Abstract
Cardiomyopathies are a leading cause of heart failure and are often caused by mutations in sarcomeric genes, resulting in contractile dysfunction and cellular damage. This may stimulate the production of a robust proinflammatory response. To determine whether myocardial inflammation is associated with cardiac dysfunction in dilated cardiomyopathy (DCM) caused by MYBPC3 mutation, we used the well-characterized cMyBP-C(t/t) mouse model of DCM at 3 months of age. Compared to wild type (WT) mice, DCM mice exhibited significantly decreased fractional shortening (36.4±2% vs. 15.5±1.0%, p < 0.0001) and significantly increased spleen weight (5.3±0.3% vs. 7.2±0.4 mm/mg, p = 0.002). Intriguingly, flow cytometry analysis revealed a significant increase in total (CD45+CD11b+Ly6C−MHCII+F480+) macrophages (6.5±1.4% vs. 14.8±1.4%, p = 0.002) and classically activated (CD45+CD11b+Ly6C−MHCII+F480+CD206−) proinflammatory (M1) macrophages (3.4±0.8% vs. 10.3±1.2%, p = 0.0009) in DCM hearts as compared with WT hearts. These results were further confirmed by immunofluorescence analysis of heart tissue sections. Splenic red pulp (CD11b+Ly6C+MHCIIlowF480hi) macrophages were significantly elevated (2.4±0.1% vs. 1.3±0.1%, p = 0.0001) in DCM compared to WT animals. Serum cytokine analysis in DCM animals exhibited a significant increase (0.65±0.2 vs. 2.175±0.5 pg/mL, p = 0.02) in interleukin (IL)-6 compared to WT animals. Furthermore, RNAseq analysis revealed the upregulation of inflammatory pathways in the DCM hearts. Together, these data indicate a robust proinflammatory response in DCM hearts, likely in response to cellular damage triggered by MYBPC3 mutation and resultant contractile dysfunction.
Keywords: Dilated Cardiomyopathy, Inflammation, Sarcomere Biology, MYBPC3, Mouse Models
1. Introduction
Heart failure (HF), the final common pathway of many cardiovascular diseases, is a global health problem that afflicts an estimated 5.8 million Americans [1, 2] and 30–50 million patients worldwide [3]. Cardiomyopathies are a leading cause of HF and are defined by a pathologically abnormal myocardium [4]. They are classified into four major categories: dilated (DCM), hypertrophic (HCM), restrictive (RCM), and arrhythmogenic right ventricular cardiomyopathy (ARVC) [4]. Genetic mutations underlie a significant proportion of cardiomyopathies [4]. For example, 75% of inherited HCMs are caused by mutations in the genes encoding myosin heavy chain (MYH7) and cardiac myosin binding protein-C (cMyBP-C, MYBPC3) [4]. Compared to HCM, DCM is a more heterogeneous disease, the causes of which may be idiopathic, familial/genetic, viral, and/or immune [5]. Intriguingly, an estimated 35%–40% of genetic DCMs are thought to arise from mutations in sarcomeric genes [4]. In genetic forms of cardiomyopathy, sarcomere protein mutations may generate severe mechanical stress as a result of contractile deficiencies that are unable to meet the cellular demands required for normal contractile function [1]. This deterioration in contractile function correlates with exacerbated cardiac myocyte damage or death, which, in turn, may trigger myocardial inflammation, further aggravating the progression of cardiomyopathy [1, 6].
Inflammation has been recognized as a hallmark of HF [7] with levels of circulating and cardiac proinflammatory cytokines indicating HF severity in humans [8], as well as contributing to many of the pathological responses during HF in animals [7, 9, 10]. Based on this inflammatory response to damaged myocardium, some researchers formulated the cytokine hypothesis, which holds that proinflammatory cytokines are produced by the damaged myocardium during chronic HF and that this inflammatory response promotes monocyte activation and the further production of cytokines, thus augmenting cardiac dysfunction [11]. However, to date, no large-scale therapeutic strategies targeting proinflammatory protein mediators in HF have been successfully translated to clinical practice, indicating a profound underappreciation of the role of inflammation in HF [7]. As such, we have recently suggested that attention be turned towards modulation of the underlying inflammatory cellular networks, including monocyte/macrophage populations, responsible for cytokine production, as well as regulation and generation of immune responses [7]. Indeed, macrophages and monocytes play a central role in inflammation and innate immunity [7, 12]. The mammalian heart has been shown to contain a population of resident macrophages that proliferate following myocardial infarction and hemodynamic stress that signals further monocyte recruitment to the heart, contributing to myocardial interstitial fibrosis and adverse cardiac remodeling [12]. However, in cardiomyopathies caused by sarcomeric protein mutation, cardiac inflammation, as it manifests relative to contractile dysfunction and cardiac remodeling, has not been elucidated.
We recently reported the use of a well-characterized mouse model of genetic DCM harboring a homozygous knock-in mutation in the MYBPC3 gene that translates into a truncated variant of cMyBP-C [13–16]. Using this mouse model (cMyBP-C(t/t)), we demonstrated that oxidative stress, which is significantly elevated in genetically induced cardiomyopathy, correlates with the level of contractile dysfunction, cellular damage and cardiac remodeling [1]. In the current study, we use the cMyBP-C(t/t) mouse model to test the hypothesis that myocardial inflammation associates with cardiac dysfunction in dilated cardiomyopathy caused by MYBPC3 mutation. Our data indicate a substantial elevation in the proinflammatory macrophages in cMyBP-C(t/t) DCM hearts and that such elevation is likely a response to cellular damage triggered by MYBPC3 mutation and the resultant contractile dysfunction.
2. Materials and Methods
2.1 Mouse model of DCM
In this study, we used a 3-month-old knock-in mouse model of DCM [17], in which a homozygous mutation in MYBPC3 causes a C′-modified cMyBP-C (cMyBP-C(t/t)) that does not incorporate into sarcomeres, resulting in a cMyBP-C null heart [16, 17], and age-matched wild type (WT) mice (FVB/N strain). Male animals were used for these studies. All animal protocols were approved by the Institutional Animal Care and Use Committee at Loyola University Chicago (LU# 205109) and were implemented in accordance with the guidelines listed in the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health. Prior to organ harvest, animals were euthanized in a carbon dioxide (CO2) chamber by slow flow of CO2 (10–30% of chamber volume per minute), followed by continued exposure for 15–30 minutes after breathing had stopped.
2.2 In vivo cardiac function by noninvasive echocardiography
Echocardiography using a Vevo 2100 (Fujifilm, Visual Sonics, Inc., Toronto, Canada) and an MS-550D 22–55 MHz transducer was performed to assess cardiac function in WT and DCM mice anesthetized with 1.5%–2% Isoflurane in 100% oxygen via inhalation. Left ventricular internal diameter (LVID), wall thickness, and contractile function were measured using short-axis M-mode imaging and the Visual Sonics Vevo 2100 analysis package as described previously [1].
2.3 Histopathological analysis of splenic architecture
To evaluate general spleen histology, spleens were isolated from WT and DCM mice and then formalin-fixed and embedded in paraffin. Spleens were sectioned at 5-μm thickness, deparaffinized, rehydrated, and stained with Masson’s trichrome as previously described [7].
2.4 Isolation of mononuclear cells from the heart
Single mononuclear cells were isolated from WT and DCM hearts as previously described [7]. Briefly, mouse hearts were completely excised; following removal of fatty tissue and aorta, they were placed in heparinized saline. Hearts were then minced into fine pieces, and blood was removed by repeated wash with saline. The tissue was digested in 10 mL of digestion buffer for 50 minutes at 37°C with constant shaking. Released cells were separated from solid tissue by filtration through a 70-μm nylon cell-strainer (Cat. No. 352350, Corning) and washed with R10 media (RPMI 1640 supplemented with 10% fetal bovine serum) on ice. The remaining solid tissue was digested for 25 minutes at 37°C and was filtered as described above. R10 media supplemented with EDTA (2mmol/L) were then added to the collected cells. Single-cell suspensions were layered on a density gradient using Histopaque-1077 (Cat. No. 10771, Sigma-Aldrich) and then centrifuged at 2000g for 20 minutes at 4°C. Following centrifugation, the upper 75% of the total volume was collected to exclude myocytes and cellular debris from the mononuclear fraction. Saline was then added to dilute the collected gradient solution, followed by centrifugation of the cell suspension at 2000g for 10 minutes at 4°C. The pelleted cells were resuspended in 400 μl of cold Flow Cytometry Staining Buffer Solution (Cat. No. 00-4222-47, eBioscience) and processed for flow cytometry analysis as described below.
2.5 Isolation of splenocytes and peripheral blood cells
Spleens were removed from WT and DCM mice and placed in a 35-mm cell culture dish (Cat. No. 353001, BD Biosciences) with 5mL DMEM culture medium (Gibco, Invitrogen). Splenocytes and peripheral blood cells were isolated from WT and DCM mice as previously described (7). Briefly, spleens were perfused with 5 mL of DMEM at multiple locations to disperse single cells. The final cell suspension was centrifuged at 150g for 5 minutes at 4°C, and the pellet was treated with 0.4 mL of RBC lysis buffer for 5 minutes at room temperature. Washed splenocytes were then resuspended in FACS staining buffer.
Peripheral blood cells were isolated from approximately 500 μl of peripheral blood collected in BD Microtainer Plastic Capillary Blood Collectors with Dipotassium EDTA (Cat. No. 02-669-38, Thermo Fisher Scientific). Erythrocytes were lysed with 1 mL of 1X RBC Lysis Buffer Solution (Cat. No. 00-4333-57, eBioscience) for 5 minutes at 37°C. The lysis reaction was neutralized with 10 mL of saline. Peripheral blood leukocytes were collected by centrifugation at 500g for 5 minutes. The resulting pellets were resuspended in 400 μl of cold Flow Cytometry Staining Buffer Solution and processed for flow cytometry analysis as described below.
2.6 Flow Cytometry
Isolated cell suspensions from WT and DCM hearts, spleens and peripheral blood (described above) were incubated for 30 minutes on ice with TruStain fcX (anti-mouse CD16/32) Antibody (Cat. No. 101319, BioLegend) to block Fc receptors. Samples were then incubated with a cocktail of fluorophore-labeled monoclonal antibodies (described below) for 1 hour at 4°C protected from light. Following a wash with cold saline, samples were centrifuged at 2000 rpm for 2 minutes at 4°C and then permeabilized for 1 hour at 4°C with 0.5% Triton-X diluted in cold saline to allow for uptake of an intracellular antibody directed against CD206. Samples were centrifuged as before and resuspended in 400 μl of cold Flow Cytometry Staining Buffer Solution. The following antibodies were used for cell labeling: anti-mouse F4/80, PerCP-Cyanine5.5, Clone BM8 (Cat. No. 45-4801-82, eBioscience); anti-mouse Ly6C, PE-Cyanine7, Clone HK1.4 (Cat. No. 25-5932-82, eBioscience); anti-mouse CD11b, Alexa Fluor 700, Clone M1/70 (Cat. No. 56-0112-82, eBioscience); anti-mouse Ly6G (GR-1), eFluor 450, Clone RB6-8C5 (Cat. No. 48-5931-82, eBioscience); anti-mouse CD45, eFluor 605NC, Clone 30-F11 (Cat. No. 93-0451-42, eBioscience); anti-mouse MHCII I-M, PE, Clone AF6-120.1 (Cat. No. 12-5320-82, eBioscience); and rat anti-mouse CD206, FITC (Cat. No. MCA2235FB, AbD Serotech). Within leukocyte (CD45+) populations isolated from mouse heart tissue, total macrophages were identified as CD45+CD11b+Ly6C−MHCII+F480+ cells. Proinflammatory or classically activated (M1) and anti-inflammatory or alternatively activated (M2) macrophages were classified from the above marker based on the absence or presence of CD206 (mannose receptor) expression, respectively. Within splenocyte samples, monocytes were defined as CD11b+Ly6C+ cells, and monocytes within the red pulp region of the spleen were identified as Ly6G−CD11b+Ly6C+F480hi cells, while red pulp macrophages were identified as CD11b+Ly6C+MHCIIlowF480hi cells. Within peripheral blood samples, monocytes were characterized as CD45+Gr1−CD11b+Ly6C+ cells. Monocytes were further subclassified as proinflammatory, CD45+Gr1−CD11b+Ly6Chi cells, and patrolling, CD45+Gr1−CD11b+Ly6Clow cells. Cells were normalized for total cell population. Data were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software, version 7.6.3.
2.7 Immunofluorescence analysis for inflammatory markers
To detect whether inflammatory markers were elevated in DCM heart tissue sections, we used confocal microscopy analysis of sectioned heart tissues. Hearts were removed and frozen in Tissue-Tek O.C.T. Compound (Cat. No. 4583, Sakura Finetek). Hearts were sectioned into 5-μm slices, fixed in 4% paraformaldehyde for 5 minutes, and washed 3 times in saline (pH 7.4) for 3 minutes. For Ly6G staining, a 1:100 dilution of BD Pharmingen PE Rat Anti-Mouse Ly-6G antibody (Cat. No. 551461, BD Biosciences) was incubated overnight with the sections at 4°C. Anti-Neutrophil antibody (NIMP-R14, Cat. No. ab2557, Abcam) was added at a 1:50 dilution for 2 hours at room temperature for NIMP-R14 staining. For CD68 staining, a 1:200 dilution of Rat anti-Mouse CD68 (Cat. No. MCA1957, AbD Serotec) was added to the sections for 1 hour at room temperature. Sections were washed as above. Donkey anti-Rat Cy3 secondary antibody (Cat. No. AP189C, Millipore) was then added at a 1:300 dilution for 1 hour at room temperature to NIMP-R14- or CD68-stained sections, followed by washing as described above. For CD68-MRC1 co-staining, MRC1-Alexa Fluor 488 (Cat. No. 141709, BioLegend) was added at a 1:100 dilution for 1 hour at room temperature, followed by washing. Nuclei were stained with a 1:1,000 dilution of DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) (Cat. No. D1306, Thermo Fisher Scientific) for 20 minutes at room temperature, followed by washing. Cover slips were mounted using a drop of Mowiol 4-88 mounting medium (Cat. No. 81381, Sigma-Aldrich).
2.8 Cytometric bead array immunoassay for serum cytokines
Serum was collected from both WT and DCM peripheral blood samples, which were allowed to clot at room temperature for 1 h and were then centrifuged at 2000g for 15 minutes at 4°C. Serum was stored at −80°C until use. The BD Cytometric Bead Array Mouse Inflammation Kit (Cat. No. 552364, BD Biosciences) was used to measure the serum concentrations of interleukin (IL)-6, monocyte chemoattractant protein (MCP)-1, interferon (IFN)-γ, IL-12p70, IL-10, and tumor necrosis factor (TNF)-α per the manufacturer’s instructions. Briefly, 50 μl of the Mouse Inflammation Capture Bead suspension were incubated for 2 h at room temperature with 50 μl of either Mouse Inflammation Standards or test samples and 50 μl of R-phycoerythrin (R-PE)-conjugated detection antibody. To remove unbound R-PE detection reagent, the samples were then washed and resuspended in 300 μl of Wash Buffer. Data were acquired on a BD LSRFortessa Cell Analyzer and analyzed using FCAP Array software.
2.9 RNA isolation and sequencing (RNA-Seq) analysis
Minced heart tissue from three WT and three DCM mice was homogenized in 1 mL of TriZol (BioRad) using a bead homogenizer, and total RNA was isolated using the Aurum Total RNA Fatty and Fibrous Tissue Kit (Cat. No. 732-6820, Bio-Rad Laboratories), according to the manufacturer’s instructions. Total RNA samples were pooled by genotype and were then used for RNA-sequencing. The quality of the total RNA was confirmed with the use of an Agilent 2100 Bioanalzyer. Using the TruSeq® Stranded mRNA Library Prep kit (Cat. No. RS-122-2201, Illumina, Inc.), mRNA in the total RNA was converted into a library of template molecules of known strand origin. Specifically, the mRNA was isolated using poly-T oligos attached to magnetic beads. The resulting mRNA was fragmented and reverse transcribed. The cDNA product was amplified, and sequencing adapters and barcodes were ligated onto the fragments for each respective sample to create cDNA libraries ready for sequencing. The samples were sequenced on the Illumina HiSeq platform rendering 100 bp paired-end reads. The reads for each sample were aligned to University of California at Santa Cruz Mus musculus mm10 reference sequence using STAR aligner. Aligned reads were assigned to genes, and counts were calculated for each sample using the Python tool HTSeq. The fold change was determined by dividing the experimental sample by the control, with any 0s found in the denominator replaced with 1s to avoid undefined values. Fold change values were then ranked, and any experimental values <32 were removed after graphing the HTSeq count distributions to empirically determine a cutoff. Fold changes <2-fold were also discarded. The resulting genes were graphed in heatmaps using the gplots v3.0.1 bioconductor package for R, version 3.3.0. Using the ToppFun application from ToppGene Suite [18], functional enrichment analyses were performed separately for the up- and downregulated gene sets. For network representation of select enriched biological processes and pathways, we used Cytoscape [19] (see Supplementary Table 1 for a complete list of enriched features in the overexpressed and underexpressed gene sets in the cMyBP-C(t/t) mouse model of DCM).
2.10 Statistical analysis
All values are reported as mean values ± SEM. Significant differences between mean values were determined using the unpaired Student’s t-test. Statistically significant differences were considered as P < 0.05.
3. Results
3.1 DCM mice exhibit cardiac contractile dysfunction and splenomegaly
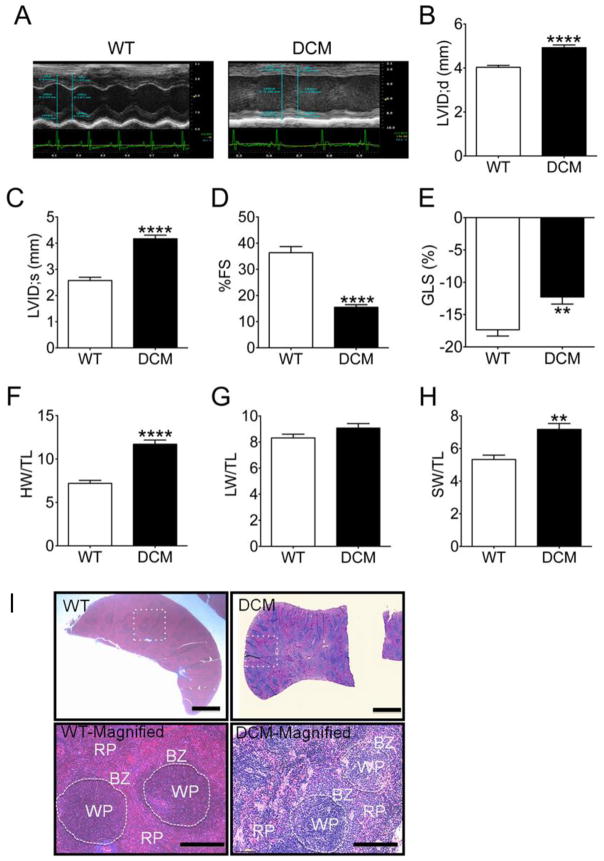
Previous work from our group and others have indicated that cMyBP-C(t/t) mice develop an increased left ventricular internal diameter (LVID), reduced fractional shortening (FS) and prominent histological abnormalities, including eccentric hypertrophy, myocardial disarray, fibrosis, and loss of structural integrity indicative of cardiac remodeling and dilation (DCM) [1, 13, 15, 20]. To confirm these observations in the animals used in this study, we implemented short-axis M-mode echocardiography to assess cardiac function in cMyBP-C(t/t) DCM animals as compared with WT animals (Fig. 1A–F). cMyBP-C(t/t) animals had an elevation in LVID (Fig. 1A) at both end-diastole (Fig. 1B) (p < 0.0001) and end-systole (Fig. 1C) (p < 0.0001), indicating LV chamber dilation and DCM. We further observed a significant reduction in FS (Fig. 1D) (p < 0.0001) and a severe deficit in percent global longitudinal strain (%GLS) (Fig. 1E) (p = 0.005) in DCM compared to WT hearts, indicating cardiac contractile dysfunction in DCM hearts. The ratio of heart weight (HW) to tibia length (TL) was significantly elevated in DCM compared to WT animals (Fig. 1F) (p < 0.0001). No difference was found in the ratio of lung weight (LW) to TL between WT and DCM animals (Fig. 1G) (p = 0.12). Intriguingly, DCM mice exhibited significantly increased ratio of spleen weight (SW) to TL (Fig. 1H) (p = 0.002) compared with WT mice, i.e., splenomegaly in DCM animals.
Fig. 1. DCM mice exhibit cardiac dysfunction and splenomegaly.
(A) Representative short-axis M-mode echocardiographic images of WT and DCM hearts. LVID at (B) peak diastole and (C) peak systole in WT (n = 7) and DCM (n = 10) hearts (****p < 0.0001). Functional measurements derived from LVID measurements showing (D) fractional shortening (FS) and global longitudinal strain (GLS) in WT (n = 7) and DCM (n = 10) hearts (**p = 0.05, ****p < 0.0001). Quantification of the ratio of (F) heart weight (HW) to tibia length (TL), (G) Lung weight (LW) to TL, (H) spleen weight (SW) to TL in WT (n = 7) and DCM (n = 10) animals (**p = 0.002, ****p<0.0001). Representative Masson’s trichrome-stained spleen sections from WT and DCM mice. The lower magnification (2X) of cross-sectioned spleens (top) shows a rounded edge in DCM spleens, which is a hallmark of splenomegaly. The high power magnification (20X) (bottom) shows less abundant splenic macrophage populations in the border zone area of DCM mouse spleens compared to WT mouse spleens. The dotted circle shows the border between red pulp (RP) and white pulp regions (WP) of the spleen. BZ= border zone.
Splenic hematopoiesis has been reported in several diseases, such as atherosclerosis [21] myocardial infarction [22] and colitis [23]. Splenic macrophages have a key role in directing lymphocyte efflux from spleen [24]. Cross and longitudinal sections of spleens from WT and DCM mice demonstrated that those from DCM mice have rounded edges (Fig. 1I), which is a hallmark of splenic enlargement or splenomegaly (Fig. 1H). Changes in the marginal zone cell population in DCM spleens were more evident compared to WT spleens, and these cells had a less abundant and somewhat patchy distribution (Fig. 1I). DCM spleens also revealed expansion and early bridging of white pulp compared to WT spleens, suggesting immune activation.
3.2 Activated M1 polarized macrophages infiltrate DCM hearts
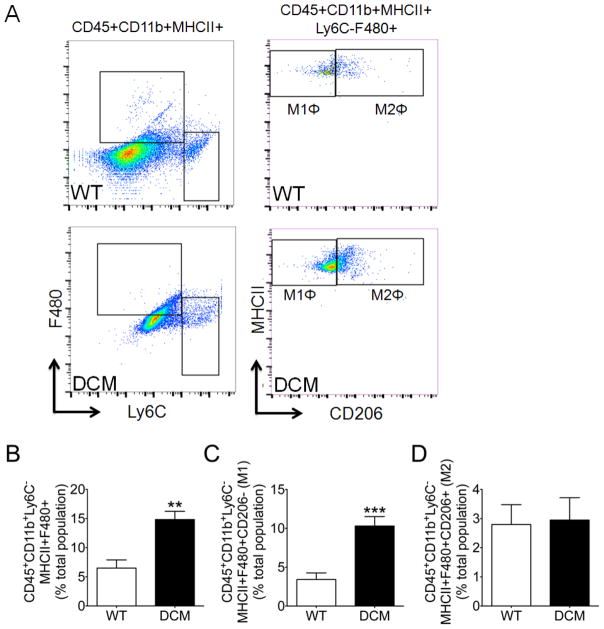
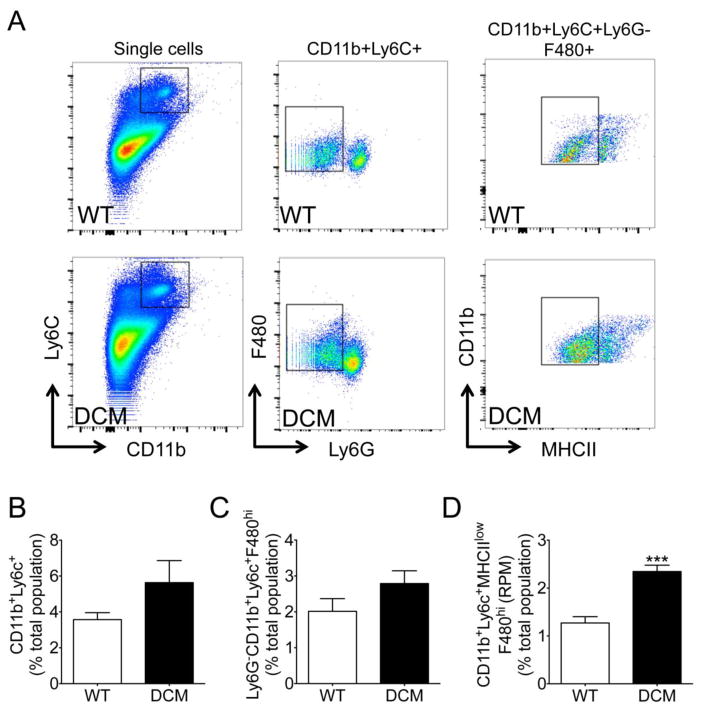
Following confirmation of cardiac dysfunction, cMyBP-C(t/t) DCM mice were used to determine whether myocardial inflammation was elevated in this form of genetically produced cardiomyopathy. Previously, we observed a greater tendency toward cellularity in DCM hearts as compared with WT hearts, suggesting cell infiltration or expansion of resident cardiac cells [1]. As such, we next sought to determine the levels and classification of leukocyte populations within the hearts of DCM and WT animals. Supplementary Figure S1 and 2A depict the gating strategy used to identify activated macrophages (CD45+CD11b+Ly6C−MHCII+F480+), proinflammatory M1 macrophages (CD45+CD11b+Ly6C−MHCII+F480+CD206−), and anti-inflammatory M2 macrophages (CD45+CD11b+Ly6C−MHCII+F480+CD206+). Strikingly, total activated macrophages (Fig. 2B) (p = 0.002) were significantly augmented in DCM compared to WT hearts. Importantly, DCM hearts exhibited a significantly greater abundance of tissue M1 macrophages (Fig. 2C) compared to WT hearts (p = 0.0009), suggesting a proinflammatory response in DCM hearts. We observed no difference in the levels of tissue M2 macrophages between DCM and WT hearts (Fig. 2D) (p = 0.88).
Fig. 2. Proinflammatory macrophages infiltrate DCM hearts.
Cardiac mononuclear cells were isolated following tissue digestion and gradient centrifugation purification. (A) Representative gating strategy and scatter plots used to identify activated macrophages (CD45+CD11b+Ly6C−MHCII+F480+), proinflammatory M1 macrophages (CD45+CD11b+Ly6C−MHCII+F480+CD206−), and anti-inflammatory M2 macrophages (CD45+CD11b+Ly6C−MHCII+F480+CD206+) in WT and DCM hearts. Flow cytometry quantification of (B) activated macrophages, (C) M1 macrophages, and (D) M2 macrophages in WT (n = 6) and DCM (n = 7–8) hearts (**p = 0.002, ***p = 0.0009).
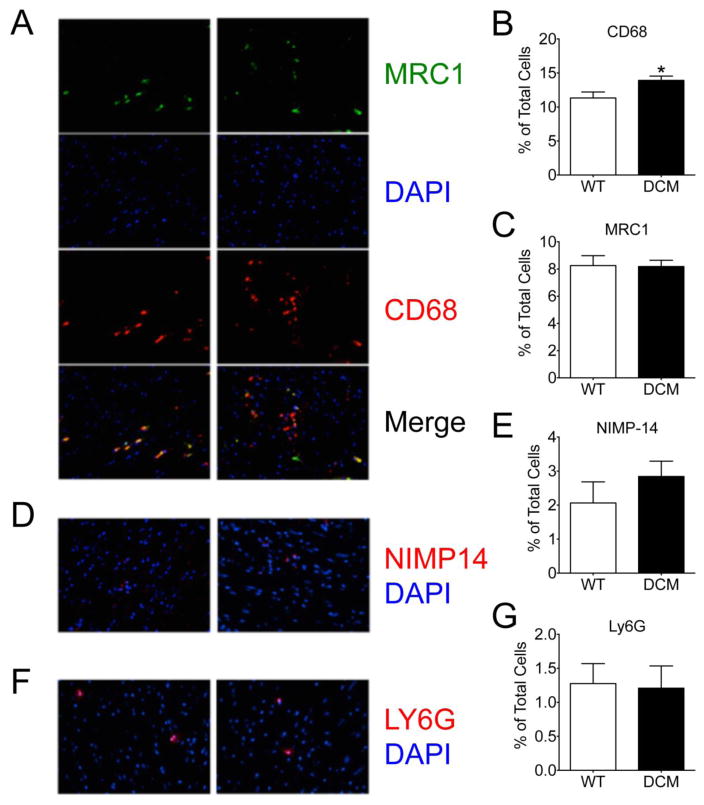
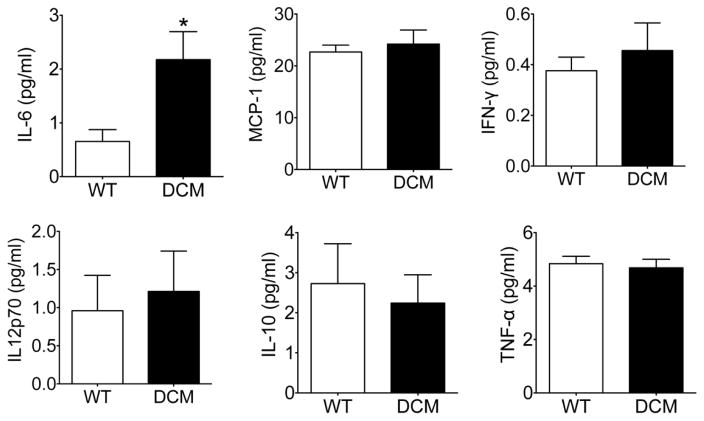
To confirm the proinflammatory environment observed in DCM compared to WT hearts, we performed immunohistochemical staining of cardiac tissue sections from these animals. In agreement with our flow cytometry data, we observed a significant increase in cells positive for CD68 (pan-macrophage marker) in DCM hearts compared to WT hearts (Fig. 3A, B) (p = 0.016). No difference was observed in cells positive for Mannose Receptor, C Type 1 (MRC1) (M2 macrophages) (Fig. 3A, C) (p 0.93) between the groups. Similarly, no difference was seen in the number of cells positive for the neutrophil markers NIMP14 (Fig. 3D, E) (p =0.31) or Ly6G (Fig. 3F, G) (p = 0.89) between DCM and WT hearts. Together, these data demonstrate that M1 proinflammatory macrophages are increased in the hearts of DCM mice. In support of this, DCM mice exhibited a significant elevation in serum levels of the circulating proinflammatory cytokine IL-6 (p = 0.02) compared to WT mice (Fig. 4A). However, no differences between groups were identified for serum levels of MCP-1, IFN-γ, IL-12p70, IL-10, and TNF-α (Fig. 4B–F).
Fig 3. Elevation in CD68+ proinflammatory macrophages in DCM heart tissue.
Representative confocal images and corresponding quantification of (A, B) CD68+ macrophages and (A, C) MRC1+ M2 anti-inflammatory macrophages (n = 58 sections from 5 WT hearts, 75 sections from 5 DCM hearts, *p = 0.016) and their merge, as well as (D, E) NIMP14+ (n = 40 sections from 4 WT hearts, 40 sections from 4 DCM hearts) and (F, G) Ly6G+ (n = 18 sections from 3 WT hearts, 29 sections from 1 DCM heart) neutrophils in WT and DCM heart tissue sections. DAPI (4′,6-diamidino-2-phenylindole).
Fig. 4. Serum IL-6 levels are elevated in DCM animals.
Serum cytokine levels of (A) IL-6, (B) MCP-1, (C) IFN-γ, (D) IL12p70, IL-10, and TNF-α in WT (n = 10) and DCM (n = 12–13) peripheral blood as determined by cytometric bead array immunoassay (*p = 0.02). IL (interleukin); tumor necrosis factor (TNF), interferon (IFN), monocyte chemoattractant protein (MCP).
3.3 Splenic red pulp macrophages (RPM) are augmented in DCM animals
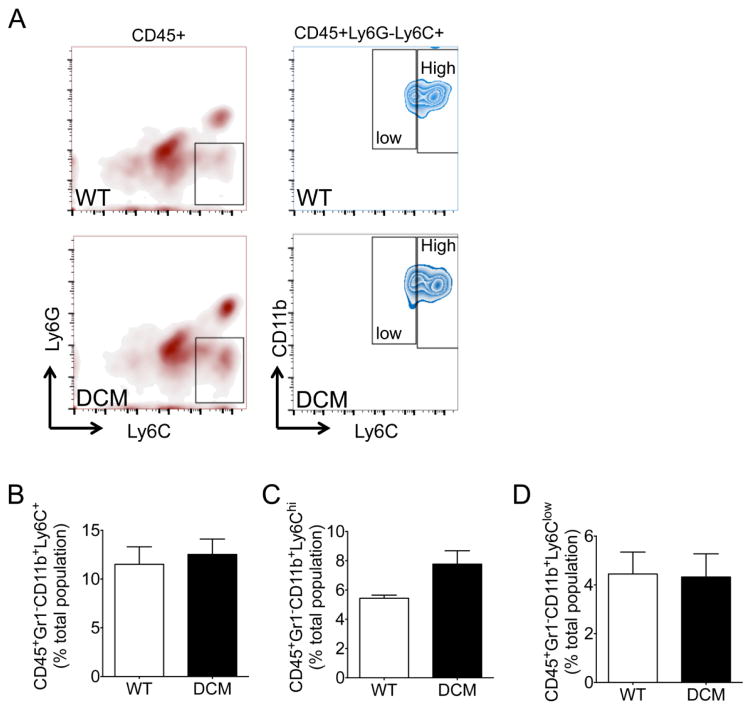
The spleen plays a critical role in regulating the progression of pathological LV remodeling in chronic heart failure [7]. Our data indicated a proinflammatory alteration in the cellular network in DCM hearts (Fig. 2–3); therefore, we next quantified monocytes/macrophages in the spleens and peripheral blood of DCM and WT animals by flow cytometry analysis. As the spleen is the largest lymphoid organ in the body and is enlarged in DCM animals (Fig. 1H), it is expected to be a primary contributor to monocyte and macrophage abundance and the progression of cardiac remodeling [7]. Figure S2 and 5A show the gating strategy used to classify splenic monocytes (CD11b+Ly6C+ cells), monocytes within the red pulp region of the spleen (Ly6G−CD11b+Ly6C+F480hi), and red pulp macrophages (CD11b+Ly6C+MHCIIlowF480hi cells). No difference was identified between splenic monocytes (Fig. 5B) (p = 0.13) and monocytes within the red pulp region of the spleen (Fig. 5C) (p = 0.15). However, red pulp macrophages were markedly increased in the spleens of DCM compared to WT mice (Fig. 5D) (p = 0.0001). Supplementary figure S3 and figure 6A depict the gating strategy used to identify total monocytes (CD45+Gr1−CD11b+Ly6C+), as well as proinflammatory (CD45+Gr1−CD11b+Ly6Chi) and anti-inflammatory (CD45+Gr1−CD11b+Ly6Clow) monocytes, in the peripheral blood samples of WT and DCM animals. The levels of all three peripheral blood monocyte populations did not differ between WT and DCM animals (Fig. 6B–D). However, a near-significant (p = 0.06) increase in proinflammatory monocytes was noted in the peripheral blood of DCM mice (Fig. 6C).
Fig. 5. Splenic red pulp macrophages (RPM) are augmented in DCM animals.
(A) Representative scatter plots used to identify splenic monocytes (CD11b+Ly6C+ cells), monocytes within the red pulp region of the spleen (Ly6G−CD11b+Ly6C+F480hi), and red pulp macrophages (CD11b+Ly6C+MHCIIlowF480hi cells) in WT and DCM spleens. Flow cytometry quantification of (B) splenic monocytes, (C) monocytes within the red pulp region of the spleen, and (D) red pulp macrophages in WT (n = 6–7) and DCM (n = 6–7) spleens (***p = 0.001).
Fig. 6. Circulating mononuclear phagocytes in DCM and WT peripheral blood.
(A) Representative gating strategy used to identify total monocytes (CD45+Gr1−CD11b+Ly6C+), proinflammatory monocytes (CD45+Gr1−CD11b+Ly6Chi), and anti-inflammatory monocytes (CD45+Gr1−CD11b+Ly6Clow) in peripheral blood samples from WT and DCM mice. Flow cytometry quantification of (B) total monocytes, (C) proinflammatory monocytes, and (D) anti-inflammatory monocytes in WT (n = 4) and DCM (n = 5) peripheral blood samples.
3.4 Gene set functional enrichment analyses and network visualization
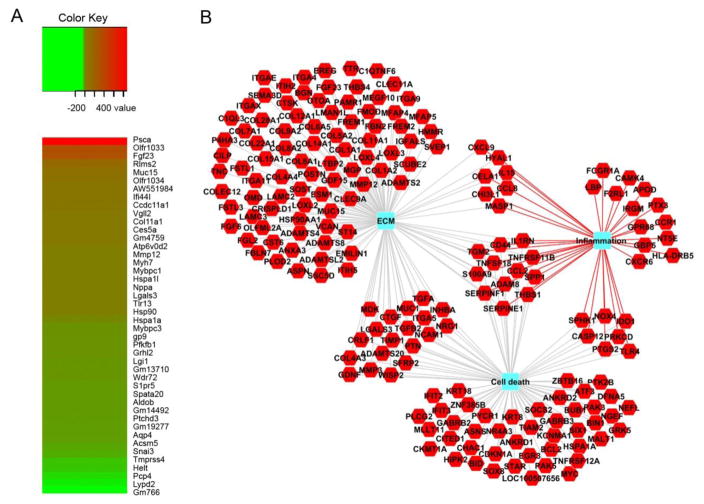
To elucidate the cellular inflammatory and damage-associated gene expression changes occurring in the hearts of DCM animals that may trigger the observed cardiac inflammatory response, we performed RNA-Seq analysis using total mRNA isolated from WT and DCM hearts (Fig. 7A). In total, we identified 635 upregulated genes and 385 downregulated genes in DCM compared with WT hearts. Interestingly, RNA-Seq revealed a significant upregulation of several alarmins, including S100A9, HSP90aa1, HSPa1l, HSPa1a, and LGALS3, as well as TLR4 and TLR13, genes encoding toll-like receptors involved in innate immune responses, in DCM compared with WT hearts. Additionally, genes encoding the monocyte chemoattractant proteins CCL8 and CCL12 were upregulated in DCM hearts. As expected, we observed an upregulation of several hypertrophy genes, including MYH7 and NPPA, in DCM hearts. Intriguingly, we further identified elevated expression of MYBPC1, which encodes the slow-skeletal isoform of MyBP-C in DCM hearts. As expected, MYBPC3 expression was markedly reduced in DCM compared to WT hearts. Additionally, functional enrichment analysis of the upregulated and downregulated gene sets using the ToppFun application of ToppGene Suite showed enrichment (p value <0.05 FDR) for several human DCM-related or -associated genes (Supplementary Table 1).
Fig. 7. Differentially regulated genes in DCM compared to WT hearts.
(A) RNA-Seq heat maps depicting clusters of genes differentially regulated in DCM versus WT hearts. Shown is a subset of the most upregulated (red) and downregulated (green) genes from the total gene set. The fold change, up or down, is represented in the keys for the respective panels. n = 3 pooled samples per genotype. (B) Network of significantly enriched gene ontology terms for upregulated genes in DCM hearts. The red hexagons denote upregulated genes in DCM compared to WT hearts. Blue rectangles represent significantly enriched (p < 0.05; FDR) biological processes using the ToppFun application of ToppGene Suite.
To understand the global interactions of genes overexpressed or underexpressed in DCM compared to WT hearts, overrepresentation analyses for biological processes and pathways were performed. Such analyses typically apply hypergeometric tests to estimate whether a biological process is represented in a given dataset more than expected by chance. Network representation of select enriched biological processes and pathways in DCM compared to WT hearts is depicted in Figure 7B, which represents genes involved in inflammation, cell death and the extracellular matrix.
4. Discussion
In the present study, we have established that 1) a proinflammatory cellular environment is present in cMyBP-C(t/t) hearts, 2) the inflammatory response is linked to the infiltration and/or expansion of proinflammatory monocytes and M1 macrophages, and 3) both contractile dysfunction and cardiac (and splenic) remodeling occur in parallel, all fundamentally triggered by mutation in the MYBPC3 gene. Cardiomyopathies are diseases of the heart muscle that cause cardiac dysfunction and can progress to HF [25]. These myocardial diseases are prominent causes of morbidity and mortality in both children and adults [26]. Taken together, it is estimated that they affect one in 390 people; however, asymptomatic individuals often go undiagnosed [25, 27]. Inherited gene defects have been recognized as key regulators of the pathogenesis of cardiomyopathies [28]. Intriguingly, several instances have been reported in which mutations within the same gene produce a different cardiac phenotype, suggesting that the type and/or location of the mutation is critical in determining this phenotype [29].
Since the discovery of the first “diseased gene” (MYH7) in HCM, over 50 genes, including many encoding proteins of the cardiac sarcomere, have been identified as generating various forms of cardiomyopathy [4, 30]. For example, mutations in MYBPC3, which encodes the contractile regulatory protein cMyBP-C, are the most common cause of HCM, accounting for nearly 50% of the identified mutations [31]. Due to the contractile abnormalities arising from such mutations, the heart undergoes a limited range of compensatory responses to maintain proper cardiac function, which can explain why many forms of cardiomyopathy demonstrate an overlapping phenotype [29]. These responses often result in changes that can lead to either systolic pump failure or a deficiency in diastolic compliance [32]. Given that their etiology lies in progressive contractile dysfunction, cardiomyopathies arising from mutations in sarcomeric genes typically worsen over time, which may promote hypertrophy and myocyte damage or death. In some cases, following prolonged hypertrophy, the addition of sarcomeres to ventricular myocytes increases myocyte length, which causes eccentric hypertrophy, ventricular dilation, and decreased force production [29]. Furthermore, myocyte damage can lead to the deposition of fibrotic lesions within the myocardium [29]. Cell damage and/or death by apoptotic and necrotic mechanisms have/has been demonstrated to occur in human HF [33, 34]. As a result of this damage, the disruption of cell integrity could trigger an inflammatory response with pathological consequences [35].
In most cases, inflammation occurs naturally in response to injury to promote tissue repair [32]. However, an overwhelming inflammatory response may trigger further tissue damage and disease [32, 36]. In DCM, it has been suggested that myocardial inflammation can lead to LV dysfunction that is mediated by cytotoxic T-lymphocytes, natural killer (NK) cells, and macrophages, as well as the release of inflammatory mediators by infiltrating lymphocytes and macrophages [32]. However, the current understanding of inflammation in cardiomyopathies holds that the progression of myocarditis to inflammatory dilated cardiomyopathy is mediated by infectious agents, most often a virus, and an autoimmune response, as diagnosed by endomyocardial biopsy [37]. According to the World Health Organization, myocarditis is classified as an inflammatory muscle disease associated with cardiac dysfunction, and DCM represents the chronic phase of the disease [38]. In addition to myocarditis-induced DCM, inflammation is a key pathomechanism underlying the progression of LV remodeling following acute myocardial infarction (AMI) to what is often referred to as ‘ischemic cardiomyopathy’ [36, 39]. Following AMI, the irreversible loss of myocardium triggered by myocyte necrosis activates biochemical intracellular signaling processes that modulate reparative changes, including cardiac dilatation, hypertrophy and the formation of a collagen scar [39]. Such processes involve migrating macrophages, monocytes and neutrophils, which are critical for local infarct zone remodeling [39, 40]. Swirski et al. previously demonstrated the rapid recruitment of monocytes from a splenic reservoir to the infarcted heart where they participated in tissue repair [40, 41]. During steady-state conditions, macrophages comprise the primary immune cells present in the heart with a minimal contribution of monocytes [12]. However, under conditions of sterile injury, such as AMI that disrupts this steady-state, cardiac macrophages are primarily derived from blood monocytes [12]. Several studies have suggested a biphasic tissue macrophage response following MI in which proinflammatory Ly6Chi monocytes and M1 macrophages regulate tissue digestion, followed by infiltration of Ly6Clow monocytes and M2 macrophages that resolve inflammation leading to scar formation [22, 40, 42, 43]. Recently, however, Hilgendorf et al. have demonstrated in Nr4a1−/− mice that this biphasic response is dominated by Ly6Chi monocytes [44], a paradigm which alters the current approach taken towards AMI in that the focus is now on modulating the polarity of macrophages to the reparative condition [40].
Our current study challenges consensus about pathomechanisms underlying the involvement of inflammation in the development of LV remodeling/cardiomyopathy. As discussed above, while cardiomyopathies are traditionally categorized as being familial (genetic), non-familial (non-genetic), and inflammatory, we suggest that inflammation may play a role in the development of cardiomyopathies outside of inflammatory DCM and ischemic cardiomyopathy to include cardiomyopathies arising from genetic defects, i.e., sarcomere gene mutations such as those caused by mutation of the MYBPC3 gene. The progressive development of cardiac inflammation may be one variable that underlies the delayed manifestations of disease observed in both animal models and humans, with overt HF becoming apparent only upon aging, and with different degrees of penetrance. To test this hypothesis, we used the cMyBP-C(t/t) mouse model of genetic DCM to demonstrate a significant elevation in total macrophages and M1 proinflammatory macrophages in the hearts of DCM animals compared to WT controls (Fig. 2–3). No difference was observed in the levels of cardiac neutrophils between DCM and WT mice (Fig. 3D–G). This could have resulted from disease severity in DCM animals at this age (Fig. 1), during which early neutrophil responses may be replaced by advanced mononuclear phagocytic responses. We further identified a significant elevation in splenic size (Fig. 1H) and remodeling (Fig. 1I) in DCM mice compared to WT controls. This may indicate activation of the cardiosplenic axis [7] in response to cardiac damage in DCM animals. Indeed, we observed a significant increase in the levels of red pulp macrophages in the spleens of DCM mice (Fig. 5D). While red pulp macrophages mainly serve as scavengers of senescent erythrocytes, they may also play a role in the induction of innate and adaptive immunity [45]. We further detected an elevation in the levels of circulating peripheral blood proinflammatory monocytes in DCM compared to WT mice, but no difference in total blood monocytes. No striking differences were found in total peripheral monocytes or between pro- and anti-inflammatory monocytes between DCM and WT animals. However, at this stage in the disease process, it is possible that many circulating proinflammatory cells have already infiltrated into damaged cardiac tissue, thus accounting for these observations. Indeed, our RNA-seq analyses demonstrate a clear upregulation in several cardiac alarmins and chemokines, which may attract monocytes/macrophages to DCM hearts. We further observed an upregulation in genes involved in cell death and inflammation in DCM hearts (Fig. 7, Supplementary Table 1). These gene changes in DCM hearts may be the result of myocardial damage and could serve as the stimulus for inflammatory cell recruitment into the cardiac tissue of these animals.
The levels of inflammatory macrophages observed in the hearts of DCM mice likely indicate a low-grade inflammatory response to cardiac myocyte damage, rather than a profound response, as observed in AMI. Indeed, Kuusisto et al. have described the presence of low-grade inflammation in the myocardium of patients who have a single HCM-causing mutation in the alpha-tropomyosin gene (TPM1-D175N) and who develop cardiac hypertrophy, myocyte disarray and myocardial fibrosis [46]. The authors provide immunohistological evidence of invading inflammatory monocytes undergoing cardiac transendothelial migration, using myocardial autopsy specimens from HCM patients. They further detected increased plasma levels of proinflammatory cytokines, including IL-6 and C-reactive protein, in HCM compared to age-matched control patients [46]. Similarly, in our study, we observed a significant elevation in IL-6 levels in the serum of DCM compared to WT animals (Fig. 4A). However, no difference was detected in the serum levels of MCP-1, IFN-γ, IL-12p70, IL-10, and TNF-α between WT and DCM mice (Fig. 4B–F). This result could be a reflection of the complexity of cytokine networks in HF [47] or a potential low-grade inflammatory response that has local cellular effects within the DCM heart, but correspondingly minimal impact on altering systemic inflammatory cytokine levels. Indeed, cardiac improvement following splenectomy in mice presenting with ischemic cardiomyopathy was not accompanied by a reduction in the serum levels of IL-6, MCP-1, IFN-γ, IL-12p70, IL-10, or TNF-α [7]. This suggests that the improved LV remodeling following splenectomy was caused by the loss in tissue infiltration of inflammatory cells in splenectomized mice rather than alterations in systemic cytokine levels [7], highlighting the importance of inflammatory cells as mediators of HF progression.
Importantly, Kuusisto et al. further described a significant association between the degree of myocardial inflammatory cell infiltration and fibrosis in their HCM patient group [46]. They concluded that a variable myocardial and systemic inflammatory response capable of modifying cardiac fibrosis is present in HCM patients having the TPM1-D175N sarcomere protein mutation [46]. Indeed, the role of inflammatory cells in fibroblast activation was recently reviewed by Hartupee et al. [48], and the responses of cardiac fibroblasts to myocardial damage-associated molecular patterns (DAMPs) was recently reviewed by Turner [49]. In their review, Hartupee et al. elegantly describe the contrasting inflammatory responses that occur in response to tissue injury, one that is required for proper tissue repair and one that may become hyperactive following tissue injury, leading to further tissue damage and pathological tissue fibrosis [48]. They go on to describe the central role fibroblast activation plays in tissue inflammation and myocardial fibrosis provoked by tissue injury [48]. As described above, we previously identified severe cardiac fibrosis in the hearts of cMyBP-C(t/t) mice. Accordingly, based on our findings here, we propose interplay among MYBPC3 mutation-induced cardiac myocyte damage, cardiac fibrosis, and myocardial inflammation in the progression of LV remodeling in genetic forms of cardiomyopathy.
Conclusions
In summary, we have demonstrated that a proinflammatory response occurs in a form of genetic dilated cardiomyopathy caused by MYBPC3 mutation. This is likely a response to cardiac myocyte damage resulting from contractile dysfunction and mechanical stress. While we deduce this conclusion from the study of one model of DCM caused by mutation of the MYBPC3 gene, the phenotypic changes observed in the hearts of these animals are representative of other forms of progressive sarcomere protein-mutated cardiomyopathies. As such, we suggest that a cardiac proinflammatory response occurs in cardiomyopathies resulting from mutations in other sarcomeric genes producing similar cardiac phenotypes. Additionally, we do recognize and appreciate that the kinetics of macrophage accumulation prior to cardiac contractile dysfunction and remodeling, as well as the elucidation of whether macrophage accumulation in DCM hearts contributes to adverse cardiac remodeling and dysfunction, remain to be determined. Therefore, our future studies will focus on a establishing a novel paradigm in which proinflammatory mononuclear phagocytes mediate myocardial autoimmune injury contributing to pathological remodeling and disease progression in these forms of genetic cardiomyopathy. Accordingly, therapeutic strategies targeted towards reduction of the proinflammatory cellular environment, or maintenance of an anti-inflammatory cellular environment, in genetic cardiomyopathies may represent a novel treatment avenue to forestall adverse cardiac remodeling and disease progression to HF.
Supplementary Material
Highlights.
M1 proinflammatory macrophages and splenic red pulp macrophages are significantly increased in cMyBP-Ct/t DCM hearts and spleens, respectively.
Myocardial inflammation is associated with contractile dysfunction and cardiac/splenic remodeling in a mouse model of genetic dilated cardiomyopathy caused by MYBPC3 mutation.
Acknowledgments
This work was supported by the National Institutes of Health [grant numbers R01s HL105826, HL130356 and K02 HL114749 to S.S.; R01 HL125735 to S.D.P.], VA Merit I01 BX002706 to S.D.P., the American Heart Association Midwest Predoctoral Fellowship [15PRE22430028 to T.L.L.IV.] and Grant-in-Aid [14GRNT20490025 to S.S]. The authors thank Christine E. Seidman and Jonathan G. Seidman, Harvard Medical School, Boston, MA 02115, USA, for providing the cMyBP-C(t/t) mouse model of genetically produced dilated cardiomyopathy and Gina Kuffel, Department of Public Health Sciences, Health Sciences Division, Loyola University Chicago, Maywood, IL 60153, USA, for data processing of RNA-seq samples.
Abbreviations
- ARVCM
arrhythmogenic right ventricular cardiomyopathy
- cMyBP-C
cardiac myosin binding protein-C
- DCM
dilated cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- HF
heart failure
- IFN
interferon
- IL
interleukin
- LV
left ventricle
- MCP
monocyte chemoattractant protein
- RCM
restrictive cardiomyopathy
- TNF
tumor necrosis factor
Footnotes
Disclosures
The authors have no conflicting interests to disclose in relation to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch TL, IV, Sivaguru M, Velayutham M, Cardounel AJ, Michels M, Barefield D, Govindan S, Dos Remedios C, van der Velden J, Sadayappan S. Oxidative Stress in Dilated Cardiomyopathy Caused by MYBPC3 Mutation. Oxid Med Cell Longev. 2015;2015:424751. doi: 10.1155/2015/424751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holzem KM, Gomez JF, Glukhov AV, Madden EJ, Koppel AC, Ewald GA, Trenor B, Efimov IR. Reduced response to I blockade and altered hERG1a/1b stoichiometry in human heart failure. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto T, Kass DA. Stressed hearts inflame the body (in a good way) Proc Natl Acad Sci USA. 2015;112:7113–7114. doi: 10.1073/pnas.1507821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caforio AL, Daliento L, Angelini A, Bottaro S, Vinci A, Dequal G, Tona F, Iliceto S, Thiene G, McKenna WJ. Autoimmune myocarditis and dilated cardiomyopathy: focus on cardiac autoantibodies. Lupus. 2005;14:652–655. doi: 10.1191/0961203305lu2193oa. [DOI] [PubMed] [Google Scholar]
- 6.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. [DOI] [PubMed] [Google Scholar]
- 7.Ismahil MA, Hamid T, Bansal SS, Patel B, Kingery JR, Prabhu SD. Remodeling of the mononuclear phagocyte network underlies chronic inflammation and disease progression in heart failure: critical importance of the cardiosplenic axis. Circ Res. 2014;114:266–282. doi: 10.1161/CIRCRESAHA.113.301720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozkurt B, Mann DL, Deswal A. Biomarkers of inflammation in heart failure. Heart Fail Rev. 2010;15:331–341. doi: 10.1007/s10741-009-9140-3. [DOI] [PubMed] [Google Scholar]
- 9.Mann DL. Inflammatory mediators and the failing heart: past, present, and the foreseeable future. Circ Res. 2002;91:988–998. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 10.Hamid T, Gu Y, Ortines RV, Bhattacharya C, Wang G, Xuan YT, Prabhu SD. Divergent tumor necrosis factor receptor-related remodeling responses in heart failure: role of nuclear factor-kappaB and inflammatory activation. Circulation. 2009;119:1386–1397. doi: 10.1161/CIRCULATIONAHA.108.802918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braunwald E. Biomarkers in heart failure. N Engl J Med. 2008;358:2148–2159. doi: 10.1056/NEJMra0800239. [DOI] [PubMed] [Google Scholar]
- 12.Hulsmans M, Sam F, Nahrendorf M. Monocyte and macrophage contributions to cardiac remodeling. J Mol Cell Cardiol. 2015;93:149–55. doi: 10.1016/j.yjmcc.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1771. doi: 10.1172/JCI7377C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McConnell BK, Fatkin D, Semsarian C, Jones KA, Georgakopoulos D, Maguire CT, Healey MJ, Mudd JO, Moskowitz IP, Conner DA, Giewat M, Wakimoto H, Berul CI, Schoen FJ, Kass DA, Seidman CE, Seidman JG. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res. 2001;88:383–389. doi: 10.1161/01.res.88.4.383. [DOI] [PubMed] [Google Scholar]
- 15.Palmer BM, Georgakopoulos D, Janssen PM, Wang Y, Alpert NR, Belardi DF, Harris SP, Moss RL, Burgon PG, Seidman CE, Seidman JG, Maughan DW, Kass DA. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ Res. 2004;94:1249–1255. doi: 10.1161/01.RES.0000126898.95550.31. [DOI] [PubMed] [Google Scholar]
- 16.Barefield D, Kumar M, de Tombe PP, Sadayappan S. Contractile dysfunction in a mouse model expressing a heterozygous MYBPC3 mutation associated with hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2014;306:H807–15. doi: 10.1152/ajpheart.00913.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer BM, McConnell BK, Li GH, Seidman CE, Seidman JG, Irving TC, Alpert NR, Maughan DW. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Mol Cell Biochem. 2004;263:73–80. doi: 10.1023/B:MCBI.0000041849.60591.45. [DOI] [PubMed] [Google Scholar]
- 21.Dutta P, Courties G, Wei Y, Leuschner F, Gorbatov R, Robbins CS, Iwamoto Y, Thompson B, Carlson AL, Heidt T, Majmudar MD, Lasitschka F, Etzrodt M, Waterman P, Waring MT, Chicoine AT, van der Laan AM, Niessen HW, Piek JJ, Rubin BB, Butany J, Stone JR, Katus HA, Murphy SA, Morrow DA, Sabatine MS, Vinegoni C, Moskowitz MA, Pittet MJ, Libby P, Lin CP, Swirski FK, Weissleder R, Nahrendorf M. Myocardial infarction accelerates atherosclerosis. Nature. 2012;487:325–329. doi: 10.1038/nature11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leuschner F, Rauch PJ, Ueno T, Gorbatov R, Marinelli B, Lee WW, Dutta P, Wei Y, Robbins C, Iwamoto Y, Sena B, Chudnovskiy A, Panizzi P, Keliher E, Higgins JM, Libby P, Moskowitz MA, Pittet MJ, Swirski FK, Weissleder R, Nahrendorf M. Rapid monocyte kinetics in acute myocardial infarction are sustained by extramedullary monocytopoiesis. J Exp Med. 2012;209:123–137. doi: 10.1084/jem.20111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griseri T, McKenzie BS, Schiering C, Powrie F. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity. 2012;37:1116–1129. doi: 10.1016/j.immuni.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons AB, Parish CR. Are murine marginal-zone macrophages the splenic white pulp analog of high endothelial venules? Eur J Immunol. 1995;25:3165–3172. doi: 10.1002/eji.1830251127. [DOI] [PubMed] [Google Scholar]
- 25.Raju H, Alberg C, Sagoo GS, Burton H, Behr ER. Inherited cardiomyopathies. BMJ. 2011;343:d6966. doi: 10.1136/bmj.d6966. [DOI] [PubMed] [Google Scholar]
- 26.Hughes SE, McKenna WJ. New insights into the pathology of inherited cardiomyopathy. Heart. 2005;91:257–264. doi: 10.1136/hrt.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahon NG, Murphy RT, MacRae CA, Caforio AL, Elliott PM, McKenna WJ. Echocardiographic evaluation in asymptomatic relatives of patients with dilated cardiomyopathy reveals preclinical disease. Ann Intern Med. 2005;143:108–115. doi: 10.7326/0003-4819-143-2-200507190-00009. [DOI] [PubMed] [Google Scholar]
- 28.Fatkin D, Graham RM. Molecular mechanisms of inherited cardiomyopathies. Physiol Rev. 2002;82:945–980. doi: 10.1152/physrev.00012.2002. [DOI] [PubMed] [Google Scholar]
- 29.Harvey PA, Leinwand LA. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194:355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–666. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 31.Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J Muscle Res Cell Motil. 2012;33:75–80. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- 32.Pankuweit S, Ruppert V, Maisch B. Inflammation in dilated cardiomyopathy. Herz. 2004;29:788–793. doi: 10.1007/s00059-004-2626-9. [DOI] [PubMed] [Google Scholar]
- 33.Galluzzi L, Maiuri MC, Vitale I, Zischka H, Castedo M, Zitvogel L, Kroemer G. Cell death modalities: classification and pathophysiological implications. Cell Death Differ. 2007;14:1237–1243. doi: 10.1038/sj.cdd.4402148. [DOI] [PubMed] [Google Scholar]
- 34.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 35.Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M, Criollo A, Nemchenko A, Hill JA, Lavandero S. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis. 2011;2:e244. doi: 10.1038/cddis.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prabhu SD, Frangogiannis NG. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calabrese F, Thiene G. Myocarditis and inflammatory cardiomyopathy: microbiological and molecular biological aspects. Cardiovasc Res. 2003;60:11–25. doi: 10.1016/s0008-6363(03)00475-9. [DOI] [PubMed] [Google Scholar]
- 38.Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 39.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 40.Prabhu SD. It takes two to tango: monocyte and macrophage duality in the infarcted heart. Circ Res. 2014;114:1558–1560. doi: 10.1161/CIRCRESAHA.114.303933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, Aikawa E, Mempel TR, Libby P, Weissleder R, Pittet MJ. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nahrendorf M, Pittet MJ, Swirski FK. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan X, Anzai A, Katsumata Y, Matsuhashi T, Ito K, Endo J, Yamamoto T, Takeshima A, Shinmura K, Shen W, Fukuda K, Sano M. Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J Mol Cell Cardiol. 2013;62:24–35. doi: 10.1016/j.yjmcc.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 44.Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer-Crosbie M, Hedrick CC, Libby P, Nahrendorf M, Weissleder R, Swirski FK. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res. 2014;114:1611–1622. doi: 10.1161/CIRCRESAHA.114.303204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borges da Silva H, Fonseca R, Pereira RM, dos Cassado AA, Alvarez JM, D’Imperio Lima MR. Splenic Macrophage Subsets and Their Function during Blood-Borne Infections. Front Immunol. 2015;6:480. doi: 10.3389/fimmu.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuusisto J, Karja V, Sipola P, Kholova I, Peuhkurinen K, Jaaskelainen P, Naukkarinen A, Yla-Herttuala S, Punnonen K, Laakso M. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart. 2012;98:1007–1013. doi: 10.1136/heartjnl-2011-300960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prabhu SD. Cytokine-induced modulation of cardiac function. Circ Res. 2004;95:1140–1153. doi: 10.1161/01.RES.0000150734.79804.92. [DOI] [PubMed] [Google Scholar]
- 48.Hartupee J, Mann DL. Role of inflammatory cells in fibroblast activation. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner NA. Inflammatory and fibrotic responses of cardiac fibroblasts to myocardial damage associated molecular patterns (DAMPs) J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.