingly, the hydrolysis of the β(1,3) linkage in GGG3G to give GGG and G via a −3→+1 binding mode can occur, although this is 20-fold less favored than −2→+2 cleavage. The −3→+1 hydrolysis mode of GGG3G had a ca. 200-fold lower specificity constant than the analogous reaction with the all-β(1,4)-linked GGGG, again demonstrating the limited capacity of VvEG16(ΔV152) to cleave β(1,3)-linkages. Thus, the heptasaccharide G3GGG3GGG was specifically hydrolyzed at the β(1,3) bond with a nearly identical specificity constant (kcat/Km = 25 M−1s−1) to the hydrolysis of GGG3G into cellotriose and glucose (kcat/Km = 28 M−1s−1, Table 1). The lack of hydrolysis of this substrate to G3GG+G3GGG, G3GGG3G+GG, or G3GGG3GG+G via the favored β(1,4) bond hydrolysis again reflects a strong bias against β(1,3)-linkages between the negative subsites. Regarding positive subsite interactions, the hydrolysis of GGG3G into GG and G3G had a ~20-fold lower specificity constant than the analogous hydrolysis of GGGG into two GG molecules (Table 1), which suggests that β(1,3) linkages are slightly disfavoured between subsites +1 and +2. Analogous to the cello-oligosaccharide data, the composite analysis of MLGO hydrolysis demonstrates clear evidence for +1 and +2 subsite binding in VvEG16, but no indication of a kinetically relevant +3 subsite.
The hydrolysis of the XyG tetradecasaccharide XXXGXXXG by at the internal, unbranched Glc residue exhibited a 20-fold higher specificity constant than XXXG-CNP, thus indicating a significant contribution of positive subsite binding to catalysis (Table 1) The kcat/Km value for XXXGXXXG hydrolysis was also similar to those of cellotetraose and cellopentaose, which further highlights the accommodation of xylosyl branches in the VvEG16 active-site. Notably, the kinetic constants for XXXGXXXG hydrolysis by VvEG16(ΔV152) were similar to those of the archetypal XEH from nasturtium (Tropaeolum majus), TmNXG1 (Baumann, et al. 2007).
In light of the defining ability of XETs to perform substrate transglycosylation, we also examined the capacity of VvEG16 to catalyze this reaction using well-defined oligosaccharide substrates. Indeed, HPLC analysis under the initial-rate conditions used to measure GGGG hydrolysis indicated that transglycosylation also occurs at a significant rate to produce cellohexaose (GGGGGG) and GG (Figure S12A, Table 1). However, similar analysis with XXXGXXXG at elevated substrate concentrations revealed no detectable formation of (XXXG)3. The potential of VvEG16 to catalyze hetero-transglycosylation was also tested (Fry et al. 2008, Hrmova et al. 2007). Using GGGG as a glycosyl donor substrate in the presence of an excess of XXXG as a potential acceptor, no transglycosylation products were observed by HPLC. On the other hand, with XXXGXXXG as a donor substrate in the presence of an excess of cellobiose as an acceptor, a small peak at a retention time between that of XXXG and XXXGXXXG slowly increased over time (Figure S12B). This peak may be due to XXXGGG, however the small amount formed precluded structural characterization. Taken together, the data suggest that significant transglycosylation by VvEG16 is only observed with linear β-glucans at elevated concentrations (>1 mM acceptor).
Three-dimensional Structures of VvEG16 variants in complex with matrix glycan oligosaccharides
To illuminate the structural basis for the unique catalytic specificity of VvEG16, we performed X-ray crystallography of variants of the enzyme in complex with representative oligosaccharide substrates.
EG16 tertiary structure
Despite extensive efforts, wild-type VvEG16 and the catalytically inactive variant VvEG16(E89A) resisted crystallization independently and in the presence of oligosaccharide substrates and products. Likewise, we were unable to crystallize VvEG16(ΔV152/E89A) in the apo form. In contrast, high-quality crystals of VvEG16(ΔV152/E89A) were obtained in the presence of the linear MLG octasaccharide GG3GGG3GGG (β-D-Glcp-(1,4)-β-D-Glcp-(1,3)-β-D-Glcp-(1,4)-β-D-Glcp-(1,4)-β-D-Glcp-(1,3)-β-D-Glcp-(1,4)-β-D-Glcp-(1,4)-β-D-Glcp) and the branched xyloglucan tetradecasaccharide XXXGXXXG (1.65 Å and 1.79 Å resolution, respectively; Table S3). Strikingly, both of these extended oligosaccharides acted to template crystallization by spanning two protein molecules in the asymmetric unit that do not have direct protein-protein contacts with one another. (Figure 5A and Figure 6A, vide infra). Two additional oligosaccharide complexes were also obtained: VvEG16(ΔV152/E89A) in complex with cellotetraose (GGGG; Figure S13) and VvEG16(ΔV152/C22S/C188S) in complex with the heptasaccharide XXXG (Figure S14; 0.97 Å and 1.59 Å resolution, respectively; Table S3). The VvEG16(ΔV152/E89A):GGGG structure appears to be the highest resolution structure determined for a GH16 member to-date (Hehemann et al. 2012, Labourel et al. 2014, Vasur et al. 2009).
Figure 5.
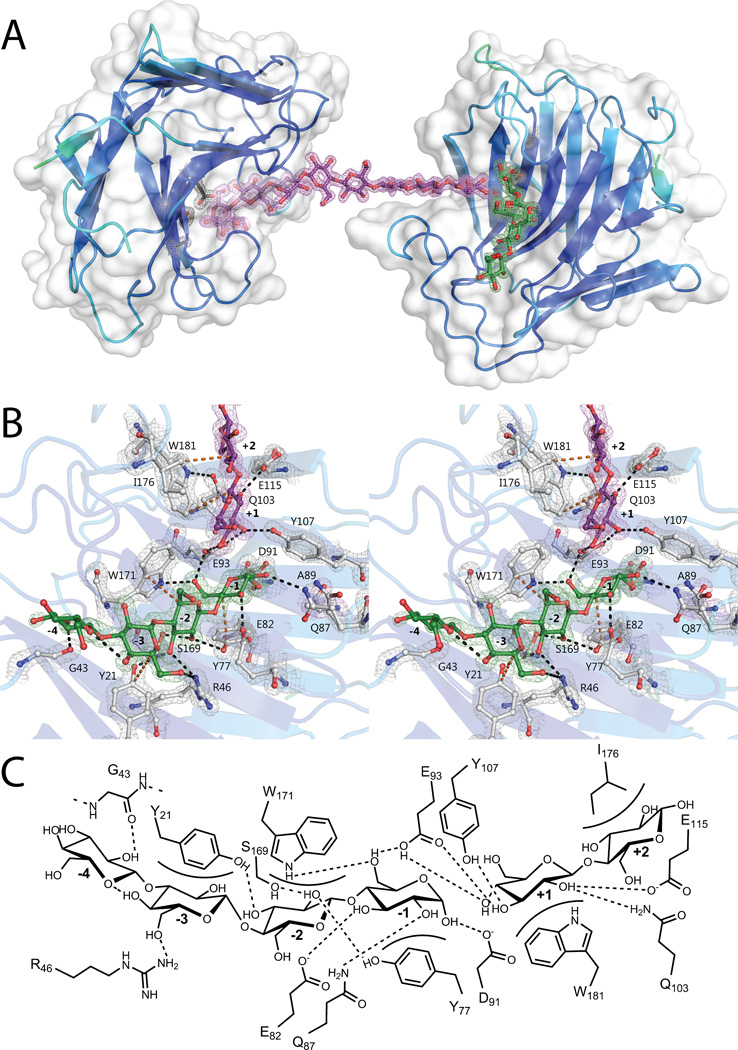
Structure of VvEG16(ΔV152,E89A) in complex with β(1,3)/β(1,4) mixed-linkage gluco-oligosaccharides: A) The asymmetric unit of VvEG16(ΔV152,E89A) contains two protein molecules, chain A (right) and chain B (left). The surface representation is shown in white and the secondary structure cartoon is coloured according to B-factors. The two protein molecules are bridged by a single molecule modelled as GG3GGG3GGG, in purple. A second oligosaccharide, modeled as G3GGG, in green, is observed in the negative subsites of chain A. A single glucose residue is modelled in the +1 subsite of chain B (hidden behind the −1 glucose residue). The active site residues EXDXE are shown in black and the 2mFo-DFc map, contoured at 1.5σ, is shown for both ligands and active site residues. B) Active-site detail shown in wall-eyed stereo, with the ligands colored as in panel A. Sidechains which interact with the ligands are shown in white with all oxygen atoms red and all nitrogen atoms blue. The 2mFo-DFc map is contoured at a 1.5σ level. Individual glucose residues are labelled according to the subsite they occupy. Amino acid numbering begins from the N-terminal glycine remaining after TEV-protease digestion. Putative hydrogen bonding interactions are indicated with black dotted lines and hydrophobic interactions are indicated with orange dotted lines C) Schematic representation of the interactions between VvEG16(ΔV152,E89A) chain A and G3GGG in the negative subsites and GG in the positive subsites. Putative hydrogen bonding interactions are shown as dotted lines and hydrophobic interactions are shown as curved surfaces. Amino acid residues are numbered as in panel B.
Figure 6.
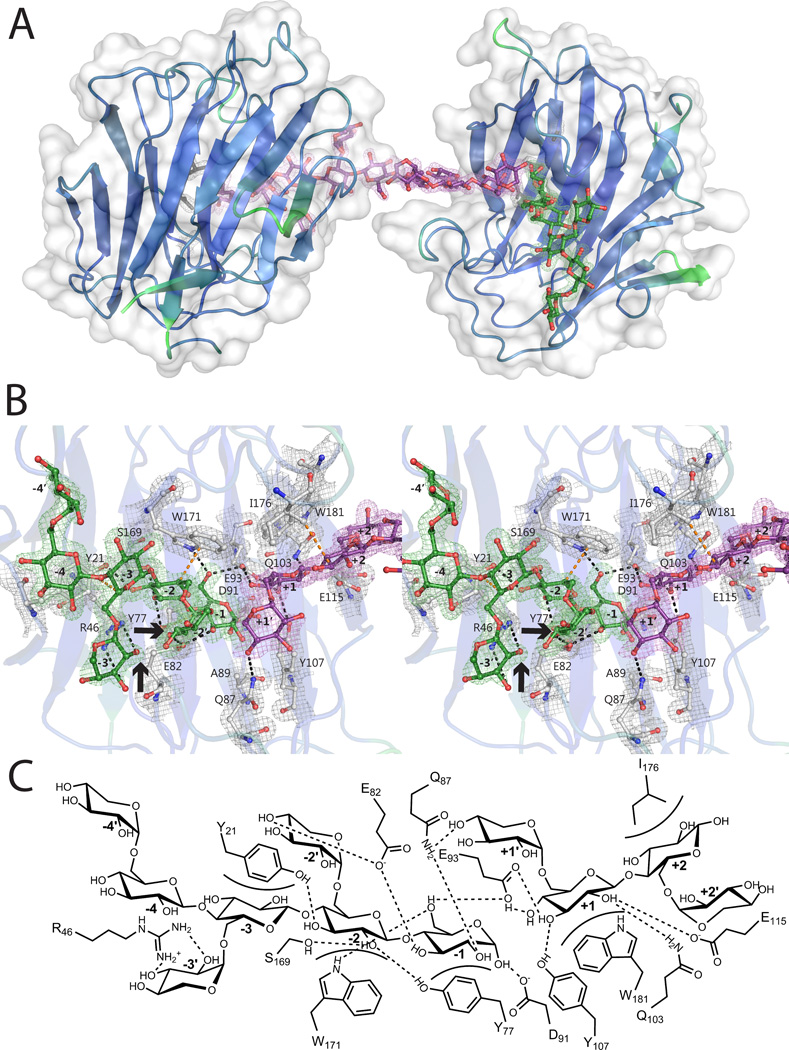
Structure of VvEG16(ΔV152,E89A) in complex with xylogluco-oligosaccharides: A) The asymmetric unit of VvEG16(ΔV152,E89A) contains two protein molecules, chain A (right) and chain B (left). The surface representation is shown in white and the secondary structure cartoon is coloured according to B-factors. The two protein molecules are bridged by a single molecule modelled as XXXGXXXG, in purple. A second oligosaccharide, modeled as XXXG, in green, is observed in the negative subsites of one protein molecule. B) Active-site detail shown in wall-eyed stereo; coloring is as in Figure 5B. Galactose attachment sites are indicated with arrows. C) Schematic representation of active-site interactions, analogous to Figure 5C.
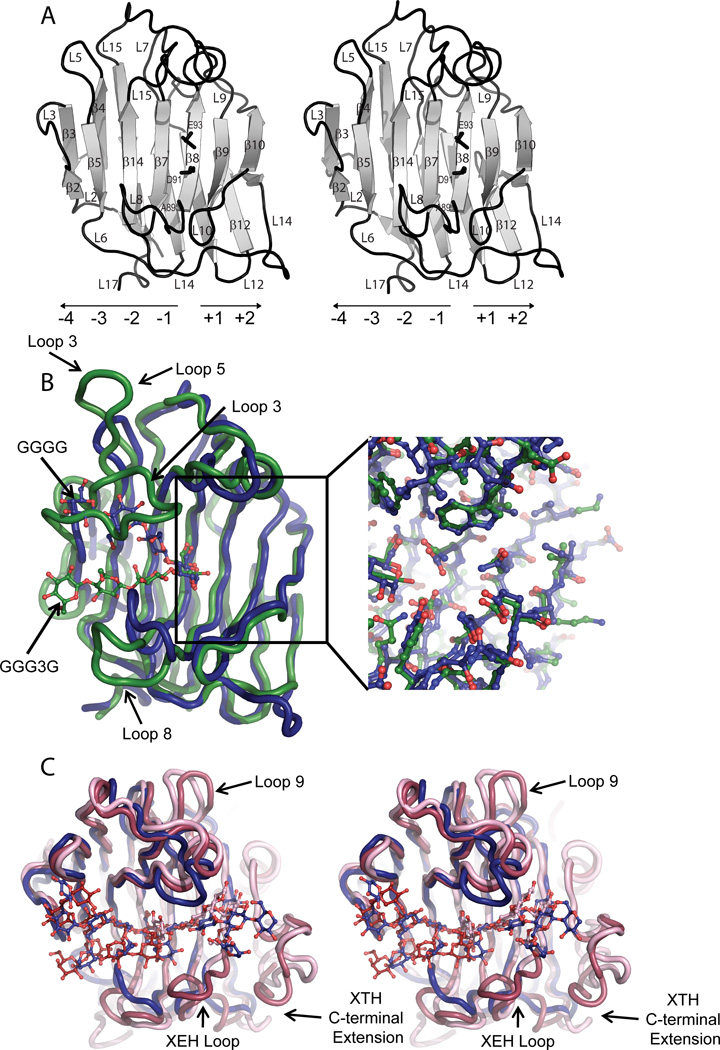
Overall, VvEG16 has a β-jelly roll fold that is typical of GH16 members (Lombard, et al. 2014) and comprises 16 β-strands and 17 loops (Figure 7A, cf. Figure S1). Superposition of all four complexes reveals that there are no major differences in protein conformation: Chain A of the bMLGO and tXyGO complexes each superpose onto the GGGG complex with all-atom RMSD values for the protein residues of 0.63 and 0.87 Å, respectively and superpose onto the XXXG complex with all-atom RMSD values of 0.70 and 0.87 Å, respectively. In the VvEG16(ΔV152/E89A) complexes, the general acid/base catalytic residue, E93, the mutated catalytic nucleophile, E89A, and the catalytic “helper” residue, D91, are found co-linearly on strand β8, with their sidechains directed into the active-site cleft. This strand delineates the positive enzyme subsites, which extend toward the reducing-end of bound saccharide substrates, from the negative subsites, which extend in the opposite direction (Figure 7A). Extending from the rigid core of β-strands, high relative B-factors, indicative of conformational flexibility, were observed in all complexes for loops 5 and 12, residues 154–159 of loop 14, and residues 177–181 of loop 15. Notably, the ΔV152 mutation is located at the beginning of Loop 14 on the convex side of the β-jelly-roll and thus is distant from the active-site cleft. This structural observation explains the insignificant effect of this mutation on catalysis (Table 1). Furthermore, our ability to crystallize the VvEG16(ΔV152) variants, but not the VvEG16(E89A) and wild-type variants, may be due to decreased disorder and more favorable packing of the shortened loop. Of the six cysteine residues found in VvEG16, C22 and C188 are surface-exposed, and C26, C64, C124 and C194 are buried. C64 and C124, which are conserved among all dicot EG16s (Figure S1), appear to be well-positioned to form a disulfide bond, yet only partial occupancy was observed in the high-resolution (0.97 Å) cellotetraose complex, possibly due to the reducing conditions used during crystallization.
Figure 7.
Tertiary structural comparison of VvEG16(ΔV152,E89A) with representative licheninase, XET, and XEH enzymes of GH16. A) Cartoon representation of VvEG16(ΔV152,E89A) in wall-eyed stereo with active site residues (A89, D91, E93) shown as black sticks (based on PDB ID: 5DZE). Loops and β-strands are labeled in order from the N-terminus to C-terminus (cf. Figure S1) and the directionality of active-site subsites indicated. The location of the V152 deletion at the beginning of loop 14 is noted with an arrow B) Superposition of the VvEG16(ΔV152,E89A):GGGG complex (dark blue, PDB ID: 5DZE) with P. macerans licheninase:GGG3G complex (green, PDB ID: 1U0A). The ligands are shown as sticks in colors corresponding to the protein structures. The inset highlights the similarity of the two enzymes in the positive subsites, in contrast to the major loop differences observed in the negative subsites (loop 3 corresponds to the “licheninase loop”). C) Wall-eyed stereo view of the superposition of the VvEG16(ΔV152,E89A):XyGO complex (dark blue, PDB ID: 5DZG) with TmNXG1:XLLG negative-subsite complex (dark pink, PDB ID 2UWA, XLLG from PDB ID: 2VH9), and PttXET16-34:XLG positive-subsite complex (light pink, PDB ID: 1UMZ).
Key features of MLG recognition by VvEG16
The 1.7 Å resolution complex of two VvEG16(ΔV152/E89A) molecules bridged by their mutual recognition of a single GG3GGG3GGG molecule provides a comprehensive view of the active-site interactions with the preferred substrate of the wild-type enzyme (Figure 5). The asymmetric unit contains two polypeptide chains, two oligosaccharides and a monosaccharide. Chain A was modelled from G1 to V207 (except missing the complete H9 residue) and Chain B was modelled from E10 to V207. Electron density consistent with the presence of the α-anomer of G3GGG in the negative subsites and the non-reducing-terminus of GG3GGG3GGG in the positive subsites of Chain A was also observed (Figure 5B & C). Likewise, electron density was observed for the α-anomer of the reducing terminus of GG3GGG3GGG in the negative subsites of Chain B. A single β-glucose was also modelled into the +1 subsite of Chain B. Strikingly, the two polypeptide chains are completely separated by solvent water molecules and the bridging oligosaccharide. To our knowledge, such crystal packing is unique among GHs, and is reminiscent of the mutual recognition of a single DNA molecule by multiple DNA-binding proteins (Fujii 1999, Murphy 1999, Shi et al. 1998),
Strongly supported by kinetic data for cello-oligosaccharide and MLGO hydrolysis (Table 1), the VvEG16(ΔV152/E89A):MLGO complex allows the clear definition of 5 glucosyl-binding subsites (−3→+2) and highlights a potential, weakly interacting −4 subsite. At the catalytic center of Chain A, the reducing-end glucose in subsite −1 interacts with strands β7 and β8 and loop 8, forming hydrogen bonds with Q87, E82, and W171, as well as an aromatic stacking interaction with Y77 (Figure 5B,C). Toward the negative subsites, the glucosyl residue in subsite −2 interacts with strands β2, β5, β7 and β14, forming hydrogen bonds with R46, S169, Y77 and Y21, as well as an aromatic stacking interaction with W171. The glucosyl residue in subsite −3 forms a hydrogen bond with R46 of strand β5 and an aromatic stacking interaction with Y21 of strand β2. The β(1,3)-linked glucosyl residue in the −4 position of chain A forms an apparent hydrogen bond with G43 of loop 5 which is not observed in any other complex. The crystallographic observation of binding a linear arrangement of β(1,4) glucosyl residues in subsites −3 to −1, and tolerance of a β(1,3) kink in subsite −4, succinctly rationalizes the kinetic data on MLGO hydrolysis, in which rigorous exclusion of Glcβ(1,3) residues from subsites −3, −2, and −1 is observed (Table 1). Notably, no hydrogen bonding interaction is observed with the unsubstituted 6-OH of the glucosyl residue in the −2 subsite. This, along with the significant twist of the glucan chain (>90° between −1 and −3) is more typical of endo-xyloglucanases than endo-glucanases that act on unbranched substrates, such as cellulases or licheninases (Gloster et al. 2007, Mark et al. 2009, McGregor et al. 2015).
In the positive subsites of Chain A of the VvEG16(ΔV152/E89A):MLGO complex, the glucosyl residue in subsite +1 interacts with strands β8 and β9 and loops 10 and 15, forming hydrogen bonds with E93, E115, Q103, N105 and Y107, and an aromatic stacking interaction with W181. Despite the kinetic importance of +2 subsite binding for β(1,3)- and β(1,4)-linked oligosaccharides (Table 1), the only apparent interaction in this position is a hydrophobic interaction with I176 of loop 15. At the same time, significant binding plasticity in the positive subsites allows the accommodation of both linkages; positioning of Glcβ(1,3) in subsite +1 is required to realize the essentially exclusive mode of cleavage of MLG (Figure 3). All protein-carbohydrate interactions through the negative and positive subsites discussed above were essentially recapitulated in Chain B.
Key features of XyG recognition by VvEG16
To understand the consequences of polysaccharide branching on substrate recognition in light of the significant activity of VvEG16 on tXyG and XXXGXXXG (Table 1), we crystallized VvEG16(ΔV152/E89A) in the presence of a mixture of variably galactosylated, Glc8-based tXyGOs. Similar to the bMLGO complex, the resulting 1.8 Å resolution structure has an asymmetric unit containing 2 protein molecules bridged only by their mutual recognition of an extended oligosaccharide spanning the positive subsites of Chain A and the negative subsites of Chain B. This bridging ligand was modelled as the tetradecasaccharide XXXGXXXG, since no significant electron density from terminal galactosyl branches was observed (Figure 6). Chain A additionally contained an apparent XXXG moiety in the negative subsites, while the positive subsites of Chain B were not occupied by a carbohydrate.
The backbone glucosyl residues of the tXyGO ligands superpose perfectly with those of the bMLGO complex across the −3 to +1 subsites (Figure S15). Analogous to the VvEG16(ΔV152/E89A):MLGO complex, the glucosyl residue of XXXG in Chain A at potential subsite −4, and especially its pendant xylosyl residue, have no apparent interaction with the protein (Figure 6B,C). The −3’ xylosyl residue participates in a hydrogen bonding interaction with R46, which displaces the amino acid sidechain from the interaction observed with 6-OH of the −3 Glc in the MLGO complex. Interestingly, this interaction also forces the −3’ xylose into an unusual 0S2 skew-boat conformation in Chain A, while a 2S0 skew-boat conformation is observed in Chain B (all other monosaccharide moieties were found in the typical 4C1 chair conformation, Table S4, as verified by Privateer (Agirre et al. 2015)). This suggests a significantly strained interaction and may partially account for the lesser activity of VvEG16 on tXyG versus bMLG (Table 1). The 4-OH of the −2’ Xyl participates in a strong hydrogen bonding interaction with E82, which remains properly positioned to form another hydrogen bond with the 3-OH of the −1 glucose.
In the positive subsites of Chain A, Glc in subsite +1 is tightly anchored, yet there is only a single potential (~3.4 Å) hydrogen bonding interaction between Q87 and the 4-OH of the +1’ Xyl. In light of the comparatively limited number of positive subsites and corresponding protein-carbohydrate interactions, this region does not appear to be a major discriminating factor for polysaccharide specificity.
Supporting GGGG and XXXG complex structures
The 0.97 Å resolution structure of VvEG16(ΔV152/E89A) in complex with cellotetraose bound in the negative subsites and glucose in the +1 subsite (Figure S13) was determined with an R/Rfree ratio of 0.136/0.150 (Table S3). These relatively high individual values can be partially attributed to density in the positive subsites which could not be modelled with confidence, due to the partial occupancy of many species, including glucose, water and possibly glycerol. In contrast, cellotetraose (GGGG) was clearly modelled in the negative subsites, and superposed perfectly with the glucan backbones in the −3 through −1 subsites of the VvEG16(ΔV152/E89A):MLGO and VvEG16(ΔV152/E89A):XyGO complexes (Figure S15). In contrast to the well-defined electron density in these subsites, the weighted 2mFo-DFc difference density for the non-reducing-terminal glucosyl residue could only be observed when the cut-off was lowered to 1.5σ, due to a high relative B-factor (21.7 Å2, compared to 6.5–8.4 Å2 for Glc residues in the −3 to −1 positions). As in the bMLGO and tXyG complexes, this suggests that the putative −4 subsite may only be weakly interacting or absent.
The surface-cysteine variant VvEG16(ΔV152/C22S/C188S) was originally produced in an unsuccessful attempt to eliminate over-labelling by a XXXG-N-bromoacetylglycosylamine inhibitor (Fenger and Brumer 2015). Fortuitous crystallization of this variant in complex with the heptasaccharide XXXG in the negative subsites revealed identical interactions to those observed in the VvEG16(ΔV152/E89A):XyGO complex (Figure S14, Table S3). Here again, strict superposition of glucosyl residues in subsites −3 to −1 was observed (Figure S15), whereas interactions in a potential weakly interacting −4 subsite were not apparent.
EG16 tertiary structure vis-à-vis GH16 licheninases and XTH gene products
The superposition of VvEG16(ΔV152/E89A) with representatives from the bacterial licheninase and plant XET/XEH clades reveals how major sequence insertions and deletions along the evolutionary trajectories of extant GH16 enzymes are manifested in their tertiary structures (Figure 7 cf. Figure 1). A key defining feature in the phylogeny of these enzymes is the presence of a regular β-strand (β8) bearing the catalytic residues. This motif is distinct from other members of GH16, including the closely related laminarinases (β(1,3)-glucanases, EC 3.2.1.39), in which an additional residue in this strand produces a “β-bulge” (Michel, et al. 2001) (Figure 1).
The superposition of the high-resolution VvEG16(ΔV152/E89A):GGGG complex with the Paenibacillus macerans licheninase:GGG3G product complex (PDB ID 1U0A) (Gaiser et al. 2006) reveals a striking structural homology and almost perfect alignment of amino acids in the positive subsites (Figure 7B). Although the glucosyl residue in the −1 subsite is similarly positioned by the same hydrogen bonding and aromatic stacking interactions in both VvEG16 and the P. macerans licheninase, the distal negative subsites in these enzymes are highly divergent. This is the result of significantly shorter Loops 3 and 5 in EG16 members than in both licheninases (Gaiser, et al. 2006) and laminarinases (Fibriansah et al. 2007) (Figure 7 cf. Figure S1). These loop differences dramatically alter the trajectory of unbranched β-glucans across the concave surface of the β-jelly-roll fold (Figure 7B). In particular, the recognition of a β(1,3) linkage between subsites −2 and −1 is a key defining feature of licheninase specificity, which arises directly from steric restriction by the extended Loops 3 and 5 in these enzymes (Planas 2000). In VvEG16, the more open active-site not only enables binding of all-β(1,4)-linked glucan chains in subsites −3 to −1, but also curiously disfavors binding of Glcβ(1,3) residues in these subsites.
Not least, this relief of steric constriction is also central to the accommodation of highly branched xyloglucan chains in the VvEG16 active-site cleft (Figure 7C). As such, the glucan backbone of the XXXG moiety in the negative subsites of the VvEG16(ΔV152/E89A):tXyGO complex closely superposes with the glucan backbone of XLLG in the TmNXG1(ΔYNIIG) complex (PDB ID 2VH9, (Mark, et al. 2009)). Despite VvEG16 and TmNXG1 displaying similar hydrolysis kinetics toward XyG and XXXGXXXG substrates, there is limited similarity in the positions of xylosyl branches in the corresponding complexes. This suggests either significant flexibility in substrate recognition or the evolution of distinct binding modes in these broad, negative-subsite clefts.
A clear distinction of plant XTH gene products in GH16 is the presence of a large C-terminal extension (InterPro Domain IPR010713, “XET_C”), as well as a lengthening of Loop 9, which significantly increase the surface area in the positive subsite region and directly enable specific recognition of branched xyloglucan substrates (Johansson, et al. 2004, Mark, et al. 2009, Mark et al. 2011). Superposition of the VvEG16(ΔV152/E89A):tXyGO complex with the Populus tremula × tremuloides XET16–34:XLG positive-subsite complex (Johansson, et al. 2004) reveals notable tertiary structural differences in this region of the active-site (Figure 7C). Firstly, a lesser number of positive subsites in VvEG16 is clearly evident, due to the lack of the “XET_C” domain. Furthermore, VvEG16 and PttXET16–34 exhibit distinct differences in the trajectory of the glucan backbone through the positive subsites, which arise from alternate conformations of Loop 14. Yet despite these differences, VvEG16 demonstrates a much higher degree of overall tertiary structural similarity with archetypal XET and XEH (excluding the C-terminal “XET_C” extension), in spite of lower sequence similarity, than with licheninases (Figure 7B,C, Figure S1). Thus, while VvEG16 exhibits overall a more open, XET/XEH-like active-site cleft, which enables recognition and hydrolysis of both MLG and XyG, residue-specific variations and the addition of the C-terminal extension appear to be key contributors to the evolution of the strict XyG-specificity observed for the majority of characterized XTH subfamily members.
DISCUSSION
Plant EG16 members represent a unique class of bi-functional matrix glycan hydrolases
In light of the widespread distribution of MLG and XyG across the plant kingdom, there is a clear and pressing need to achieve a comprehensive understanding of the enzymology of the biosynthesis and biodegradation of these key matrix glycans (Attia and Brumer 2016, Burton, et al. 2010, Fincher 2009, Pauly and Keegstra 2016). Endogenous endo-hydrolysis of MLG, the key initial step in glucose mobilization, was previously thought to be catalyzed exclusively by plant Glycoside Hydrolase Family 17 (GH17) members (Fincher 2009). On the other hand, endogenous endo-hydrolysis of the XyG backbone was only known among a small subset of GH16 members, the XEHs, which are encoded by Group III-A XTH genes (Baumann, et al. 2007, Eklöf and Brumer 2010).
Building upon our previous work on a Populus trichocarpa homolog (Eklöf, et al. 2013), we demonstrated here that members of the EG16 clade within GH16, which exclusively comprises plant homologs (Figure 1), uniquely catalyze the endo-hydrolysis of both MLG and XyG with comparable efficiency. Importantly, the kinetic analysis of VvEG16 presented here represents the first quantitative demonstration that predominant mixed-linkage endo-glucanase activity is found among plant enzymes that are phylogenetically and structurally distinct from canonical GH17 members. Moreover, these EG16 members have notably broader substrate specificity than both bacterial licheninases (Planas 2000) and plant XEHs (Eklöf and Brumer 2010) of GH16.
EG16 members represent extant transitional enzymes linking the evolution of bacterial licheninases and plant XTH gene products
Molecular phylogeny in combination with detailed structural enzymology of VvEG16 has allowed us to elucidate key protein features that give rise to the bi-functionality of EG16 members. Moreover, this composite analysis allows us to propose that EG16 members are extant “transitional enzymes” (by analogy with transitional fossils in paleontology) in the evolution of GH16 members. Specifically, the experimentally determined crystal complexes of VvEG16 directly reveal key, stepwise structural changes that give rise to the functional diversification of extant bacterial licheninases, plant EG16s, and plant XETs and XEHs.
Our global phylogenetic analysis of GH16 β-jellyroll domains across species and activities demonstrates a clear delineation of licheninases, EG16 members, and XTH gene products into distinct, well-supported clades (Figure 1A). This phylogeny essentially recapitulates that first presented by Barbeyron, Michel, et al. (Barbeyron, et al. 1998, Michel, et al. 2001), but with the key insertion of the EG16 clade intermediate between the licheninases and XTH gene products. In consideration of the early evolutionary origin of bacteria and key sequence features present in all bacterial GH16 enzymes which have been lost in EG16 enzymes (such as the extended loop 3), it is most likely that a licheninase-like protein served as the ancestor to both extant bacterial licheninases and plant EG16 enzymes. In addition to the common β-jellyroll fold, composed of 13 core β-strands and a regular β8-strand bearing the catalytic EXDXE motif, this ancestral protein also would have included an extended version of Loop 3 (referred to here as the “Licheninase Loop”), as found in extant bacterial mixed-linkage endo-glucanases (Figure S1).
Subsequent truncation of Loop 3 in the divergence of EG16 members (Figure 1B) resulted in the widening of the active-site cleft, thereby allowing these enzymes to address both the defining licheninase substrate, MLG, as well as highly branched XyG chains (Figure 7). It is presently unclear in which kingdom this mutation may have occurred; just as there are no EG16-like (short Loop 3) proteins among known bacterial sequences, there are no licheninase-like (long Loop 3) sequences among known plant sequences. However, EG16 members are found in the genomes of early-diverging plants, currently represented by the mosses Physcomitrella patens and Selaginella moellendorffi (Figure 1B), which highlights the ancient evolution of this class of enzymes.
In turn, we propose that the broadened active-site cleft of an ancestral EG16 member poised this protein scaffold for further evolution into extant xyloglucan endo-transglycosylases (Group I, II, and III-B XTH gene products (Eklöf and Brumer 2010)) by addition of the distinguishing “XET_C” C-terminal domain extension (Figure 1B). This is the most parsimonious explanation in light of the existence of EG16 members in plants, vis-à-vis a conceivable tandem event directly giving rise to a XET-like homolog via the simultaneous truncation of Loop 3 and gain of the XET_C extension in an ancestral licheninase. Notably, no proteins are currently known that comprise both a long, licheninase-like Loop 3 and a XET_C extension, which supports the proposed evolution of XETs from an EG16-like (short Loop 3) ancestor. Collectively, the truncated Loop 3 and XET_C extension, together with various active-site point mutations, has resulted in the high specificity of XETs for XyG over MLG. In this context, it is interesting to note that a recently discovered Equisetum β-glucan:XyG heterotransglycosylase does not represent an evolutionary intermediate between licheninases and XETs, but appears to represent a later functional divergence in Group I/II XTH gene products (Simmons, et al. 2015).
Finally, we have previously provided phylogenetic, biochemical, and tertiary structural evidence that the comparatively fewer number of predominant XEHs (Group III-A XTH gene products) evolved subsequently from strict XETs by the introduction of a short, 5 amino-acid loop insert on the active-site cleft, immediately preceding strand β10 (Figure S1, “XEH loop”; Figure 1B) (Baumann, et al. 2007). These XyG hydrolases have apparently evolved to meet needs for seed-storage XyG mobilization and fruit ripening in select species (reviewed in (Baumann, et al. 2007)), and also appear to be active in vegetative tissues of Arabidopsis (Kaewthai, et al. 2013).
Thus, the structural enzymology of VvEG16 vis-à-vis other GH16 members provides compelling evidence for the following evolutionary trajectory: ancestral licheninase→ancestral EG16→ancestral XET→ancestral XEH. Through evolution, the ancestral bacterial licheninase-encoding genes have expanded greatly across species (Lombard, et al. 2014), while ancestral plant XTH genes have also seen massive expansion to comprise 20–50 members in individual bryophytes, lycophytes, and angiosperms (reviewed in (Eklöf and Brumer 2010, Eklöf, et al. 2013)). Strikingly, EG16 members appear to be restricted to one member in each plant genome (Figure 1B), with many prominent dicots, including the model species Arabidopsis thaliana, lacking EG16 representatives altogether (Eklöf, et al. 2013). Indeed, EG16 members have been largely overlooked, or in some cases have been mistakenly classified as XTH gene products (Eklöf, et al. 2013, Geisler-Lee et al. 2006, Yokoyama et al. 2010), for this reason. The now clear delineation of EG16 will significantly enable future bioinformatics analysis, as well as consideration of functional data in an updated phylogenetic context (Hatfield and Nevins 1987).
In vivo roles of EG16 members
Despite their limited distribution, transcriptional analysis suggests that EG16 members are not simply vestigial, but are likely to play functional roles in plants. In particular, VvEG16 was first identified in mRNA extracted from each of root, leaf and flower of V. vinifera (Peng, et al. 2007). Also in retrospect, EG16 members have been found in EST libraries from flowers, leaves, and roots of a variety of plants at different developmental stages, including citrus trees and cotton (Arpat et al. 2004, Forment et al. 2005). In the absence of systematic studies, it is unclear under which conditions EG16-encoding genes might be specifically upregulated. Curiously, however, all EG16 members identified thus far lack a trafficking signal peptide, lack N-glycosylation sites, and contain numerous unpaired cysteine residues. These characteristics argue against apoplastic role for EG16 members (unless secretion occurs via a non-classical pathway (Rose and Lee 2010)) This is in sharp contrast to other plant XyG- and MLG-active enzymes, namely the XTH gene products of GH16 (Eklöf and Brumer 2010) and the mixed-linkage β-glucanases of GH17 (Fincher 2009). As such, we tentatively suggest that EG16 members may not be not involved in wall polysaccharide remodelling in the classical sense, but may function in housekeeping or modifying roles within the confines of the cell membrane, or after cell death. Certainly, the unique MLG cleavage specificity of VvEG16, which leads to polysaccharide gelation in vitro, is worthy of consideration in light of potential wall-modifying roles and may also have biotechnological application.
Although the biological role(s) of the EG16 members presently remain enigmatic, the present enzyme structure-function analysis sets the stage for future genome mining, transcriptomic analysis, cellular localization, and forward- and reverse-genetic analyses across plant species. It is especially intriguing that mosses, grasses, and trees all encode EG16 members. Elucidating to what extent EG16 function in vivo is conserved or specialized among species with such intrinsically diverse cell wall compositions and growth habits now remains to be resolved.
EXPERIMENTAL PROCEDURES
All chemicals and resins were obtained from Sigma-Aldrich unless otherwise specified. UV/Vis spectroscopy was run on a Cary 60 (Agilent) equipped with a single-cell temperature controller. Chromogenic substrate degradation was monitored on a Cary 300 Bio (Agilent) with a temperature-controlled 8-cell dual-beam sample holder. SDS-PAGE was run using Bio-Rad mini Protean TGX 4–20% gels. Gels were imaged in a Bio-Rad Gel Doc XR+ imager. Michaelis-Menten parameters (Km, kcat) were determined by non-linear fit using the Michaelis-Menten model with or without substrate inhibition in OriginPro 9.1. Sequence alignments were displayed using ESPript 3.0 (Robert and Gouet 2014).
Bioinformatic Analysis
Amino acid sequences of GH16 enzymes with confirmed activities were extracted from the CAZy database, trimmed using ScanProsite (Castro et al. 2006) and aligned using the Expresso method (Armougom et al. 2006). The resulting alignments were manually refined using the alignment explorer in MEGA6 v6.06. A phylogenetic tree was derived from the resulting sequence alignment using the maximum likelihood method in MEGA6. The reliability of the tree was tested by bootstrap analysis using 100 resamplings of the data set. Five cellulases from GH7 were used as an outgroup to root the tree. Where apparent substrate-specific clades could be identified, branches were collapsed and displayed as triangles (see Table S5 for accession codes).
EG16 protein sequences were identified through BLAST searches of the NCBI GenBank and JGI Phytozome databases. Bioinformatic analysis was performed on all 33 of the EG16 protein sequences found to date (accession numbers and species of origin can be found in Figure 1). Protein sequence alignments were performed using the MUSCLE algorithm in MEGA6 v6.06 (Tamura et al. 2013) with the UPGMB clustering method. The resulting alignment was manually refined using the alignment explorer in MEGA6 on the basis of the secondary structure of VvEG16 (this work). A phylogenetic tree was derived from the resulting sequence alignment using the maximum likelihood method in MEGA6. Two licheninases, from B. subtilis and B. licheniformis as well as two XTHs, TmNXG1 and PttXET16–34 were used as outgroups after removal of all signal peptides and the XTH C-terminal extensions. The reliability of the tree was tested by bootstrap analysis using 100 resamplings of the data set.
Cloning
10G Hi-Control and BL21(DE3) Hi-Control E.coli were obtained from an Expresso T7 cloning kit (Lucigen). pET24a vector was purchased from EMD Millipore. T5 exonuclease, Q5 High-Fidelity DNA Polymerase and Phusion High-Fidelity DNA Polymerase were purchased from New England Biolabs (Ipswich, MA). Taq DNA ligase was purchased from MCLAB (San Francisco, CA). Nucleotides were purchased from Amresco. Oligonucleotide primers were purchased from Integrated DNA Technologies (IDT, San Diego, CA) (Table S6). PCR reactions were run in a Bio-Rad S1000 Thermal Cycler. Genes of interest were purchased from IDT as GBlocks and amplified by PCR using Q5 DNA polymerase (New England Biolabs). All cloning was done using Gibson Assembly (Gibson et al. 2009) and primers for Gibson assembly were designed using the NEBuilder web tool (New England Biolabs). Site-directed mutagenesis was performed using the QuikChange method (Stratagene).
Protein Expression and Purification
BL21(DE3) Hi-Control bearing VvEG16 (NCBI refseq: XP_002273975.1), VvEG16(ΔV152) or VvEG16 (ΔV152/E89A) expression plasmids were grown at 37°C in Studier media (1× YT supplemented with 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 2 mM MgSO4, 5 mM Na2SO4, 0.05% glucose, and 0.5% glycerol) to an OD600 of 1.8–2.0 prior to induction with 0.2 mM IPTG overnight (16–18 h) at 16°C. The cells were collected by centrifugation and resuspended in buffer A (300 mM NaCl, 20 mM imidazole, 20 mM NaPi, pH 7.5) supplemented with 1 mM EDTA (to prevent proteolysis). The cells were lysed by a single pass through a French press. The lysate was clarified by centrifugation before protein purification using a HisTrap FF crude (GE Life Sciences) column. After elution with a 10 CV linear gradient from 20–500 mM imidazole, the protein was desalted into SEC buffer (20 mM MOPS, 1 mM EDTA, pH 7.5), supplemented with 1 mM DTT and concentrated to 5 mg/mL using a 10 kDa Centricon (EMP millipore) maintained at 4°C. The protein was then cleaved by TEV protease (1 mg TEV protease/50 mg VvEG16, 16h, 4°C), run over a freshly-charged HisTrap column, concentrated down to 10 mg/mL and purified over an XK 16/100 column (GE Life Sciences) packed with Superdex 75 (GE Life Sciences) run with SEC buffer at 1 mL/min. The pure protein was then concentrated down to 20–50 mg/mL and flash-frozen in LN2. All purified proteins were analyzed for sequence correctness and post-translational modification by intact mass spectrometry using a Waters Nanoacquity UPLC coupled to a Xevo G2-S QTof as previously described (Sundqvist et al. 2007).
Wild-type PpXG5 (Gloster, et al. 2007) was generated by site-directed mutagenesis from a clone of PpXG5 (E323G) in pET21a with a 6×–C-terminal polyhistidine tag (Spadiut et al. 2011). This was desalted into 20 mM NH4HCO3 pH 7.5 prior to concentration to ~10 mg/mL using a 30 kDa Centricon. Sucrose was then added (9 mg/mg PpXG5) and the solution was frozen in LN2 and lyophilized. CjBgl35A and PtEG16 were expressed as described (Eklöf, et al. 2013, Larsbrink et al. 2011). TEV protease was prepared as described (Tropea et al. 2009) using BL21(DE3) Hi-Control cells bearing the pRARE2 plasmid for expression.
Carbohydrate Analysis
HPAEC-PAD was performed on a Dionex ICS-5000 system equipped with an AS-AP auto-sampler with a temperature-controlled sample tray run in a sequential injection configuration using Chromeleon 7 control software. The injection volume was 10 µL unless otherwise specified. A 3×250 mm Dionex CarboPac PA200 column and a 3×50mm guard column were used for all HPAEC separations. Separations were run using gradients A-C previously specified (Larsbrink, et al. 2011).
MALDI-TOF analysis of oligosaccharides was performed on a Bruker Autoflex system (Bruker Daltonics) operated in reflectron mode. Oligosaccharide samples (0.1–10 mg/mL) were mixed 1:1 with 10 mg/mL 2,5-dihydroxybenzoic acid in 1:1 H2O:MeOH directly on a Bruker MTP 384 ground steel MALDI plate and allowed to dry under ambient conditions.
Liquid chromatography-mass spectrometry was performed on a Waters Xevo Q-TOF with a nanoACQUITY UPLC system. Samples were separated on a 0.32×150 mm Hypercarb KAPPA column packed with 3 µm porous graphitized carbon particles run at 8 µL/min. Buffer A was 25 mM formic acid in 95:5 H2O:MeCN, raised to pH 5 with ammonium hydroxide, buffer B was 25 mM formic acid in 5:95 H2O:MeCN with equivalent ammonium hydroxide added. Mass spectrometry was run in positive mode, optimized for resolution, with a 2 second scan time. MS/MS was performed by CID using 15 V collision energy. Following the injection of a 1 µL of sample, the gradient was run at 30°C: 0–5 min, 90% A, 10% B; 5–25 min, linear gradient to 30% B; 25–25.5 min, linear gradient back to 10% B; 25.5–30 min, equilibration with 10% B.
Substrates
Tamarind xyloglucan (tXyG), barley β-glucan (bMLG), konjac glucomannan (kGM), carboxymethyl cellulose (CMC), wheat arabinoxylan, mixed-linkage glucan oligosaccharides (MLGOs) and cellooligosaccharides (G3–6) were purchased from Megazyme International. Hydroxyethyl cellulose (HEC) was purchased from Fluka. Guar gum (gGM) was purchased from West Point Naturals, a local store in Vancouver, Canada.
tXyGOs were prepared essentially as described in (Eklöf et al. 2012) using 5 U of His6-PpXG5 per gram of tXyG in 10 mM NH4OAc, pH 5.5 incubated at 30°C, instead of T. reesei cellulase. XXXG and XXXGXXXG were subsequently produced by incubation with CjBgl35A as described (McGregor, et al. 2015). [β-D-Glcp-(1◊3)-β-D-Glcp-(1◊4)-β-D-Glcp-(1◊4)-β-D-Glcp] (G3GGG) and [β-D-Glcp-(1◊4)-β-D-Glcp-(1◊3)-β-D-Glcp-(1◊4)-β-D-Glcp-(1◊4)-β-D-Glcp-(1◊3)-β-D-Glcp-(1◊4)-β-D-Glcp-(1◊4)-β-D-Glcp] (GG3GGG3GGG) were prepared as described (McGregor, et al. 2015).
4-nitrophenyl-β-glucoside (G-PNP) and 4-nitrophenyl-β-cellobioside (GG-PNP) were purchased from Sigma-Aldrich. 4-nitrophenyl-β-cellotrioside (GGG-PNP), 2,4-dinitrophenyl-β-cellotrioside (GGG-DNP) and 2,4-dinitrophenyl-β-cellobioside (GG-DNP) were kind gifts from the Withers lab (UBC). 2-chloro-4-nitrophenyl-β-XXXG (XXXG-CNP) was prepared in-house as previously described (Ibatullin et al. 2008). 2-chloro-4-nitrophenyl-β-cellobioside (GG-CNP) and 2-chloro-4-nitrophenyl-β-cellotrioside (GGG-CNP) were purchased from Megazyme International.
Enzyme Activity Determination
The bicinchoninic acid-copper (BCA) assay was performed as previously described (Arnal et al. 2016). The pH-activity optimum of VvEG16(ΔV152) was determined by incubating 1 mg/mL tXyG with 5 µg/mL VvEG16(ΔV152) for 15 minutes at room temperature in 50 mM buffer containing 1 mM EDTA. The buffers used were: sodium citrate (pH 3.75–5.5), sodium phosphate (pH 6.1–8.3) and glycine-HCl (pH 8.6–9.9). The temperature optimum was determined in 20 mM citrate, pH 6.0 with 1 µg/mL VvEG16(ΔV152) and 1 mg/mL tXyG incubated for 15 minutes. Chromogenic substrate hydrolysis was quantified essentially as described (McGregor, et al. 2015) using 20 mM sodium citrate, pH 6, incubated at 30°C. Incubation times and enzyme concentration were minimized for GGG-PNP due to cleavage of GGG-PNP into GG and G-PNP. To monitor polysaccharide hydrolysis by HPAEC-PAD, polysaccharides (tXyG or bMLG, 0.1 mg/mL final) were mixed with an appropriate amount of enzyme in a 1 mL reaction containing 20 mM sodium citrate, pH 6.0, and incubated at 30°C. 100 µL samples were diluted into 400 µL of 0.1 M Na2CO3 and run on HPAEC-PAD using gradient C. XXXGXXXG, MLGO and cellooligosaccharide kinetics were determined as previously described (McGregor et al. 2016). The regiospecificity of cellooligosaccharide hydrolysis was determined using the H218O labelling method previously described (McGregor, et al. 2015).
Co-crystallization of VvEG16 variants in complex with oligosaccharides
Crystals were grown using the sitting-drop method at 4°C; 10 mg/mL VvEG16(ΔV152/E89A) in SEC buffer supplemented with 5 mM DTT was co-crystallized with 2 mM cellotetraose in 0.8 M NaH2PO4 + 1.2 M K2HPO4 yielding large hexagonal prismic needle clusters. The same enzyme stock was co-crystallized with 5 mM G3GGG3GGG (potentially contaminated by ~0.5 mM of putative GG3GGG3GGG according to HPAEC-PAD) in 0.1 M NaCl, 0.1 M HEPES pH 7.5, 1.6 M (NH4)2SO4 to yield small rectangular prisms with rounded ends. Co-crystallization with 10 mg/mL tXyGO2 in 20% PEG 6000 in 100 mM pH 6.0 MES buffer yielded small tetragonal bipyramidal crystals. 10 mg/mL VvEG16(ΔV152/C22S/C188S) in SEC buffer was co-crystallized with 5 mM XXXG in 30% PEG 6000 in 0.1 M Bicine at pH 9.0 and 4°C yielding large hexagonal prismic needle clusters. All crystals were cryoprotected with 25% (v/v) glycerol in well solution and flash-frozen in liquid N2.
Data collection and refinement
Diffraction experiments were performed at the Canadian Light Source (Saskatoon) on beamline 08ID-1 run with MxDC for the XXXG, cellotetraose and GG3GGG3GGG crystals. Diffraction experiments were performed at the Stanford Synchrotron Radiation Lightsource (Menlo Park) on beamline 7-1 run with Blu-Ice (McPhillips et al. 2002) for the tXyGO2 complex. Datasets were processed using XDS (Kabsch 2010). For initial phasing, a search model was generated from PttXET16–34 in complex with a tXyGO (PDBID: 1UMZ) using chainsaw (Stein 2008). The initial structure (VvEG16(ΔV152/E89A)-tXyGO2) was phased by molecular replacement using phaser-2.5.0 (McCoy et al. 2007) yielding poor initial phases. This initial model was improved via auto-building in ARP/wARP 7.4 (Langer et al. 2008) in CCP4 (Winn et al. 2011), which improved the phases, followed by manual adjustment using Coot (Emsley and Cowtan 2004) and refinement with Refmac5 (Murshudov et al. 1997, Pannu et al. 1998, Vagin et al. 2004). Final refinement was completed with simulated annealing in Phenix (Adams et al. 2010, Afonine et al. 2012, Chen et al. 2010). Subsequent structures were solved using VvEG16(ΔV152/E89A)-tXyGO2 as the search model and refined using Phenix.refine. The cellotetraose complex was refined with anisotropic B-factors for all non-hydrogen atoms and the GG3GGG3GGG complex was refined with anisotropic B-Factors for all non-hydrogen and non-water atoms. Models were validated using the Phenix Molprobity and Coot validation tools along with the PDB Adit server and Privateer (Agirre, et al. 2015). No residues were found in the disallowed region of the Ramachandran plot for the tXyGO2 or GG3GGG3GGG complexes. However, H41 of the GGGG complex was identified as a Ramachandran outlier by the wwPDB X-Ray validation server (Table S3) due to a minor alternate conformation observed for the backbone of D40 which could not be likewise modelled for the backbone of H41. All structure figures were prepared using PyMOL (Schrödinger).
Supplementary Material
Acknowledgments
Mr. McGregor thanks the Natural Sciences and Engineering Research Council of Canada (NSERC) for an Alexander Graham Bell Canada Graduate Doctoral Scholarship. This work was supported by NSERC (Discovery Grant), the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund. Waters Corporation is gratefully acknowledged for the provision and skilled maintenance of the LC-MS system used in this study. The authors acknowledge the kind contributions of Agrobacterium sp. β-glucosidase, and chromogenic substrates from the Withers group at UBC. HB thanks Prof. Lacey Samuels (UBC Botany) for stimulating discussions. The authors thank A. Gonzalez and Y. Tsai at the Stanford Synchrotron Radiation Lightsource for the autoxds script. We thank the Canadian Macromolecular Crystallography Facility staff for assistance with data collection.
Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Research described in this paper was performed using beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research. We thank the CMCF staff for assistance with data collection.
Footnotes
ACCESSION NUMBERS
Coordinates for all complexes are available in the RCSB Protein Data Bank (PDB) with identifiers 5DZE (GGGG, Glc complex), 5DZF (bMLGO complex), 5DZG (tXyGO2 complex), and 5SV8 (XXXG complex).
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
NM performed phylogenetic analysis, selected and cloned genes, expressed proteins, generated mutants, produced oligosaccharides, co-crystallized VvEG16 variants with all oligosaccharides, collected diffraction data, solved protein structures, collected and analyzed all activity data and prepared all figures. VY collected and analyzed all protein intact mass spectra and worked with NM on enzyme stability experiments. CCT designed crystal screening experiments and provided valuable guidance and advice for all aspects of crystal preparation prior to diffraction. FVP and HB provided project guidance and analyzed data. NM and HB co-wrote the manuscript, with input from the other co-authors.
SUPPORTING INFORMATION LEGENDS
Figure S1: Protein sequence alignment of EG16s with Licheninases and XTH gene products.
Figure S2: Analysis of purified VvEG16 proteins.
Figure S3: Comparison of wild-type VvEG16 and VvEG16 (ΔV152) activities.
Figure S4: Dependence of VvEG16(ΔV152) activity on pH and temperature.
Figure S5: Identification of oligosaccharide series produced by action of VvEG16(ΔV152) on bMLG.
Figure S6: Identification of the MLGO produced by the action of VvEG16(ΔV152) on bMLG (Glc4-MLGO) by HPAEC-PAD.
Figure S7: Identification of the MLGO produced by the action of VvEG16(ΔV152) on bMLG (Glc4-MLGO) by LC-MS.
Figure S8: Time-dependent MLG hydrolysis by VvEG16(ΔV152).
Figure S9: Kinetic data and Michaelis-Menten curves used to determine the kcat and KM values for the hydrolysis of various chromogenic substrates by VvEG16(ΔV152).
Figure S10: Kinetic data and Michaelis-Menten curves used to determine the kcat and KM values for the hydrolysis of various oligosaccharide substrates by VvEG16(ΔV152).
Figure S11: Mass spectra of products produced by the hydrolysis of cello-oligosaccharides by VvEG16(ΔV152) in H218O.
Figure S12: Homo- and hetero-transglycosylation of oligosaccharides by VvEG16(ΔV152).
Figure S13: Structure of VvEG16(ΔV152,E89A) in complex with cellooligosaccharides.
Figure S14: Structure of VvEG16(ΔV152,C22S,C188S) in complex with a xyloglucan oligosaccharide.
Figure S15: Superimposition of the carbohydrate ligands bound within the active site of each experimentally determined structure of VvEG16.
Table S1: EG16 expression constructs screened for protein production.
Table S2: 18O-labeling of each product from cellooligosaccharide hydrolysis.
Table S3: X-ray diffraction data statistics.
Table S4: Privateer validation results.
Table S5: Sequences used for generation of the GH16 phylogenetic tree.
Table S6: Primer sequences used for cloning and mutagenesis.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre J, Iglesias-Fernández J, Rovira C, Davies GJ, Wilson KS, Cowtan KD. Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant Cell Walls. New York, NY: Garland Science; 2010. [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D–Coffee. Nucl. Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal G, Attia M, Asohan J, Brumer H. Methods in Molecular Biology, Protein-Carbohydrate Interactions. Springer; 2016. A low-volume, parallel copper-bicinchoninic acid (BCA) assay for glycoside hydrolases. (in press) [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. Functional genomics of cell elongation in developing cotton fibers. Plant. Mol. Biol. 2004;54:911–929. doi: 10.1007/s11103-004-0392-y. [DOI] [PubMed] [Google Scholar]
- Attia MA, Brumer H. Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Current opinion in structural biology. 2016;40:43–53. doi: 10.1016/j.sbi.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Barbeyron T, Gerard A, Potin P, Henrissat B, Kloareg B. The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol Biol Evol. 1998;15:528–537. doi: 10.1093/oxfordjournals.molbev.a025952. [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklof JM, Michel G, Kallas AM, Teeri TT, Czjzek M, Brumer H. Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism. Plant Cell. 2007;19:1947–1963. doi: 10.1105/tpc.107.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 2006;61:451–467. doi: 10.1007/s11103-006-0021-z. [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell. 2002;14:3073–3088. doi: 10.1105/tpc.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan M, Burton RA, Dhugga KS, Rafalski AJ, Tingey SV, Shirley NJ, Fincher GB. Endo-(1,4)-beta-Glucanase gene families in the grasses: temporal and spatial Co-transcription of orthologous genes. Bmc Plant biology. 2012;12:19. doi: 10.1186/1471-2229-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB. Heterogeneity in the chemistry, structure and function of plant cell walls. Nature Chemical biology. 2010;6:724–732. doi: 10.1038/nchembio.439. [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC. Biochemistry and Molecular Biology of Plants. Somerset, NJ: John Wiley & Sons, Inc.; 2000. The cell wall; pp. 55–108. [Google Scholar]
- Castro Ed, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucl. Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochemical Journal. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. The XTH Gene Family: An Update on Enzyme Structure, Function, and Phylogeny in Xyloglucan Remodeling. Plant Physiol. 2010;153:456–466. doi: 10.1104/pp.110.156844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklöf JM, Ruda MC, Brumer H. Distinguishing xyloglucanase activity in endo-β(1→4)glucanases. Meth. Enzymol. 2012;510:97–120. doi: 10.1016/B978-0-12-415931-0.00006-9. [DOI] [PubMed] [Google Scholar]
- Eklöf JM, Shojania S, Okon M, McIntosh LP, Brumer H. Structure-Function Analysis of a Broad Specificity Populus trichocarpa Endo-β-glucanase Reveals an Evolutionary Link between Bacterial Licheninases and Plant XTH Gene Products. J. Biol. Chem. 2013;288:15786–15799. doi: 10.1074/jbc.M113.462887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fenger TH, Brumer H. Synthesis and Analysis of Specific Covalent Inhibitors of endo-Xyloglucanases. ChemBioChem. 2015;16:575–583. doi: 10.1002/cbic.201402663. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Masuda S, Koizumi N, Nakamura S, Kumasaka T. The 1.3 Å crystal structure of a novel endo-β-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96. Proteins. 2007;69:683–690. doi: 10.1002/prot.21589. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Exploring the evolution of (1,3;1,4)-β-D-glucans in plant cell walls: comparative genomics can help! Current Opinion in Plant biology. 2009;12:140–147. doi: 10.1016/j.pbi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Forment J, Gadea J, Huerta L, Abizanda L, Agusti J, Alamar S, Alos E, Andres F, Arribas R, Beltran JP, Berbel A, Blazquez MA, Brumos J, Canas LA, Cercos M, Colmenero-Flores JM, Conesa A, Estables B, Gandia M, Garcia-Martinez JL, Gimeno J, Gisbert A, Gomez G, Gonzalez-Candelas L, Granell A, Guerri J, Lafuente MT, Madueno F, Marcos JF, Marques MC, Martinez F, Martinez-Godoy MA, Miralles S, Moreno P, Navarro L, Pallas V, Perez-Amador MA, Perez-Valle J, Pons C, Rodrigo I, Rodriguez PL, Royo C, Serrano R, Soler G, Tadeo F, Talon M, Terol J, Trenor M, Vaello L, Vicente O, Vidal C, Zacarias L, Conejero V. Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol. Biol. 2005;57:375–391. doi: 10.1007/s11103-004-7926-1. [DOI] [PubMed] [Google Scholar]
- Frankova L, Fry SC. Biochemistry and physiological roles of enzymes that cut and paste plant cell-wall polysaccharides. J. Exp. Bot. 2013;64:3519–3550. doi: 10.1093/jxb/ert201. [DOI] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR. Mixed-linkage (1→3,1→4)-β-d-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytologist. 2008;179:104–115. doi: 10.1111/j.1469-8137.2008.02435.x. [DOI] [PubMed] [Google Scholar]
- Fujii Y. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. The EMBO Journal. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser OJ, Piotukh K, Ponnuswamy MN, Planas A, Borriss R, Heinemann U. Structural Basis for the Substrate Specificity of a Bacillus 1,3-1,4-β-Glucanase. Journal of Molecular biology. 2006;357:1211–1225. doi: 10.1016/j.jmb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, Sundberg B, Karpinski S, Teeri TT, Kleczkowski LA, Henrissat B, Mellerowicz EJ. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006;140:946–962. doi: 10.1104/pp.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerttula S, Zinkgraf M, Muday GK, Lewis DR, Ibatullin FM, Brumer H, Hart F, Mansfield SD, Filkov V, Groover A. Transcriptional and Hormonal Regulation of Gravitropism of Woody Stems in Populus. Plant Cell. 2015;27:2800–2813. doi: 10.1105/tpc.15.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gloster TM, Ibatullin FM, Macauley K, Eklöf JM, Roberts S, Turkenburg JP, Bjørnvad ME, Jørgensen PL, Danielsen S, Johansen KS, Borchert TV, Wilson KS, Brumer H, Davies GJ. Characterization and Three-dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families GH5 and GH12. J. Biol. Chem. 2007;282:19177–19189. doi: 10.1074/jbc.M700224200. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucl. Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer DH, Weston C. Heat stress affects flowering, berry growth, sugar accumulation and photosynthesis of Vitis vinifera cv. Semillon grapevines grown in a controlled environment. Functional Plant biology. 2010;37:206–214. [Google Scholar]
- Guillon F, Bouchet B, Jamme F, Robert P, Quéméner B, Barron C, Larré C, Dumas P, Saulnier L. Brachypodium distachyon grain: characterization of endosperm cell walls. J. Exp. Bot. 2011;62:1001–1015. doi: 10.1093/jxb/erq332. [DOI] [PubMed] [Google Scholar]
- Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K. Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann Bot. 2014;114:1309–1318. doi: 10.1093/aob/mct292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield RD, Nevins DJ. Hydrolytic Activity and Substrate Specificity of an Endoglucanase from Zea mays Seedling Cell Walls. Plant Physiol. 1987;83:203–207. doi: 10.1104/pp.83.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J-H, Correc G, Thomas F, Bernard T, Barbeyron T, Jam M, Helbert W, Michel G, Czjzek M. Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia galactanivorans. J Biol Chem. 2012;287:30571–30584. doi: 10.1074/jbc.M112.377184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson K, Teleman A, Suortti T, Reinikainen T, Jaskari J, Teleman O, Poutanen K. Hydrolysis of barley (1→3), (1→4)-β-d-glucan by a cellobiohydrolase II preparation from Trichoderma reesei. Carbohydrate Polymers. 1995;26:109–119. [Google Scholar]
- Hrmova M, Farkas V, Lahnstein J, Fincher GB. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-beta-D-glucans. J. Biol. Chem. 2007;282:12951–12962. doi: 10.1074/jbc.M611487200. [DOI] [PubMed] [Google Scholar]
- Ibatullin FM, Baumann MJ, Greffe L, Brumer H. Kinetic Analyses of Retaining endo-(Xylo)glucanases from Plant and Microbial Sources Using New Chromogenic Xylogluco-Oligosaccharide Aryl Glycosides. Biochemistry. 2008;47:7762–7769. doi: 10.1021/bi8009168. [DOI] [PubMed] [Google Scholar]
- Johansson P, Brumer H, Baumann MJ, Kallas AM, Henriksson H, Denman SE, Teeri TT, Jones TA. Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding. Plant Cell. 2004;16:874–886. doi: 10.1105/tpc.020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallographica Section D Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewthai N, Gendre D, Eklof JM, Ibatullin FM, Ezcurra I, Bhalerao RP, Brumer H. Group III-A XTH Genes of Arabidopsis Encode Predominant Xyloglucan Endohydrolases That Are Dispensable for Normal Growth. Plant Physiol. 2013;161:440–454. doi: 10.1104/pp.112.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemle SN, Zhang X, Esker AR, Toriz G, Gatenholm P, Cosgrove DJ. Role of (1,3)(1,4)-β-Glucan in Cell Walls: Interaction with Cellulose. Biomacromolecules. 2014;15:1727–1736. doi: 10.1021/bm5001247. [DOI] [PubMed] [Google Scholar]
- Kong Y, Peña MJ, Renna L, Avci U, Pattathil S, Tuomivaara ST, Li X, Reiter W-D, Brandizzi F, Hahn MG, Darvill AG, York WS, O'Neill MA. Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 2015;167:1296–1306. doi: 10.1104/pp.114.255943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE. Stereochemistry and the Mechanism of Enzymatic Reactions. Biological Reviews. 1953;28:416–436. [Google Scholar]
- Labourel A, Jam M, Jeudy A, Hehemann J-H, Czjzek M, Michel G. The β-Glucanase ZgLamA from Zobellia galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin. J. Biol. Chem. 2014;289:2027–2042. doi: 10.1074/jbc.M113.538843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Izumi A, Ibatullin, Farid M, Nakhai A, Gilbert, Harry J, Davies, Gideon J, Brumer H. Structural and enzymatic characterization of a glycoside hydrolase family 31 α-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochemical Journal. 2011;436:567–580. doi: 10.1042/BJ20110299. [DOI] [PubMed] [Google Scholar]
- Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD. Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta. 2010;232:1191–1205. doi: 10.1007/s00425-010-1246-2. [DOI] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucl. Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casado G, Urbanowicz BR, Damasceno CMB, Rose JKC. Plant glycosyl hydrolases and biofuels: a natural marriage. Current Opinion in Plant biology. 2008;11:329–337. doi: 10.1016/j.pbi.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Malet C, Jimenez-Barbero J, Bernabe M, Brosa C, Planas A. Stereochemical course and structure of the products of the enzymic action of endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase from Bacillus licheniformis. Biochemical Journal. 1993;296:753–758. doi: 10.1042/bj2960753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Kaewthai N, Eklof JM, Miller JG, Brumer H, Fry SC, Verbelen JP, Vissenberg K. Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis thaliana. J. Exp. Bot. 2011;62:261–271. doi: 10.1093/jxb/erq263. [DOI] [PubMed] [Google Scholar]
- Mark P, Baumann MJ, Eklöf JM, Gullfot F, Michel G, Kallas ÅM, Teeri TT, Brumer H, Czjzek M. Analysis of nasturtium TmNXG1 complexes by crystallography and molecular dynamics provides detailed insight into substrate recognition by family GH16 xyloglucan endo-transglycosylases and endo-hydrolases. Proteins. 2009;75:820–836. doi: 10.1002/prot.22291. [DOI] [PubMed] [Google Scholar]
- Mark P, Zhang Q, Czjzek M, Brumer H, Agren H. Molecular dynamics simulations of a branched tetradecasaccharide substrate in the active site of a xyloglucan endo-transglycosylase. Molecular Simulation. 2011;37:1001–1013. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor N, Arnal G, Brumer H. Methods in Molecular Biology, Protein-Carbohydrate Interactions. Springer; 2016. Quantitative Kinetic Characterization of Glycoside Hydrolases Using High-Performance Anion-Exchange Chromatography (HPAEC) (in press) [DOI] [PubMed] [Google Scholar]
- McGregor N, Morar M, Fenger TH, Stogios P, Lenfant N, Yin V, Xu X, Evdokimova E, Cui H, Henrissat B, Savchenko A, Brumer H. Structure-function analysis of a mixed-linkage Beta-glucanase/xyloglucanase from key ruminal Bacteroidetes Prevotella bryantii B14. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.691659. jbc.M115.691659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips TM, McPhillips SE, Chiu H-J, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- Meier H, Reid JSG. Reserve Polysaccharides Other Than Starch in Higher Plants. In: Loewus PDFA, Tanner PDW, editors. Plant Carbohydrates I. Berlin Heidelberg: Springer; 1982. pp. 418–471. [Google Scholar]
- Mewalal R, Mizrachi E, Mansfield SD, Myburg AA. Cell Wall-Related Proteins of Unknown Function: Missing Links in Plant Cell Wall Development. Plant Cell Physiol. 2014;55:1031–1043. doi: 10.1093/pcp/pcu050. [DOI] [PubMed] [Google Scholar]
- Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, Dideberg O. The k-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of clan-B glycoside hydrolases. Structure. 2001;9:513–525. doi: 10.1016/s0969-2126(01)00612-8. [DOI] [PubMed] [Google Scholar]
- Minic Z, Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiology and Biochemistry. 2006;44:435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Murphy FV. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. The EMBO Journal. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of Prior Phase Information Strengthens Maximum-Likelihood Structure Refinement. Acta Crystallographica Section D Biological Crystallography. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. Xyloglucan and its Interactions with Other Components of the Growing Cell Wall. Plant Cell Physiol. 2015;56:180–194. doi: 10.1093/pcp/pcu204. [DOI] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu LF, Mansoori N, de Souza A, Schultink A, Xiong GY. Hemicellulose biosynthesis. Planta. 2013;238:627–642. doi: 10.1007/s00425-013-1921-1. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Biosynthesis of the Plant Cell Wall Matrix Polysaccharide Xyloglucan. In: Merchant SS, editor. Annual Review of Plant Biology. Vol. 67. Palo Alto: Annual Reviews; 2016. pp. 235–259. [DOI] [PubMed] [Google Scholar]
- Peng FY, Reid KE, Liao N, Schlosser J, Lijavetzky D, Holt R, Martínez Zapater JM, Jones S, Marra M, Bohlmann J, Lund ST. Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development. Gene. 2007;402:40–50. doi: 10.1016/j.gene.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Planas A. Bacterial 1,3-1,4-β-glucanases: structure, function and protein engineering. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2000;1543:361–382. doi: 10.1016/s0167-4838(00)00231-4. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Herve C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual review of plant biology. 2011;62:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Lee SJ. Straying off the Highway: Trafficking of Secreted Plant Proteins and Complexity in the Plant Cell Wall Proteome. Plant Physiol. 2010;153:433–436. doi: 10.1104/pp.110.154872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Gianzo C, Iglesias N, Guitian E, Revilla G, Zarra I. AtBGAL10 Is the Main Xyloglucan beta-Galactosidase in Arabidopsis, and Its Absence Results in Unusual Xyloglucan Subunits and Growth Defects. Plant Physiol. 2012;158:1146–1157. doi: 10.1104/pp.111.192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Pauly M. Glycosyltransferases and cell wall biosynthesis: novel players and insights. Current Opinion in Plant biology. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. In: Merchant S, Briggs WR, Ort D, editors. Annual Review of Plant Biology. Vol. 61. Palo Alto: Annual Reviews; 2010. pp. 263–289. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal Structure of a Smad MH1 Domain Bound to DNA: Insights on DNA Binding in TGF-β Signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Simmons TJ, Mohler KE, Holland C, Goubet F, Franková L, Houston DR, Hudson AD, Meulewaeter F, Fry SC. Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalises cellulose. Plant J. 2015 doi: 10.1111/tpj.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadiut O, Ibatullin FM, Peart J, Gullfot F, Martinez-Fleites C, Ruda M, Xu C, Sundqvist G, Davies GJ, Brumer H. Building Custom Polysaccharides in Vitro with an Efficient, Broad-Specificity Xyloglucan Glycosynthase and a Fucosyltransferase. J Am Chem Soc. 2011;133:10892–10900. doi: 10.1021/ja202788q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. Journal of Applied Crystallography. 2008;41:641–643. [Google Scholar]
- Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. A general, robust method for the quality control of intact proteins using LC-ESI-MS. Journal of Chromatography B. 2007;852:188–194. doi: 10.1016/j.jchromb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea JE, Cherry S, Waugh DS. Expression and Purification of Soluble His6-Tagged TEV Protease. In: Doyle SA, editor. High Throughput Protein Expression and Purification. Humana Press; 2009. pp. 297–307. [DOI] [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O’Neill MA, York WS. Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydrate Research. 2015;402:56–66. doi: 10.1016/j.carres.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Tyler L, Bragg JN, Wu JJ, Yang XH, Tuskan GA, Vogel JP. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. Bmc Genomics. 2010;11:21. doi: 10.1186/1471-2164-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallographica Section D Biological Crystallography. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Vasur J, Kawai R, Andersson E, Igarashi K, Sandgren M, Samejima M, Ståhlberg J. X-ray crystal structures of Phanerochaete chrysosporium Laminarinase 16A in complex with products from lichenin and laminarin hydrolysis. FEBS Journal. 2009;276:3858–3869. doi: 10.1111/j.1742-4658.2009.07099.x. [DOI] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Verhertbruggen Y, Scheller HV, Ronald PC. Abundance of mixed linkage glucan in mature tissues and secondary cell walls of grasses. Plant Signal Behav. 2013:8. doi: 10.4161/psb.23143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakarchuk WW, Kilburn DGRCM, Jr, Warren RAJ. The molecular cloning and expression of a cellobiase gene from an Agrobacterium in Escherichia coli. Molec. Gen. Genet. 1986;205:146–152. [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D Biological Crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Qinggang Z, Zhengke Z, Kun M, Yali H, Qiuyan B, Jiangtao S, Jingping R. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) Genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS ONE. 2015;10:e0123668. doi: 10.1371/journal.pone.0123668. [0123620pp.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Yuan SH, Guo H, Chen F, Tuskan GA, Cheng ZM. Evolution and divergence in the coding and promoter regions of the Populus gene family encoding xyloglucan endotransglycosylase/hydrolases. Tree Genetics & Genomes. 2012;8:177–194. [Google Scholar]
- Yokoyama R, Uwagaki Y, Sasaki H, Harada T, Hiwatashi Y, Hasebe M, Nishitani K. Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. Plant Journal. 2010;64:645–656. doi: 10.1111/j.1365-313X.2010.04351.x. [DOI] [PubMed] [Google Scholar]
- York WS, van Halbeek H, Darvill AG, Albersheim P. Structural analysis of xyloglucan oligosaccharides by 1H-n.m.r. spectroscopy and fast-atom-bombardment mass spectrometry. Carbohydrate Research. 1990;200:9–31. doi: 10.1016/0008-6215(90)84179-x. [DOI] [PubMed] [Google Scholar]
- Zverlov VV, Velikodvorskaya GA. Cloning the Clostridium thermocellum thermostable laminarinase gene in Escherichia coli: The properties of the enzyme thus produced. Biotechnol Lett. 1990;12:811–816. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.