Abstract
Niemann-Pick disease type C (NPC) is a fatal, neurodegenerative, cholesterol storage disorder. With new therapeutics in clinical trials, it is imperative to improve diagnostics and facilitate early intervention. We used metabolomic profiling to identify potential markers and discovered three unknown bile acids that were increased in plasma from NPC but not control subjects. The bile acids most elevated in the NPC subjects were identified as 3β,5α,6β-trihydroxycholanic acid and its glycine conjugate, both of which were shown to be metabolites of cholestane-3β,5α,6β-triol, an oxysterol elevated in NPC. A high-throughput, mass spectrometry-based method was developed and validated to measure the glycine-conjugated bile acid in dried blood spots. Analysis of dried blood spots from 4992 controls, 134 NPC carriers, and 44 NPC subjects provided 100% sensitivity and specificity in the study samples. Quantification of the bile acid in dried blood spots, therefore, provides the basis for a newborn screen for NPC that is ready for piloting in newborn screening programs.
INTRODUCTION
Niemann-Pick C (NPC) disease is a rare, progressive, neurodegenerative, cholesterol storage disorder. Mutations in NPC1 (95% of NPC cases) and NPC2 are responsible for NPC in humans. NPC1, a multi-spanning transmembrane protein, and NPC2, a soluble cholesterol-binding protein, both reside in late endosome/lysosomes (1). Loss of function in either protein results in lysosomal cholesterol and sphingolipid accumulation, accompanied by hallmarks of cellular oxidative stress, including increased amounts of reactive oxygen species (ROS) and marked increase in cholesterol oxidation products (2–4). The complex lipid storage in NPC, as in other sphingolipidoses, is associated with alterations in neuronal morphology and neuronal cell loss that underlie the neuroinflammatory and neurodegenerative disease pathology (5).
NPC disease encompasses a broad spectrum of systemic, neurological, and psychiatric signs and symptoms (1, 6). Affected individuals often present in early childhood with ataxia, seizures, and progressive impairment of motor and intellectual function, and usually die in adolescence, though increasingly NPC disease is being recognized among adults with cognitive defects (7, 8). Approximately half of NPC patients initially present with visceral symptoms (enlarged liver or spleen or both) or neonatal cholestasis. Diagnosis of this disorder, however, has been challenging because of the non-specific symptoms and the lack of a rapid diagnostic assay. As a result, the disease frequently goes unrecognized or misdiagnosed, with diagnostic delays averaging 4–5 years, during which time neurological symptoms inexorably progress and opportunities for intervention are lost. Because therapies targeting the lipid storage have been shown to be effective in pre-clinical models (9–14) and clinical trials (15), there is a pressing need for a sensitive and specific diagnostic test that can be effectively applied to newborn populations.
Current approaches for diagnosing NPC are not well suited for newborn screening. Cholesterol staining of fibroblasts using filipin, which has been the diagnostic standard, requires an invasive biopsy, takes up to three months, and is frequently indeterminate (8). DNA diagnostics are not practical because of the high preponderance of private mutations causing NPC1 disease and the large number of variants of uncertain functional consequence in the NPC1 gene (16). In contrast to many other lysosomal storage disorders (LSDs), NPC1 is not caused by mutations in a soluble enzyme, but a structural lysosomal protein. Thus, newborn screening based upon enzyme activity assays is not feasible. Recently, cholestane-3β,5α,6β-triol – a cholesterol oxidation product – was shown to be a sensitive and specific marker for Niemann-Pick disease (4), and a liquid chromatography-tandem mass spectrometry (LC-MS/MS) assay that measures the plasma concentration of this metabolite has emerged as a diagnostic tool (17). An oxysterol-based screen, however, is limited by the requirement for derivatization and the co-migration of an interference peak that derives from the newborn screening cards and cannot be adequately resolved during a short liquid chromatography run.
To identify a more suitable marker, we performed unbiased metabolomic profiling of plasma from NPC subjects, focusing on bile acids because of the known perturbations in sterol metabolism, the reports of unusual urinary bile acids in NPC (18, 19), and the favorable chemical properties of bile acids for mass spectrometry detection. Through profiling of bile acid species in the plasma from NPC subjects, we discovered an unusual β-hydroxysteroid bile acid and its glycine and taurine conjugates, which were strikingly elevated in the plasma and dried blood spots of NPC1 subjects as compared with controls. Here, we describe elucidation of the structures of these bile acids, development of LC-MS/MS methodology for the analysis of bile acids in dried blood spots, and establishment and validation of a newborn screen based on quantification of the conjugated bile acid. This dried blood spot-based assay provides the basis for a screen for NPC that is ready for piloting in newborn screening programs.
RESULTS
Profiling of bile acids in NPC1 and control plasma samples
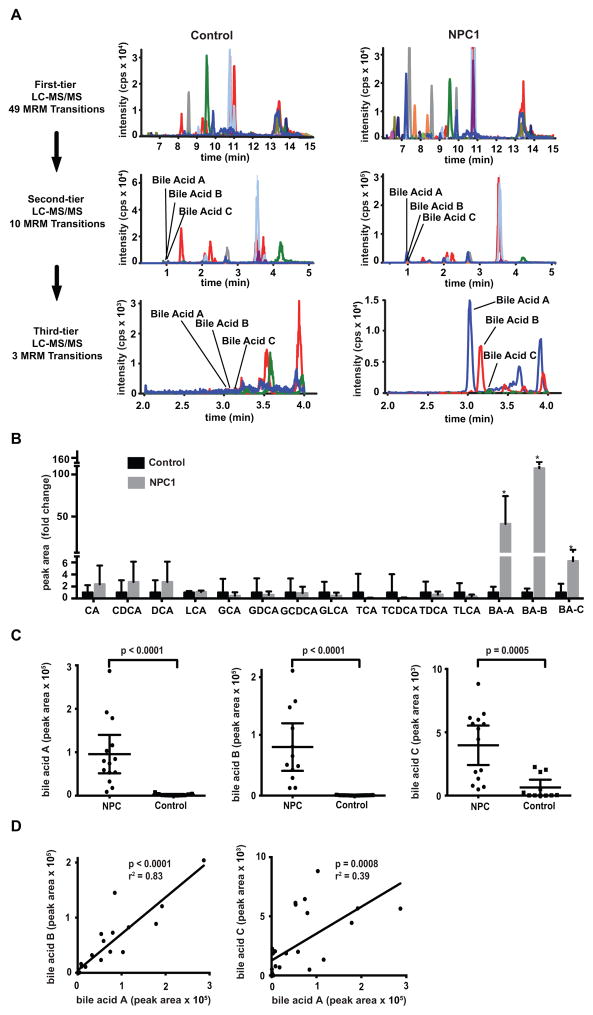
To identify potential markers, we profiled bile acids in NPC1 and control plasmas using a three-tier targeted metabolomics strategy based on LC-MS/MS operated in multiple reaction monitoring (MRM) mode (Fig. 1A). The first-tier assay included 49 MRM transitions with 17 min run time to broadly detect possible bile acids. For the second tier, 10 MRM transitions and 7.5 min run time were used to detect only those bile acids with signal-to-noise ratio greater than five. Only three MRM transitions and six min run time were used in the third-tier assay to confirm the three candidate bile acid species (referred to as bile acids A, B, and C) identified by second-tier assay. In contrast to other bile acids monitored, these three unknown species were markedly elevated in the NPC1 but not control plasma samples (Fig. 1B). The unknown bile acids were detected in same MRM transitions as cholic acid, glycocholic acid, and taurocholic acid, respectively, but their retention times differed, suggesting that they might be isomers of these bile acids. Bile acids A and B were increased 41-fold and 144-fold, respectively, in NPC1 plasma, and were able to completely discriminate between NPC1 and control plasma samples (Fig. 1C). Plasma bile acid C was elevated 6-fold in NPC1 samples (p = 0.0005), but there was partial overlap with control samples. The high correlation between the three bile acids’ plasma concentrations suggested that they are related, possibly within the same metabolic pathway (Fig. 1D). Because the bile acid C signal was 20-fold lower than those of bile acids A and B and prone to interferences from plasma, we did not pursue bile acid C further.
Fig. 1. NPC1 biomarker screening.
(A) Three-tier targeted metabolomics strategy for identification of bile acid biomarkers. First-tier screen includes 49 multiple reaction monitoring (MRM) transition (17 min run time). Second tier includes 10 MRM transitions (7.5 min run time) to characterize peaks with signal-to-noise ratio greater than five. Third tier (6 min run time) quantifies unknown bile acid peaks (A, B and C) that are elevated in NPC1 compared to control. (B) Comparison of bile acid concentration in NPC1 (n = 12) versus control (n = 11) samples obtained from second-tier profiling. Data are presented as mean fold-change + SD normalized to control. *P = 0.0005, 0.0003, 0.0007 for bile acid A, B, C in NPC1 versus controls, respectively. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; GCA, glycocholic acid; GDCA, glycodeoxycholic acid; GCDCA, glycochenodeoxycholic acid; GLCA, glycolithocholic acid; TCA, taurocholic acid; TDCA, taurodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; TLCA, taurolithocholic acid; BA-A, bile acid A; BA-B, bile acid B; BA-C, bile acid C. (C) Comparison of bile acids A, B and C in NPC1 (n = 12) and control (n = 11) plasma samples obtained from third-tier profiling. Data are presented as mean ± 95% CI peak area. P < 0.0001 for bile acids A and B in NPC1 versus controls. P = 0.0005 for bile acid C in NPC1 versus controls. (D) Correlation between bile acids A and B in NPC plasma samples, r2 = 0.83, P < 0.0001; correlation between bile acids A and C in NPC plasma samples, r2 = 0.39, P = 0.0008.
Elucidation of the structure of the bile acids
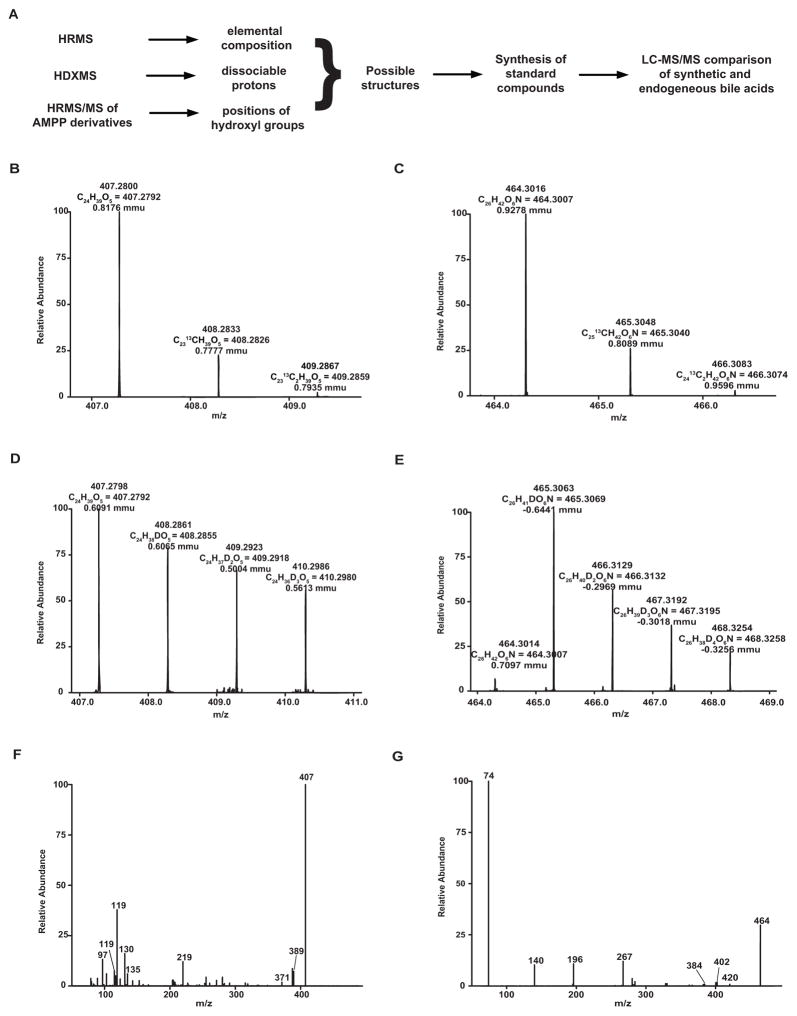
Our strategy for identification of bile acid structures is outlined in Fig. 2A. High resolution mass spectrometric analysis of bile acids A and B on LTQ-Orbitrap mass spectrometer in the negative mode showed accurate m/z values of 407.2800 and 464.3016, respectively, for [M–H]− corresponding to formulas C24H39O5 (calculated mass, 407.2803) and C26H42NO6 (calculated mass, 464.3018) with mass errors less than 1 mDa, respectively (Fig. 2B and C). Hydrogen/deuterium (H/D) exchange experiments indicated that there are four (3 OH, COOH) and five (3 OH, NH, COOH) exchangeable hydrogens in bile acids A and B (Fig. 2D and E), respectively. The higher energy collisional dissociation (HCD) spectrum of bile B contains an abundant ion at m/z 74.0256, corresponding to deprotonated glycine (C2H4O2N; calculated mass: 74.0248), confirming that bile acid B is a glycine conjugate. However, unambiguous assignment of structures for bile acids A and B was hindered by lack of interpretable fragments from steroid skeletons in HCD spectra (Fig. 2F and G). Therefore, we converted isolated bile acids A and B into their N-(4-aminomethylphenyl) pyridinium (AMPP) amides, which produced informative charge-remote fragmentation for structure identification in HCD and identified the key fragments that can differentiate the positions of hydroxylation (Supplemental Materials, tables S1 and S2). To aid interpretation of the product ion spectra of derivatized bile acids A and B, we studied the fragmentation patterns of the AMPP derivatives of a series of bile acids and analogs 1–10 (fig. S1–S12; tables S3–S5) and identified the key fragments that can differentiate the positions of hydroxylation (Supplemental Materials, fig. S13). On the basis of these fragmentation patterns, we assigned the hydroxyl groups to bile acids A and B. The structures of bile acids A and B were preliminarily proposed as 5α-cholanic acid-3β,5α,6β-triol and 5α-cholanic acid-3β,5α,6β-triol N-(carboxymethyl)-amide, respectively (Supplemental Materials, fig. S14). These structures were then confirmed by the synthesis of the reference compounds and comparative LC-MS/MS analysis of endogenous and synthesized compounds (Supplemental Materials, fig. S13 and S15).
Fig. 2. Identification and confirmation of structure of unknown bile acids.
(A) Strategy for identification of bile acid structures. HRMS, high-resolution mass spectrometry; HDXMS, hydrogen/deuterium exchange mass spectrometry; HRMS/MS, high-resolution tandem mass spectrometry; AMPP, N-(4-aminomethylphenyl) pyridinium. High-resolution mass spectra are shown for bile acid A (B) and bile acid B (C). H/D exchange mass spectra are shown for bile acid A (D) and bile acid B (E). Higher energy collisional dissociation (HCD) mass spectra is shown for bile acid A (F) and bile acid B (G).
Biosynthesis of the bile acids
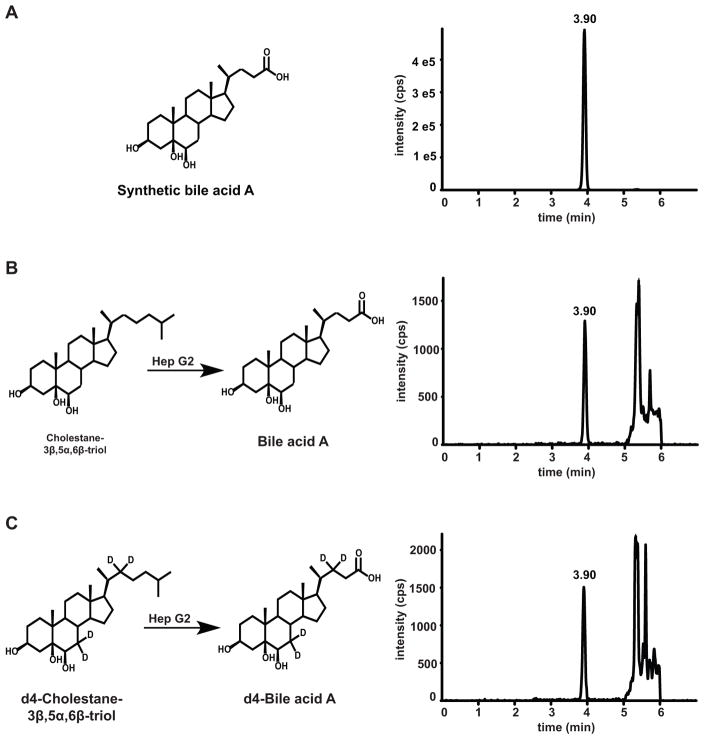
Although bile acid A has been reported as a major metabolite of cholestane-3β,5α,6β-triol in rat (20), the biosynthesis of bile acids A and B in humans has not previously been described. To explore the biosynthetic route of the bile acids, we incubated the human hepatoblastoma-derived cell line Hep G2 with cholestane-3β,5α,6β-triol and 7,7,22,22-d4-cholestane-3β,5α,6β-triol. We found that bile acid A and 7,7,22,22-d4-bile acid A were produced, thus confirming that bile acid A was a product of cholestane-3β,5α,6β-triol metabolism (Fig. 3). No bile acid B was found, consistent with the known defects in synthesis of conjugated bile acids in Hep G cells (21), presumably due to deficiency of bile acid CoA:amino acid N-acyltransferase (BAAT).
Fig. 3. Biosynthesis of bile acid A from cholestane-3β,5α,6β-triol.
(A) Synthetic bile acid A detected by MRM transition m/z 407 → 407. (B) Bile acid A generated from cholestane-3β,5α,6β-triol in HepG2 cells and detected by MRM transition m/z 407 → 407. (C) d4-Bile acid A generated from d4-cholestane-3β,5α,6β-triol in HepG2 cells and detected by MRM transition m/z 411 → 411.
Selection of bile acid marker for newborn screening of NPC1 disease
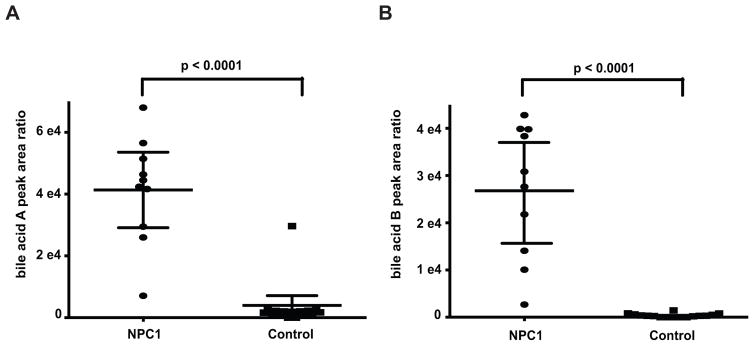
To explore the ability of bile acids A and B to serve as markers for newborn screening of NPC1 disease, we measured these metabolites in 10 NPC and 16 control dried blood spots. Bile acids A and B were elevated 12- and 101-fold, respectively, in NPC1 subjects compared to the control group (Fig. 4). Whereas bile acid A could separate all but one control subject from NPC subjects, bile acid B could unambiguously discriminate NPC1 subjects from controls, indicating that bile acid B may be the more specific marker for purposes of a newborn screening application.
Fig. 4. Detection of bile acid biomarkers in dried blood spots.
(A) Bile acid A in NPC1 (n = 10) and control (n = 16) dried blood spot samples. Data are presented as mean ± 95% CI peak area. P < 0.0001 for NPC1 versus controls. (B) Bile acid B in NPC1 (n = 10) and control (n = 16) dried blood spots sample. Data are presented as mean ± 95% CI peak area. P < 0.0001 for NPC1 versus controls.
Development and validation of two-tiered LC-MS/MS assay for bile acid B in dried blood spots
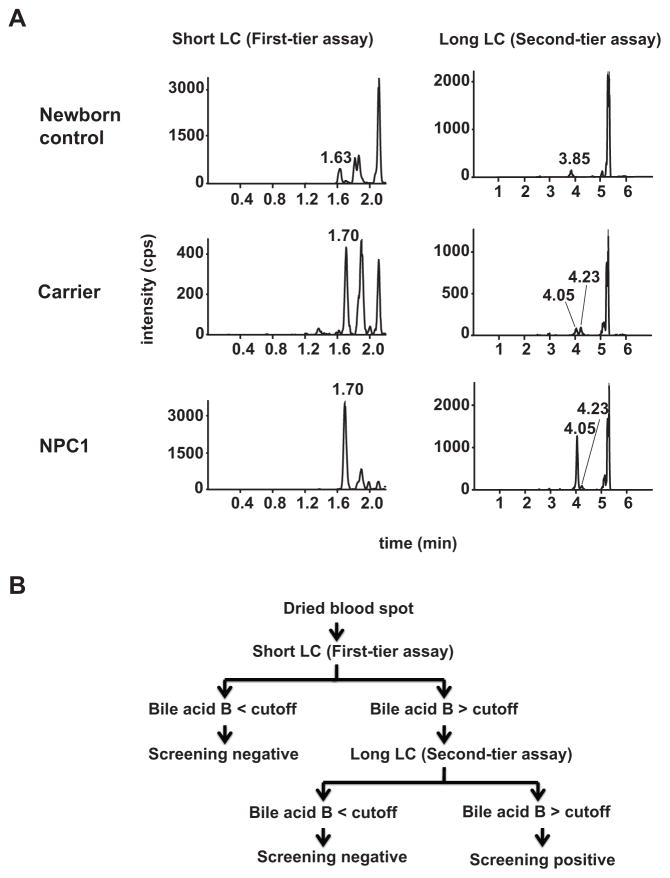
Our goal was to develop a short LC-MS/MS method (~2 min) that would allow sufficient throughput to meet a general requirement for a newborn screening assay. A major challenge for development of a high throughput LC-MS/MS method is separation of interferences from bile acid B within the short LC run time. A long LC run time (7 min) was initially developed, which separated all the interferences from bile acid B. Two interference peaks eluted closely to bile acid B (retention time at 4.05 min, Fig. 5A). Most newborn dried blood spots only showed an interference peak that eluted at 3.85 min, whereas dried blood spots from NPC1 subjects and carriers showed an interference peak that eluted at 4.23 min. We next developed a short LC condition with a sample analysis time of 2.2 min, in which bile acid B (retention time at 1.7 min) was baseline resolved from the major interference peak (retention time at 1.63 min) in normal newborn dried blood spots. However, under the short LC condition the bile acid B could not be separated from the interference peak in NPC1 and carrier dried blood spots. Therefore, a two-tier assay strategy was adopted, in which a short (2.2 minutes) and long (7 minutes) LC conditions were used as first and second tier assays, respectively (Fig. 5B). Thus, using the first-tier method, more than 500 samples/day can be analyzed. Samples with bile acid B values above the cut-off value and inability to separate from the second interference peak could then be submitted to the highly selective second-tier assay, permitting adjudication of the false positives from the first-tier assay. Together, this tiered strategy can serve as the basis of a screen for NPC.
Fig. 5. Two-tier newborn screen for NPC1 disease.
(A) Chromatograms of bile acid B in dried blood spots from a newborn control, adult NPC1 carrier, and NPC1 patient, as resolved with short LC (first-tier assay) and long LC conditions (second-tier assay). The bile acid B was eluted at 1.7 and 4.05 min under short and long LC conditions, respectively. There are two interferences eluted close to bile acid B. An interference peak presents in most newborn dried blood spots was baseline resolved from bile acid B under both short (1.63 min) and long LC (3.85 min) conditions. The dried blood spots from NPC1 subjects and carriers showed an interference peak that was co-eluted with bile acid B at 1.7 min under short LC condition, but baseline separated from bile acid B under long LC condition at 4.23 min. (B) Algorithm for two-tier newborn screening of NPC1 disease.
Detailed method development is described in Supplemental Materials. The method was validated according to commonly accepted criteria (22, 23) for sensitivity, selectivity, accuracy, precision, linearity, carry-over, recovery, matrix effect, effect of spotting volumes, effect of hematocrit, effect of punch location, and stabilities in whole blood, dried blood spots, processed samples, and stock solutions (Supplemental Materials, table S8). Bile acid B was stable in dried blood spots in newborn screening cards for up to 66 days at room temperature storage.
Establishment and validation of cut-off value for newborn screen
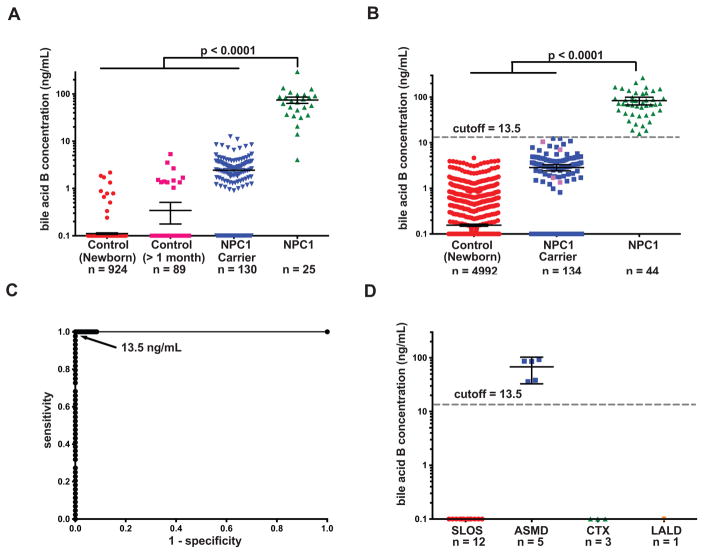
We used the validated bile acid B dried blood spot assay to establish the cut-off value for NPC1 newborn screening. For this training set, we analyzed dried blood spot samples from 1013 normal subjects (including 924 newborns and 89 subjects at other ages), 130 NPC1 carriers, and 25 NPC1 subjects. NPC1 and NPC1 carrier dried blood spots were obtained from previously diagnosed patients and obligate heterozygotes, respectively. The reference ranges for control, NPC1 carrier, and NPC1 subjects were <5 - 5.34, <5 - 12.5, and 5.45 - 294 ng/mL, respectively (Fig. 6A). The only NPC1 subject with bile acid B concentration <10 ng/mL was neurologically asymptomatic and was identified because of a sibling with splenomegaly who was diagnosed with NPC1, who carried a high frequency variant (c.665A>C, p.N222S) (16). A cut-off at 13.5 ng/mL allowed nearly complete discrimination of NPC1 subjects from controls and NPC1 carriers.
Fig. 6. Establishment and validation of cut-off for NPC1 newborn screen.
(A) Bile acid B concentrations in dried blood spots from newborn control, control at other age (> 1 month old), NPC1 carrier, and NPC1 patients in a training set. Bile acid B concentrations below the LLOQ (5 ng/mL) were quantifiable though the %CV and %RE for these samples were above acceptance criteria for the validated assay. Data presented on semi-log plots are shown as mean ± 95% CI. Samples with no detectable bile acid B peak were assigned as 0.1 ng/ml for purposes of plotting. P < 0.0001 for NPC1 versus controls and NPC1 carriers. (B) Determination of bile acid B concentrations and application of the 13.5 ng/mL cut-off to a test sample set consisting of newborn control, NPC1 carrier, and NPC1 dried blood spots. All newborn control and NPC1 samples are new samples, and carriers denoted by pink symbols are new samples. Samples were coded, randomized and the operator blinded to group assignment, thus reducing bias and noise/variance in the results and enabling unbiased statistical analysis of the data. Data are presented on semi-log plots and are shown as mean ± 95% CI. Samples with no detectable bile acid B peak were assigned as 0.1 ng/ml for purposes of plotting. P < 0.0001 for NPC1 versus controls and NPC1 carriers. (C) Application of cut-off value of 13.5 ng/mL yields sensitivity and specificity of 100%, and ROC area under the curve of 1.0. The NPC1 carriers in blue were analyzed in training set and re-analyzed in validation set. (D) Bile acid B concentrations in SLOS, ASMD, CTX, and LALD dried blood spots from cut-off validation sample set. Data are presented on semi-log plots and are shown as mean ± 95% CI. Samples with no detectable bile acid B peak were assigned as 0.1 ng/ml for purposes of plotting. P < 0.0001 for ASMD versus controls.
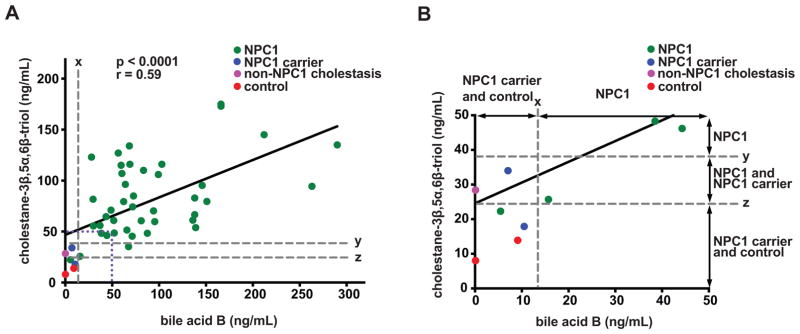
The cut-off was validated by analysis of a second (“test”) set of dried blood spot samples including 4992 normal newborns, 134 NPC1 carriers (130 carriers analyzed in the training set plus four new carriers), and 44 NPC1 patients. The range of bile acid B concentrations in normal newborns, NPC1 carriers, and NPC1 patients was <5, <5 -12.4, 15.7 – 266 ng/mL, respectively (Fig. 6B). A cut-off of 13.5 ng/mL provided 100% sensitivity and specificity for detection of NPC1 subjects in the test set, yielding a receiver operator characteristic (ROC) area under the curve of 1.0 (Fig. 6C). Only 1 normal newborn sample (0.017%) needed to be resolved using the second-tier assay. Among NPC1 subjects, there was no significant correlation between bile acid B concentration and age or disease severity (Supplemental Materials, fig. S16). The mean bile acid B level for female NPC1 subjects was 35% lower than male subjects (P < 0.05). All the normal newborns and NPC1 carriers were below the cut-off, while all the NPC1 samples were above the cut-off. To assess specificity of the assay, we examined samples from patients with other disorders of sterol metabolism [acid-sphingomyelinase deficiency (ASMD), cerebrotendinous xanthomatosis (CTX), lysosomal acid lipase deficiency (LALD), and Smith-Lemli-Opitz Syndrome (SLOS)]. Of these, only ASMD subjects demonstrated bile acid B concentrations (36 - 92.8 ng/mL) above the cut-off (Fig. 6D). The new screen was “benchmarked” by comparing bile acid B concentrations in dried blood spots with the plasma concentration of cholestane-3β,5α,6β-triol, the current diagnostic standard (Fig. 7). As anticipated, there was significant correlation between the bile acid and its precursor (r = 0.59), and both assays were equally sensitive in detecting NPC1 subjects. However, the bile acid B marker was more specific, completely discriminating NPC1 subjects from NPC1 carriers and permitting proper assignment of non-NPC1 cholestasis.
Fig. 7. Comparison of bile acid B in dried blood spots with cholestane-3β,5α,6β-triol in plasma.
(A) Correlation of bile acid B in dried blood spots with cholestane-3β,5α,6β-triol in plasma (n = 47), r = 0.59, P < 0.0001. x, bile acid cutoff for NPC1 carriers and controls; y, cholestane-3β,5α,6β-triol cutoff for NPC1 carriers; z, cholestane-3β,5α,6β-triol cutoff for controls. (B) The inset bounded by the dotted purple line in panel (A) has been expanded to more clearly compare assay specificity.
DISCUSSION
A major barrier to delivery of effective treatment for NPC disease is the profound diagnostic delay caused by the non-specific nature of early symptoms, genetic heterogeneity, and lack of a biochemical marker that can be applied to a dried blood spot-based newborn screen. Preclinical studies clearly demonstrate that intervention with effective treatments at this early stage, before the onset of neurological symptoms, can modify the course of the disease and extend lifespan (9, 13), underscoring the need for new diagnostic algorithms. To address this unmet need, we performed targeted metabolomic profiling, identifying disease markers that were strikingly elevated in NPC1 subjects. The metabolite structures were identified by mass spectrometric techniques and confirmed as 3β,5α,6β-trihydroxycholanic acid and its glycine conjugate. We developed and validated a high-throughput, mass spectrometry (MS)-based method to quantify the bile acid glycine conjugate in dried blood spots, which provides the basis for a highly sensitive and specific newborn screen for NPC that is ready for piloting in newborn screening laboratories.
The bile acids identified by our profiling strategy are unusual in that they have not been reported in human plasma and possess a 3β-hydroxyl-5α-hydroxyl-cholanic acid rather than the 3α-hydroxyl-5β-cholanic acid characteristic of the common circulating bile acids (24). Although low amounts of 3β-hydroxy bile acids have been reported in human fecal contents and urine, their steroid skeletons are different from the bile acids identified in our study (18, 19, 25). These observations suggest that the 3β-hydroxy bile acids are likely specific for distinguishing Niemann-Pick disease from other causes of cholestasis. Using stable isotope labeling, we demonstrated that 3β,5α,6β-trihydroxycholanic acid (bile acid A) is converted from cholestane-3β,5α,6β-triol in human hepatoma cells, suggesting that 3β,5α,6β-trihydroxycholanic acid may be formed in vivo in humans. Although generation of the glycine conjugate could not be tested in the cell culture model, 3β,5α,6β-trihydroxycholanic acid is likely converted in vivo by bile acid CoA:amino acid acyltransferase into its more polar glycine conjugate (bile acid B). Bile acid C is very likely a taurine conjugate of 3β,5α,6β-trihydroxycholanic acid (fig. S14). The marked elevation of these unusual bile acids in the plasma of the NPC1 subjects is entirely consistent with the known increases in cholestane-3β,5α,6β-triol, the metabolic precursor of these bile acids, in the plasma of NPC patients. In contrast to cholestane-3β,5α,6β-triol, which is also elevated in other disorders of sterol metabolism (ASMD, CTX, and LALD) (26), bile acid B was elevated only in NPC and ASMD subjects. Thus, elevation of these bile acid markers appears to be a downstream consequence of both the tissue oxidative stress that is a prominent feature of inborn errors of sterol metabolism (2, 3, 27, 28) and the altered disposal of bile acids in Niemann-Pick disease (29).
Detection of the bile acid markers in dried blood spots from newborn screening cards enabled development of a high throughput newborn screen for NPC. The screen further incorporates a second-tier LC run, a well-accepted strategy in newborn screens to increase specificity without sacrificing sensitivity. The exceptional performance of the screen – 100% sensitivity and specificity for discrimination of NPC1 subjects from controls and NPC1 carriers coupled with a 0.017% re-assay rate – should minimize false positives caused by the ~1% carrier frequency in the population (16). As compared to cholestane-3β,5α,6β-triol, which is the current diagnostic standard, bile acid B eliminates the ~25% overlap of NPC1 carriers that are in the NPC1 range (17), as well as false positives among cholestatic neonates caused by interference peaks that cannot be separated in short LC runs (30). A potential source of bias in this retrospective study, however, is that non-neonatal NPC1 patients were used as surrogates for assay validation because of the lack of authentic dried blood spots from NPC1 neonates (the youngest patient in the study was 4 months old and neurologically asymptomatic). Nonetheless, in light of case reports demonstrating dramatic elevation of the bile acid precursor cholestane-3β,5α,6β-triol in NPC1 and NPC2 neonates (31), it seems probable that the bile acids will be elevated in NPC patients during the neonatal period. Moreover, bile acid B concentrations were independent of age and severity of disease, suggesting that the screening test will be universally applicable to all NPC1 subjects, including neonates.
Advances in mass spectrometry have expanded the scope of newborn screening to more than 50 inborn errors of metabolism. Tandem MS is considered the method of choice for screening of metabolic disorders, and it is widely used to screen for amino acid, organic acid, and fatty acid disorders. Recently, a multiplex tandem MS assay, which is based upon use of a panel of substrates to quantify enzymatic activity, was introduced to screen for five LSDs: Fabry, Gaucher, Krabbe, Niemann-Pick A/B (ASMD), and Pompe diseases (32). Newborn screening using this method is mandated to begin in four US states (33). There are several reasons to believe that NPC disease is also an excellent candidate for newborn screening. The frequency of the disorder (1:100,000) (1, 6) is comparable to the five currently screened LSDs (34) and at least twice as frequent as many commonly screened metabolic disorders (35), and disease-modifying therapies are available (miglustat) or entering into late stage clinical trials (2-hydroxypropyl-β-cyclodextrin). Moreover, as with other LSDs, it is anticipated that neonatal screening will reduce long-term morbidity, and the screening and treatment will cost less than long-term care and lost productivity (36). Finally, coupling of the bile acid assay, which has exceptional receiver operating characteristics, with a cut-off that will exclude all carriers and controls, could yield a newborn screen capable of detecting the vast majority of NPC1 cases. Determination of the positive predictive value of the assay will await prospective validation. In the future, modifications of this screen, such as elimination of liquid chromatography or shifting to positive ion mode detection, could facilitate incorporation of the NPC screen into a multiplexed panel.
At present, none of the disease conditions included in the Recommended Universal Screening Panel, which is the newborn screening standard, target a disorder involving an integral membrane (in other words, non-luminal enzyme) lysosomal protein or an inborn error of sterol metabolism and trafficking. Thus, this bile acid-based NPC screen represents an important advance in neonatal screening. Broad implementation of newborn screening for NPC would eliminate the diagnostic delay and shift diagnosis of the disease to the newborn period before the onset of neurological symptoms. Drug intervention and non-pharmacological supportive care during this asymptomatic period have the potential to markedly delay disease progression and extend life.
MATERIALS AND METHODS
Study Design
The study was a retrospective case-control design. Dried blood spot samples were obtained from four groups: (1) newborn controls, (2) controls > 30 days of age, (3) NPC1 carriers (obligate heterozygotes who are parents of affected NPC1 subjects), and (4) previously diagnosed NPC1 subjects. The goal of the study was to evaluate the performance of a dried blood spot-based newborn screen for NPC1. For this study, an assay was developed to measure bile acid B in the dried blood, and this assay met FDA guidance criteria with respect to precision, accuracy, and replication (22). Samples were coded and randomized, and the operator was blinded to group assignment to reduce bias and noise/variance in the results. The assay cut-off was established using a training set of samples. The cut-off was then applied in a second (“test”) assay using new, independent samples except that NPC1 carriers analyzed in the training set were re-assayed together with new carriers. Data are presented on semi-log plots and are shown as mean ± 95% CI. Statistical differences between the groups were determined using Mann-Whitney U test. The performance of the assay is presented on an ROC plot.
Chemicals and reagents
Deoxycholic acid (DCA), chenodeoxycholic acid (CDCA), cholic acid (CA), α-muricholic acid, β-muricholic acid, glycodeoxycholic acid (GDCA), glycochenodeoxycholic acid (GCDCA), and glycocholic acid (GCA) were obtained from Steraloids, Inc. Bile acid structures are shown in fig. S1. Bile acid A (37), 5-cholanic acid-3α,4β,7α-triol (7)(38), and AMPP (39) were synthesized according to published procedures. N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), 4-(dimethylamino)pyridine (DMAP), diethylamine, N,N-dimethylformamide, trisodium citrate, sodium hydroxide (NaOH), chloroform, Dulbecco’s modified Eagle’s medium, fetal calf serum, and penicillin G and streptomycin sulfate were obtained from Sigma–Aldrich. HPLC solvents (methanol and acetonitrile) were HPLC grade and were purchased from EMD Chemicals. Milli-Q ultrapure water was prepared in-house with a Milli-Q Integral Water Purification System.
Plasma and dried blood spot sample collection
NPC1 dried blood spot samples were obtained from NIH, Rush University Medical Center, Universitätsklinikum Münster, and University of Heidelberg. NIH also provided NPC1 plasma, NPC1 carrier, and SLOS dried blood spot samples. All NPC1 and NPC1 carrier dried blood spots were obtained from previously diagnosed patients and obligate heterozygotes, respectively. Normal plasmas and dried blood spots were obtained from anonymized residual samples at St. Louis Children’s Hospital and New York State Newborn Screening Program. Genzyme provided ASMD dried blood spot samples. Hospital de Clínicas de Porto Alegre provided ASMD, CTX, and LALD dried blood spot samples. All plasma samples were collected in ethylenediamine tetraacetic acid dipotassium salt (EDTA-K2) containing tubes. The collection and analysis of de-identified human samples was approved by the Human Studies Committee at Washington University.
Bile acid marker screening and identification
Sample preparation for plasma and dried blood spots
Plasma samples (50 μL) were aliquoted into 2 mL polypropylene tubes (VWR). Methanol (150 μL) was added to each tube. The samples were vortexed for 3 min, centrifuged for 10 min at 3000 g, and the supernatants were transferred to clean glass inserts in 1.5 mL HPLC vials for LC-MS/MS assay. A 3 mm disc was punched from each dried blood spot using a Harris Micro-Punch (Thermo Fisher Scientific). Each punch was transferred to a clean 2 mL polypropylene tube (VWR). An aliquot of 50 μl of water was added to each punch, and the mixture was vortexed for 10 min. Methanol (150 μL) was added to each tube. The samples were vortexed for 3 min, centrifuged for 10 min at 3000 g, and the supernatants were transferred to clean glass inserts in 1.5 mL HPLC vials for LC-MS/MS assay.
First-tier LC-MS/MS bile acid marker screening in plasma
The NPC1 and control samples were randomized so that each group was evenly distributed in the run order. In this way, bias and noise/variance in the results caused by instrument fluctuation were reduced, enabling subsequent unbiased statistical analysis of the data. LC–MS/MS analysis was conducted on a Shimadzu Prominence UFLC system coupled with an Applied Biosystems/MDS Sciex 4000QTRAP mass spectrometer using multiple reaction monitoring (MRM). Separation of bile acids was carried out at 50 °C using a Waters XBridge C18 analytical column (4.6×50 mm, 2.5 μm) connected to a Phenomenex SecurityGuard C18 guard column (4 × 3 mm) at a flow rate of 1 mL/min. The mobile phase consisted of 10 mM ammonium acetate and 0.1% ammonium hydroxide in water (solvent A), and acetonitrile-methanol (1:4) (solvent B). The step gradient was as follows: 0–0.1 min, 10%; 0.1–10 min, 10 to 75% solvent B; 10–12 min, 75% solvent B; 12–12.1 min, 75 to 100% solvent B; 12.1–15 min, 100%; 15–15.1 min, 100 to 10% solvent B; 15.1–17 min, 10% solvent B. The effluent was directed into the mass spectrometer for data acquisition within the 13-min time window (2 – 15 min); elsewhere, effluent was sent to waste to minimize source contamination. The injection volume was 5 μL, and the total run time was 17 min. The ESI source temperature was 500 °C; the ESI needle was −4500 V; the entrance potential was −10 V; and the collision cell exit potential was −10 V. The collision and curtain gas were set at medium and 20, respectively. Both desolvation gas and nebulizing gas were set at 35 L/min. The MRM transitions, declustering potentials, and the collision energies are listed in table S6. Unconjugated bile acids such as CA, CDCA, DCA, UDCA, HDCA, HDCA, and LCA are extremely difficult to fragment in collision-induced dissociation and were therefore monitored by the MRM transitions from their precursor ions to product ions with the same m/z values. The dwell time was set at 20 ms for each MRM transition. Data were acquired and analyzed by Analyst software (version 1.5.2). In a separate LC run, fractions containing the bile acids of interest were collected for further structure analysis.
Second-tier LC-MS/MS bile acid marker screening in plasma
The same LC-MS/MS system and column as those in the first-tier bile acid marker screening were used for the second-tier screening. The chromatography was carried out at 50 °C. The mobile phase consisted of 0.1% ammonium hydroxide in water (solvent A) and acetonitrile-methanol (1:4) (solvent B). The step gradient was as follows: 0–5 min, 45 to 75% solvent B; 5–5.1 min, 75 to 100% solvent B; 5.1–6 min, 100% solvent B; 6–6.1 min, 100 to 45% solvent B; 6.1–7.5 min, 45% solvent B. The effluent was directed into the mass spectrometer for data acquisition from 0.9 to – 6 min; elsewhere, effluent was sent to waste to minimize source contamination. The injection volume was 5 μL, and the total run time was 7.5 min. The ESI source temperature was 500 °C; the ESI needle was −4500 V; the entrance potential was −10 V; and the collision cell exit potential was −10 V. The collision and curtain gas were set at medium and 20, respectively. Both desolvation gas and nebulizing gas were set at 35 L/min. The MRM transitions, declustering potentials, and the collision energies are listed in table S7. The dwell time was set at 20 ms for each MRM transition. Data were acquired and analyzed by Analyst software (version 1.5.2).
Third-tier LC-MS/MS bile acid marker screening in plasma and analysis of bile acids A and B in dried blood spots
The same LC-MS/MS system as that in the first- and second-tier bile acid marker screening was used for the third-tier screening. Separation of bile acids A and B was carried out at 50 °C using a Waters XBridge C18 analytical column (4.6×100 mm, 3.5 μm) connected to a Phenomenex SecurityGuard C18 guard column (4 × 3 mm) at a flow rate of 1 mL/min. The mobile phase consisted of 2.9 mM diethylamine in water (solvent A) and acetonitrile-methanol (1:9) (solvent B). The step gradient was as follows: 0–3 min, 40% to 55% solvent B; 3–3.1 min, 55% to 100% solvent B; 3.1–4 min, 100% solvent B; 4–4.1 min, 100% to 40% solvent B; 4.1–6 min, 40% solvent B. The effluent was directed into the mass spectrometer for data acquisition within the 2-min time window (2 – 4 min) in which bile acids A and B were eluted; elsewhere, effluent was sent to waste to minimize source contamination. The total run time was 6 min. The injection volume was 2 μL for plasma samples and 20 μL for dried blood spot samples. The ESI source temperature was 500 °C; the ESI needle was −4500 V; the declustering potential was −120 V for bile acids A and B, and −150 V for bile acid C; the entrance potential was −10 V; and the collision cell exit potential was −10 V. The collision and curtain gas were set at medium and 20, respectively. Both desolvation gas and nebulizing gas were set at 35 L/min, and the collision energies were −35 and −75eV for bile acids A and B, respectively. For MRM for third-tier LC-MS/MS bile acid marker screening, the dwell time was set at 50 ms for each of the signals from transitions of m/z 407 to 407 (bile acid A), m/z 464 to 74 (bile acid B), m/z 514 to 80 (bile acid C). For MRM for analysis of bile acids in dried blood spots, the dwell time was set at 50 ms for each of the signals from transitions of m/z 407 to 407 (bile acid A) and m/z 464 to 74 (bile acid B). Data were acquired and analyzed by Analyst software (version 1.5.2). In a separate LC run, fractions containing the bile acids of interest were collected from plasma samples for further structure analysis.
High resolution mass spectrometry and H/D exchange experiment of underivatized bile acids A and B
The bile acids A and B isolated from third tier screening were dissolved in methanol and directly infused into a LTQ Orbitrap Velos ETD mass spectrometer (Thermo Fisher Scientific) via Harvard syringe pump at 5 μL/min. The full FT-MS scan (m/z 250–800) and HCD MS/MS scans were performed with precursor isolation width of 1 m/z. Full scan and HCD MS/MS mass spectra were recorded at a resolution of 100,000 at m/z 400. Automatic gain control (AGC) was used to accumulate sufficient ions. For survey scans, AGC target was 1 × 106 (maximum injection time 1 s). For HCD, AGC target was 1 × 105 (maximum inject time 100 and 25 ms, respectively). HCD was performed at normalized collision energy of 95%. Helium was used as the buffer and collision gas at a pressure of 1 × 10−3 mbar (0.75 mTorr). Data acquisition was controlled by Xcalibur 2.1 software package. Spray voltage was set to −4 kV and temperature of the heated transfer capillary was 300 °C. The vaporizer heating was off. Sheath and auxiliary nitrogen gas were both applied at a flow rate of 0 arbitrary units (AU). The exchange of the labile hydrogen atoms in bile acids A and B for deuterium atoms was carried out by preparing solutions of the analytes in deuterated methanol. The final samples were immediately infused directly into the ESI LTQ Orbitrap Velos ETD mass spectrometer.
Derivatization of bile acids with AMPP
10 μL of 5 mg/mL AMPP suspension in acetonitrile, 10 μL of 1 M EDC/1 M DMAP in chloroform, and 10 μL of N,N-dimethylformamide were added to the dried bile acids in a 1.2 mL glass insert in a μL plate (VWR) to derivatize the samples. Mixtures were capped, vortexed, and heated for 1 hour at 50 °C. The mixture was dried with nitrogen stream at 50 °C and reconstituted with 200 μL of methanol-water (1:1).
LC-HRMS analysis of bile acid AMPP derivatives
The separation was performed on a Shimadzu 10A HPLC system (Shimadzu Scientific Instruments) coupled with the LTQ Orbitrap Velos ETD mass spectrometer, operating with ESI source in positive mode. A MAC-MOD ACE 3 C18 (2×50 mm, 3 μm) connected to a Phenomenex SecurityGuard C18 guard column (4 × 3 mm) was used for the chromatographic separation, and it was maintained at room temperature. The mobile phase consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile-methanol (1:4) (solvent B). The step gradient was as follows: 0–2.5 min, 20% to 100% solvent B; 2.5–6 min, 100% solvent B; 6–6.1 min, 100% to 20% solvent B; 6.1–7 min, 20% solvent B. The effluent was directed into the mass spectrometer for data acquisition within the 4-min time window (2 – 6 min) in which bile acids A and B were eluted; elsewhere, effluent was sent to waste to minimize source contamination. The mass spectrometer performed a full FT-MS scan (m/z 250–800), and HCD MS/MS scan precursor isolation width was 1 m/z. Full scan and HCD MS/MS mass spectra were recorded at a resolution of 100,000 at m/z 400. Automatic gain control (AGC) was used to accumulate sufficient ions. For survey scans, AGC target was 1 × 106 (maximum injection time 1 s). For HCD, AGC target was 1 × 105 (maximum inject time 100 and 25 ms, respectively). Helium was used as the buffer and collision gas at a pressure of 1 × 10−3 mbar (0.75 mTorr). HCD was performed at normalized collision energy of 80%. Data acquisition was controlled by Xcalibur 2.1 software package. Spray voltage was set to 4.5 kV. The vaporizer temperature and temperature of the heated transfer capillary were 380 and 250°C, respectively. Sheath and auxiliary nitrogen gas were applied at a flow rate of 60 and 20 arbitrary units (AU), respectively.
Biosynthesis of bile acid from cholestane-3β,5α,6β-triol in Hep G2 cells
Hep G2 cells (1 × 106 cells/well) were seeded in a 6-well plate in triplicate and maintained in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal calf serum and 100 U of penicillin G/mL and 100 μg streptomycin sulfate at 37 °C in a humidified atmosphere with 5% CO2. Hep G2 cells were treated when they were 80% confluent. For the treatment, stock solutions were made in DMSO for triol and d4-triol. DMSO concentration never exceeded 1% in the culture medium. Cells were exposed to the test compounds (2.5 μg/mL cholestane-3β,5α,6β-triol or 7,7,22,22,22-d4-cholestane-3β,5α,6β-triol) or solvent control (1% DMSO) for 24 h. The conditioned medium (50 μL) was removed and transferred to a new 2 mL polypropylene tube, to which methanol (200 μl) was added. The samples were centrifuged at 9391g for 10 minutes at room temperature. The supernatant was transferred to a glass HPLC insert and analyzed immediately after preparation by LC-MS/MS.
LC–MS/MS analysis was conducted on a Shimadzu Prominence UFLC system coupled with an Applied Biosystems/MDS Sciex 4000QTRAP mass spectrometer using multiple reaction monitoring (MRM). The ESI source temperature was 550 °C; the ESI needle was −4500 V; the declustering potential was −120 V for bile acid A and d4-bile acid A and −140 V for bile acid B and d4-bile acid B; the collision energy were −35 eV for bile acid A and d4-bile acid A and −72 eV for bile acid B and d4-bile acid B; the entrance potential was −10 V for bile acid A, d4-bile acid A, bile acid B, and d4-bile acid B; and the collision cell exit potential was −11 V for bile acid A, d4-bile acid A, bile acid B, and d4-bile acid B. The collision and curtain gas were set at medium and 20, respectively. The desolvation gas and nebulizing gas were set at 60 and 35 L/min, respectively. For MRM, the dwell time was set at 50 ms for the transitions of m/z 407 to 407 (bile acid A), m/z 411 to 411 (d4-bile acid A), m/z 464 to 74 (bile acid B), and m/z 468 to 74 (d4-bile acid B). Data were acquired and analyzed by Analyst software (version 1.5.2). The liquid chromatography was carried out at ambient temperature using an ACE Excel 3 Super C18 column (4.6 × 100 mm, 3 μm; MAC-MOD Analytical) connected to a Phenomenex SecurityGuard Gemini C18 guard column (4 × 3 mm). The solvent gradient using 2.9 mM diethylamine and 20 mM hexafluoro-2-propanol in water (phase A) and acetonitrile/methanol (1:4) (phase B) at a flow rate of 1 ml/min was as follows: 0 – 3.5 min 50 – 60% B, 3.5 – 3.6 min 60 – 100% D, 3.6 – 5.0 min 100% B, 5.0 – 5.1 min 100 – 50% B, and 5.1 – 7.0 min 50% B. The effluent was directed to waste at 0 – 2.5 and 5 – 7 min, and to the mass spectrometer at 2.5 – 6 min. The injection volume was 2 μL.
Two-tier assay for NPC1 newborn screening
Stock solution preparation
All the stock solutions (1 mg/mL) and working solutions (10 μg/mL) of bile acid B and bile acid B-[13C2,15N] were prepared in acetonitrile-water (1:1). A working solution containing 25 μg/mL of bile acid B was prepared by the dilution of the stock solution with methanol. The internal standard working solution (12 ng/mL of bile acid B-[13C2,15N]) was prepared in 1% SDS and 50 mM trisodium citrate in water, pH 12.
Standard curve and quality control samples
A freshly collected adult blood unit was washed with three portions of saline to remove anticoagulants and the buffy coat. After centrifugation and removal of the last saline wash, the combined red cells were reconstituted to a hematocrit of 55% ± 0.5% with pooled human cord plasma (blood bank at Barnes-Jewish Hospital) that had been verified to have undetectable bile acid B. The calibration standards (5, 10, 20, 50, 100, 200, 250, 500 ng/mL), lower limit of quantification (LLOQ, 5 ng/mL), lower limit (LLQC, 10 ng/mL), low (LQC, 30 ng/mL), middle (MQC, 150 ng/mL), and high (HQC, 300 ng/mL) quality control samples were prepared by serial dilution after bile acid B working solution was spiked into reconstituted blood. All blood samples were spotted onto Whatman 903 newborn screening cards in 50 μL aliquots, then dried for at least 3 hours at room temperature and stored at −20 °C in airtight bags with desiccant to minimize moisture accumulation. To evaluate the effect of spotting volume, LQC and HQC were also spotted in 75 and 100 μL aliquots onto Whatman 903 newborn screening cards. To evaluate the effect of hematocrit, LQC and HQC were prepared in blood at five hematocrit levels (40%, 50%, 55%, 60%, and 70%), and spotted onto Whatman 903 newborn screening cards in 50 μL aliquots.
Sample preparation for dried blood spots
A 3 mm disc was punched from each dried blood spot using a Harris Micro-Punch (Thermo Fisher Scientific) and placed into a 96-well plate (1 mL/well). An aliquot of 50 μl of internal standard working solution was added to each punch, and the mixture was vortexed for 10 min. Acetonitrile (150 μL) was added to each tube. The samples were vortexed for 3 min and centrifuged for 10 min at 3000 g. Supernatant was transferred to a clean 96-well plate (1 mL/well) and evaporated to dryness under a stream of nitrogen. The residue was reconstituted with 150 μL of water.
LC-MS/MS analysis of bile acid B in dried blood spot samples
LC–MS/MS analysis was conducted on a Shimadzu Prominence UFLC system coupled with an Applied Biosystems/MDS Sciex 4000QTRAP mass spectrometer using multiple reaction monitoring (MRM). The ESI source temperature was 550 °C; the ESI needle was −4500 V; the declustering potential was −140 V; the collision energy was −72 eV; the entrance potential was −10 V; and the collision cell exit potential was −11 V. The collision and curtain gas were set at medium and 20, respectively. The desolvation gas and nebulizing gas were set at 60 and 35 L/min, respectively. For MRM, the dwell time was set at 200 and 50 ms for the transition of m/z 464 to 74 (bile acid B) and m/z 467 to 77 (internal standard), respectively. Data were acquired and analyzed by Analyst software (version 1.5.2).
In the first-tier assay, the chromatography was performed using an ACE Excel 3 Super C18 column (4.6 × 50 mm, 3 μm; MAC-MOD Analytical) connected to a Phenomenex SecurityGuard Gemini C18 guard column (4 × 3 mm) at ambient temperature. The compartment of the autosampler was set at 4 °C. The mobile phase A (2.9 mM diethylamine and 20 mM hexafluoro-2-propanol in water) and mobile phase B (methanol) were operated with a gradient elution as follows: 0 – 1.3 min 50 – 80% B, 1.3 – 1.4 min 80 – 100% B, 1.4 – 1.6 min 100% B, 1.6 – 1.7 min 100 – 50% B, and 1.7 – 2.2 min 50% B at a flow rate of 1 ml/min. The effluent was directed to waste at 0 – 1.2 min, and to the mass spectrometer at 1.2 – 2.2 min. The injection volume was 20 μL.
The liquid chromatography in the second-tier assay was carried out at ambient temperature using an ACE Excel 3 Super C18 column (4.6 × 100 mm, 3 μm; MAC-MOD Analytical) connected to a Phenomenex SecurityGuard Gemini C18 guard column (4 × 3 mm). The solvent gradient using 2.9 mM diethylamine and 20 mM hexafluoro-2-propanol in water (phase A) and acetonitrile/methanol (1:4) (phase B) at a flow rate of 1 ml/min was as follows: 0 – 3.5 min 50 – 60% B, 3.5 – 3.6 min 60 – 100% D, 3.6 – 5.0 min 100% B, 5.0 – 5.1 min 100 – 50% B, and 5.1 – 7.0 min 50% B. The effluent was directed to waste at 0 – 2.5 and 5 – 7 min, and to the mass spectrometer at 2.5 – 5 min. The injection volume was 20 μL.
Linearity, precision and accuracy
Calibration curves were constructed with Analyst software (version 1.5.2) by plotting the corresponding peak area ratios of analyte/internal standard versus the corresponding analyte concentrations using weighted (1/x2) least squares regression analysis. The linearity response of bile acid B was assessed over the respective calibration range from three batches of analytical runs. The precision and accuracy of the assay were determined at LLOQ, LLQC, LQC, MQC, and HQC concentrations over the three batch runs. For each QC concentration, analysis was performed for six replicates in each batch. Precision and accuracy are denoted by percent coefficient of variance (%CV) and percent relative error (%RE), respectively. The accuracy and precision were required to be within ± 15%RE of the nominal concentration and ≤15%CV, respectively, for LLQC, LQC, MQC, and HQC samples. The accuracy and precision were required to be within ± 20% RE of the nominal concentration and ≤20% CV for LLOQ samples in the intra-batch and inter-batch assays (22).
Sample stability
The storage stability of dried blood spots and processed sample stabilities in the autosampler were determined at the LQC and HQC concentrations (n = 3). Storage stability of bile acid B in dried blood spots was tested at −20 °C and room temperature for 66 days, and at 37 °C for 90 hours. In the autosampler, stability was tested over seven days by injecting the first batch of the validation samples. The LQC and HQC in whole blood were placed on the benchtop at room temperature for 27 hours and then spotted onto newborn screening cards to examine stability of bile acid B in whole blood. Stock solution stability was established by quantification of samples from dilution of two stock solutions that have been stored at −20 °C for 91 days and at room temperature on the bench for 22 hours, respectively, to the final solution (500 ng/mL in water). The storage stability of internal standard working solution was tested at room temperature for 12 days. A fresh standard curve was established each time.
Analysis of clinical dried blood spot samples
All the clinical samples were first submitted to first-tier assay. Samples consisting of calibration standards in duplicate, a blank, a blank with internal standard, QC samples (LQC, MQC, and HQC), and unknown clinical samples were analyzed. The clinical samples with bile acid B above the LLOQ in the first tier assay together with calibration standards, blank, blank with internal standard, and QC samples in the same batch were re-assayed with the second-tier assay. The LC-MS/MS acceptance criteria were as indicated in FDA recommendations (22).
Statistical analysis
The GraphPad Prism version 6.0 (GraphPad Software) was used to perform Mann-Whitney U test, receiver-operator curve (ROC) analysis, and Pearson correlations. Microsoft Excel was used for calculations of percent coefficient of variance (%CV) and percent relative error (%RE). Mann-Whitney U test was applied to calculate differences between NPC1 and normal (control or control and NPC1 carrier) groups, and between ASMD and control groups. All presented P values are two-sided, and P < 0.05 was considered to be statistically significant. Bile acid A and B correlation was analyzed using Pearson correlations because the data showed a normal distribution.
Supplementary Material
Fig. S1. Structures of bile acids and analog that were used to study fragmentation patterns of the AMPP derivatives.
Fig. S2. HCD mass spectra of AMPP-derivatized 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), 6 (F), 7 (G), 8 (H), 9 (I), 10 (J).
Fig. S3. Proposed fragmentation pathway to fragments A – H in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S4. Proposed fragmentation pathway to fragments I – K in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S5. Proposed fragmentation pathway to fragments L (L′)– M (M′) in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S6. Proposed fragmentation pathway to fragments N – P in AMPP derivatives of unconjugated bile acids 1 – 6.
Fig. S7. Proposed fragmentation pathway to fragments Q – T in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S8. Proposed fragmentation pathway to fragments GA – GE, GH in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S9. Proposed fragmentation pathway to fragments GF, GG, GI in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S10. Proposed fragmentation pathway to fragments GJ - GM in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S11. Proposed fragmentation pathway to fragments GN - GW in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S12. Proposed fragmentation pathway to fragments GX – GZ, GAA - GAB in glycine conjugated bile acid AMPP derivatives.
Fig. S13. Comparison of chromatograms and HCD mass spectra of AMPP derivatives of bile acids A and B in NPC plasma and solutions of synthetic compounds.
Fig. S14. Structures of bile acid biomarkers for NPC.
Fig. S15. Comparison of chromatograms of bile acids A and B in NPC1 plasma and solutions of synthetic compounds.
Fig. S16. Correlation of bile acid B concentrations with patient parameters
Table S1. Accurate masses and calculated elemental composition of fragment ions of 21,26,27-trinorcholestan-25-oic acid-3β,5α,6β-triol and bile acid A AMPP derivatives.
Table S2. Accurate masses and calculated elemental composition of fragment ions of bile acid B AMPP derivative.
Table S3. Accurate masses and calculated elemental composition of fragment ions of deoxycholic acid, chenodeoxycholic acid, and 5β-cholanic acid-3α,4β,7α-triol AMPP derivatives.
Table S4. Accurate masses and calculated elemental composition of fragment ions of cholic acid, α-muricholic acid, and β- muricholic acid AMPP derivatives.
Table S5. Accurate masses and calculated elemental composition of fragment ions of glycocholic acid, glycochenodeoxycholic acid, and glycodeoxycholic acid AMPP derivatives.
Table S6. MRM transitions and MS parameters for bile acids in the first-tier biomarker screening.
Table S7. MRM transitions and MS parameters for bile acids in the second-tier biomarker screening.
Table S8. Accuracy and precision of QC samples.
Acknowledgments
We are grateful to the National Niemann-Pick Disease Foundation for their assistance in obtaining samples from NPC1 and NPC1 carrier subjects. The authors express their appreciation to the families and patients who participated in this study.
Funding: This work was supported by grants from the National Niemann-Pick Disease Foundation (X.J.), Dana’s Angels Research Trust (D.S.O. and N.Y.F.), Ara Parseghian Medical Research Foundation (D.S.O. and N.Y.F.), Support Of Accelerated Research for NPC Disease (D.S.O.), NIH T32 HL07275 (L.M.), and by NIH Grant R01 NS081985 (D.S.O. and J.E.S.). This study was also supported by the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (F.D.P.) and a Bench to Bedside award from the Office of Rare Diseases (F.D.P. and D.S.O.). This work was performed in the Metabolomics Facility at Washington University (NIH P30 DK020579).
Footnotes
Author contributions: X.J. performed mass spectrometry experiments, synthesized precursors for biosynthesis of novel bile acids, analyzed and interpreted the data and wrote the manuscript. R.S. performed sample preparation for development and validation of newborn screening. L.M. under the direction of D.F.C. synthesized standard compounds of novel bile acids and deuterated internal standard. F-F.H. and D.F.C. contributed to structural identification of novel bile acids and preparation of the manuscript. D.E.S. helped in experiments for structural identification of novel bile acids. B.E. and S.G. determined biosynthesis of novel bile acids. N.Y.F., D.J.D., J.J.O., E.B-K., H.R., X.Z., J.R., T.M., and R.G. collected blood spots and contributed to preparation of the manuscript. F.D.P., J.E.S. and D.S.O. planned the studies and wrote the manuscript.
Competing interests: D.S.O. and X.J. are named as co-inventors on a patent application pertaining to use of bile acid biomarkers in NPC disease.
Data and materials availability: All reasonable requests for chemical compounds and assay protocols described in this work will be fulfilled via an MTA or licensing agreements with Washington University.
REFERENCES AND NOTES
- 1.Vanier MT. Niemann-Pick disease type C. Orphanet J Rare Dis. 2010;516 doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tint G, Pentchev P, Xu G, Batta A, Shiefer S, Salen G, Honda A. Cholesterol and oxygenated cholesterol concentrations are markedly elevated in peripheral tissue but not in brain from mice with Niemann-Pick type C phenotype. J Inher Metab Dis. 1998;21:853–863. doi: 10.1023/a:1005474803278. [DOI] [PubMed] [Google Scholar]
- 3.Zhang JR, Coleman T, Langmade SJ, Scherrer DE, Lane L, Lanier MH, Feng C, Sands MS, Schaffer JE, Semenkovich CF, Ory DS. Niemann-Pick C1 protects against atherosclerosis in mice via regulation of macrophage intracellular cholesterol trafficking. J Clin Invest. 2008;118:2281–2290. doi: 10.1172/JCI32561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter FD, Scherrer DE, Lanier MH, Langmade SJ, Molugu V, Gale SE, Olzeski D, Sidhu R, Dietzen DJ, Fu R, Wassif CA, Yanjanin NM, Marso SP, House J, Vite C, Schaffer JE, Ory DS. Cholesterol oxidation products are sensitive and specific blood-based biomarkers for Niemann-Pick C1 disease. Sci Transl Med. 2010;2:56ra81. doi: 10.1126/scitranslmed.3001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walkley SU, Suzuki K. Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim Biophys Acta. 2004;1685:48–62. doi: 10.1016/j.bbalip.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Patterson MC, Hendriksz CJ, Walterfang M, Sedel F, Vanier MT, Wijburg F, Group NCGW. Recommendations for the diagnosis and management of Niemann-Pick disease type C: an update. Mol Genet Metab. 2012;106:330–344. doi: 10.1016/j.ymgme.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Bauer P, Balding DJ, Klunemann HH, Linden DE, Ory DS, Pineda M, Priller J, Sedel F, Muller A, Chadha-Boreham H, Welford RW, Strasser DS, Patterson MC. Genetic screening for Niemann-Pick disease type C in adults with neurological and psychiatric symptoms: findings from the ZOOM study. Hum Mol Genet. 2013;22:4349–4356. doi: 10.1093/hmg/ddt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stampfer M, Theiss S, Amraoui Y, Jiang X, Keller S, Ory DS, Mengel E, Fischer C, Runz H. Niemann-Pick disease type C clinical database: cognitive and coordination deficits are early disease indicators. Orphanet J Rare Dis. 2013;835 doi: 10.1186/1750-1172-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson CD, Ali NF, Micsenyi MC, Stephney G, Renault S, Dobrenis K, Ory DS, Vanier MT, Walkley SU. Chronic cyclodextrin treatment of murine Niemann-Pick C disease ameliorates neuronal cholesterol and glycosphingolipid storage and disease progression. PLoS One. 2009;4:e6951. doi: 10.1371/journal.pone.0006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Li H, Repa JJ, Turley SD, Dietschy JM. Genetic variations and treatments that affect the lifespan of the NPC1 mouse. J Lipid Res. 2008;49:663–669. doi: 10.1194/jlr.M700525-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Ramirez CM, Miller AM, Repa JJ, Turley SD, Dietschy JM. Cyclodextrin overcomes the transport defect in nearly every organ of NPC1 mice leading to excretion of sequestered cholesterol as bile acid. J Lipid Res. 2010;51:933–944. doi: 10.1194/jlr.M000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Turley SD, Burns DK, Miller AM, Repa JJ, Dietschy JM. Reversal of defective lysosomal transport in NPC disease ameliorates liver dysfunction and neurodegeneration in the npc1−/− mouse. Proc Natl Acad Sci U S A. 2009;106:2377–2382. doi: 10.1073/pnas.0810895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vite CH, Bagel JH, Swain GP, Prociuk M, Sikora TU, Stein VM, O’Donnell P, Ruane T, Ward S, Crooks A, Li S, Mauldin E, Stellar S, De Meulder M, Kao ML, Ory DS, Davidson C, Vanier MT, Walkley SU. Intracisternal cyclodextrin prevents cerebellar dysfunction and Purkinje cell death in feline Niemann-Pick type C1 disease. Sci Transl Med. 2015;7:276ra226. doi: 10.1126/scitranslmed.3010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zervas M, Somers KL, Thrall MA, Walkley SU. Critical role for glycosphingolipids in Niemann-Pick disease type C. Curr Biol. 2001;11:1283–1287. doi: 10.1016/s0960-9822(01)00396-7. [DOI] [PubMed] [Google Scholar]
- 15.Patterson MC, Vecchio D, Prady H, Abel L, Wraith JE. Miglustat for treatment of Niemann-Pick C disease: a randomised controlled study. Lancet Neurol. 2007;6:765–772. doi: 10.1016/S1474-4422(07)70194-1. [DOI] [PubMed] [Google Scholar]
- 16.Wassif CA, Cross JL, Iben J, Sanchez-Pulido L, Cougnoux A, Platt FM, Ory DS, Ponting CP, Bailey-Wilson JE, Biesecker LG, Porter FD. High incidence of unrecognized visceral/neurological late-onset Niemann-Pick disease, type C1, predicted by analysis of massively parallel sequencing data sets. Genet Med. 2016;18:41–48. doi: 10.1038/gim.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Sidhu R, Porter FD, Yanjanin NM, Speak AO, te Vruchte DT, Platt FM, Fujiwara H, Scherrer DE, Zhang J, Dietzen DJ, Schaffer JE, Ory DS. A sensitive and specific LC-MS/MS method for rapid diagnosis of Niemann-Pick C1 disease from human plasma. J Lipid Res. 2011;52:1435–1445. doi: 10.1194/jlr.D015735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvelius G, Hjalmarson O, Griffiths WJ, Bjorkhem I, Sjovall J. Identification of unusual 7-oxygenated bile acid sulfates in a patient with Niemann-Pick disease, type C. J Lipid Res. 2001;42:1571–1577. [PubMed] [Google Scholar]
- 19.Maekawa M, Misawa Y, Sotoura A, Yamaguchi H, Togawa M, Ohno K, Nittono H, Kakiyama G, Iida T, Hofmann AF, Goto J, Shimada M, Mano N. LC/ESI-MS/MS analysis of urinary 3beta-sulfooxy-7beta-N-acetylglucosaminyl-5-cholen-24-oic acid and its amides: new biomarkers for the detection of Niemann-Pick type C disease. Steroids. 2013;78:967–972. doi: 10.1016/j.steroids.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi S, Imai Y, Suzuoki Z, Matsu T, Noguchi S. Biologic studies of cholestane-3-beta,5-alpha,6-beta-triol and its derivatives. 3. The metabolic fate and metabolites of cholestane-3-beta,5-alpha,6-beta-triol in animals. J Pharmacol Exp Ther. 1968;159:399–408. [PubMed] [Google Scholar]
- 21.Everson GT, Polokoff MA. HepG2. A human hepatoblastoma cell line exhibiting defects in bile acid synthesis and conjugation. J Biol Chem. 1986;261:2197–2201. [PubMed] [Google Scholar]
- 22.Guidance for Industry: Bioanalytical Method Validations. 2001 May; http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf.
- 23.Spooner N, Lad R, Barfield M. Dried blood spots as a sample collection technique for the determination of pharmacokinetics in clinical studies: considerations for the validation of a quantitative bioanalytical method. Anal Chem. 2009;81:1557–1563. doi: 10.1021/ac8022839. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res. 2014;55:1553–1595. doi: 10.1194/jlr.R049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann AF, Loening-Baucke V, Lavine JE, Hagey LR, Steinbach JH, Packard CA, Griffin TL, Chatfield DA. Altered bile acid metabolism in childhood functional constipation: inactivation of secretory bile acids by sulfation in a subset of patients. J Pediatr Gastroenterol Nutr. 2008;47:598–606. doi: 10.1097/MPG.0b013e31816920a6. [DOI] [PubMed] [Google Scholar]
- 26.Pajares S, Arias A, Garcia-Villoria J, Macias-Vidal J, Ros E, de las Heras J, Giros M, Coll MJ, Ribes A. Cholestane-3beta,5alpha,6beta-triol: high levels in Niemann-Pick type C, cerebrotendinous xanthomatosis, and lysosomal acid lipase deficiency. J Lipid Res. 2015;56:1926–1935. doi: 10.1194/jlr.M060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Cuyar LF, Hunter B, Harris PL, Perry G, Smith MA, Castellani RJ. Cerebrotendinous xanthomatosis: case report with evidence of oxidative stress. Redox report : communications in free radical research. 2007;12:119–124. doi: 10.1179/135100007X200173. [DOI] [PubMed] [Google Scholar]
- 28.Korade Z, Xu L, Mirnics K, Porter NA. Lipid biomarkers of oxidative stress in a genetic mouse model of Smith-Lemli-Opitz syndrome. Journal of inherited metabolic disease. 2013;36:113–122. doi: 10.1007/s10545-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amigo L, Mendoza H, Castro J, Quinones V, Miquel JF, Zanlungo S. Relevance of Niemann-Pick type C1 protein expression in controlling plasma cholesterol and biliary lipid secretion in mice. Hepatology. 2002;36:819–828. doi: 10.1053/jhep.2002.35617. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Ory DS. Towards a New Diagnostic Standard for Niemann-Pick C Disease. EBioMedicine. 2016;4:18–19. doi: 10.1016/j.ebiom.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reunert J, Lotz-Havla AS, Polo G, Kannenberg F, Fobker M, Griese M, Mengel E, Muntau AC, Schnabel P, Sommerburg O, Borggraefe I, Dardis A, Burlina AP, Mall MA, Ciana G, Bembi B, Burlina AB, Marquardt T. Niemann-Pick Type C-2 Disease: Identification by Analysis of Plasma Cholestane-3beta,5alpha,6beta-Triol and Further Insight into the Clinical Phenotype. JIMD Rep. 2015;23:17–26. doi: 10.1007/8904_2015_423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Scott CR, Chamoles NA, Ghavami A, Pinto BM, Turecek F, Gelb MH. Direct multiplex assay of lysosomal enzymes in dried blood spots for newborn screening. Clin Chem. 2004;50:1785–1796. doi: 10.1373/clinchem.2004.035907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsden D, Levy H. Newborn screening of lysosomal storage disorders. Clin Chem. 2010;56:1071–1079. doi: 10.1373/clinchem.2009.141622. [DOI] [PubMed] [Google Scholar]
- 34.Wang RY, Bodamer OA, Watson MS, Wilcox WR ACMG Work Group on Diagnostic Confirmation of Lysosomal Storage Diseases. Lysosomal storage diseases: diagnostic confirmation and management of presymptomatic individuals. Genet Med. 2011;13:457–484. doi: 10.1097/GIM.0b013e318211a7e1. [DOI] [PubMed] [Google Scholar]
- 35.Burke W, Tarini B, Press NA, Evans JP. Genetic screening. Epidemiol Rev. 2011;33:148–164. doi: 10.1093/epirev/mxr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Karnebeek CD, Mohammadi T, Tsao N, Sinclair G, Sirrs S, Stockler S, Marra C. Health economic evaluation of plasma oxysterol screening in the diagnosis of Niemann-Pick Type C disease among intellectually disabled using discrete event simulation. Mol Genet Metab. 2015;114:226–232. doi: 10.1016/j.ymgme.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Kawanami J. Bile acids and steroids. XX. Hog bile acids. 4. Synthesis of 3β,6β-dihydroxy-5β-cholanic acid and 3β,6α-dihydroxy-5α-cholanic acid. Bull Chem Soc Jpn. 1961;34:509–513. [Google Scholar]
- 38.Yoshimura T, Mahara R, Kurosawa T, Ikegawa S, Tohma M. An efficient synthesis of 4 beta- and 6 alpha-hydroxylated bile acids. Steroids. 1993;58:52–58. doi: 10.1016/0039-128x(93)90052-o. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Han RH, Han X. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal Chem. 2013;85:9312–9320. doi: 10.1021/ac402078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciuffreda P, Casati S, Alessandrini L, Terraneo G, Santaniello E. Synthesis of deuterated isotopomers of 7alpha- and (25R,S)-26-hydroxycholesterol, internal standards for in vivo determination of the two biosynthetic pathways of bile acids. Steroids. 2003;68:733–738. doi: 10.1016/s0039-128x(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 41.Jopling J, Henry E, Wiedmeier SE, Christensen RD. Reference ranges for hematocrit and blood hemoglobin concentration during the neonatal period: data from a multihospital health care system. Pediatrics. 2009;123:e333–337. doi: 10.1542/peds.2008-2654. [DOI] [PubMed] [Google Scholar]
- 42.Yang K, Dilthey BG, Gross RW. Identification and quantitation of fatty acid double bond positional isomers: a shotgun lipidomics approach using charge-switch derivatization. Anal Chem. 2013;85:9742–9750. doi: 10.1021/ac402104u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Structures of bile acids and analog that were used to study fragmentation patterns of the AMPP derivatives.
Fig. S2. HCD mass spectra of AMPP-derivatized 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), 6 (F), 7 (G), 8 (H), 9 (I), 10 (J).
Fig. S3. Proposed fragmentation pathway to fragments A – H in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S4. Proposed fragmentation pathway to fragments I – K in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S5. Proposed fragmentation pathway to fragments L (L′)– M (M′) in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S6. Proposed fragmentation pathway to fragments N – P in AMPP derivatives of unconjugated bile acids 1 – 6.
Fig. S7. Proposed fragmentation pathway to fragments Q – T in AMPP derivatives of unconjugated bile acids 1 – 7.
Fig. S8. Proposed fragmentation pathway to fragments GA – GE, GH in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S9. Proposed fragmentation pathway to fragments GF, GG, GI in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S10. Proposed fragmentation pathway to fragments GJ - GM in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S11. Proposed fragmentation pathway to fragments GN - GW in AMPP derivatives of glycine conjugated bile acids 8 – 10.
Fig. S12. Proposed fragmentation pathway to fragments GX – GZ, GAA - GAB in glycine conjugated bile acid AMPP derivatives.
Fig. S13. Comparison of chromatograms and HCD mass spectra of AMPP derivatives of bile acids A and B in NPC plasma and solutions of synthetic compounds.
Fig. S14. Structures of bile acid biomarkers for NPC.
Fig. S15. Comparison of chromatograms of bile acids A and B in NPC1 plasma and solutions of synthetic compounds.
Fig. S16. Correlation of bile acid B concentrations with patient parameters
Table S1. Accurate masses and calculated elemental composition of fragment ions of 21,26,27-trinorcholestan-25-oic acid-3β,5α,6β-triol and bile acid A AMPP derivatives.
Table S2. Accurate masses and calculated elemental composition of fragment ions of bile acid B AMPP derivative.
Table S3. Accurate masses and calculated elemental composition of fragment ions of deoxycholic acid, chenodeoxycholic acid, and 5β-cholanic acid-3α,4β,7α-triol AMPP derivatives.
Table S4. Accurate masses and calculated elemental composition of fragment ions of cholic acid, α-muricholic acid, and β- muricholic acid AMPP derivatives.
Table S5. Accurate masses and calculated elemental composition of fragment ions of glycocholic acid, glycochenodeoxycholic acid, and glycodeoxycholic acid AMPP derivatives.
Table S6. MRM transitions and MS parameters for bile acids in the first-tier biomarker screening.
Table S7. MRM transitions and MS parameters for bile acids in the second-tier biomarker screening.
Table S8. Accuracy and precision of QC samples.