Abstract
The neural connectome is a critical determinant of brain function. Circuits of precisely wired neurons, and the features of transmission at the synapses connecting them, are thought to dictate information processing in the brain. While recent technological advances now allow to define the anatomical and functional neural connectome at unprecedented resolution, the elucidation of the molecular mechanisms that establish the precise patterns of connectivity and the functional characteristics of synapses has remained challenging. Here, we describe the power and limitations of genetic approaches in the analysis of mechanisms that control synaptic connectivity and function, and discuss how recent methodological developments in proteomics might be used to elucidate the molecular synaptic connectome that is at the basis of the neural connectome.
Introduction
The proper function of the nervous system is defined by neuronal circuits, in which individual neurons are precisely connected via synapses with specific properties. Recent methodological developments, such as optogenetics, in vivo electrophysiology and imaging, virus-based tracing, tissue-clearing and light sheet microscopy, and three-dimensional reconstruction of circuits by serial electron microscopic imaging, have led to substantial progress in defining the core anatomical connectivity of the mammalian brain. However, it is unclear how this anatomical connectivity is established at the molecular and cellular level [1,2], and how the synapses within such circuits are endowed with synapse type-specific functional features.
For over 50 years, since Roger W. Sperry formulated the 'chemoaffinity hypothesis', posing that individual cells expose 'identification tags' by which they are distinguished to the level of single neurons and implying that such tags determine the specificity of synaptogenesis [3], it has been assumed that cell type-specific surface receptors and adhesion proteins determine the specificity of connectivity. Subsequently, many candidate proteins that might act as such identification tags have been identified, but the question as to whether and how these molecules define the connectivity and function of specific synapses has remained largely unresolved [4].
Two main problems account for this lack of progress. First, multiple synaptic adhesion proteins appear to operate in concert or even in parallel and redundantly, rather than individually, to control synapse formation and define synapse-specific functional features [5,6]. Accordingly, obtaining detailed insights into the mechanism of specific synapse and circuit formation from analyses of individual adhesion proteins or even of entire protein families has proven challenging. Second, it has long been impossible to systematically tackle the problem of synapse type-specific protein composition and function, beyond mere localization and characterization of individual protein species and their interactors.
From a biochemical point of view, synapses can be considered supramolecular protein machines that are assembled from the repertoire of synaptic adhesion proteins, scaffold proteins, receptors, ion channels, and the components of the synaptic signaling machinery available in the connected neurons [7]. The qualitative and quantitative features of the composition and stoichiometry of the synaptic protein machinery subsequently determine the specificity and unique properties of a given synaptic connection. In turn, the mutation of genes encoding synaptic protein components causes qualitative or quantitative changes in the composition of defined synapses, triggering perturbations in synapse and network function that are at the basis of multiple neurological and psychiatric diseases.
In view of the major role that the protein composition of synapses plays in brain function, the elucidation of the composition of synaptic protein complexes and an understanding of the molecular mechanisms that control synaptic specificity and function have immense importance. Here, we first describe the power and limitations of genetic approaches in the analysis of mechanisms that control the specific generation and composition of synapses. We then consider recent methodological developments in cell subcompartment-specific proteomics, and discuss how these approaches might be used to determine the molecular synaptic connectome that is at the basis of the neural connectome.
Genetic dissection of the mechanisms controlling synapse-specific organization and function
Synaptic adhesion molecules play an important role in synapse development, by mediating cell-cell recognition and by linking pre- and postsynaptic partners [8]. A subset of synaptic adhesion molecules has the capacity to induce the differentiation of initial cell-cell contacts into pre- or postsynaptic specializations. This was first demonstrated for the postsynaptic adhesion molecules of the neuroligin family, which induce presynaptic differentiation when presented to contacting axons in cell culture [9]. Conversely, the presynaptic binding partners of neuroligins, the neurexin family of alternatively spliced adhesion molecules, induce postsynaptic differentiation [10]. The formation of a neurexin-neuroligin complex across the synaptic cleft induces pre- and postsynaptic differentiation by recruiting key components of the synaptic machinery, such as scaffolding proteins and neurotransmitter receptors [10–12] (Fig. 1A), thereby organizing synaptic protein composition [13]. A host of additional synaptic ligands for neurexins that expand or modulate the repertoire of neurexin interactions have since been identified. Among these are postsynaptic adhesion molecules of the leucine-rich repeat transmembrane neuronal protein (LRRTM) family, which also form trans-synaptic complexes with neurexins [14–16]. In addition, secreted proteins can bridge neurexins and postsynaptic receptors into tri-partite complexes. The astrocyte-derived secreted protein Hevin for example facilitates binding between neurexin and neuroligin splice variants that normally have weak affinity for each other [17], modulating the neurexin repertoire. Cerebellin-1 (Cbln1), a secreted glycoprotein of the C1q/tumor necrosis factor (TNF) superfamily, binds neurexins and the GluD2 glutamate receptor [18,19], whereas the related C1q-like proteins C1ql2 and C1ql3 couple neurexin-3 to postsynaptic kainate receptors [20], thus expanding the repertoire of neurexin interactions to directly organize postsynaptic neurotransmitter receptor composition at excitatory synapses. At inhibitory synapses, the extracellular matrix protein Punctin/MADD-4 was recently identified as a secreted synaptic organizer that binds both neurexin and neuroligin and regulates postsynaptic GABAA receptor clustering [21,22].
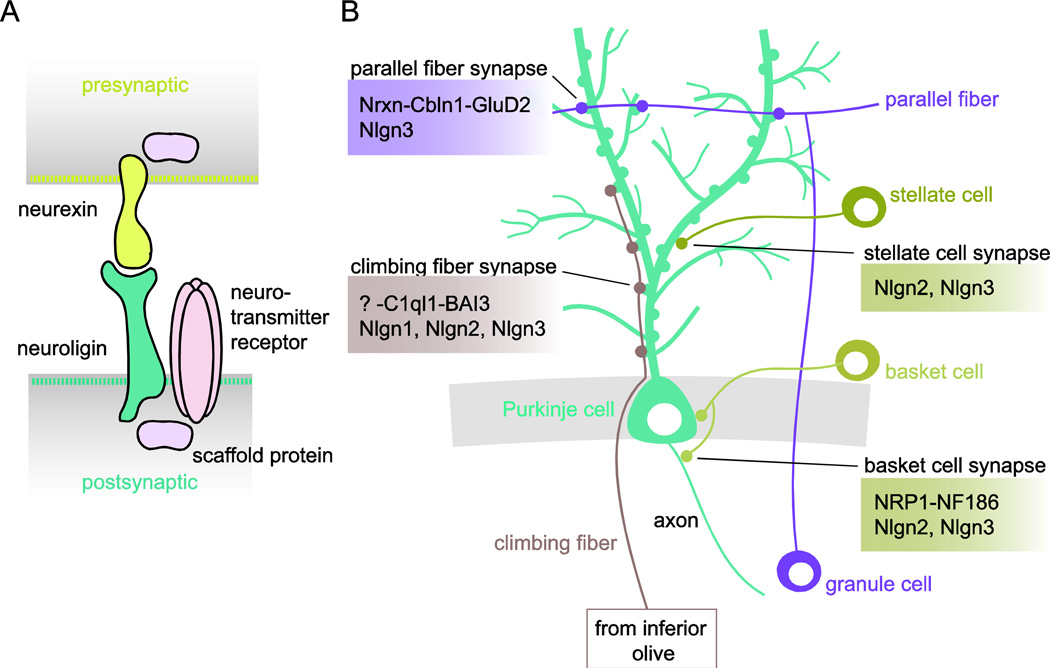
Figure 1. Genetic dissection of Purkinje cell connectivity in the cerebellum.
(A) Presynaptic neurexin molecules bind to postsynaptic neuroligin molecules to form a trans-synaptic adhesive complex that organizes the pre- and postsynaptic machinery by recruiting scaffolding proteins and neurotransmitter receptors. (B) Purkinje cells (PCs) receive excitatory input from climbing fibers (CFs) originating from the inferior olivary nucleus and from parallel fibers (PFs) originating from cerebellar granule cells. Local interneurons, stellate and basket cells, provide inhibitory input. At excitatory PF-PC synapses, Cbln1 is secreted from PFs and forms a tri-partite complex with presynaptic neurexin (Nrxn) isoforms and the postsynaptic GluD2 neurotransmitter receptor to regulate synaptogenesis. Neuroligin-3 (Nlgn3) is not required for synapse formation, but is required for long-term depression of PF-PC synapses, although this is under debate. At excitatory CF-PC synapses, C1ql1 is secreted from CFs and binds the postsynaptic adhesion GPCR BAI3 to regulate CF synapse formation. Whether C1ql1 also binds a presynaptic receptor at CF-PC synapses to form a tri-partite complex remains to be determined. Nlgn1, Nlgn2, and Nlgn3 all contribute to the specification of CF-PC synapse functional properties. At inhibitory basket cell-PC synapses, Semaphorin 3A is secreted from PCs to attract basket cell axons via its receptor Neuropilin-1 (NRP1) on these axons. NRP1 then binds in trans to Neurofascin 186 (NF186), which is expressed in a gradient on the PC soma and the axon initial segment. Both Nlgn2 and Nlgn3 contribute to stellate/basket cell inhibitory synapse function.
In addition to these neurexin-based adhesive complexes, other synaptic adhesion molecules capable of inducing synaptic differentiation have been identified. These include members of the LRR family of synaptic adhesion molecules [23], and the homophilic adhesion molecule SynCAM1 [24], which contributes to the patterning of the synaptic cleft into adhesive subcompartments, with different adhesion receptors occupying distinct regions of the cleft [25]. Of note, this summary is far from exhaustive, and is only meant to highlight the increasingly complex adhesive interactions that regulate synapse organization, formation and function. Other molecularly diverse families of adhesion molecules with important roles in regulating connectivity or synaptic function have been identified [4], and recent single-cell profiling studies are beginning to propose the functional relevance of cell type-specific repertoires of adhesion molecules [26,27]. Determining the role of cell type-specific signatures of adhesion molecules in synapse formation and the specification of synapse type-specific functional features constitutes a major challenge. Here we focus on the genetic dissection of adhesion molecules in the development of cerebellar Purkinje cell (PC) connectivity as an example of a well-characterized neural circuit. PC dendrites receive two main types of excitatory input: parallel fibers (PF) from cerebellar granule cells, which terminate on distal dendrites, and climbing fibers (CF) from the inferior olive, which terminate on proximal dendrites (Fig. 1B). In addition to these excitatory inputs, PCs also receive inhibitory input from various types of interneurons, stellate and basket cells (Fig. 1B).
A major synaptic organizer of excitatory PF-PC synapses is Cbln1, which is secreted from cerebellar granule cell axons and binds the postsynaptic GluD2 receptor on PC dendrites. Both are required for the formation and plasticity of this synapse [28]. Cbln1 also binds presynaptic neurexin, and the tri-partite neurexin-Cbln1-GluD2 complex is required for PF-PC synapse formation and plasticity [18,19] (Fig. 1B). C1ql1, which is secreted from climbing fibers, is a major organizer of excitatory CF-PC synapses. C1ql1 binds to the adhesion G protein-coupled receptor BAI3 on PC dendrites. Both C1ql1 and BAI3 are required for the formation and maintenance of the CF-PC synapse [29,30] (Fig. 1B). Whether C1ql1 also binds neurexins at CF-PC synapses to form a tri-partite complex, analogous to the interaction of C1ql2/3 with neurexin-3 at hippocampal mossy fiber synapses [20], or interacts with a different presynaptic receptor remains to be determined. Together, these studies show that secreted cues, released from distinct presynaptic inputs and through differential interactions with postsynaptic receptors, play a critical role in establishing specific excitatory synaptic connectivity in the cerebellum.
A systematic analysis of neuroligin function in cerebellar PCs revealed that all three neuroligins expressed in cerebellum, (Nlgn1, -2, and -3), are required for specifying the functional properties, but not the formation, of CF-PC synapses [31]. Combinatorial PC-specific loss of Nlgn1 and Nlgn3, which localize to excitatory synapses, decreases CF-PC synapse size and strength. A similar phenotype is found in Nlgn1/2/3 knockout mice, but here the additional deletion of Nlgn2, which localizes to inhibitory synapses, also results in a loss of distal CF synapses, indicating that Nlgn2 indirectly contributes to CF-PC synapse development through a poorly understood mechanism [31] (Fig. 1B). The contributions of all three neuroligins at PF-PC synapses appeared dispensable in this study, but an independent study showed that Nlgn3 immunoreactivity mainly localizes to PF-PC synapses, and found that loss of Nlgn3 impairs long-term depression (LTD) at PF-PC synapses and causes an ectopic expansion of CF synapses onto distal PC dendrites [32].
The development of inhibitory synapses onto PCs is also regulated by several adhesion molecules. A subcellular gradient of Neurofascin-186 (NF186) on the PC soma and axon initial segment (AIS) directs the formation of pinceau synapses by basket cells (BCs) [33]. The secreted axon guidance cue Semaphorin 3A (SEMA3A), released by PCs, attracts BC axons via the SEMA3A receptor Neuropilin-1 expressed on BC axons. Neuropilin-1 then interacts in trans with NF186 on the AIS to form pinceau synapses [34], revealing an interplay between guidance and adhesive mechanisms to establish specific inhibitory synaptic connectivity (Fig. 1B). Neuroligins on the other hand contribute to specifying the functional properties of inhibitory synapses onto PCs, but are not required for the formation of inhibitory stellate/basket cell synapses. Loss of Nlgn2 impairs basket/stellate cell synaptic function, whereas loss of Nlgn3, which localizes to both excitatory and inhibitory synapses, has no effect. Combined loss of Nlgn2 and Nlgn3 however impairs inhibitory synaptic transmission more strongly than Nlgn2 deletion alone [31], indicating some redundancy between Nlgn isoforms at stellate/basket cell synapses, with Nlgn2 having a more important contribution (Fig. 1B).
Taken together, the emerging picture from this work is that many different cell-surface cues, axon guidance and adhesive, secreted and membrane-bound, act at specific types of inputs and subcellular compartments to construct precisely wired neural circuits.
Uncovering the molecular synaptic connectome: towards compartment-specific proteomics
The genetic dissection of the cerebellar PC circuit illustrates that different molecules act at distinct types of synapses. Systematically and comprehensively identifying synapse type-specific protein compositions has long been challenging. Novel proteomics-based approaches are rapidly changing this, enabling new insight into the mechanisms that define the connectivity and function of specific synaptic connections.
Most attempts to elucidate the composition of synaptic protein complexes have been based on the biochemical isolation of synaptic subfractions, such as synaptosomes, synaptic membranes, or postsynaptic densities. In combination with mass spectrometric analyses, over 2000 (potential) synaptic proteins were identified [35–42]. However, a key limitation in this context has been that whole brains or brain regions containing complex mixtures of different types of neurons, glial cells and synapses were typically used as input material.
Complementing classical subfractionation approaches, attempts were made to isolate more specific synaptic protein complexes, using purely biochemical techniques, antibody-based affinity purification of synaptic proteins, or mouse genetics to tag synaptic proteins in order to provide an additional level of specificity biochemical approaches alone could not afford. Examples include glutamatergic synaptosomes [43] (BOX 1), components of GABAergic or glutamatergic synapses [44,45], components of the pre- and postsynaptic constituents of synaptic protein complexes [46], or neurotransmitter receptor complexes [47–49]. To date, only a single study, employing transgenic cell type-specific expression of the GFP-tagged GluD2 receptor that localizes to parallel fiber inputs on Purkinje cells, isolated and characterized synaptic protein complexes originating from a defined population of neurons and synapses [50].
BOX 1. Fluorescent sorting of specific synaptosomes.
Synaptosomes are isolated, functional synaptic particles consisting of a resealed presynaptic compartment and partial postsynaptic element. A major limitation of conventional synaptosome preparations is that they contain a mixture of many synapse types together with neuronal and non-neuronal contaminants [73–75]. Fluorescence Activated Synaptosome Sorting (FASS) of a subset of glutamatergic synapses purified from a VGLUT1VENUS (vesicular glutamate transporter 1) knock-in mouse allows to deplete most contaminants and to enrich for VGLUT1VENUS synapses to near homogeneity [43,76]. Recent improvements on this technique include the fluorescent sorting of EGFP-labeled terminal fields of AAV transduced cortical mouse neurons (unpublished), enabling the analysis of synaptic proteomes of genetically identified afferents. Even though the amount of material recovered from these sorts is extremely low, current mass spectrometry technologies can confidently identify and quantify thousands of proteins from only a few micrograms of proteins and excel when analyzing samples with reduced protein complexity. FASS-based proteomics thus has the potential to contribute to unraveling the protein networks of distinct synapse populations.
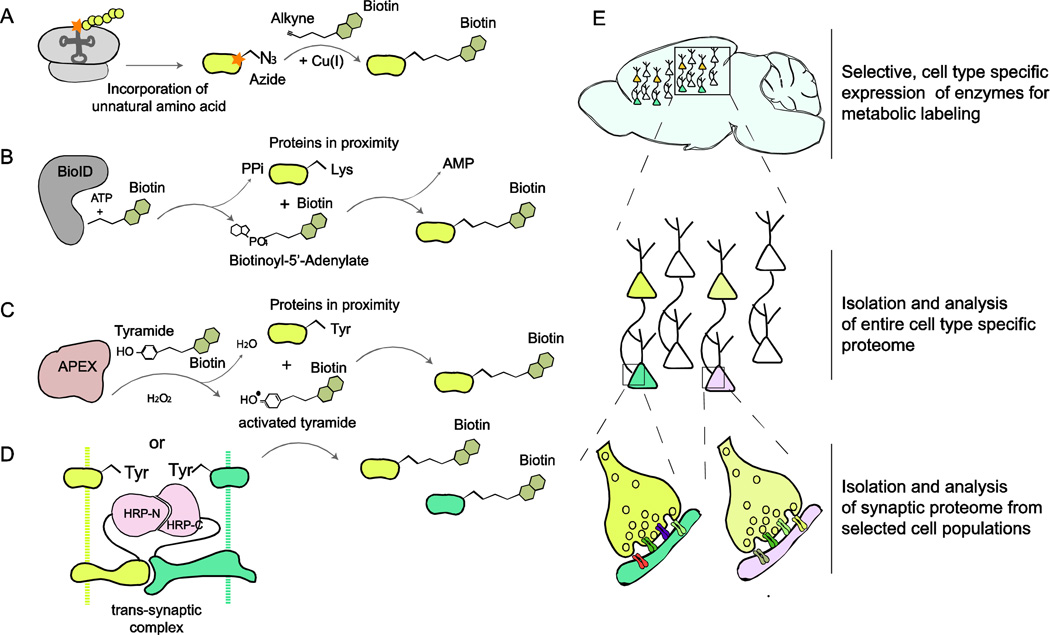
Recent developments in the metabolic labeling of proteins (BOX 2) and the subsequent isolation of proteins from defined cell populations or subcellular compartments open new avenues for the analysis of specific synaptic proteomes. Bio-orthogonal non-canonical amino-acid-tagging (BONCAT), for instance, is based on the incorporation of non-canonical amino acids, such as azidonorleucine (ANL), instead of methionine into newly synthesized proteins [51] (Fig. 2A). In cells expressing an unnatural aminoacyl-tRNA-synthetase, translated proteins incorporate ANL and can subsequently be labeled with a functional group for affinity purification, for example by coupling the azide group of ANL to alkyne-bearing tags (e.g. biotin) using so-called 'click chemistry'. Several published studies utilized this methodology for the labeling and comparative proteome analysis of newly synthetized proteins in primary neuronal cultures or in brain slices after stimulation with neuronal receptor agonists or as response to neuronal activity [52–54]. However, initially developed and widely used for applications in cell culture, this methodology has recently been applied successfully for labeling of whole cell type-specific proteomes in Drosophila [55] and C. elegans [56]. So far, BONCAT is mostly limited to applications in cell cultures and small animals. The delivery of non-canonical amino acids to the brain in vivo and possible side-effects caused by the expression of unnatural aminoacyl-tRNA-synthetases or the presence of ANL [55], still pose major barriers for applications in mammals.
BOX 2. Quantitative proteomic analysis of mammalian synapses.
Metabolic labeling of rodents with a modified diet highly enriched in stable, heavy isotopes (such as nitrogen-15) facilitates the relative quantitation of thousands of synaptic proteins. The benefit of this strategy compared to label-free proteomic analysis is that mixing “light” and “heavy” brain extracts prior to biochemical enrichment of synaptic compartments provides experimental control over the inherent variability of these procedures [77]. Isotopic labeling in combination with gene knockout allows the analysis of the effect of loss of a single protein on synaptic protein composition [78]. Isotopic labeling in combination with modulation of sensory stimulation (e.g. whisker trimming, noise exposure, or eye lid suture) allows in-depth proteomic characterization of the barrel, auditory, or visual cortex [79]. Further, pulse-chase labeling of rodent brains with stable isotopes enables the analysis of synaptic proteome dynamics and identified an extremely long-lived neuronal adhesion protein [80,81]. While isotope-based strategies ensure accurate protein quantitation, they still lack the spatial resolution needed to resolve changes in protein levels of specific synapses. Isotopic labeling in combination with synapse-specific proximity-labeling has the potential to resolve this issue.
Figure 2. Overview of chemical labeling techniques and their application for targeted proteome analysis.
(A) Bioorthogonal noncanonical amino acid tagging (BONCAT): the approach is based on the in vivo incorporation of non-canonical, azide containing amino acids such as L-azidohomoalanine (AHA) into newly synthesized proteins. These proteins can be subsequently labeled with biotin by ‘click-chemistry’ and isolated for further analysis. (B) In vivo proximity protein biotinylation by promiscuous biotin-ligase (BioID): biotinylation occurs through Biotinoyl-5’-Adenylate which is released by a mutated variant of E.coli biotin ligase BirA. Biotinoyl-5’-Adenylate is a highly reactive compound that quickly reacts with lysines of proximal proteins. (C) Biotin-labeling by ascorbate-peroxidase (APEX) and (D) by split-horseradish-peroxidase: both techniques utilize the ability of peroxidase enzymes to generate highly reactive species from tyramide derived compounds such as biotin-phenol. These react quickly with aromatic groups (usually tyrosine and tryptophan, but also histidine and cytosine) of proteins in close proximity. (E) Application of metabolic labeling for analysis of cell-type/compartment-specific proteomes: Selective expression of biotin-labeling enzymes in cells of interest (e.g. utilizing the Cre-Lox-system) allows isolation of proteins expressed in identified cells. Additionally, targeting these enzymes to synaptic compartments provides an opportunity for the labeling and subsequent determination of synapse-type specific protein composition in desired cell populations.
Biotin proximity labeling by the promiscuous biotin ligase BioID [57] represents a second approach with potential suitability for cell-selective metabolic labeling of proteins with biotin (Fig. 2B). Compared to BONCAT, this approach allows not only for labeling of entire cellular proteomes, but also of subcellular nano-proteomes, by targeting BioID fusion proteins to defined subcellular compartments. Numerous studies have demonstrated the applicability of this method for the identification of components of different protein complexes, including protein complexes that mediate nonsense-mediated mRNA decay [58], control centrosome organization [59], or constitute the nuclear pore [60]. To date, the BioID methodology has only been applied in cell culture. Efficient biotinylation by BioID requires high, non-physiological biotin concentrations, and has relatively slow kinetics. A new and improved version of BioID requires lower biotin concentrations [61]. The application of this methodology in the brain would require an external supply of biotin by injections, which is feasible as biotin is efficiently transported across the blood-brain barrier [62,63]. Hence, BioID approaches have substantial potential for applications of selective biotin labeling of proteins in vivo, by expressing BioID in defined neuronal cell populations and/or by targeting BioID to defined subcellular compartments (e.g. postsynaptic spines or presynaptic terminals).
A BioID-related approach for proximity labeling employs ascorbate-peroxidase (APEX) [64]. This technique utilizes the enzymatic activation of tyramides, such as biotin-phenol, by ascorbate-peroxidase (APEX) in the presence of hydrogen peroxide [65,66] (Fig. 2C). Activated tyramides are highly reactive and conjugate rapidly with proteins in the proximity by covalent attachment to aromatic amino acids (preferentially tyrosines). Several recent publications demonstrated that the APEX approach is a powerful way to specifically isolate proteomes of selected cell types or subcellular compartments and organelles [64,67–69]. So far, the requirement of tyramides and hydrogen peroxide for protein labeling has limited the application of this technique to cell cultures or small animals. However, the fast kinetics of the reaction provides an avenue for the use of the APEX technique in situ, e.g. in acute brain slices from animals that express APEX in specific neuronal populations and/or subcellular compartments.
A recent publication demonstrated the feasibility of biotin labeling of synapses in vivo using split horseradish peroxidase [70]. Here, enzymatic activity of non-functional fragments of horseradish peroxidase fused to pre- and postsynaptic adhesion proteins, was restored upon formation of trans-synaptic adhesion complexes and association of the peroxidase fragments (Fig. 2D). This allowed for proximity biotin labeling and visualization of synapses in the intact mouse brain. While the initial use of this approach was limited to the visualization of synaptic contacts, it is also potentially applicable for the isolation and proteomic analysis of specific synaptic complexes in the intact brain.
During the review of the manuscript two studies with a special importance for the discussed issues were published [71,72]. In the study published by Loh and colleagues, cultured neurons were infected with lentiviral constructs encoding for horseradish-peroxidase (HRP) targeted to the synaptic cleft of inhibitory or excitatory synapses, respectively, by fusion to the transmembrane and cytoplasmic part of synaptic adhesion proteins known to be localized at the respective type of synapses. Biotinylation of proteins localized to the synaptic cleft in the proximity of HRP was achieved by addition of biotin-phenol and hydrogen peroxide (see Fig. 2). Uezu and colleagues utilized the BioID approach to label components of excitatory and inhibitory postsynaptic densities (PSD) in vivo. BioID constructs targeted to inhibitory or excitatory PSDs by fusion to gephyrin or PSD95, respectively, were delivered by AAV injections into neonatal animals. To achieve efficient protein biotinylation by BioID, biotin was delivered exogenously by subcutaneous injections. In both studies specific biotinylation and subsequent isolation of proteins corresponding to inhibitory or excitatory synapses, respectively, could be achieved. The mass spectrometric analysis of isolated proteins identified known synaptic proteins as well as new components of inhibitory and excitatory synaptic complexes. Thus, both studies demonstrated the feasibility and power of the protein labeling technologies for synaptic proteome analysis. Further development of these approaches and their extension for the applications in defined neuronal populations will significantly contribute to our understanding of the molecular organization of neuronal synapses.
Conclusion
In summary, proximity-labeling methods represent fascinating new tools for the analysis of specific neuronal and synaptic proteomes (Fig. 2E). Depending on the specific targeting of the enzymatic agents, analyses can be very focused, even down to specific synapses. In combination with evolving mass spectrometric analysis, including conventional ‘shotgun’ proteomics as well as targeted and quantitative approaches, these techniques hold the promise to drive substantial progress in our understanding of the molecular determinants of synaptic specificity and function. In combination with sophisticated combinatorial genetic approaches, we can begin to understand how the molecular synaptic connectome controls the anatomical and functional connectivity of neural circuits.
Highlights.
The CNS represents a precisely organized synaptic network formed by diverse neurons
Neuronal adhesion proteins substantially contribute to synaptic complex organization
Molecular compositions of synaptic complexes define their functional capabilities
New techniques will help to gain insight in the molecular organization of synapses
Acknowledgments
We apologize to those authors whose work we could not include in this review, due to space limitations and the emphasis on recent publications in Current Opinion articles. Work in the authors’ laboratories is supported by NIH award R00DC013805-02 and Hartwell Individual Biomedical Research Award (to J.N.S.); the French Agence Nationale de la Recherche (ANR-12-JSV4-0005-01 VGLUTIQ, and ANR-10-LABX-43 BRAIN) and the French Fondation pour la Recherche Medicale (ING20150532192) (to E.H.); the Max Planck Society, the European Commission (EU-AIMS FP7-115300) and the German Research Foundation (CNMPB) (to N.B.); European Research Council (ERC) Starting Grant (#311083) and Research Organization Flanders (FWO) Odysseus Grant and Project Grant (#G094016N) (to J.d.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
* of special interest; ** of outstanding interest
- 1.Yogev S, Shen K. Cellular and molecular mechanisms of synaptic specificity. Annu Rev Cell Dev Biol. 2014;30:417–437. doi: 10.1146/annurev-cellbio-100913-012953. [DOI] [PubMed] [Google Scholar]
- 2.Sanes JR, Yamagata M. Many paths to synaptic specificity. Annu Rev Cell Dev Biol. 2009;25:161–195. doi: 10.1146/annurev.cellbio.24.110707.175402. [DOI] [PubMed] [Google Scholar]
- 3.Sperry RW. Chemoaffinity in the Orderly Growth of Nerve Fiber Patterns and Connections. Proc Natl Acad Sci U A. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:4. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- 5.Brose N. Synaptogenic proteins and synaptic organizers: “many hands make light work”. Neuron. 2009;61:650–652. doi: 10.1016/j.neuron.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Brose N. Why we need more synaptogenic cell-adhesion proteins. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3717–3718. doi: 10.1073/pnas.1300505110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emes RD, Grant SG. Evolution of synapse complexity and diversity. Annu Rev Neurosci. 2012;35:111–131. doi: 10.1146/annurev-neuro-062111-150433. [DOI] [PubMed] [Google Scholar]
- 8.Giagtzoglou N, Ly CV, Bellen HJ. Cell adhesion, the backbone of the synapse: “vertebrate” and “invertebrate” perspectives. Cold Spring Harb Perspect Biol. 2009;1:a003079. doi: 10.1101/cshperspect.a003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 10.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budreck EC, Kwon OB, Jung JH, Baudouin S, Thommen A, Kim HS, Fukazawa Y, Harada H, Tabuchi K, Shigemoto R, et al. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc Natl Acad Sci U A. 2013;110:725–730. doi: 10.1073/pnas.1214718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui TJ, Craig AM. Synaptic organizing complexes [Internet] Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30:7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz L-J, Kim IH, Manhaes AC, Rodrigues WS, Pamukcu A, et al. Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1α and NL1 via Hevin. Cell. 2016;164:183–196. doi: 10.1016/j.cell.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Elegheert J, Kakegawa W, Clay JE, Shanks NF, Behiels E, Matsuda K, Kohda K, Miura E, Rossmann M, Mitakidis N, et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science. 2016;353:295–299. doi: 10.1126/science.aae0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuda K, Budisantoso T, Mitakidis N, Sugaya Y, Miura E, Kakegawa W, Yamasaki M, Konno K, Uchigashima M, Abe M, et al. Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. ** Matsuda and colleagues show that the C1q-like proteins C1ql2 and C1ql3 are released from hippocampal mossy fibers and bridge a complex of a presynaptic neurexin 3 isoform and postsynaptic kainate receptors to trans-synaptically regulate kainate receptor localization and function.
- 21.Maro GS, Gao S, Olechwier AM, Hung WL, Liu M, Özkan E, Zhen M, Shen K. MADD-4/Punctin and Neurexin Organize C. elegans GABAergic Postsynapses through Neuroligin. Neuron. 2015;86:1420–1432. doi: 10.1016/j.neuron.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu H, Pinan-Lucarré B, Ji T, Jospin M, Bessereau J-L. C. elegans Punctin Clusters GABA(A) Receptors via Neuroligin Binding and UNC-40/DCC Recruitment. Neuron. 2015;86:1407–1419. doi: 10.1016/j.neuron.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 23.de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 2014;37:539–550. doi: 10.1016/j.tins.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 25. Perez de Arce K, Schrod N, Metzbower SW, Allgeyer E, Kong GK, Tang AH, Krupp AJ, Stein V, Liu X, Bewersdorf J, et al. Topographic Mapping of the Synaptic Cleft into Adhesive Nanodomains. Neuron. 2015;88:1165–1172. doi: 10.1016/j.neuron.2015.11.011. * Using cryoelectron tomography and super-resolution imaging, Perez de Arce and colleagues show that the synaptic cleft is structurally organized and that the adhesion molecule SynCAM1 shapes the edge of the synaptic cleft. Different synaptic receptors, EphB2 and SynCAM1, occupy distinct subsynaptic compartments, indicating that the synaptic cleft is organized in adhesive nanodomains.
- 26.Fuccillo MV, Foldy C, Gokce O, Rothwell PE, Sun GL, Malenka RC, Sudhof TC. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Földy C, Darmanis S, Aoto J, Malenka RC, Quake SR, Südhof TC. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- 29. Sigoillot SM, Iyer K, Binda F, Gonzalez-Calvo I, Talleur M, Vodjdani G, Isope P, Selimi F. The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells [Internet] Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.034. Together with Ref #30, this study shows that C1ql1, secreted from cerebellar climbing fibers, binds to the adhesion GPCR BAI3 on Purkinje cells to control climbing fiber synaptogenesis and territory on Purkinje cells.
- 30. Kakegawa W, Mitakidis N, Miura E, Abe M, Matsuda K, Takeo YH, Kohda K, Motohashi J, Takahashi A, Nagao S, et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron. 2015;85:316–329. doi: 10.1016/j.neuron.2014.12.020. ** Together with Ref #29, this paper shows that C1ql1 binds BAI3 at climbing fiber-Purkinje cell synapses. C1ql1 and BAI3 are required to control the selection and maintenance of a single-winner in synaptic competing of climbing fibers during development, and are also essential for cerebellar synaptic plasticity in a motor learning task.
- 31. Zhang B, Chen LY, Liu X, Maxeiner S, Lee S-J, Gokce O, Südhof TC. Neuroligins Sculpt Cerebellar Purkinje-Cell Circuits by Differential Control of Distinct Classes of Synapses. Neuron. 2015;87:781–796. doi: 10.1016/j.neuron.2015.07.020. ** Using conditional deletion of all three neuroligins specifically in cerebellar Purkinje cells, the authors demonstrate that neuroligins are not required for synapse formation, but are selectively important for postsynaptic function, and differentially contribute to climbing fiber and stellate /basket cell inputs on Purkinje cells.
- 32.Baudouin SJ, Gaudias J, Gerharz S, Hatstatt L, Zhou K, Punnakkal P, Tanaka KF, Spooren W, Hen R, De Zeeuw CI, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- 33.Ango F, di Cristo G, Higashiyama H, Bennett V, Wu P, Huang ZJ. Ankyrin-based subcellular gradient of neurofascin, an immunoglobulin family protein, directs GABAergic innervation at purkinje axon initial segment. Cell. 2004;119:257–272. doi: 10.1016/j.cell.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 34. Telley L, Cadilhac C, Cioni J-M, Saywell V, Jahannault-Talignani C, Huettl RE, Sarrailh-Faivre C, Dayer A, Huber AB, Ango F. Dual Function of NRP1 in Axon Guidance and Subcellular Target Recognition in Cerebellum. Neuron. 2016;91:1276–1291. doi: 10.1016/j.neuron.2016.08.015. * Telley and colleagues show that the formation of precisely targeted, local GABAergic circuits in the cerebellum is controlled by an interplay between axon guidance and cell adhesion mechanisms. Semaphorin 3A, secreted by Purkinje cells, binds to Neuropilin-1 on inhibitory basket cell axons to attract them to the Purkinje cell axon initial segment, where Neuropilin-1 interacts in trans with the adhesion molecule Neurofascin-186 to form basket cell-Purkinje cell synapses.
- 35.Collins MO, Husi H, Yu L, Brandon JM, Anderson CNG, Blackstock WP, Choudhary JS, Grant SGN. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 36.Distler U, Schmeisser MJ, Pelosi A, Reim D, Kuharev J, Weiczner R, Baumgart J, Boeckers TM, Nitsch R, Vogt J, et al. In-depth protein profiling of the postsynaptic density from mouse hippocampus using data-independent acquisition proteomics. Proteomics. 2014;14:2607–2613. doi: 10.1002/pmic.201300520. [DOI] [PubMed] [Google Scholar]
- 37.Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla K-H, et al. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 2004;279:987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 38.Liu S-H, Cheng H-H, Huang S-Y, Yiu P-C, Chang Y-C. Studying the protein organization of the postsynaptic density by a novel solid phase- and chemical cross-linking-based technology. Mol. Cell. Proteomics MCP. 2006;5:1019–1032. doi: 10.1074/mcp.M500299-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- 40.Walikonis RS, Jensen ON, Mann M, Provance DW, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. Off. J. Soc. Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm BG, Mandad S, Truckenbrodt S, Kröhnert K, Schäfer C, Rammner B, Koo SJ, Claßen GA, Krauss M, Haucke V, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88:759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 43.Biesemann C, Gronborg M, Luquet E, Wichert SP, Bernard V, Bungers SR, Cooper B, Varoqueaux F, Li L, Byrne JA, et al. Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting. EMBO J. 2014;33:157–170. doi: 10.1002/embj.201386120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Serwanski DR, Miralles CP, Bahr BA, De Blas AL. Two pools of Triton X-100-insoluble GABA(A) receptors are present in the brain, one associated to lipid rafts and another one to the post-synaptic GABAergic complex. J. Neurochem. 2007;102:1329–1345. doi: 10.1111/j.1471-4159.2007.04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heller EA, Zhang W, Selimi F, Earnheart JC, Ślimak MA, Santos-Torres J, Ibañez-Tallon I, Aoki C, Chait BT, Heintz N. The biochemical anatomy of cortical inhibitory synapses. PloS One. 2012;7:e39572. doi: 10.1371/journal.pone.0039572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berninghausen O, Rahman MA, Silva JP, Davletov B, Hopkins C, Ushkaryov YA. Neurexin Ibeta and neuroligin are localized on opposite membranes in mature central synapses. J Neurochem. 2007;103:1855–1863. doi: 10.1111/j.1471-4159.2007.04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 48.Schwenk J, Pérez-Garci E, Schneider A, Kollewe A, Gauthier-Kemper A, Fritzius T, Raveh A, Dinamarca MC, Hanuschkin A, Bildl W, et al. Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat. Neurosci. 2016;19:233–242. doi: 10.1038/nn.4198. [DOI] [PubMed] [Google Scholar]
- 49.Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR, 3rd, et al. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 2012;1:590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 2009;7:e83. doi: 10.1371/journal.pbio.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc. Natl. Acad. Sci. U. S. A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schanzenbächer CT, Sambandan S, Langer JD, Schuman EM. Nascent Proteome Remodeling following Homeostatic Scaling at Hippocampal Synapses. Neuron. 2016;92:358–371. doi: 10.1016/j.neuron.2016.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowling H, Bhattacharya A, Zhang G, Lebowitz JZ, Alam D, Smith PT, Kirshenbaum K, Neubert TA, Vogel C, Chao MV, et al. BONLAC: A combinatorial proteomic technique to measure stimulus-induced translational profiles in brain slices. Neuropharmacology. 2016;100:76–89. doi: 10.1016/j.neuropharm.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodas JJL, Nehring A, Höche N, Sweredoski MJ, Pielot R, Hess S, Tirrell DA, Dieterich DC, Schuman EM. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;12:2464–2476. doi: 10.1002/pmic.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Erdmann I, Marter K, Kobler O, Niehues S, Abele J, Müller A, Bussmann J, Storkebaum E, Ziv T, Thomas U, et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat. Commun. 2015;6:7521. doi: 10.1038/ncomms8521. * Using cell selective expression of mutated methionyl-tRNA synthetase, which enables incorporation of the non-canonical amino acid azidonorleucine (ANL) instead of methionine, the authors demonstrate applicability of the BONCAT technique for labeling and isolation of cell type specific proteome in vivo.
- 56.Yuet KP, Doma MK, Ngo JT, Sweredoski MJ, Graham RLJ, Moradian A, Hess S, Schuman EM, Sternberg PW, Tirrell DA. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2705–2710. doi: 10.1073/pnas.1421567112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. This work describes for the first time the development and application of proximal biotin labeling by a promiscuous biotin ligase (BioID).
- 58.Schweingruber C, Soffientini P, Ruepp M-D, Bachi A, Mühlemann O. Identification of Interactions in the NMD Complex Using Proximity-Dependent Biotinylation (BioID) PloS One. 2016;11:e0150239. doi: 10.1371/journal.pone.0150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spector R, Mock D. Biotin transport through the blood-brain barrier. J. Neurochem. 1987;48:400–404. doi: 10.1111/j.1471-4159.1987.tb04107.x. [DOI] [PubMed] [Google Scholar]
- 63.Spector R, Mock DM. Biotin transport and metabolism in the central nervous system. Neurochem. Res. 1988;13:213–219. doi: 10.1007/BF00971535. [DOI] [PubMed] [Google Scholar]
- 64.Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen C-L, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12093–12098. doi: 10.1073/pnas.1515623112. * Applying cell type-specific expression of ascorbate peroxidase (APEX) targeted to defined subcellular compartments in Drosophila melanogaster, the authors demonstrate the applicability of APEX-based protein labeling for characterization subcellular proteomes in vivo.
- 68.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of Primary Cilia by Proximity Labeling. Dev. Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, Ting AY. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 2016 doi: 10.1038/nbt.3563. ** Using a split-horseradish peroxidase (HRP) approach, the authors demonstrate for the first time applicability of the peroxidase-based protein proximity biotinylation in selected neurons in the intact mouse brain. N- and C-terminal HRP fragments, which have no enzymatic activity on their own, were fused to the synaptic adhesion proteins neurexin and neuroligin, respectively. Upon trans-synaptic interaction between neuroligin and neurexin, enzymatic activity of HRP is reconstituted, and thus enables biotinylation of proximal proteins in the synaptic cleft. The method was used for visualization of synapses between specific sets of neurons, however, could also be adapted for isolation and analysis of synaptic proteins from desired cell populations.
- 71. Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell. 2016;166:1295.e21–1307.e21. doi: 10.1016/j.cell.2016.07.041. Utilizing a proximity biotinylation approach by horseradish peroxidase (HRP) targeted to the synaptic cleft of glutamatergic or GABAergic synapses, respectively, Loh and colleagues were able to label and isolate proteins localized at synaptic cleft at both major types of neuronal synapses in primary neuronal culture.
- 72. Uezu A, Kanak DJ, Bradshaw TWA, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353:1123–1129. doi: 10.1126/science.aag0821. ** Uezu and collegues could demonstrate for the first time the feasibility of the use of proximity biotinylation by promiscous biotin ligase (BioID) in the mouse brain in vivo. BioID constructs targeted to the postsynaptic densities (PSD) of inhibitory or exitatory synapses, respectively, were delivered via AAV injection. The analysis of isolated biotinylated proteins identified not only known components of synaptic complexes, but also new important constituents of the inhibitory PSD.
- 73.Cotman CW, Matthews DA. Synaptic plasma membranes from rat brain synaptosomes: isolation and partial characterization. Biochim. Biophys. Acta. 1971;249:380–394. doi: 10.1016/0005-2736(71)90117-9. [DOI] [PubMed] [Google Scholar]
- 74.Henn FA, Anderson DJ, Rustad DG. Glial contamination of synaptosomal fractions. Brain Res. 1976;101:341–344. doi: 10.1016/0006-8993(76)90274-2. [DOI] [PubMed] [Google Scholar]
- 75.Dodd P, Hardy JA, Oakley AE, Strong AJ. Synaptosomes prepared from fresh human cerebral cortex; morphology, respiration and release of transmitter amino acids. Brain Res. 1981;224:419–425. doi: 10.1016/0006-8993(81)90871-4. [DOI] [PubMed] [Google Scholar]
- 76.Luquet E, Biesemann C, Munier A, Herzog E. Purification of Synaptosome Populations Using Fluorescence Activated Synaptosome Sorting [Internet] In: Poulopoulos A, editor. Synapse Development Methods Mol Biol. Springer US; 2017. [DOI] [PubMed] [Google Scholar]
- 77.McClatchy DB, Liao L, Lee JH, Park SK, Yates JR. Dynamics of subcellular proteomes during brain development. J. Proteome Res. 2012;11:2467–2479. doi: 10.1021/pr201176v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savas JN, Ribeiro LF, Wierda KD, Wright R, DeNardo-Wilke LA, Rice HC, Chamma I, Wang YZ, Zemla R, Lavallee-Adam M, et al. The Sorting Receptor SorCS1 Regulates Trafficking of Neurexin and AMPA Receptors. Neuron. 2015;87:764–780. doi: 10.1016/j.neuron.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butko MT, Savas JN, Friedman B, Delahunty C, Ebner F, Yates JR, 3rd, Tsien RY. In vivo quantitative proteomics of somatosensory cortical synapses shows which protein levels are modulated by sensory deprivation. Proc Natl Acad Sci U A. 2013;110:E726–E735. doi: 10.1073/pnas.1300424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]