Abstract
During development a limited number of progenitors generate diverse cell types that comprise the nervous system. Neuronal diversity, which arises largely at the level of neural stem cells, is critical for brain function. Often these cells exhibit temporal patterning: they sequentially produce neurons of distinct cell fates as a consequence of intrinsic and/or extrinsic cues. Here, we review recent advances in temporal patterning during neuronal specification, focusing on conserved players and mechanisms in invertebrate and vertebrate models. These studies underscore temporal patterning as an evolutionarily conserved strategy to generate neuronal diversity. Understanding the general principles governing temporal patterning and the molecular players involved will improve our ability to direct neural progenitors towards specific neuronal fates for brain repair.
Introduction
Our brains are comprised of thousands of complex processing networks, each made up of multiple neuron types with unique morphological and functional properties. Neuronal diversity underlies the ability to process sensory information and perform complex tasks. The production of diverse types of neurons can often be traced back to decisions made at the level of neural stem cells (called neuroblasts in Drosophila; See Box 1). Indeed, prior to establishing neural circuitry, neural stem cells must divide, know when to stop dividing, and their progeny must adopt distinct neuronal fates in appropriate numbers and stoichiometry. Altering neuronal diversity, numbers or stoichiometry, all have profound impacts on circuit composition and ultimately brain function.
Box 1. Essential terms.
Neural progenitor: A multipotent stem cell, called a neuroblast in Drosophila, that is responsible for producing neurons and/or glia during development.
Competence: The ability of a neural stem cell to produce specific neuron types. Spatial and temporal patterning influence progenitor competence (e.g. temporally patterned progenitors can lose the ability to produce early-born neuronal fates late in life).
Symmetric division: A mitotic division type that produces daughters of the same fate. Progenitors can divide symmetrically to produce either two progenitor daughters or two neuronal progeny.
Asymmetric cell division: A mitotic division that produces two progeny of different fates.
Ganglion Mother Cell: A cell type generated when a Drosophila type I neuroblast (Figure 1) or INP undergoes an asymmetric self-renewing division. GMCs typically divide to generate two post-mitotic neuronal progeny.
Intermediate Neural Progenitor: Neurogenic progeny generated when a Drosophila type II neuroblast (Figure 1B) undergoes an asymmetric self-renewing division. These cells amplify a progenitor lineage by undergoing multiple rounds of asymmetric divisions to self-renew and produce GMCs.
Intermediate progenitor cells (IPCs): Neurogenic progeny generated by cortical progenitors, which can self-renew or generate neurons directly (Figure 2C). Additional IPC-like cells with greater proliferative potential, called outer radial glia, exist in mammals in varying proportions. For example, primates have far more outer radial glia than rodents. The large variability in mammalian neocortex sizes is attributed to these cells.
Spatial patterning: The generation of distinct neuronal progeny based on positional cues that restrict progenitor competence.
Temporal patterning: The generation of distinct neuronal progeny based on time. Extrinsic or intrinsic timing cues restrict progenitor competence to produce specific cell types as development progresses, resulting in the ordered birth of different cell types.
Temporal transcription factor: A transcription factor that is expressed for a fixed period in progenitors to mediate temporal patterning. These factors are necessary and sufficient to dictate progenitor competence.
Neuroepithelium: A pseudostratified epithelium that expands via symmetric divisions before being converted into neural progenitors.
An obvious contribution to neuronal diversity comes from spatial patterning, where regional compartments along spatial axes produce different neuronal subtypes [reviewed in 1–4]. For instance, spatially segregated neural stem cells along the length of the spinal cord produce motor neurons that adopt unique fates to innervate either limbs or abdominal muscles [reviewed in 4]. A less intuitive source of neuronal diversity arises from temporal patterning, whereby the same neural stem cell produces distinct neuronal cell-types over time [reviewed in 5]. Both spatial and temporal patterning mechanisms are evolutionarily conserved modes by which neuronal diversity is generated. Due to limited space we do not provide an exhaustive review of the literature but instead focus mainly on temporal patterning at the level of neural stem cells. We summarize recent findings in invertebrate and vertebrate systems, highlighting conserved players and temporal patterning mechanisms.
Temporal patterning during Drosophila neurogenesis
The genetically tractable fruit fly, Drosophila melanogaster, with its stereotyped nervous system, has been invaluable in advancing our understanding of how neuroblasts change over time to generate different neuronal progeny. Indeed, temporal patterning of neuroblasts was first described in the Drosophila ventral nerve cord (VNC) [6–8].
The Drosophila VNC is analogous to the vertebrate spinal cord. During embryogenesis, neuroblasts with unique spatial identities, defined by their segmentally repeated bilateral organization, give rise to all neurons and glia of the VNC [9]. These type I (Figure 1A) neuroblasts pass through several rounds of self-renewing asymmetric divisions. Each division produces a ganglion mother cell (GMC), which divides once more to generate a pair of neurons or glia [reviewed in 5]. Progeny produced in early divisions are displaced by those produced by later ones, which results in a layering that reflects neuronal birth order (Figure 1D) [6].
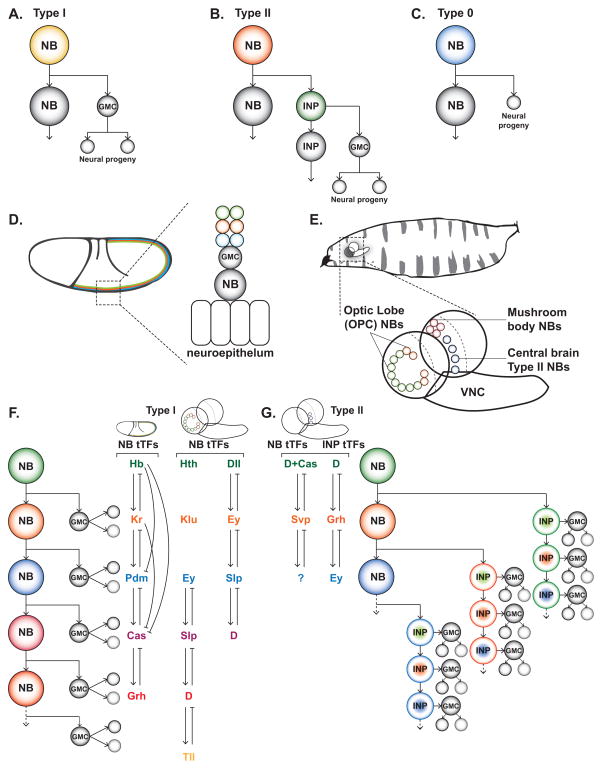
Figure 1. Neuroblast division modes and mechanisms of temporal patterning in Drosophila.
(A–C) The three main division modes used during Drosophila neurogenesis. Type I is the result of self-renewing asymmetric divisions that produce a neuroblast (NB) and an intermediate precursor called a ganglion mother cell (GMC), which divides only once to produce two cells. Type II is a self-renewing division that produces a NB and intermediate neural progenitor (INP), which then asymmetrically divides multiple times to give rise to GMCs that only divide once. Finally, type 0 is the result of self-renewing asymmetric divisions that produce a neuroblast (NB) and a single neuron.
(D) Neuroblasts delaminate from the embryonic, single-layered epithelium. Neuroblasts then divide asymmetrically (type I mode depicted here) throughout development. After each new GMC divides, earlier born neurons are displaced deeper into the embryo, resulting in a laminar organization with the earliest born neurons (green layer) occupying the deepest layer and the youngest neurons (blue layer) positioned closest to the surface.
(E) The larval brain is located in the anterior of the animal and will grow throughout development as new neurons are added from NBs. At this stage, the brain can be split into three main sections, the optic lobe, the central brain, and the ventral nerve cord (VNC), each containing populations of NBs. Depicted here, optic lobe NBs from the outer proliferation center (OPC), which will give rise to medulla neurons, central brain type II NBs, which give rise to a diverse set of central brain neurons, and mushroom body NBs, which produce the intrinsic neurons of the mushroom body.
(F) Embryonic and larval OPC NBs transition through three independent temporal series to dictate temporal fate. The result of this type of patterning is the ordered birth of different neuron types from single progenitors. In the embryonic VNC, most NBs transition through Hb → Kr → Pdm → Cas → Grh. In the optic lobe, NBs transition through either Hth → Klu → Ey → Slp → D → Tll or Dll → Ey → Slp → D. Temporal transcription factors do not specify a single cell fate but rather define early to late temporal identities in multiple lineages. Importantly, these factors can regulate each other, offering a model for progression through the temporal series.
(G) In the type II NBs of the central brain, an additional layer of temporal patterning is added in INPs. Neuroblasts transition through D+Cas → Svp → unknown factor, while INPs progress through D → Grh → Ey. Combinatorial temporal pattering allows for each temporal transcription factor in the NB to give rise to multiple neuron types without extending the NB lineage.
As they age, individual neuroblasts of the VNC sequentially express a series of transcription factors (known as temporal Transcription Factors or tTFs). The temporally restricted expression of these transcription factors defines unique identity windows during which different neurons are generated. Although the neuroblast itself changes tTF expression over time, neurons born within a given tTF window usually maintain expression of that tTF. Most VNC neuroblasts express the tTF series: Hunchback (Hb) → Krüppel (Kr) → POU domain proteins 1 and 2 (referred to as ‘Pdm’) → Castor (Cas) → Grainyhead (Grh) (Figure 1F) [6,7]. Neurons adopt distinct fates based on the tTF window in which they were born. Importantly, these tTFs differ from classical cell-fate determinants, since though they are used in most VNC neuroblast lineages, progeny fates differ between lineages [5]. Together, spatial and temporal inputs define unique neuronal identities.
Additional tTF series were later described in neuroblasts derived from the Drosophila Outer Proliferation Center (OPC) neuroepithelium, which produce neurons for the medulla neuropil of the visual system. The majority of OPC neuroblasts progress through the tTF series: Homothorax (Hth) → Klumpfuss (Klu) → Eyeless (Ey) → Sloppy-paired 1 and 2 (Slp) → Dichaete (D) → Tailless (Tll) [10,11], while a smaller population of neuroblasts localized at the tips of the OPC, which produce neurons for the medulla, lobula and lobula plate neuropils, progress through a different tTF series: Distal-less (Dll) → Ey → Slp → D (Figure 1F) [12]. Thus, similar to VNC neuroblasts, OPC neuroblasts in each region progress through the same tTF series but can give rise to different lineages. Once again highlighting that tTFs do not act as classical cell-fate determinants. Interestingly, during the Dll temporal window, neuroblasts divide asymmetrically to self-renew and generate a neuron directly (type 0, Figure 1C) before switching to type I neurogenesis for the remainder of the temporal series. Together with the VNC, these studies underscore temporal patterning as a clear strategy for expanding neuronal diversity, which can be further augmented by spatial patterning.
Likewise, recent work has demonstrated that overlaying multiple tiers of temporal patterning can be used to enrich neuronal diversity. Unlike VNC and optic lobe type I neuroblasts that generate GMCs, type II neuroblasts in the larval central brain generate intermediate neural progenitors (INPs), ganglion motherlike cells with increased proliferative potential [13]. Each INP undergoes several asymmetric divisions, and each division generates a GMC that produces neurons or glia (Figure 1B). Here, both neuroblasts and INPs progress through their own tTF series in parallel: D+Cas → Seven-up (Svp) → unknown factor(s) in the neuroblasts, and D → Grh → Ey in all INPs; together they combinatorially specify neuronal fate (Figure 1G) [13]. Interestingly, similar to type II neuroblasts in Drosophila, human outer subventricular zone neural progenitors also generate INP-like cells [reviewed in 14]. Thus, combinatorial temporal patterning may be a conserved strategy for expanding neuronal diversity within a lineage.
Mechanisms of transitions between tTFs
What are the mechanisms that mediate temporal transitions? Studies in VNC and optic lobe neuroblasts, as well as central brain INPs, indicate that transcriptional cross-regulation among tTFs is a common theme for regulating temporal transitions. For the most part, optic lobe neuroblasts and central brain INPs appear to follow a simple ‘feedforward activation/feedback repression’ mechanism: each factor in the series promotes expression of the next tTF, while repressing the previous tTF (Figure 1F) [reviewed in 15]. However, this mechanism is likely more complicated, as has been shown for VNC neuroblasts. For instance, a given tTF can repress the tTF that is next-plus-one in the series [6,7]. In addition, yet-to-be-identified independent mechanisms also promote sequential tTF expression in VNC neuroblasts since loss of Hb or Kr does not affect later-born fates [16]. The transition from Hb→Kr, unlike other transitions in VNC neuroblasts, is coupled to cytokinesis [16] and requires the orphan nuclear receptor Svp [17,18]. Svp acts as a ‘switch’ factor to facilitate the Hb→Kr transition by repressing hb. Notably, export of svp mRNA from the nucleus for translation is coupled to cytokinesis [17], which may explain the dependence of the Hb→Kr transition on cell division.
How is the length of a temporal window regulated? Are transitions governed by cell intrinsic or extrinsic cues? In vitro cultured VNC neuroblasts progress through the normal temporal series, arguing in favor of cell intrinsic mechanisms [16]. However, since neuroblasts in culture produce neurons, these experiments cannot rule out the possibility that neuronal progeny provide extrinsic feedback to regulate temporal transitions in neuroblasts. Indeed, a recent study reported that abdominal VNC neuroblasts undergo apoptosis in response to cues from progeny neurons [19]. Heterochronic transplants of older neuroblasts into younger hosts (or vice versa), and in vitro cultures of isolated neuroblasts, where cell-contacts are prevented, will clarify whether intrinsic programs dominate over extrinsic cues.
These findings contrast with those of the Drosophila mushroom body neuroblasts in the central brain. While a tTF series has not yet been identified in mushroom body neuroblasts, they are likely temporally patterned since they sequentially produce three main classes of neurons from embryogenesis through pupation [20]. Unlike temporal windows in VNC and optic lobe neuroblasts, which last for a handful of cell cycles, temporal windows in mushroom body neuroblasts span up to seventy-five cell cycles, and the mechanisms used to switch between each temporal window do not appear to be intrinsically fixed [reviewed in 21]. The timing of each transition in the mushroom body, and therefore the length of each temporal window, varies in response to extrinsic cues such as hormonal signals (ecdysone) and nutrition [22,23]. Importantly, while neuronal numbers can vary when extrinsic factors are modified, neuronal diversity is maintained. While in vitro culturing and transplantation experiments remain to be performed for optic lobe neuroblasts, recent work has demonstrated that once optic lobe neuroblasts initiate the temporal series, they are insensitive to nutrient conditions [24]. Indeed, in nutrient deprived conditions, the neuroepithelium, and therefore the initial pool of available neuroblasts, is reduced but temporal transitions are unaffected. Thus, although the overall size of the brain is smaller, neuronal diversity and the number of neurons produced by each optic lobe neuroblast are invariant [24].
Studies in the mushroom body have also highlighted the importance of post-transcriptional regulation for temporal patterning. Transcriptional profiling of mushroom body neuroblasts at defined developmental time points identified IGF-II mRNA-binding protein (imp) and Syncrip (syp) as two differentially expressed RNA binding proteins through time [25]. Imp is highly expressed at early time points and decreases at late time points and vice versa for Syp expression. Imp promotes and Syp inhibits translation of chronologically inappropriate morphogenesis (chinmo), whose high-to-low temporal expression level in neurons determines early versus late neuronal fate [26]. chinmo mRNA is also regulated by Let-7, a conserved miRNA [22], well-studied as a heterochronic gene that regulates the larval to adult transition in C. elegans [reviewed in 27]. Thus, not surprisingly, temporal patterning and transitions from one window to the next are regulated at multiple levels.
Temporal patterning of vertebrate neural progenitors
The sequential birth of different cell types is a hallmark of temporal patterning not just in invertebrates but also in vertebrates (in the retina, cortex, spinal cord and hindbrain), thus highlighting temporal patterning as an evolutionarily conserved strategy for generating neuronal diversity [reviewed in 14,28,29]. Temporal patterning mechanisms in vertebrates parallel those identified in Drosophila and in some striking examples even show conservation down to the level of specific tTFs. Here we review a small subset of recent studies in the vertebrate retina and cortex, as well as in vitro studies, which offer insights into how temporal transitions occur in vivo. While we mainly limit our discussion to temporal patterning at the level of neural stem cells, we refer our readers to some excellent recent reviews, which provide a more comprehensive discussion of vertebrate neurogenesis [1,3–5,14,27–29].
Temporal patterning in the retina
The vertebrate retina develops from multipotent retinal progenitor cells (RPCs), which, as a population, give rise to distinct cell types in a characteristic yet highly overlapping birth order: ganglion, horizontal, cone and amacrine cells are born early whereas bipolar cells, rods and Müller glia are born late (Figure 2A) [reviewed in 28]. Clonal analysis of zebrafish and rat RPCs revealed that they do not have fixed lineages. Instead they vary considerably in the number and the composition of their progeny, and make stochastic decisions for self-renewing or differentiating, and for the order in which they produce different retinal cell types [30,31]. These individual stochastic choices are balanced at the level of the whole tissue to produce retinal cells in characteristic numbers and order (Figure 2B).
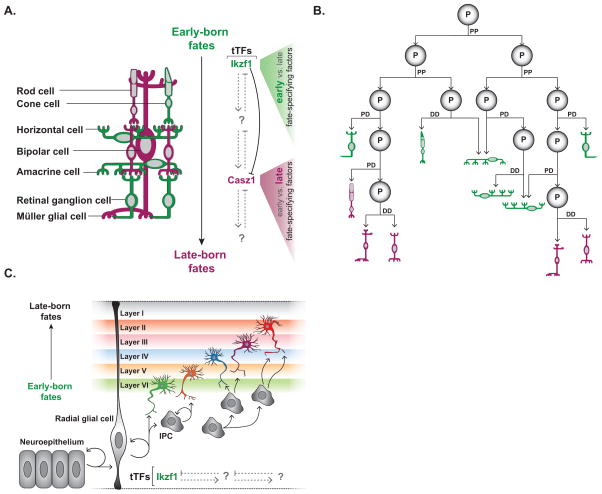
Figure 2. Temporal patterning in the vertebrate retina and cortex.
(A) There are seven main cell types in the adult retina: ganglion cells, horizontal cells, cone cells, amacrine cells, bipolar cells, rod cells and Müller glia. Although the birth order of retinal cell types overlaps highly, and individual retinal progenitor cells (RPCs) produce variable clone sizes, cone cells, horizontal cells, amacrine cells, and retinal ganglion cells are born early (green cells) whereas rod cells, bipolar cells and Muller glial cells are born late (purple cells). Early-fate specifying transcription factors are biased to be expressed early, perhaps as a result of the temporal transcription factor Ikzf1, which is expressed in RPCs early in development. In contrast, late-fate specifying transcription factors are biased to be expressed late, perhaps as a result of RPCs expressing the temporal transcription factor Casz1 late in development. The cross-regulatory interaction between Ikzf1 and Casz1 is reminiscent of the interaction between temporal transcription factors in Drosophila.
(B) Initially, RPCs tend to undergo PP divisions to expand the progenitor population, before switching to PD and DD divisions (See Box 2). Although progenitors are capable of producing any cell type in any temporal window, they are biased to produce specific fates as they age (See Box 2). Interestingly, cell fate correlates with division mode: retinal ganglion cells are born from PD divisions, amacrine cells arise from both PD and DD divisions, whereas horizontal, bipolar, rod, and cone cells tend to be born from DD divisions.
(C) The vertebrate cortex is built in an inside-out fashion, where early born cells reside in the deepest layer and later born cells migrate past their earlier born siblings. Neuroepithelial cells initially undergo PP division to expand the progenitor population before giving rise to radial glial cells, which are multipotent cortical progenitors. Radial glia divide asymmetrically to give rise to cells with limited proliferative potential called intermediate precursor cells (IPCs). Early in development, cortical progenitors express the transcription factor Ikzf1, a candidate temporal transcription factor associated with early-born neuronal fates, which is also expressed in early progenitors in the mammalian retina.
Recent studies have linked the production of specific retinal cell fates with the expression of unique transcription factors (fate determinants) in RPCs [reviewed in 28]. Quantitative modeling of vertebrate retinal clones has argued that although expression of fate determinants in an individual RPCs may be stochastic, at the tissue-level early-fate determinant expression is biased to precede late-fate determinant expression, which can account for the known birth order and number of retinal cell types [32]. This raises the question of how fate determinant expression is biased through time? Some recent studies suggest a model similar to Drosophila neurogenesis: Two transcription factors were identified that are expressed in all RPCs in non-overlapping temporal windows, indicative of a tTF series: Ikzf1, the ortholog of Drosophila Hb (an early VNC tTF), is expressed in young RPCs and promotes specification of early-born cell types [33], whereas Casz1, the ortholog of Drosophila Cas (a late VNC tTF), is expressed in older RPCs and promotes late-born fates [34]. An emerging view is that tTFs bias fate determinant expression during their temporal windows. Thus, progression through the tTF series dictates the characteristic birth-order of different neuronal types. Importantly, whereas expression of Drosophila tTFs deterministically define specific early or late fates, vertebrate tTFs appear to only bias RPC competence towards early or late fates via the regulation of specific cell fate determinants. Interestingly, as with Hb repression of Cas in Drosophila VNC neuroblasts, Ikzf1 inhibits Casz1 expression, providing evidence that cross regulation between tTFs is an evolutionarily conserved strategy to help regulate temporal transitions and ensure sequential expression of tTFs (Figure 2A) [34].
It is important to note that deterministic and stochastic mechanisms can operate together in the context of retinal neurogenesis. For example, mouse RPCs that express Cdh6 produce all retinal cell types in a stochastic manner. However, divisions from this RPC that are destined to produce RGCs are restricted to generate only a single sub-type of ganglion cell [35]. In addition to this example, the outcome of some RPC terminal divisions are deterministic, such as for the early and late Olig2 expressing populations, which are biased to give rise to particular retinal neurons [36]. These terminally dividing RPCs are similar to Drosophila GMCs, which are also terminally dividing cells that are limited to generate a specific set of neurons depending on the tTF they inherit.
Temporal patterning in the cortex
The mammalian cerebral cortex is a laminar structure, a feature that is intimately linked with temporal patterning. Early-born neurons settle in deep layers and later-born neurons climb past them to settle in more superficial layers, thus building six distinct layers from the inside out (Figure 2C) [reviewed in 14]. Here too, Ikzf1 is expressed early in cortical progenitors (radial glia) and appears to bias neurogenesis in favor of early fates, suggestive of its role as a tTF (Figure 2C) [37]. Although tTF series remain largely unknown in the cortex, in vivo and in vitro studies of corticogenesis have shed light on the relative contributions of intrinsic versus extrinsic cues to the temporal program [reviewed in 5,14].
How do radial glia sequentially give rise to neurons with different laminar identities through time? Pioneering work using heterochronic transplantation assays determined that progenitors lose competence to produce early born neurons (deep laminar fate) as they age [38–40]. Interestingly, it was also shown that young cortical progenitors transplanted into an older host could follow the host program and produce late born neurons (superficial laminar fate) [39]. This demonstrates that young progenitors are competent to produce late born neurons and that environmental conditions can extrinsically determine late born fate. The role of extrinsic signaling in regulating cell fate has long been recognized [reviewed in 41] and is well documented for regulating the switch from neurogenesis to gliogenesis in the cerebral cortex [42]. Additionally, it is now known that post-mitotic neurons can regulate progenitor competence within the neurogenic phase by secreting neurotrophins [43].
Similar to in vivo corticogenesis, in vitro corticogenesis proceeds with remarkably robust temporal patterning [44,45]. However, depending on culture conditions (medium and density), neurons of the 6 layers are produced in proportions that differ from those observed in vivo [reviewed in 46]. Progenitors present in neurospheres, which are clustered progenitors and thus subject to signals from surrounding cells, change their gene expression profiles progressively over time, in a manner similar to that observed in vivo [reviewed in 46]. In contrast, cell-contact isolated progenitors (i.e. sparsely plated progenitors lacking cell-to-cell contact as compared to neurospheres) are more limited in their pattern of gene expression transitions [47]. Moreover, while pluripotent stem-cell-derived cortical progenitors cultured in minimal medium have limited neurogenic potential for upper-layer neuronal fates, they display increased potential to generate these fates when transplanted into a newborn host cortex [48]. Taken together, the in vivo and in vitro data demonstrate that extrinsic cues act together with intrinsic programs to convey time dependent neuronal specification in the cortex. Identifying additional sources of extrinsic cues (which could come from cortical progenitors, neurons, or other sources such as the meninges, cerebrospinal fluid, or the vascular niche) will be important for enabling in vitro corticogenesis systems to more accurately recapitulate in vivo developmental processes. It will be particularly interesting to continue to determine how extrinsic cues influence temporal patterning: Do these cues modify progenitor competence? Do they affect division mode (production of neurons or INPs, for example)? Do they regulate the time spent in a given tTF window? Perhaps the best example of the type of answers we can expect to these questions was recently shown using hindbrain progenitors both in vivo and in vitro, where extrinsic signaling through Tgfβ acts as a switch to restrict progenitor competence, suppress early fate, and promote late born identity [49].
Although corticogenesis is conserved among mammals, there are several divergent features, most notably size and neuronal diversity, which can be traced to species-specific progenitor properties [reviewed in 50]. Cortical progenitors arise from a neuroepithelium, which expands through symmetric divisions. Progenitors initially self-renew and generate neurons directly, similar to type 0 neurogenesis in Drosophila. They subsequently generate intermediate precursor cells (IPCs), which divide symmetrically to produce two identical progenitors or two neurons. In primates, both neuroepithelial expansion and neuronal production are drawn-out over weeks and months compared to days in mice [reviewed in 50]. Interestingly, these species-specific temporal qualities are observed in vitro under the same culture conditions, arguing for intrinsic species-specific tuning of the cell cycle, division mode, etc. in progenitors [reviewed in 50]. Increased cortical size in primates has also been attributed to a class of progenitor called Outer Radial Glia, which are IPC-like cells derived from radial glia, but which have greater proliferative potential than standard radial glia-derived IPCs [51]. As with type II neuroblasts in the Drosophila central brain, the presence of IPC-like cells in the cortex may provide opportunities for layering temporal patterning programs to increase neuronal diversity and numbers.
Conclusions and future perspectives
Neuronal specification relies on a complex interplay between temporal and spatial patterning, division mode and competence at the level of the progenitor. The work highlighted here indicates that these considerations need not be independent but instead can influence and bias each other. While still in its infancy, our growing understanding of how neural progenitor temporal identity and competence are regulated during development will help increase the efficiency and accuracy of somatic cell reprogramming and directed differentiation. Interdisciplinary studies that combine in vivo and in vitro approaches, as well as cross-species comparisons, are likely to identify fundamental governing principles while pointing out species-specific peculiarities.
Box 2. Temporal regulation of modes of division and its influence on neuronal specification.
Neural progenitors can divide in three different ways: self-expanding symmetric divisions that generate 2 progenitors (PP), self-replacing asymmetric divisions that generate a progenitor and a differentiating daughter (PD), or self-consuming symmetric divisions that produce 2 differentiating daughters (DD) [52]. Progenitor modes of division are temporally regulated during development [30,53]. Initially, retinal progenitor divisions are predominantly PP, then PD, followed by DD (Figure 2B). Intriguingly, neuronal fate specification appears to correlate with division mode: retinal ganglion cells are born from PD divisions, amacrine cells arise from both PD and DD divisions, whereas horizontal, bipolar, rod, and cone cells tend to be born from DD divisions (Figure 2B) [30]. While it is unclear how progenitors switch between modes of division over time, tTFs might bias the generation of different neuronal fates by regulating progenitor division mode. The DrosophilatTF Castor and the spatial factor Antennapedia converge to drive embryonic neuroblasts in the thoracic region to switch from producing two neurons through an intermediate GMC to producing a neuron directly (type 1 → type 0 neurogenesis, Figure 1C) [54]. Future studies will need to establish whether modes of division are a driving force behind cell fate specification or whether they are controlled in parallel by upstream factors that regulate both fate and division mode.
Highlights.
Temporal patterning of neural progenitors is a conserved strategy to generate cell diversity.
In Drosophila, tTFs deterministically regulate cell fate.
In vertebrates, tTFs bias stochastic cell-fate decisions towards specific fates.
Intrinsic and extrinsic cues can regulate temporal transitions.
Acknowledgments
We thank Claire Bertet, Marc Amoyel and members of the Desplan laboratory for helpful input and suggestions. Research of the authors is supported by the National Institutes of Health grants R21 NS095288-01 and R01 EY13012 to CD. VMF is supported by a Natural Sciences and Engineering Research Council of Canada fellowship and AMR by funding from NIH 5T32HD007520-17. We apologize to all of our colleagues whose work we did not cite due to limited space but has been invaluable to our understanding of neurogenesis.
Footnotes
Conflict of interest
Nothing declared
Conflict of interest
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Azzarelli R, Hardwick LJA, Philpott A. Emergence of neuronal diversity from patterning of telencephalic progenitors. Wiley Interdiscip Rev Dev Biol. 2015;4:197–214. doi: 10.1002/wdev.174. [DOI] [PubMed] [Google Scholar]
- 2.Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays. 2004;26:739–51. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- 3.Alfano C, Studer M. Neocortical arealization: Evolution, mechanisms, and open questions. Dev Neurobiol. 2013;73:411–447. doi: 10.1002/dneu.22067. [DOI] [PubMed] [Google Scholar]
- 4.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 5.Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody T, Odenwald WF. Programmed Transformations in Neuroblast Gene Expression during Drosophila CNS Lineage Development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- 7.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 8.Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 10*.Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498:456–462. doi: 10.1038/nature12319. Together with [11] this study demonstrated that optic lobe neuroblasts undergo temporal patterning. Together they were the first studies to identify a temporal series outside of the VNC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol. 2013;380:12–24. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal Patterning of Neuroblasts Controls Notch-Mediated Cell Survival through Regulation of Hid or Reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. This study showed that layered temporal patterning in type II neuroblasts, which produce INPs, is used to further amplify neuronal diversity in the central brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodato S, Arlotta P. Generating Neuronal Diversity in the Mammalian Cerebral Cortex. Annu Rev Cell Dev Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Chen Z, Desplan C. Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol. 2013;105:69–96. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of Temporal Identity Transitions in Drosophila Neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Mettler U, Vogler G, Urban J. Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–37. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- 18.Kanai MI, Okabe M, Hiromi Y. Seven-up controls switching of transcription factors that specify temporal identities of drosophila neuroblasts. Dev Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Arya R, Sarkissian T, Tan Y, White K. Neural stem cell progeny regulate stem cell death in a Notch and Hox dependent manner. Cell Death Differ. 2015;22:1–10. doi: 10.1038/cdd.2014.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 21.Yasugi T, Nishimura T. Temporal regulation of the generation of neuronal diversity in Drosophila. Dev Growth Differ. 2015 doi: 10.1111/dgd.12245. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y-C, Chen C-H, Mercer A, Sokol NS. let-7-Complex MicroRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via chinmo. Dev Cell. 2012;23:202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Lin S, Marin EC, Yang C-P, Kao C-F, Apenteng BA, Huang Y, O’Connor MB, Truman JW, Lee T. Extremes of Lineage Plasticity in the Drosophila Brain. Curr Biol. 2013;23:1908–1913. doi: 10.1016/j.cub.2013.07.074. This study addressed the relative contribution of intrinsic vs. extrinsic cues during temporal patterning in mushroom body neuroblasts, which have prolonged temporal windows. It showed that unknown extrinsic signals regulate temporal transitions in the mushroom body, which is in contrast to antenal lobe lineages where similar environmental manipulations did not alter the numbers or types of progeny produced by a neuroblast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Lanet E, Gould AP, Maurange C. Protection of Neuronal Diversity at the Expense of Neuronal Numbers during Nutrient Restriction in the Drosophila Visual System. Cell Rep. 2013;3:587–594. doi: 10.1016/j.celrep.2013.02.006. This study showed that in the optic lobes while neuroepithelial proliferation is sensitive to nutrient conditions, neuroblast proliferation is not, thus neuronal diversity is preserved under nutrient-poor conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Liu Z, Yang C-P, Sugino K, Fu C-C, Liu L-Y, Yao X, Lee LP, Lee T. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science (80-) 2015;350:317–320. doi: 10.1126/science.aad1886. This study showed that the high-to-low and low-to-high gradients of the RNA-binding proteins Imp and Syp, respectively, regulate temporal identity by regulating Chinmo expression. Thus other factors, which affect post-transcriptional regulation, and not just tTFs are important for temporal fate specification. [DOI] [PubMed] [Google Scholar]
- 26.Zhu S, Lin S, Kao C-F, Awasaki T, Chiang A-S, Lee T. Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 27.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- 29.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–47. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 30.He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How Variable Clones Build an Invariant Retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes FLAF, Zhang G, Carbonell F, Correa Ja, Harris WA, Simons BD, Cayouette M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138:227–235. doi: 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Boije H, Rulands S, Dudczig S, Simons BD, Harris WA. The Independent Probabilistic Firing of Transcription Factors: A Paradigm for Clonal Variability in the Zebrafish Retina. Dev Cell. 2015;34:532–543. doi: 10.1016/j.devcel.2015.08.011. This study presents a simple computational model based on the independent and probabilistic expression of key fate-specifying factors in the retina, which can quantitatively account for the variance associated with reintal clones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros Confers Early Temporal Competence to Mouse Retinal Progenitor Cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 34**.Mattar P, Ericson J, Blackshaw S, Cayouette M. A Conserved Regulatory Logic Controls Temporal Identity in Mouse Neural Progenitors. Neuron. 2015;85:497–504. doi: 10.1016/j.neuron.2014.12.052. This study demonstrated a temporal series in the vertebrate retina by showing that Casz1, the ortholog to Drosophila Cas, is expressed in mid-/late retinal progenitors and is necessary for thier specification. They also showed transcriptional cross-regulation between these factors similar to that observed in flies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De la Huerta I, Kim IJ, Voinescu PE, Sanes JR. Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci U S A. 2012;109:17663–17668. doi: 10.1073/pnas.1215806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafler BP, Surzenko N, Beier KT, Punzo C, Trimarchi JM, Kong JH, Cepko CL. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci. 2012;109:7882–7887. doi: 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Alsio JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc Natl Acad Sci. 2013;110:E716–E725. doi: 10.1073/pnas.1215707110. This study shows that as in Drosophila VNC neuroblasts and in the vertebrate retina, Ikzf1 may act as a tTF early in a temporal series in the cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 39.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 40.Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- 41.Toma K, Hanashima C. Switching modes in corticogenesis: mechanisms of neuronal subtype transitions and integration in the cerebral cortex. Front Neurosci. 2015;9:274. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnabé-Heider F, Wasylnka Ja, Fernandes KJL, Porsche C, Sendtner M, Kaplan DR, Miller FD. Evidence that Embryonic Neurons Regulate the Onset of Cortical Gliogenesis via Cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 43.Parthasarathy S, Srivatsa S, Nityanandam A, Tarabykin V. Ntf3 acts downstream of Sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development. 2014;141:3324–3330. doi: 10.1242/dev.114173. [DOI] [PubMed] [Google Scholar]
- 44.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van Den Ameele J, Espuny-Camacho I, Passante L, Herpoel A, Schiffmann SN, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 45.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 46.Gaspard N, Gaillard A, Vanderhaeghen P. Making cortex in a dish: In vitro corticopoiesis from embryonic stem cells. Cell Cycle. 2009;8:2491–2496. doi: 10.4161/cc.8.16.9276. [DOI] [PubMed] [Google Scholar]
- 47*.Okamoto M, Miyata T, Konno D, Ueda HR, Kasukawa T, Hashimoto M, Matsuzaki F, Kawaguchi A. Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat Commun. 2016;7:11349. doi: 10.1038/ncomms11349. This study identified a subset of genes whose expression changes over time in cortical progenitors independent of their differentiation status, and that intrinsic and extrinsic cues are needed for these gene expression changes to occur normally, though they occur in cell-cycle arrested progenitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, et al. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Dias JM, Alekseenko Z, Applequist JM, Ericson J. Tgfβ Signaling Regulates Temporal Neurogenesis and Potency of Neural Stem Cells in the CNS. Neuron. 2014;84:927–939. doi: 10.1016/j.neuron.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 50.Van den Ameele J, Tiberi L, Vanderhaeghen P, Espuny-Camacho I. Thinking out of the dish: What to learn about cortical development using pluripotent stem cells. Trends Neurosci. 2014;37:334–342. doi: 10.1016/j.tins.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 52.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 53.Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic Progenitor Behavior and Unitary Production of Neurons in the Neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurange C, Cheng L, Gould AP. Temporal Transcription Factors and Their Targets Schedule the End of Neural Proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]